Abstract
Background
Schistosomiasis is a chronic neglected tropical disease that is characterized by continued inflammatory challenges to the exposed population and it has been established as a possible risk factor in the aetiology of bladder cancer. Improved diagnosis of schistosomiasis and its associated pathology is possible through mass spectrometry to identify biomarkers among the infected population, which will influence early detection of the disease and its subtle morbidity.
Methodology
A high-throughput proteomic approach was used to analyse human urine samples for 49 volunteers from Eggua, a schistosomiasis endemic community in South-West, Nigeria. The individuals were previously screened for Schistosoma haematobium and structural bladder pathologies via microscopy and ultrasonography respectively. Samples were categorised into schistosomiasis, schistosomiasis with bladder pathology, bladder pathology, and a normal healthy control group. These samples were analysed to identify potential protein biomarkers.
Results
A total of 1306 proteins and 9701 unique peptides were observed in this study (FDR = 0.01). Fifty-four human proteins were found to be potential biomarkers for schistosomiasis and bladder pathologies due to schistosomiasis by label-free quantitative comparison between groups. Thirty-six (36) parasite-derived potential biomarkers were also identified, which include some existing putative schistosomiasis biomarkers that have been previously reported. Some of these proteins include Elongation factor 1 alpha, phosphopyruvate hydratase, histone H4 and heat shock proteins (HSP 60, HSP 70).
Conclusion
These findings provide an in-depth analysis of potential schistosoma and human host protein biomarkers for diagnosis of chronic schistosomiasis caused by Schistosoma haematobium and its pathogenesis.
Author summary
Schistosomiasis, caused by S. haematobium, causes inflammation in the bladder and is common in tropical areas such as Nigeria. Undetected schistosomiasis can lead to inflammation in the bladder which may lead to bladder cancer. Diagnosis of bladder cancer in areas with common urinary schistosomiasis is difficult because the two diseases share many common symptoms. It is therefore important to identify biomarkers that could be used for early diagnosis of both schistosomiasis and schistosomiasis-associated bladder cancer. We chose a proteomic approach to identify these candidate biomarkers in a clinical cohort of urine samples obtained in Nigeria. We found several parasite- and host-specific protein biomarkers that could have diagnostic potential in a urine-based test for schistosomiasis, and schistosomiasis-associated bladder cancer. These include proteins that may also have application in drug or vaccine development. This study is one of the first to catalogue the urinary proteomes of people affected by this neglected tropical disease.
Introduction
Urinary schistosomiasis, caused by the parasite Schistosoma haematobium, is of public health significance in tropical and sub-tropical areas, with an estimated 732 million persons being vulnerable to infection worldwide in well-defined transmission areas [1]. In 2008, 17.5 million people were treated globally for schistosomiasis, 11.7 million of those from sub-Saharan Africa [2]. Schistosomiasis is considered to be endemic in Nigeria [1,3,4], where about 20 million people are affected by chronic schistosomiasis [1,5]. Schistosoma haematobium infection is reported to be more widespread than Schistosoma mansoni infection [6]. Schistosomiasis is characterized by continued health threat and inflammatory challenges in people who are exposed to long-term daily risk of infection [7]. Chronic infection with S. haematobium has been reported as a possible risk factor in the aetiology of bladder cancer [8,9]. Several studies have recorded increased urinary tract pathology conditions among populations infected with S. haematobium [3,10]. Histopathologists have also associated S. haematobium infection with the development of squamous cell carcinoma of the bladder [11]. S. haematobium has been associated with a two- to ten-fold increase in the risk of bladder squamous cell carcinoma, as well as being a potential cause of kidney damage. Hence, the parasite is considered as a group 1 carcinogen [12]. In some regions where S. haematobium is endemic, bladder cancer is the most common cancer in men and the second most common in women, just behind breast cancer, accounting for as much as 30% of all cancer cases [13].
Early disease detection of bladder cancer would significantly benefit people living in S. haematobium-endemic areas, because bladder cancer is otherwise unlikely to be recognized, as the obvious urinary tract symptoms (intermittent haematuria, dysuria, increased frequency, urgency and pain with micturition) are so commonly associated with urinary schistosomiasis that when the cancer manifests the patient is not likely to receive adequate diagnosis and may become severely debilitated with poor disease prognosis [10].
Detecting bladder cancer at the population level is challenging because direct proof requires detailed histopathological study, but invasive examinations are restricted to tertiary hospitals [14]. The detection of cancer-associated biomarkers, preferably isolated from urine and blood, has therefore become important. Such biomarkers are now being developed and will provide tools that could be useful to evaluate the specific effects of long-term exposure to S. haematobium [15].
Demonstration of schistosome-associated bladder damage by ultrasound examination is valuable and useful; however, it cannot be used to construe a diagnosis of cancer. Cancer-specific urine biomarkers may therefore play an important role in people with long-term S. haematobium infections. In addition, considering the fact that treatment of schistosomiasis relies on a single drug, praziquantel, which raises fears of resistance, there is a need to acquire a deeper understanding of the communication between the parasite and the mammalian host, with a view to identifying new methods of controlling schistosomiasis and schistosomiasis-associated bladder cancer.
One potential approach to investigating the developing relationship between the parasite and its host is proteomics. Biological fluids are promising sources of diagnostic, prognostic and treatment based biomarkers, due to their easy accessibility [16, 17]. Biological fluids are associated with tissues that release protein components into them and the disease-altered state could change either the constituents or the amount of such proteins. Biofluid-derived proteins could be parasite or host associated biomarkers.
Proteomics has been successfully employed for human-based studies of disease, where it has been a valuable approach for distinguishing diseases and generating candidate biomarkers to determine pathological state [16, 18]. Mass spectrometry (MS) based analysis of a small number of exposed and unexposed subjects has been found to reveal altered expression of proteins that may be identified as intermediate biomarkers of early disease effects [19]. In particular, the potential of the urinary proteome as a non-invasive means to identify biomarkers for carcinogen exposure and metabolism of toxic chemicals has been demonstrated by Moore et al. [19].
Several schistosome-oriented proteomics studies have focused on the parasites [12,16]. However, more information on the changes that manifest in the host proteome during active schistosomiasis is required [14]. The goal of the present study is thus to identify candidate biomarkers for the diagnosis of schistosomiasis and schistosomiasis-associated bladder cancer from adults in a rural population in south-west Nigeria, an area which is endemic for urinary schistosomiasis.
Materials and methods
Urine sample collection
Human urine samples were collected from volunteers living in Eggua, Ogun State, a schistosomiasis endemic community in South-west, Nigeria. Eggua lies between latitude 7° 6ʹ4.811ʺ N and longitude 2° 52ʹ 43.776ʺ E in a derived savanna zone. The area is largely dominated by Yoruba speaking people. The volunteers were screened for the presence of Schistosoma haematobium infection by a combination of microscopy (Fig 1), detection of macro and microhaematuria (urinalysis) and rapid diagnostic test (RDT) for schistosomiasis (Table 1). The urine samples were collected between 10:00 and 14:00 to ensure maximum egg yield and 10 mL of sample were processed for microscopic examination and egg count. The eggs were quantified by counting and classified as light infection if there were ≤50 (1–49) eggs/10 mL urine and heavy infection if there were >50 eggs/10 mL urine [10] and urinary structural bladder pathology was examined using ultrasonography, which has been published elsewhere [10]. Sample size power calculation was carried out, indicating that 44 individual samples (N) were required for a statistical power of 0.9 at significance level 0.05.
Fig 1. Schistosome eggs (arrows) in the urine of an S. haematobium infected participant as shown by light microscopy (magnification x10).
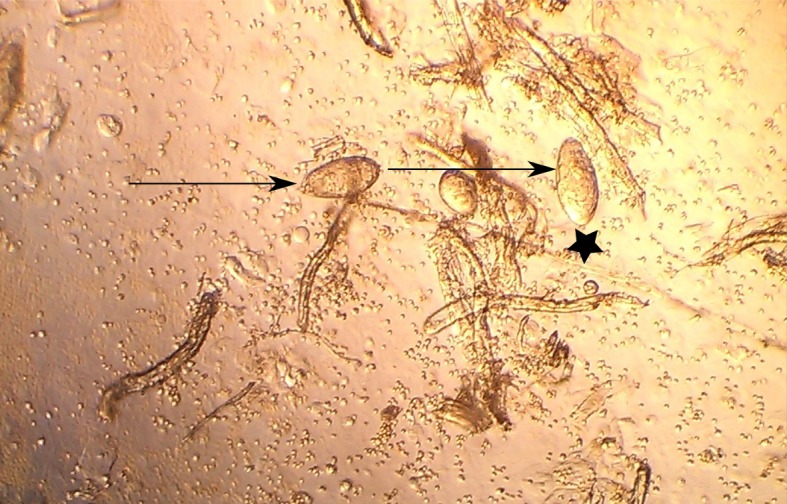
A terminal spine of the egg is indicated by a black star.
