Abstract
Methionine adenosyltransferases (MATs) are essential for cell survival because they catalyze the biosynthesis of the biological methyl donor S-adenosylmethionine (SAMe) from methionine and adenosine triphosphate (ATP). Mammalian cells express two genes, MAT1A and MAT2A, which encode two MAT catalytic subunits, α1 and α2, respectively. The α1 subunit organizes into dimers (MATIII) or tetramers (MATI). The α2 subunit is found in the MATII isoform. A third gene MAT2B, encodes a regulatory subunit β, that regulates the activity of MATII by lowering the inhibition constant (Ki) for SAMe and the Michaelis constant (Km) for methionine. MAT1A expressed mainly in hepatocytes maintains the differentiated state of these cells whereas MAT2A and MAT2B are expressed in non-parenchymal cells of the liver (hepatic stellate cells [HSCs] and Kupffer cells) and extrahepatic tissues. A switch from the liver-specific MAT1A to MAT2A has been observed during conditions of active liver growth and de-differentiation. Liver injury, fibrosis, and cancer are associated with MAT1A silencing and MAT2A/MAT2B induction. Even though both MAT1A and MAT2A are involved in SAMe biosynthesis, they exhibit distinct molecular interactions in liver cells. This review provides an update on MAT genes and their roles in liver pathologies.
Keywords: Methionine adenosyltransferases, S-adenosylmethionine, Liver injury, Hepatocellular carcinoma
1.Introduction
Methionine adenosyltransferase (MAT) enzymes are essential for the biosynthesis of S-adenosylmethionine (SAMe), a biological methyl donor required for the methylation of nucleic acids, phospholipids, histones, biogenic amines, and proteins. SAMe biosynthesis is impaired in patients with chronic liver disease because of the inactivation of hepatic MAT. Experiments in mice showed that chronic hepatic SAMe depletion induced the spontaneous development of steatohepatitis and hepatocellular carcinoma (HCC). More recent research has also shown that MAT proteins have distinct interactomes and altered MAT gene expression can contribute to liver injury and cancer. In this review, we provide a detailed overview of MAT genes and enzymes, as well as their regulation and dysregulation that occur in normal and diseased livers.
2. Methionine adenosyltransferases
2.1.MAT expression and structure
SAMe is a methyl donor required for most methylation reactions in mammalian cells.1 MAT (E.C.2.5.1.6) catalyzes reactions involving transfer of the adenosyl moiety of adenosine triphosphate (ATP) to methionine to form SAMe.2 Three distinct MAT genes, MAT1A, MAT2A, and MAT2B encode the protein products, MATα1, MATα2, and MATβ respectively.3 MATα1 is a catalytic subunit of MAT isoenzymes that is mainly expressed in liver (mostly hepatocytes) and pancreatic acinar cells.4 It is a 396 amino acid protein that forms wither a homodimer (MATIII) or a homotetramer (MATI).3 The MATα2 catalytic subunit (395 amino acids) and the MATβ regulatory subunit (334 amino acids) are expressed in extrahepatic tissues but are also expressed within the liver in hepatic stellate cells (HSCs) and Kupffer cells.5,6 MATα2 and MATβ interact with each other to form the MATII isoenzyme. Isothermal titration calorimetry analysis and crystallography showed that the MATα2 and β subunits interacted in a 2:1 stoichiometric ratio either as [MAT(α2)2(β)1] or [MAT(α2)4(β)2] complexes.7,8 MATβ regulates activity of the MATII complex by lowering the inhibition constant (Ki) for the product, SAMe, and the Michaelis constant (Km) for the substrate, methionine.9 Of the four MAT2B mRNA variants described, the two main splicing forms are MAT2B-V1 and MAT2B-V2. V1 is the same as MATβ while the V2-encoded protein differs from V1 in the first 20 amino acids at the N-terminus.10
2.2.MAT subcellular localization