Table 1. Pathology and infection status of sampled volunteers.
| Sample Group | Gender (n = 49) | S. haematobium Infection by egg count | Microhaematuria | Infection by RDT | Bladder Pathology | |
|---|---|---|---|---|---|---|
| PS | Male | 8 | Present | Absent | Absent | Present |
| Female | 7 | |||||
| SH | Male | 5 | Present | Present | Present | Absent |
| Female | 7 | |||||
| PT | Male | 5 | Absent | Absent | Absent | Present |
| Female | 7 | |||||
| NPS | Male | 4 | Absent | Absent | Absent | Absent |
| Female | 6 | |||||
Ethical consideration
Ethical approval was obtained from the University of Ibadan and University College Hospital (UI/UCH) Ethical Committee and Ogun State Ministry of Health. All participants gave informed consent; all participants were adults and were able to decide for themselves. The informed consent document was written in both English and Yoruba languages, the latter being the language of the Nigerian communities. For those participants who could read and write, written informed consent was obtained. For those participants who could not read and write, the informed consent form was read to them in their language. All participants enrolled in the study voluntarily. The informed consent was signed by all participants and those who could not sign provided a thumb print on the informed consent form. This approach to informed consent for those who could not read and write was approved by the UI/UCH Ethical Committee.
Sample preparation and in solution protein digestion
A total of 49 individual urine samples were placed into four different categories, namely 12 schistosomiasis cases (SH), 12 bladder pathology cases (PT), 15 combined pathology and schistosomiasis (PS) cases and 10 controls with no pathology or schistosomiasis (NPS). All samples were processed on the same day using the same batches of all reagents, including trypsin, to minimise batch effects and sample to sample technical variation. An aliquot of 4 ml of urine per individual was subjected to methanol-chloroform protein precipitation followed by in solution tryptic digest prior to MS analysis. Precipitated protein was resuspended in denaturation buffer (6 M urea, 2 M thiourea, 10 mM Tris buffer, pH 8.0), and then a Bradford assay was carried out to determine protein concentration [17]. For each sample, 100 μg of protein was then further reduced by incubation at room temperature for 1 hour in reduction buffer (1 M dithiothreitol (DTT); 50 mM ammonium bicarbonate (ABC). An alkylating buffer (550 mM iodoacetamide (IAA) in 50 mM ABC) was then added and incubated in the dark at room temperature for an hour. The sample was then diluted with 4 volumes of 50 mM ABC and proteolysed overnight for 16 hours at 37°C using Trypsin-Ultra (mass spectrometry grade; New England BioLabs) according to the manufacturer’s instructions. An equivalent of 10 μg of the peptide solution was then transferred to in-house prepared stage tips for off-line solid-phase extraction, desalting and clean-up of sample, as described in previous studies [17, 20], and the desalted peptides were then dried in a refrigerated speedy vac (SPD 111v-230 Speed VAC, Thermo Savant, New York, USA).
Ultra-high performance liquid chromatography
Peptide samples were resuspended by diluting the desalted, dried peptides to 200 ng/μL using 2% acetonitrile (ACN) in HPLC grade water containing 0.1% v/v formic acid (FA) before MS analysis. Nanoflow ultra-HPLC was carried out on each sample, without pre-fractionation, using a Dionex UltiMate 3500RSnano UPLC system (Thermo Fisher, San Jose, CA, USA) equipped with a reverse phase (RP) pre-column trap (100 μm × 2 cm; 5 μm; 100 Å; C18) and analytical column (70 μm × 20 cm; 5 μm; 100 Å; C18). Equal 400 ng injections of each sample were eluted by gradient chromatography at 23°C with a flow rate of 300 nL/min, and a 6–40% gradient of water–ACN from 0 to 120 min. The binary mobile phase system used was as follows: buffer A contained water and 0.1% FA, while buffer B contained ACN and 0.1% FA. The elution gradient for peptides was 6% B from 10 min to 40% B at 60 min, then increasing to 80% B for 10 min before returning to 2% B for equilibration. The same pre-column trap and analytical column was used for all samples.
Mass Spectrometry (MS)
Discovery proteomic analysis of each sample was carried out on a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher). Analysis of samples introduced from the in-line HPLC system was achieved with the following system settings: Data-dependent automated full scan cycles were performed with automatic switching between MS/MS and MS scans at a scan range of 300–1650 m/z. The top ten most abundant precursor ions selected by the quadrupole during the initial MS scan were subjected to fragmentation using high-energy collision dissociation with normalized collision energy at a pressure of 1.2 mTorr and a dynamic exclusion time of 30 s. The abundance threshold for ion selection was 0.001 with charge exclusion of z = 1 ions. Acquisition of mass spectra was done at a resolution of 70,000 with a maximum injection time of 250 ms or a target automatic gain control value of 3×106. High-energy collision dissociation and normalized collision energy set at 27 were used for peptide fragmentation. Continuous tandem mass spectra acquisition resolution was set at 17,500 at a maximum injection time of 120 ms or target AGC of 2 × 105. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006438.
MS data processing and statistical analysis
All raw MS data was processed with MaxQuant software (version 1.5.3.12) and its built-in search engine, Andromeda, for peptide identification and protein inference, using the default settings and the human and S. mansoni databases (www.uniprot.org), as described in detail elsewhere [17] (S1 Fig). FDRs were set at 1% at both peptide and protein level. Peptide identifications were transferred to unidentified features in other LC-MS runs based on matching masses and re-calibrated retention times between runs (“match between runs” option in MaxQuant). Normalisation of data between LC-MS runs was carried out in MaxQuant, based on summation of the total peptide ion signals per sample and using a global Levenberg-Marquardt optimisation procedure that aimed to minimise the overall changes for all peptides across all samples [21]. Label-free quantification (LFQ) was then carried out using MaxQuant, based on determination of the median pair-wise common peptide ratios between samples, requiring a minimum of two peptide ratios per identified protein [21].
The LFQ values from MaxQuant were imported into Perseus software (version 1.5.3.1) for differential expression statistical analysis and visualization by hierarchical clustering and Principal Component Analysis (PCA), with a Benjamini-Hochberg multiple testing correction cut off set at FDR 0.05. Three separate independent t-tests were carried out using LFQ data to compare NPS versus SH, NPS versus PT, and NPS versus PS. One-way ANOVA was also carried out to statistically validate the differentially expressed potential biomarkers. Venn diagrams were plotted using VennDIS (version 1.01). Proteins which were determined to be significantly differentially expressed between groups were further subjected to a GO-enrichment analysis using Blast2GO [16,22]. Uniquely identified human proteins were subjected to pathway and protein-protein interaction analysis using the Reactome database (v60; www.reactome.org) and String DB (www.string-db.org), respectively.
Results
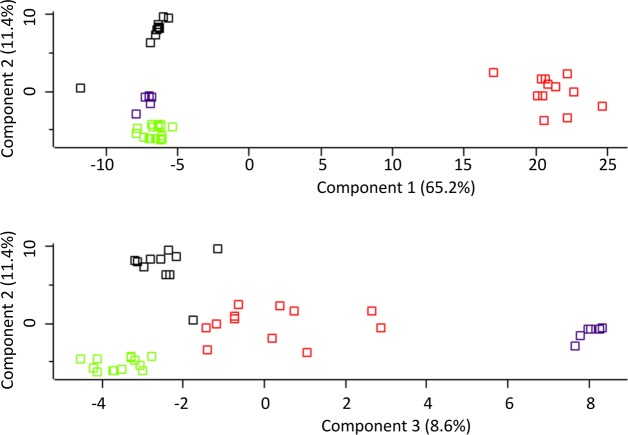
Urine samples from 49 individuals distributed across the four disease groups (NPS, SH, PT and PS) were analysed to identify candidate biomarkers for schistosomiasis and its associated pathologies (Table 1). High levels of correlation between the urinary protein components of these sample groups was demonstrated by scatterplots, hierarchical clustering (heatmap) and principal component analysis (PCA) (Figs 2 and 3). The dimensions in Fig 3 account for 76.6% and 20% of the observed variation, respectively. Hierarchical clustering of protein groups identified in SH, PT and PS and NPS samples showed clear molecular differences between groups. As expected, differences in the proteomic signatures were observed between the control group (NPS) and all disease groups, as all three disease groups clustered distinctly, with the SH and PS group being more closely related to one another as compared to the PT group (Figs 2 and 3), although a few samples of the pathology group clustered proximally to the control group (Fig 3).
Fig 2. A hierarchical heatmap showing distinct clustering of each sample group.
The colour bars above the heatmap depict the following clusters: Schistosomiasis, SH—red; Bladder Pathology, PT—blue; Pathology and Schistosomiasis, PS—green; No Pathology or Schistosomiasis, NPS—yellow. Representative protein identifiers that drive the clustering are also shown.
Fig 3. Individual sample comparison between NPS, SH, PS and PT.
Similar patterns with minor overlap were seen by multivariate testing using Principal Component Analysis (PCA). The abbreviations represent different sample groups namely: SH—S. haematobium infected group (red); PT—bladder pathology group (black); PS—group with combination of pathology and S. haematobium infection (green); and NPS—No Pathology or Schistosomiasis (blue).
A total of 209,923 tandem mass spectra were identified and used to assign peptides and unique protein group identities, leading to the identification of 9,701 non-redundant peptides and 1,306 protein groups at a false discovery rate of 1%. The majority (66.3%) of peptides identified had no missed cleavages, while 27.5% had one missed cleavage and 6.2% had two missed cleavages (S4 Fig), which is within the expected range for in solution digest of complex protein mixtures. The number of peptide sequences identified and % MS/MS spectra identified per sample are shown in S2 and S3 Figs, respectively.
Prior to quantitative analysis of differential abundance, samples from each individual were normalised at the peptide level before mass spectrometric analysis. The intensity data for all identified peptides per sample was then further normalised in MaxQuant, based on the underlying assumption that the majority of the proteome should not change significantly between any two conditions and that the average behaviour can therefore be used as a relative standard [21]. In other words, normalisation between samples was based solely on the distribution of the peptide-level data obtained, without addition of external standards or reliance on any set of housekeeping proteins that are assumed to be stably expressed, as is now commonplace in label-free proteomics. Subsequent label-free differential protein abundance analysis was carried out at the protein level using MaxQuant and volcano plots of pairwise comparisons demonstrated the effectiveness of the normalisation procedures, with the majority of proteins found to be not significantly differentially expressed between the various comparisons after permutation-based FDR truncation (FDR = 0.05; S5 Fig).
A total of 36 Schistosoma proteins were identified in the host urine when the MS output was searched against a combination of human and Schistosoma databases (Table 2). These 36 identities were considered to be confident identities due to the relatively small size of the Schistosoma database compared to the human database in the combined database. The GO terms for the molecular function of the identified human and Schistosoma-derived proteins are summarized in Figs 4 and 5. More (124) parasite protein groups were identified when the MS output was searched against the Schistosoma DB only, but only 31 Schistosoma proteins were differentially abundant between groups by ANOVA using label-fee quantification (LFQ). Some Schistosoma specific proteins were found in samples from individuals earlier diagnosed and classified as negative for S. haematobium infection by microscopy (Figs 6 and 7).
Table 2. Identified Schistosoma-derived proteins across all urine sample groups, and their predicted functions.
A posterior error probability (PEP) score cut-off of <0.01 was applied to all protein identities in order to ensure confident protein assignments.
| Protein ID | Identified Schistosoma Protein | PEP Score | Location | Predicted Functions |
|---|---|---|---|---|
| C4Q4S5 | Tubulin alpha chain | 1.86E-18 | Cytoskeleton | Structural/GTPase activity |
| C4Q5I7 | Calreticulin autoantigen homolog | 0.00034 | Mitochondria | Binding |
| C4QBN1 | Histone H4 | 0.002522 | Cytosol /Nucleus | Binding |
| G4LWI2 | Heat shock protein HSP60 | 0.001447 | Cytoplasm | Heat Shock protein |
| G4LYN4 | ADP-ribosylation factor, arf | 0.000271 | Membrane | Transporter |
| G4M1M0 | DNA polymerase | 0.000801 | Nucleus | Binding and catalytic |
| G4V6R4 | Putative rab9 | 6.42E-05 | Membrane | Binding |
| G4V8L4 | Putative heat shock protein 70 | 3.61E-43 | Cytosol /Nucleus | Binding/Heat Shock protein |
| G4V910 | Putative heat shock protein 70 (Hsp70) | 0.001976 | Cytosol /Nucleus | Binding/Heat Shock protein |
| G4V8L4 | Putative heat shock protein 70 | 2.60E-29 | Cytosol /Nucleus | Binding/Heat Shock protein |
| G4VAC9 | Putative uncharacterized protein | 0.001085 | Unknown | Unknown |
| G4VAD2 | Elongation factor 1-alpha | 1.72E-36 | Cytoplasm | Binding/GTPase activity |
| G4VAW0 | Serine/threonine kinase | 4.72E-05 | Nucleus | Binding |
| G4VB75 | cytoplasmic dynein light chain | 0.000763 | Cytoskeleton | Structural/Motor |
| G4VB79 | Voltage-gated potassium channel, KCNQ | 0.001114 | Membrane | Transmembrane Transporter |
| G4VDD2 | Eukaryotic translation initiation factor 5A | 0.000224 | Ribosome | Binding |
| G4VG19 | Phosphoglycerate kinase | 1.25E-43 | Mitochondria | Kinase |
| G4VG20 | Phosphoglycerate kinase | 2.68E-07 | Mitochondria | Kinase |
| G4VGA0 | Sodium/potassium-transporting ATPase subunit alpha | 0.000264 | Membrane | Transporter |
| G4VH98 | Putative fimbrin/plastin | 0.002 | Unknown | Binding |
| G4VHN3 | ATP synthase subunit beta | 7.63E-34 | Membrane | Transporter/ Bindng |
| G4VIM7 | Camp-response element binding protein-related | 0.000471 | Nucleus | Binding |
| G4VKT8 | Putative atp synthase alpha subunit vacuolar | 0.000308 | Membrane | Transporter/ Binding |
| G4VLJ0 | ATP synthase subunit alpha | 0.000104 | Membrane | Transporter/ Binding |
| G4VLJ8 | Fidgetin like-1 | 0.000756 | Nucleus | Binding and catalytic |
| G4VLN5 | Putative uncharacterized protein | 3.70E-70 | Unknown | Binding |
| G4VLW1 | Putative actin | 6.14E-19 | Membrane | Binding |
| G4VLW2 | Putative actin-1 | 2.35E-33 | Membrane | Binding |
| G4VM26 | Putative zinc finger protein | 0.000636 | Nucleus | Binding |
| G4VMG4 | Venom allergen-like (VAL) 3 protein | 0.001866 | Membrane | Unknown |
| G4VMT3 | Adapter-related protein complex 3, beta subunit | 0.001906 | Membrane | Transporter |
| G4VP51 | Putative ADP,ATP carrier protein | 0.000161 | Membrane | Structural/ Transporter |
| G4VPE8 | Putative inorganic pyrophosphatase | 6.22E-23 | Cytoplasm | Binding |
| G4VPU8 | Putative cytoplasmic dynein intermediate chain 2 | 9.23E-05 | Cytoskeleton | Structural/Motor |
| G4VQ01 | Uncharacterized protein | 0.00185 | Membrane | Unknown |
| G4VQ58 | Phosphopyruvate hydratase | 8.38E-06 | Cytoplasm | Binding |
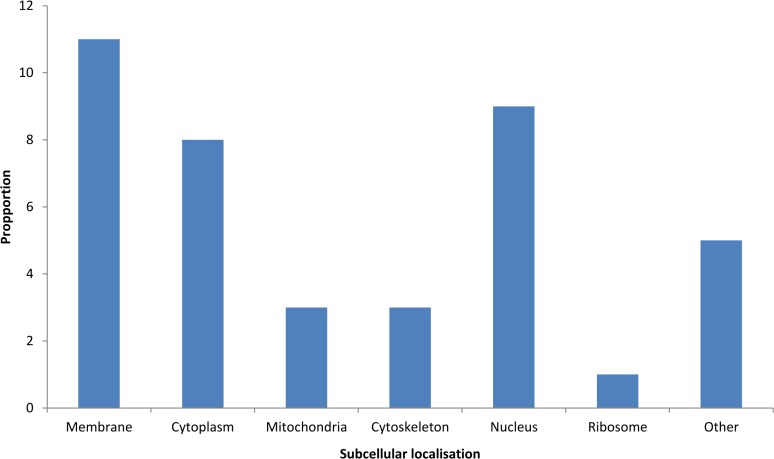
Fig 4. Allocation of observed Schistosoma-derived proteins into various subcellular locations according to Blast2GO analysis.