Originally MAT enzymes were considered cytosolic proteins and SAMe produced from MAT activity was thought to be delivered to specific compartments such as the nucleus for DNA and histone methylation reactions.11 However, the demonstration of tetrameric forms of MATI/III isoenzymes in the nucleus of hepatocytes suggested that MAT activity in this compartment provided a local source of SAMe for specific methylation reactions.12 Localization of active MATα1 protein in the nucleus correlated with the induction of histone H3K27 trimethylation, a known epigenetic modification that causes DNA methylation and gene repression.12 Monomeric MATα1 is also found in nuclei and may be involved in other interactions.12,13 Both MATα2 and MATβ subunits of the MATII isoenzyme have been detected in the cytoplasm and nucleus of mammalian cells.14 Distinct functional interactions of these proteins exist based on their subcellular localization.15
2.3.MAT transcriptional and post-transcriptional regulation
2.3.1.MAT1A transcriptional/epigenetic control
The MAT1A promoter contains consensus binding sites for glucocorticoid response elements (GRE), hepatocyte nuclear factor (HNF), interleukin-6 (IL-6), activator protein 1 (AP-1), CCAAT enhancer binding protein (C/EBP) and one or more sites for cyclic AMP response element binding protein (CREBP), E2F, signal transducers and activators of transcription (STAT), c-Myc and v-Myb.16 Even though some of these factors, such as HNF and C/EBP, are determinants of liver-specific gene expression and C/EBP controls MAT1A expression by promoter regulation,16,17 the MAT1A promoter appears to be active in non-liver cell lines such as the Chinese hamster ovary cell line indicating that its liver-specific expression is not controlled by these transcription factors and other mechanisms of transcriptional control exist for this gene.18
In normal liver, the MAT1A gene is epigenetically upregulated by hyperacetylation and cytosine hypomethylation. HepG2 cells treated with 5-aza-2′-deoxycytidine, a demethylating agent or with a histone deacetylase inhibitor exhibited enhanced MAT1A expression. A 750 base pair (bp) region upstream of the transcriptional start site of MAT1A is a site for these epigenetic modifications.19 Hypermethylation of the Mat1a promoter leading to gene silencing was observed under conditions of hepatocellular damage or abnormal proliferation such as chemically induced-liver cirrhosis and in the livers of F344 rats genetically predisposed to hepatocarcinogenesis.19,20 Coding region methylation at sites +10 and +88 relative to the transcription start site were also reported to downregulate MAT1A transcription in human HCC cell lines.21 Importantly, lower MAT1A mRNA levels and hypermethylation of the MAT1A promoter and coding regions were reported in patients with advanced non-alcoholic fatty liver disease (NAFLD with fibrosis score 3-4).22
2.3.2.MAT1A post-transcriptional control
The stability of Mat1a mRNA is negatively regulated by the binding of AU-rich RNA binding factor (AUF1) to its 3′-untranslated region. In differentiated rat hepatocytes, low levels of AUF1 are associated with increased Mat1a expression and the de-differentiation of hepatocytes in culture increases AUF1 levels with a concomitant decrease in Mat1a mRNA levels.23 MAT1A mRNA levels are also regulated by microRNA (miR) in HCC.24,25 Preneoplastic liver lesions induced by 2-acetylaminofluorene injection in rats induced miR-22 and mir-29b that inhibited Mat1a mRNA expression.24 MicroRNAs miR-485-3p, miR-495, and miR-664 are induced in HCC and induce the LIN28B component of the LIN28B/Let-7 axis (where LIN28B indicates lin-28 homolog B [Caenorhabditis elegans], a protein over-expressed in HCC that represses the tumor suppressor, Let-7).25 These microRNAs downregulate MAT1A expression, resulting in lower nuclear SAMe levels, hypomethylation of the LIN28B promoter region and increased LIN28B expression.25 Blocking the expression of these miRs recovered MAT1A expression, inhibited growth, induced apoptosis in HCC cell lines, and inhibited HCC growth in vivo.25
2.3.3.MAT2A/MAT2B transcriptional/epigenetic control