Fig 5. Summarized molecular function of the identified human and Schistosoma proteins as predicted by Blast2GO.
The abbreviations represent different sample groups namely: SH—S. haematobium infected groups (A); PT—bladder pathology group (B); PS—group with combination of pathology and S. haematobium infection (C); and Schistosoma proteins (D).
Fig 6. Abundance of Schistosoma-derived proteins and their LFQ intensity among individual samples.
SH—Schistosomiasis; PT—Bladder Pathology; PS—Pathology and Schistosomiasis; NPS—No Pathology or Schistosomiasis.
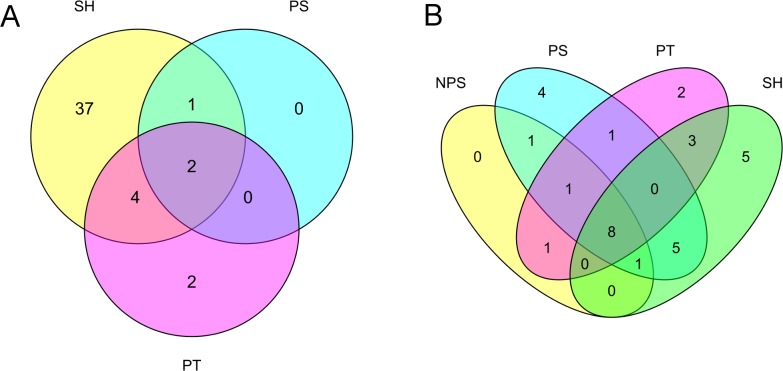
Fig 7.
Venn diagrams showing (A) overlap in Schistosoma proteins identified across different clinical groups and (B) overlap in the statistically significant human proteins identified across clinical groups. SH—S. haematobium infected groups; PT—Bladder Pathology group; PS—group with combination of pathology and S. haematobium infection; NPS—No Pathology or Schistosomiasis.
Venn diagrams were generated to identify proteins unique to each group. Out of the 36 total Schistosoma protein groups confidently identified, 5 (15.6%), 4 (12.6%) and 2 (6.3%) proteins were unique to SH, PS and PT group respectively while only 8 proteins (25%) were found common to all study groups (Fig 7A). Heat shock protein 70, elongation factor 1-alpha, camp-response element binding proteins-related, histone H4 and venom allergen-like (VAL) 3 proteins were found to be unique to SH group while tubulin alpha chain, calreticulin autoantigen homolog, heat shock protein HSP 60 and putative ADP,ATP carrier protein were found only in PS group. 2 potential biomarkers unique to the PT group include cytoplasmic dynein light chain and putative actin 1. 13 (36.1%) of the predicted Schistosoma protein were membrane-associated, 8 (22.2%) nuclear based, 4 (11%) cytoplasmic and 3 (8.3%) cytoskeletal and mitochondrial, 1 (2.8%) ribosomal and 3 (8.3%) unknown.
The result of the three independent t-tests performed using LFQ values for “NPS versus SH”, “NPS versus PT” and “NPS versus PS” revealed a total of 54 candidate human protein biomarkers for schistosomiasis and bladder pathology. The proteins are distributed into 43, 8 and 7 for “NPS versus SH” (Table 3), “NPS versus PT” and “NPS versus PS” (Table 4) respectively. 37 and 2 proteins were unique to SH and PT groups, respectively, while none were unique to the PS group (Fig 7B). A search for possible marker overlap across study groups showed that cathepsin B (P07858) was shared by all disease groups; arylsulfatase A (A0A0C4DFZ2) and phosphatidylethanolamine-binding protein 4 (Q96S96) were shared by PS and SH group (Table 5); and PT and SH groups were found to have 4 proteins in common, namely transthyretin (P02766), plasma retinol-binding protein (Q5VY30), phosphatidylcholine-sterol acyltransferase (P04180) and cartilage intermediate layer protein 2 (K7EPJ4). The majority of the human proteins identified were predicted to be membrane associated and perform “binding” molecular activities. The human proteins that were identified as differentially abundant in the SH, PT and PS groups were subjected to pathway analysis using the reactome tool and string DB. Reactome did not identify any statistically significantly overexpressed human pathways in any of the groups, as the majority of identified proteins did not have mapped identifiers. Using string DB, we found that the cellular component ‘extracellular exosomes’ (GO:0070062) was significantly functionally enriched in the SH group compared to that of the NPS group, with a FDR of 1.09e-14. Furthermore, the Kegg pathway ‘lysosome' (ID 04142) was functionally enriched with an FDR of 0.00685. The number of differentially abundant proteins unique to the PT and PS groups was too few to identify functionally enriched pathways or GO terms.
Table 3. Identified differentially abundant human proteins and their predicted functions in individuals infected with Schistosoma haematobium (SH) vs the NPS group.
| Protein ID | Identified Human Protein | PEP Scores | Location | Predicted Functions |
|---|---|---|---|---|
| Q99519 | Sialidase-1 | 2.68E+08 | Membrane | Binding |
| A0A075B6I5 | Ig lambda chain V-I region NIG-64;Ig lambda chain V-I region BL2 | 9.46E+08 | Membrane Associated | Binding/Immune |
| Q9BQ51 | Programmed cell death 1 ligand 2 | 4.76E+08 | Membrane | Binding/Immune |
| P09467 | Fructose-1,6-bisphosphatase 1 | 4.59E+08 | Cytosol/Nucleus | Binding |
| Q96L35 | Receptor protein-tyrosine kinase;Ephrin type-B receptor 4 | 4.73E+08 | Cytoplasm/Mitochondria | Kinases/Binding |
| Q08174 | Protocadherin-1 | 6.57E+08 | Membrane | Binding |
| P21796 | Voltage-dependent anion-selective channel protein 1 | 4.55E+08 | Membrane/Mitochondria | Transport/Binding |
| A0A087WXM8 | Basal cell adhesion molecule | 6.27E+08 | Membrane Associated | Binding |
| A0A087X2B5 | Basigin | 5.07E+08 | Membrane | |
| Q12794 | Hyaluronidase-1 | 3.94E+08 | Lysosome | Other Enzymatic |
| Q13332 | Receptor-type tyrosine-protein phosphatase S;Protein-tyrosine-phosphatase | 6.35E+08 | Membrane | Other Enzymatic |
| A6NNI4 | Tetraspanin;CD9 antigen | 4.59E+08 | Membrane | Binding/Immune |
| Q86UN3 | Reticulon-4 receptor-like 2 | 4.69E+08 | Membrane | Binding/Immune |
| P01701 | Ig lambda chain V-I region NEW | 8.4E+08 | Membrane | Binding/Immune |
| Q99988 | Growth/differentiation factor 15 | 1.17E+09 | Exosome | Binding/Immune |
| P07858 | Cathepsin B;Cathepsin B light chain;Cathepsin B heavy chain | 1.19E+09 | Membrane | Binding |
| Q15113 | Procollagen C-endopeptidase enhancer 1 | 2.59E+09 | Membrane | Binding |
| P53634 | Dipeptidyl peptidase 1;Dipeptidyl peptidase 1 exclusion domain chain;Dipeptidyl peptidase 1 heavy chain;Dipeptidyl peptidase 1 light chain | 1.43E+09 | Endoplasmic Recticulum/Golgi Apparatus | Binding/Enzymatic |
| Q96NY8 | Nectin-4;Processed poliovirus receptor-related protein 4 | 1.98E+09 | Cytoskeleton/Membrane | Binding |
| Q9HCN6 | Platelet glycoprotein VI | 1.91E+09 | Membrane | Binding |
| Q9NZH0 | G-protein coupled receptor family C group 5 member B | 7.23E+08 | Membrane/Nucleus | Binding |
| O00560 | Syntenin-1 | 1.24E+09 | Membrane | Binding |
| Q92520 | Protein FAM3C | 9.39E+08 | Membrane Associated | Immune |
| Q8NBJ4 | Golgi membrane protein 1 | 8.67E+08 | Membrane Associated | |
| P04180 | Phosphatidylcholine-sterol acyltransferase | 1.19E+09 | Extracellular | Enzymatic |
| K7EPJ4 | Cartilage intermediate layer protein 2;Cartilage intermediate layer protein 2 C1;Cartilage intermediate layer protein 2 C2 | 1.04E+09 | Membrane | Unknown |
| P05109 | Protein S100-A8;Protein S100-A8, N-terminally processed | 1.19E+10 | Membrane /cytosol | Binding |
| O94919 | Endonuclease domain-containing 1 protein | 1.24E+10 | Membrane | Binding/Enzymatic |
| P02766 | Transthyretin | 4.96E+09 | Cytoplasm | Binding |
| Q16777 | Histone H2A type 2-C;Histone H2A type 2-A | 8.63E+09 | Nucleus | Binding |