MAT2A transcription is upregulated during liver regeneration and in HCC.26-28 Transcription factors, specificity protein 1 (Sp1), c-Myb, and E2F upregulate MAT2A promoter activity.26 Tumor necrosis factor-a (TNF-a) induced the MAT2A promoter via nuclear factor-KB (NF-κB) and AP-1 elements present in the MAT2A promoter.29 A hypoxic tumor environment can also induce MAT2A expression because hypoxia-inducible factor-1α (HIF-1α) binds to a consensus binding site in the MAT2A promoter and activates its transcription in hepatoma cells.30 The Mat2a promoter upstream regulatory region contains several PPAR response elements (PPRE) that bind to nuclear receptors including peroxisome-proliferator activated receptors (PPAR) in rat HSCs.31 PPARγ is a marker of HSC quiescence in the normal liver and PPARβ is induced in activated HSCs during liver fibrogenesis.32,33 Both PPARγ and PPARβ occupy the same binding site on the Mat2a promoter.31 In quiescent HSCs, PPARγ acts as a negative regulator of Mat2a transcription by binding to the PPRE. However, during HSC activation, a dramatic reduction in PPARγ expression and activity allows the positive regulator, PPARβ to bind to the Mat2a PPRE and induce the expression of this gene.31 The Mat2a promoter also exhibits epigenetic regulation— it is hypomethylated in HCC, hypermethylated in the normal liver, and histone acetylation favors MAT2A expression in HCC.27
2.3.4.MAT2A/MAT2B post-transcriptional control
MAT2A mRNA stability is influenced by the binding of human RNA-binding (HuR) protein and its methylated form, methyl-HuR.23 HuR is an mRNA stabilizer whereas methyl-HuR destabilizes target mRNAs.23 During HCC and hepatocyte de-differentiation, HuR induction is associated with a decline in methyl-HuR resulting in a higher HuR/methyl-HuR ratio. HuR binding to MAT2A mRNA stabilizes its expression in HCC and in de-differentiated hepatocytes.23
In human HepG2 cells, MAT2B-V1 mRNA but not MAT2B-V2 mRNA is transcriptionally upregulated by TNF-α through an AP-1 and NF-KB-dependent mechanism.10 MAT2B transcription is also induced by an NAD+-dependent deacetylase called Sirtuin 1.34 Similar to MAT2A, HuR also stabilizes MAT2B mRNA in liver cancer cells.34
In HepG2 cells, MAT2A and MAT2B mRNA stability is also controlled by drug-induced miRs such as mir-21-3p. The over-expression of mir-21-3p and its induction by the anticancer drug, berberine, induced apoptosis and inhibited growth by down-regulating MAT2A and MAT2B.35
2.4. MAT post-translational modifications
Post-translational modifications of MAT enzymes such as nitrosylation, phosphorylation, and sumoylation influence their activity and stability. Nitrosylation or oxidation of the cysteine 121 residue in a flexible loop over the active site cleft of MATα1 protein inactivates the enzyme.36,37 Antioxidants such as glutathione and other thiol-reducing agents can prevent this inactivation.38 The phosphorylation of MATI/III by protein kinase C at threonine 342 was described over 20 years ago.39 This post-translational modification does not alter the kinetic parameters of the enzyme. However, dephosphorylation of the T-342 site by alkaline phosphatase lowers the activity of both MATI and MATIII.39
Post-translational modifications in MATα2 and MATβ were recently reported during HSC activation. HSCs are vitamin-A storing cells of the liver that become activated and proliferative during liver injury and fibrogenesis.40 The phosphorylation of both MATα2 and MATβ is strongly induced during HSC activation.41 Phospho-MATα2 and MATβ proteins are highly stable and the mutation of specific phosphorylation sites (Y371/Y374 in MATα2 and T257/Y259 in MATβ) inhibits HSC activation.41 Chemical inhibitors, gene silencing and in vitro kinase assays have shown that mitogen-activated protein kinase/ERK kinase (MEK) might be involved in MATα2 phosphorylation whereas extracellular signal-regulated kinase (ERK) might phosphorylate the MATβ protein.41
Sumoylation is a post-translational modification that involves the conjugation of proteins with a small ubiquitin modifier (SUMO) leading to alterations in protein stability, activity, and localization.42 SUMO-1 conjugation of proteins is generally associated with protein stabilization.42 Three SUMO-1 modifications of MATα2 at K340, K372, and K394 were recently shown to enhance its stability and its interactions with oncoproteins such as B-Cell CLL/lymphoma 2 (BCL-2).43
P300 (E1A binding protein) acetylates MATα2 at the K81 residue causing its destabilization, whereas a lack of this modification stabilizes the protein and is associated with HCC development.44
Table 1 summarizes the known regulators of MAT genes and proteins.