| A0A087WV17 | Osteoclast-associated immunoglobulin-like receptor | 5.67E+09 | Membrane | Binding/Immune |
| Q5VY30 | Retinol-binding protein 4;Plasma retinol-binding protein(1–182);Plasma retinol-binding protein(1–181);Plasma retinol-binding protein(1–179);Plasma retinol-binding protein(1–176) | 2.34E+09 | Membrane | Transporter/Immune |
| H0Y755 | Low affinity immunoglobulin gamma Fc region receptor III-A | 3.96E+09 | Membrane/cytoskeleton | Transporter/Immune |
| Q8NFZ8 | Cell adhesion molecule 4 | 2.09E+09 | Membrane | Binding |
| A0A087WZR4 | Low affinity immunoglobulin gamma Fc region receptor III-B | 2.11E+09 | Membrane/cytoskeleton | Transporter/Immune |
| Q9H8L6 | Multimerin-2 | 2.72E+09 | ||
| A0A0C4DFZ2 | Arylsulfatase A;Arylsulfatase A component B;Arylsulfatase A component C | 1.16E+09 | Membrane/ER | Binding |
| F6X2W2 | Neuronal growth regulator 1 | 3.44E+09 | Membrane | Transporter/Binding |
| P09603 | Macrophage colony-stimulating factor 1;Processed macrophage colony-stimulating factor 1 | 6.79E+08 | Membrane/Cytoplasm | Binding/Immune |
| Q96S96 | Phosphatidylethanolamine-binding protein 4 | 1.9E+08 | Cytoplasm/Mitochondria | Binding |
| P31946 | 14-3-3 protein beta/alpha;14-3-3 protein beta/alpha, N-terminally processed | 3.77E+08 | Membrane | Binding |
| A0A087X0D5 | Pro-cathepsin H;Cathepsin H mini chain;Cathepsin H;Cathepsin H heavy chain;Cathepsin H light chain | 5.06E+08 | Cytoplasm/Nucleus | Enzymatic |
| P09619 | Platelet-derived growth factor receptor beta | 2.68E+08 | Membrane | Binding |
SH—Schistosomiasis; PEP—Posterior error probability; Protein IDs are Uniprot accession numbers
Table 4. Identified differentially abundant human proteins and their predicted functions from PT and PS groups vs the NPS group.
| Protein ID | Identified Human Protein | PEP Scores | Location | Predicted Functions |
|---|---|---|---|---|
| A0A0G2JM94 | Leukocyte-associated immunoglobulin-like receptor 1 | 1.1E+10 | Membrane | Binding/Immune |
| P02766 | Transthyretin | 1.24E+10 | Membrane | Binding/Immune |
| Q5VY30 | Retinol-binding protein 4;Plasma retinol-binding protein(1–182);Plasma retinol-binding protein(1–181);Plasma retinol-binding protein(1–179);Plasma retinol-binding protein(1–176) | 5.67E+09 | Cytoplasm | Binding |
| A0A0C4DFZ2 | Arylsulfatase A;Arylsulfatase A component B;Arylsulfatase A component C | 2.72E+09 | Nucleus | Enzymatic |
| P07858 | Cathepsin B;Cathepsin B light chain;Cathepsin B heavy chain | 1.19E+09 | Membrane | Binding |
| P04180 | Phosphatidylcholine-sterol acyltransferase | 1.19E+09 | ||
| K7EPJ4 | Cartilage intermediate layer protein 2;Cartilage intermediate layer protein 2 C1;Cartilage intermediate layer protein 2 C2 | 1.04E+09 | Membrane | Unknown |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein;Endorepellin;LG3 peptide | 9.34E+10 | Membrane | Binding |
| Q96S96 | Phosphatidylethanolamine-binding protein 4 | 6.79E+08 | Membrane Associated/ Lysosome | Binding |
PT—Bladder Pathology; PS—Pathology and Schistosomiasis; PEP—Posterior error probability
Table 5. Identified differentially abundant human proteins and their predicted functions that were shared between individuals with combined structural bladder pathology and Schistosoma infection (PS) and Schistosoma infected individuals (SH).
| Protein ID | Identified Human Protein | PEP Scores | Location | Predicted Functions |
|---|---|---|---|---|
| A0A0C4DFZ2 | Arylsulfatase A;Arylsulfatase A component B;Arylsulfatase A Component C | 2.72E+09 | Membrane/ER | Binding |
| Q96S96 | Phosphatidylethanolamine-binding protein 4 | 6.79E+08 | Membrane Associated/ Lysosome | Binding |
| P07858 | Cathepsin B;Cathepsin B light chain;Cathepsin B heavy chain | 1.19E+09 | Membrane | Binding |
PS—Pathology and Schistosomiasis; PEP—Posterior error probability; ID—Identification
Discussion
A total of over 2,000 proteins are estimated to be present in normal human urine [23], and the highest number of human proteins identified in a proteomic study thus far is 1,823 [24]. In the present study, we observed 1,306 proteins in human urine by unfractionated MS analysis. Sample clustering analyses by PCA and heatmap placed all sample groups into clear-cut strata based on the LFQ intensities of the identified proteins, with little interference between groups, thereby indicating a distinct difference in proteomic signatures between groups.
Some of the potential Schistosoma biomarkers identified in this study are clear targets for the generation of new vaccines and drug targets against schistosomiasis. The majority of these proteins appear to be involved in binding activity according to GO-enrichment analysis. This observation is similar to the report of Sotillo et al. [16]. The parasite markers include four heat shock proteins (HSPs) which are known as highly conserved stress-induced proteins found in many trematodes and nematodes including Schistosoma specific study [25]. HSP expression in the earliest stages of intra-mammalian schistosomula development has been reported and was suggested to be as a result of thermal changes in the parasite niche/environment i.e. changes between freshwater and the human body [16, 26]. Venom allergen-like protein (VAL)-3 was identified in the present study. Sotillo et al. [16] reported downregulation of VAL-4 and -6 on the maturing schistosomula tergument and the upregulated expression of VAL-6 in cercaria and adults worm has also been reported [27]. The Schistosomas VALs comprise at least 29 members, subdivided into two major groupings, with group 1 including SmVAL1–5, 7–10, 12, 14–15, & 18–29 which have signal peptides and conserved cysteines positioned for disulphide bond formation. VAL proteins are associated with excretion/secretion products and extracellular environment of the parasite [28] and have been used as a trial vaccine against hookworm infections in humans [29]. Changes in the val gene and the resultant protein expression denotes its functions in different aspects of host-parasite biology, which include snail invasion by miracidium, intra-molluscan sporocyst development, and cercarial development and host penetration [28]. The VAL protein family are abundant in different helminth species including gastrointestinal nematodes, where they are known to carry out several roles in the infective activities of parasites [16, 30].
Actin 1 protein, as reported in this study, has been identified as a possible drug target for the treatment of schistosomiasis. Strong association between actin and S. mansoni adult worm surface membranes has been confirmed [25, 31]. Studies have described the role of actin in enhancing the activity of praziquantel (PQZ) treatment of schistosomiasis. It is suggested that PQZ intercalates in the surface membrane lipid bilayers, thereby inducing tegumental changes that leads to antigen exposure, including actin [31, 32].
Elongation factor 1-alpha, phosphopyruvate hydrase and histone-4 were all identified as potential Schistosoma biomarkers in this report, which parallels the results of deWalick et al. [25], where these proteins were identified in purified eggshell fragments of Schistosoma mansoni. The proteins identified as part of the eggshell protein skeleton are known schistosome antigens and may induce cellular or antibody responses [25]. These eggshell markers may be very useful schistosomiasis diagnostic candidates rather than vaccine candidates, since such a vaccine would be likely to target the eggs and further encourage granuloma formation and pathology rather than priming the immune system against the parasite. The significantly regulated parasite proteins were mostly predicted to be membrane-associated, when classified according to their predicted subcellular location [16, 25]. The expression of some membrane associated proteins was earlier proposed as possible vaccine antigens in different Schistosoma spp [16, 33]. Jossic et al. [34] reported membrane proteins as one of the most interesting classes of proteins among disease biomarker candidates due to their localization on the surface cells and organelles. The identification of a large number of membrane and membrane-associated proteins in the present study strongly suggests that these proteins are abundant in the urine of schistosomiasis patients and would therefore be reasonable targets for drug or vaccine development. These proteins are likely present in the urine as a result of their accessibility to the host immune system via the parasitic tegument, which constitutes the host-parasite interface between S haematobium and the human immune system. Despite some recent clarification regarding common transmembrane tegument proteins in S. mansoni [35], it remains unclear what proteins are abundantly present at this important interface in S. haematobium. An ideal vaccine candidate would present a large extracellular domain, such as the tetraspanin family of proteins, which are a promising candidate for S. mansoni vaccines [36]. The present study therefore offers some insight in to potential membrane protein targets for vaccine development in S. haematobium.
Arylsulfatase A and phosphatidylethanolamine-binding protein 4 were both found in the SH and PS sampled group. Arylsulfatases A, B, and C (arylsulfo-hydrolases) are a group of hydrolytic enzymes that occur in various tissues and fluids [37]. An increase in the activities of arylsulfatase B (ASB) has been reported in bladder tumours [38]. Also, arylsulfatase A (ASA) in the livers of Schistosoma infected mice displayed a non- significant decrease in expression vs the control, while the expression of hepatic ASB was significantly increased in Schistosma infected mice in similar study [37]. Aminophospholipids, such as phosphatidylserine and phosphatidylethanolamine are described as specific, accessible and stable markers of the luminal surface of tumour blood vessels [39]. There has already been some development of aminophospholipid-targeted diagnostic and therapeutic constructs for use in tumour intervention. Antibody-therapeutic agent conjugates and constructs that bind to aminophospholipids, including methods that specifically deliver therapeutic agents, such as toxins and coagulants, to the constitutively-expressed aminophospholipids of tumour blood vessels, thereby inducing thrombosis, necrosis and tumour regression, are particularly promising [39].
One of the four proteins shared by PT and SH samples, plasma retinol-binding protein (RBP), is a circulating plasma protein produced in the liver and adipose tissue that transports active natural metabolites of Vitamin A as retinol around the body [40]. Retinol acts pharmacologically to restore differentiation and inhibit growth in some premalignant and malignant cell both in vivo and in vitro (including bladder cancer cases) and also modulates cell proliferation, malignant transformation, apoptosis and the immune system [40, 41]. A recent study revealed that individuals with HIV and S. mansoni coinfection have significantly lower blood RBP levels when compared to participants with HIV and S. haematobium coinfection [40]. Transthyretin has also been identified by Yi-Ting et al., [42] as a potential urine-derived biomarker for bladder cancer.
The programmed cell death 1 (PD-1) surface receptor binds to two ligands, PD-L1 and PD-L2. PD-1–PD-L interaction is known to control the induction and maintenance of peripheral T cell tolerance. PD-1 and its ligands have been exploited by a variety of microorganisms to reduce the effect of antimicrobial immunity, thereby facilitating chronic infection [43]. The findings of Alvarez et al. [44] on the role played by PD-1 in innate immunity against Mycobacterium tuberculosis also showed that PD-1 signalling might be modulating host innate immunity by inhibiting natural killer (NK) cell responses to the pathogen, contributing to avoidance of immune-mediated pathology caused by excessive host response to the infection. Understanding the functions of PD-1 and its ligands in regulating antimicrobial and self-reactive T cell responses and the possibility of manipulating this pathway may eventually reveal its therapeutic potential in chronic schistosomiasis.
Human Growth/differentiation factor 15 (GDF15) could be a useful diagnostic marker for chronic urinary schistosomiasis. GDF15 is a divergent member of the transforming growth factor β family found in a broad range of cells [45]. Corre et al. [45] reported that GDF15 could be an integrative signal in pathological conditions and provide may information on the severity of disease. Expression and secretion of GDF15 is heightened in many malignant tissue and cancer cell lines (prostate, colorectal, pancreatic, gastric and oral squamous carcinoma) as compared with their normal tissues or cells [46, 47, 48, 49, 50, 51, 52].
Human sialidase protein was identified in the SH group and is known for its immunological role in regulating phagocytosis in macrophages cells [53]. Amith et al. [54] reported Neu1 sialidase as a complex with Toll like receptor (TLR)-2, -3 and -4, which is induced upon ligand binding to either receptor. Activated Neu1 sialidase hydrolyzes sialyl α-2, 3-linked β-galactosyl residues distant from ligand binding to remove steric hindrance to TLR-4 dimerization, MyD88/TLR4 complex recruitment, NFkB activation and pro-inflammatory cell responses [54].
We identified differentially abundant human proteins in each clinical group that may contribute further specificity to a panel of biomarkers. We found a total of 54 proteins that were differentially abundant compared to the control group, with 43 that were specific for schistosomiasis, 8 that were specific to bladder pathology, and 7 that were specific to those patients with bladder pathology and schistosomiasis. Of these, only 37 and 2 proteins were uniquely identified in the SH and PT groups, respectively, while none were unique to the PS group. This implies that the combined pathology of schistosomiasis and bladder pathology may not have uniquely identifiable characteristics, but rather has features common to both contributing diseases. On the other hand, the proteins that are unique to Schistosoma infected individuals or those with bladder pathology are of interest in identifying the molecular mechanisms underlying the pathology of these diseases and may serve as differential biomarkers for diagnostic purposes. Due to the relatively small numbers of unique differentially abundant proteins identified in each clinical group, pathway analysis is not especially informative. The Reactome database identified no statistically significantly dysregulated pathways, although String analysis identified that ‘extracellular exosomes’ were enriched as a cellular component, as well as the Kegg pathway relating to ‘lysosomes’. This suggests that exosomes may have a role in host/pathogen protein trafficking in the urine, which has been of recent interest in other diseases [55].
This study is currently at the discovery phase of identifying schistosome and human based biomarkers for urinary schistosomiasis and its associated pathologies. Actin 1, elongation factor 1-alpha, phosphopyruvate hydrase, heat shock protein, histone-4 and other schistosome-derived proteins identified in this study could be considered as markers for the diagnosis of urinary schistosomiasis. The presence of venom allergen-like protein-3 in the present study confirms its potential as a promising vaccine biomarker against the parasite. The human programmed cell death 1 and Growth/differentiation factor 15 proteins could also be promising markers for the diagnosis of chronic urinary schistosomiasis, a condition that is difficult to identify by microscopy. The consistency of detection of the human proteins arylsufatase and cathepsin across all disease groups (SH, PT and PS) suggests that these markers may be useful in identifying links between schistosomiasis and the development of urinary bladder cancer. We propose that a panel of biomarkers derived from both human and Schistosoma may achieve the best clinical sensitivity and specificity, and this study goes some way to identifying putative candidates for a further quantitative clinical study on a large number of blinded samples.
The reduced complexity of the protein content of urine and its non-invasive sample collection renders urine a valuable source for diagnostic biomarkers, especially for urinary tract diseases. In addition, urine is an ideal biofluid for biomarker discovery by mass spectrometry-based proteomics due to the abundant availability of urine samples and the relative stability of urine proteins. With the use of integrated high throughput technologies, we can begin to elucidate how S. haematobium and human host systems interact during infection. The momentous challenge we face is the possibility of parasite resistance to the only known drug, Praziquantel, and the ongoing problem of continual re-infection within at-risk populations. The comparative proteomics approach undertaken in this study has generated promising hypotheses regarding the mechanisms of pathogenesis that can be tested through manipulation of the host and parasite during infection. This study demonstrates that urinary proteomics is a viable approach to discovering candidate biomarkers for schistosomiasis and its associated pathology, but the results presented here require validation in a larger cohort before clinical applications can be considered.
Supporting information
The study was carried out in two phases (field sampling and laboratory experiments). The abbreviations represent different sample groups namely: SH- S. haematobium infected groups, PT- bladder pathology group, PS- group with combination of pathology and S. haematobium infection and NPS- no pathology and schistosomiasis (control group)
(TIF)
Sample names correspond to the following clinical classifications: TC1-15 = PS, TC16-27 = SH, TC28-39 = PT, and TC40-49 = NPS. SH- S. haematobium infected groups, PT- bladder pathology group, PS- group with combination of pathology and S. haematobium infection and NPS- no pathology and schistosomiasis (control group)
(TIF)
Sample names correspond to the following clinical classifications: TC1-15 = PS, TC16-27 = SH, TC28-39 = PT, and TC40-49 = NPS. SH- S. haematobium infected groups, PT- bladder pathology group, PS- group with combination of pathology and S. haematobium infection and NPS- no pathology and schistosomiasis (control group)
(TIF)
(TIF)
Permutation-based FDR truncation was set at FDR 0.05.
(PPTX)
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006438.
Funding Statement
JMB thanks the South African National Research Foundation (http://www.nrf.ac.za/) for a Research Chair grant. NCS thanks the South African Medical Research Council (http://www.mrc.ac.za/) for a Junior Research Fellowship. BC received a fellowship from the South African Medical Research Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Adenowo AF, Oyinloye BE, Ogunyinka BI, Kappo AP. Impact of human schistosomiasis in sub-Saharan Africa. Braz. J. Infect. Dis. 2015; 19: 196–205. doi: 10.1016/j.bjid.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W.H.O. World Health Organization schistosomiasis fact sheet. Available from: http://www.who.int/mediacentre/factsheets/fs115/en[accessed 2014;10.04.14].
- 3.Nmorsi OPG, Ukwandu NCD, Ogoinja S, Blackie HOT, Odike MAC. Urinary tract pathology in some Schistosoma haematobium infected Nigerians. Afr. J. Biotechnol. 2007; 6:123–127. [PubMed] [Google Scholar]
- 4.Agere IJ. Istifanus WA, Kela SL. Water usage and transmission of schistosoma haematobium in Jalingo and Ardokola Local Government Areas of Taraba State, Nigeria. Nig J Sci Tech and Environ Edu (NIJOSTEE). 2010; 3: 1. [Google Scholar]
- 5.Chitsulo L, Engels D, Montresor A, Savioli L. The global status of schistosomiasis and its control. Acta Trop. 2000; 77:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agbolade OM, Odaibo A. Schistosoma Haematobium Infection Among Pupils, and Snail Intermediate Hosts in Ago- Iwoye; Ogun State, Nigeria. Nig. J. Parasitol. 1996; 17: 1721. [Google Scholar]
- 7.Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 2008; 118:1311–1321. doi: 10.1172/JCI34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritsche HA, Barton HG, Seth PL, Ihor. National Academy of Clinical Biochemistry Guidelines for the Use of Tumor Markers in Bladder Cancer; NACB: Practice Guidelines And Recommendations For Use Of Tumor Markers In The Clinic Bladder Cancer (3H) from pTa tumours. BJU Int. 2006; 90:846–852. [Google Scholar]
- 9.European Urology Association. EAU Guidelines on muscle-invasive and mestastic bladder cancer. 2016
- 10.Onile OS, Awobode HO, Oladele VS, Agunloye AM, Anumudu CI. Detection of urinary tract pathology in some Schistosoma haematobium infected Nigerian Adults. J. Trop. Med. 2016; Article ID 5405207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mostafa MH, Sheweita SA, O’Connor PJ. Relationship between schistosomiasis and bladder cancer. Clin Microbiol Rev. 1999; 12:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driguez P, McManus DP, Gobert GN. Clinical implications of recent findings in schistosome proteomics, Exp. Rev. Prot. 2016. 13:19–33. [DOI] [PubMed] [Google Scholar]
- 13.Botelho M, Ferreia AC, Olivieira MJ, Domingues A, Machado JC, de Costa JM. Schistosoma haematobium total antigen and decreased apoptosis of normal epithelial cells. Int. J. Parasitol. 2010; 39:1083–91. [DOI] [PubMed] [Google Scholar]
- 14.Shiff C, Robert V, Jean N, Joseph Q, Joseph O, William A, et al. Ultrasound verification of bladder damage is associated with known biomarkers of bladder cancer in adults chronically infected with Schistosoma haematobium in Ghana; Trans. Royal Soc. Trop. Med. and Hyg. 2006;100:847–854. [DOI] [PubMed] [Google Scholar]
- 15.Konety BR, Nguyen T, Brenes G, Sholder A, Bastacky S, Potter D, et al. Clinical usefulness of the novel marker BLCA-4 for the detection of bladder cancer. J. Urol. 2000;164:634–639. [DOI] [PubMed] [Google Scholar]
- 16.Sotillo J, Pearson M, Becker L, Mulvenna J, Loukas A. A quantitative proteomic analysis of the tegumental proteins from Schistosoma mansoni schistosomula reveals novel potential therapeutic targets. Int. J. Parasitol. 2015; 45:505–516. doi: 10.1016/j.ijpara.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 17.Adeola HA, Soares NC, Paccez JD, Kaestner L, Blackburn JM, Zerbini LF. Discovery of novel candidate urinary protein biomarkers for prostate cancer in a multiethnic cohort of South African patients via label-free mass spectrometry. Proteomics Clin. Appl. 2015; 9: 597–609. doi: 10.1002/prca.201400197 [DOI] [PubMed] [Google Scholar]
- 18.Theodorescu D, Stefan Wi, Mark MR, Michael W, Mark C, Ingo J, et al. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006; 7:230–240. doi: 10.1016/S1470-2045(06)70584-8 [DOI] [PubMed] [Google Scholar]
- 19.Moore LE, Wiencke JK, Bates MN, Zheng S, Rey OA, Smith AH. Investigation off genetic polymorphisms and smoking in a bladder cancer case-control study in Argentina. Cancer Letters. 2004; 211:199–207. doi: 10.1016/j.canlet.2004.04.011 [DOI] [PubMed] [Google Scholar]
- 20.Rappsilber J, Mann M, Ishihama Y. Protocol for micropurification, enrichment, pre-fractionation and storage of peptides for proteomics using Stage Tips. Nat. Protocols. 2007; 2:1896–1906. doi: 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 21.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014; 13(9): 2513–26. doi: 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005; 21:3674–3676. doi: 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 23.Kalantari S, Jafari, Moradpoor R, Ghasemi E, Khalkhal E. Human Urine Proteomics: Analytical Techniques and Clinical Applications in Renal Diseases. Int. J. Prot. 2015; 782798, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marimuthu A, Meally RNO, Chaerkady R. A comprehensive map of the human urinary proteome. J. Prot. Res. 2011; 10:2734–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.deWalick S, Bexkens ML, van Balkom BWM, Wu Y, Smit CH, Hokke CH, de Groot PG, Heck AJR., Tielens AGM, van Hellemond JJ. The proteome of the insoluble Schistosoma mansoni eggshell skeleton. Int. J. Parasitol. 2011; 41:523–532. doi: 10.1016/j.ijpara.2010.12.005 [DOI] [PubMed] [Google Scholar]
- 26.Devaney E. Thermoregulation in the life cycle of nematodes. Int. J. Parasitol. 2006; 36:641–649. doi: 10.1016/j.ijpara.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 27.Rofatto HK, Parker-Manuel SJ, Barbosa TC, Tararam CA, Alan WR, Leite LC, Farias LP. Tissue expression patterns of Schistosoma mansoni Venom Allergen-Like proteins 6 and 7. Int. J. Parasitol. 2012; 42:613–620. doi: 10.1016/j.ijpara.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 28.Yoshino TP, Brown M, Wu XJ, Jackson CJ, Ocadiz-Ruiz R, Chalmers IW, et al. Excreted/secreted Schistosoma mansoni venom allergen-like 9 (SmVAL9) modulates host extracellular matrix remodelling gene expression. Int. J. Parasitol. 2014; 44:551–563. doi: 10.1016/j.ijpara.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diemert DJ, Pinto AG, Freire J, Jariwala A, Santiago H, Hamilton RG, et al. Generalized urticaria induced by the Na-ASP-2 hookworm vaccine: implications for the development of vaccines against helminths. J. Allergy Clin. Immunol. 2012; 130:e166. [DOI] [PubMed] [Google Scholar]
- 30.Tribolet L, Cantacessi C, Pickering DA, Navarro S, Doolan DL, Trieu A, et al. Probing of a human proteome microarray with a recombinant pathogen protein reveals a novel mechanism by which hookworms suppress B-cell receptor signaling. J. Infect. Dis. 2015; 211:416–425. doi: 10.1093/infdis/jiu451 [DOI] [PubMed] [Google Scholar]
- 31.Hatem T, Rashika E. Praziquantel binds Schistosoma mansoni adult worm actin. Int. J. Antimicrob. Agents. 2007; 29:570–575. doi: 10.1016/j.ijantimicag.2006.12.018 [DOI] [PubMed] [Google Scholar]
- 32.Linder E, Thors C. Schistosoma mansoni: praziquantel induced tegumental lesion exposes actin of surface spines and allows binding of actin depolymerizing factor, gelsolin. Parasitology. 1992; 105:71–9. [DOI] [PubMed] [Google Scholar]
- 33.Da’dara AA, Faghiri Z, Krautz-Peterson G, Bhardwaj R, Skelly PJ,). Schistosome Na, K-ATPase as a therapeutic target. Trans. R. Soc. Trop. Med. Hyg. 2013; 107:74–82. doi: 10.1093/trstmh/trs020 [DOI] [PubMed] [Google Scholar]
- 34.Josic D, Clifton JG, Kovac S, Hixson DC. Membrane proteins as diagnostic biomarkers and targets for new therapies. Curr Opin Mol Ther. 2008; 10:116–23. [PubMed] [Google Scholar]
- 35.Braschi S, Curwen RS, Ashton PD, Verjovski-Almeida S, Wilson A. The tegument surface membranes of the human blood parasite Schistosoma mansoni: a proteomic analysis after differential extraction. Proteomics. 2006; 6:1471–82. doi: 10.1002/pmic.200500368 [DOI] [PubMed] [Google Scholar]
- 36.Loukas A, Tran M, Pearson MS. Schistosome membrane proteins as vaccines. Int. J. Parasitol. 2007; 37:257–263. doi: 10.1016/j.ijpara.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 37.Balbaa M, El-Kersh M, Mansour H, Yacout G, Ismail M, Malky A, et al. Activity of Some Hepatic Enzymes in Schistosomiasis and Concomitant Alteration of Arylsulfatase B. J. Biochem. Mol. Biol. 2004; 37:223–228. [DOI] [PubMed] [Google Scholar]
- 38.Poys LE, Morgan IR. Urine enzyme activities in patients with transitional cell carcinoma of the bladder. Clin. Chim. Acta. 1977; 74:7–10. [DOI] [PubMed] [Google Scholar]
- 39.Thorpe, P. E., Ran, S. and Seattle, B. Methods for imaging tumour vasculature using conjugates that bind to aminophospholipids. United State Patent. 2009; US007550141B2.
- 40.Kotze SR, Zinyama-Gutsire R, Per K, Benn CS, Gomo E, Gerstoft J et al. HIV and schistosomiasis in rural Zimbabwe: the association of Retinol-binding protein with disease progression, inflammation and mortality. Int. J. Infect. Dis. 2015; 33:159–164. doi: 10.1016/j.ijid.2015.01.015 [DOI] [PubMed] [Google Scholar]
- 41.Hameed DA, El-Metwally TH. The effectiveness of retinoic acid treatment in bladder cancer-Impact on recurrence, survival and TGFα and VEGF as end-point biomarkers. Cancer Biology and Therapy. 2008; 7:92-100. [DOI] [PubMed]
- 42.Yi-Ting C, Hsiao-Wei C, Dominik D, Derek SS, Kung-Hao L, Chih-Ching W, et al. Multiplexed quantification of 63 proteins in human urine by multiple reaction monitoring-based mass spectrometry for discovery of potential bladder cancer biomarkers. J. Prot. 2012; 75:3529–3545. [DOI] [PubMed] [Google Scholar]
- 43.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ.The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nature Immunology. 2007; 8:239–245. doi: 10.1038/ni1443 [DOI] [PubMed] [Google Scholar]
- 44.Alvarez IB, Pasquinelli V, Jurado JO, Abbate E, Musella RM, de la Barrera SS, et al. Role Played by the Programmed Death 1– Programmed Death Ligand Pathway during Innate Immunity against Mycobacterium tuberculosis. J. Infect. Dis. 2010; 202: 524–532. doi: 10.1086/654932 [DOI] [PubMed] [Google Scholar]
- 45.Corre J, Hébraud B, Bourin P. Concise Review: Growth Differentiation Factor 15 in Pathology: A Clinical Role? Stem Cells Translational Medicine. 2013; 2:946–952. doi: 10.5966/sctm.2013-0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welsh JB, Sapinoso LM, Su AI. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001; 61:5974–5978. [PubMed] [Google Scholar]
- 47.Lee DH, Yang Y, Lee SJ, et al. ,. Macrophage inhibitory cytokine-1 induces the invasiveness of gastric cancer cells by up-regulating the urokinase-type plasminogen activator system. Cancer Res. 2003; 63:4648–4655. [PubMed] [Google Scholar]
- 48.Buckhaults P, Rago C, St Croix B. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001; 61:6996–7001. [PubMed] [Google Scholar]
- 49.Koopmann J, Zhang Z, White N, et al. Serum diagnosis of pancreatic adenocarcinoma using surface-enhanced laser desorption and ionization mass spectrometry. Clin Cancer Res. 2004; 10:860–868. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Yang X, Pan HY). Expression of growth differentiation factor 15 is positively correlated with histopathological malignant grade and in vitro cell proliferation in oral squamous cell carcinoma. Oral Oncol. 2009; 45:627–632. doi: 10.1016/j.oraloncology.2008.07.017 [DOI] [PubMed] [Google Scholar]
- 51.Park YJ, Lee H, Lee JH). Macrophage inhibitory cytokine-1 transactivates ErbB family receptors via the activation of Src in SK-BR-3 human breast cancer cells. BMB Rep. 2010; 43:91–96. [DOI] [PubMed] [Google Scholar]
- 52.Rothman N, Garcia-Closas M, Chatterjee N. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 2010; 42:978–984. doi: 10.1038/ng.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seyrantepe V. Regulation of Phagocytosis in Macrophages by Neuraminidase 1. J. Biol. Chem. 2010; 285:206–215. doi: 10.1074/jbc.M109.055475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amith SR, Jayanth P, Finlay T, Franchuk S, Gilmour A, Abdulkhalek S, et al. Detection of Neu1 Sialidase Activity in Regulating TOLL-like Receptor Activation. J. Vis. Exp. 2010; 43: e2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hosseini HM, Fooladi AA, Nourani MR, Ghanezadeh F. The role of exosomes in infectious diseases. Inflamm. Allergy Drug Targets. 2013; 12:29–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The study was carried out in two phases (field sampling and laboratory experiments). The abbreviations represent different sample groups namely: SH- S. haematobium infected groups, PT- bladder pathology group, PS- group with combination of pathology and S. haematobium infection and NPS- no pathology and schistosomiasis (control group)
(TIF)
Sample names correspond to the following clinical classifications: TC1-15 = PS, TC16-27 = SH, TC28-39 = PT, and TC40-49 = NPS. SH- S. haematobium infected groups, PT- bladder pathology group, PS- group with combination of pathology and S. haematobium infection and NPS- no pathology and schistosomiasis (control group)
(TIF)
Sample names correspond to the following clinical classifications: TC1-15 = PS, TC16-27 = SH, TC28-39 = PT, and TC40-49 = NPS. SH- S. haematobium infected groups, PT- bladder pathology group, PS- group with combination of pathology and S. haematobium infection and NPS- no pathology and schistosomiasis (control group)
(TIF)
(TIF)
Permutation-based FDR truncation was set at FDR 0.05.
(PPTX)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006438.