Table 1. MAT gene/protein regulators.
| MAT gene | MAT subunit encoded | Transcriptional regulators | Post-transcriptional regulators | Post-translational modifications |
|---|---|---|---|---|
| MAT1A | MATα1 | C/EBP | AUF1, miR-22, miR-29b, miR-485-3p, miR-495, miR-664 | Nitrosylation Phosphorylation |
| MAT2A | MATα2 | Sp1, c-Myb, E2F, TNF-α, HIF-1α, PPARγ, PPARβ | HuR, methyl-HuR, miR-21-3p | Sumoylation Phosphorylation Acetylation |
| MAT2B | MATβ | TNF-α, Sirtuin 1 | HuR, miR-21-3p | Phosphorylation |
MAT, methionine adenosyltransferase; C/EBP, CCAAT enhancer binding protein; AUF1, AU-rich RNA binding factor; miR, microRNA; Sp1, specificity protein 1; TNF-α, tumor necrosis factor-α; HIF-1α, hypoxia-inducible factor-1α; PPAR, peroxisome-proliferator activated receptors; HuR, human RNA-binding
2.5.MAT activity and liver function
MAT isoenzymes have different kinetic and regulatory properties. The Km for the substrate, methionine, is lowest for MATII, followed by MATI and is highest for MATIII.45 The product of MAT activity, SAMe, is a feedback inhibitor of certain MAT isoenzymes at specific concentrations.46 MATII has high sensitivity to SAMe inhibition, with a 50% inhibitory concentration (IC50) of 60 μM, the normal physiological level of SAMe in the liver. However, MATI is minimally inhibited by SAMe (IC50 = 400 μM) and MATIII is stimulated at high SAMe levels (eight-fold at 500μM SAMe levels).46 Therefore, MATI/III isoenzymes maintain high SAMe levels in the liver (6-8 g/day compared with MATII that insignificantly contributes to this SAMe pool).18 A switch from MAT1A to MAT2A is associated with liver de-differentiation, reduced SAMe biosynthesis, and favoring proliferative signaling in the liver.15 This is because high SAMe levels inhibit the mitogenic effect of growth factors.45 A summary of the MAT structure and association with liver dysfunction is shown in Table 2.
Table 2. MAT enzyme structure, properties and associations with liver dysfunction.
| MAT gene | MAT enzyme complex | Catalytic subunit | Regulatory subunit | Feedback inhibition by SAMe | Effect on cell growth | Liver de-differentiation | HCC |
|---|---|---|---|---|---|---|---|
| MAT1A | MATI | MATα1 | None | Minimal (IC50=400μM) | Inhibitor | Low | Down-regulated |
| MAT1A | MATIII | MATα1 | None | None | Inhibitor | Low | Down-regulated |
| MAT2A/MAT2B | MATII | MATα2 | MATβ | High (IC50=60μM) | Activator | High | Induced |
MAT, methionine adenosyltransferase
Deregulation of MAT1A and MAT2A genes alters SAMe homeostasis and is an important determinant of liver injury, fibrosis, and HCC. However, apart from alterations in SAMe metabolism, the MAT catalytic (MATα1 and MATα2) and regulatory (MATβ) subunit proteins were recently shown to exhibit distinct interactions with key signaling molecules in normal and diseased liver. These interactions are described below.
3.MATs and liver dysfunction
3.1.Liver injury/fatty liver/fibrosis/cirrhosis
3.1.1.Epigenetic changes in MAT1A during liver cirrhosis and NAFLD
Methionine metabolism is impaired in patients with chronic liver disease and patients with hepatic cirrhosis exhibited reduced MAT1A expression and MATI/III activity, as well as impaired methionine clearance.47,48 Hypermethylation of the MAT1A promoter might be responsible for the reduced expression of MAT1A during cirrhosis.48 Patients with advanced NAFLD exhibit MAT1A hypermethylation and lower MAT1A mRNA levels compared to patients with mild NAFLD and normal subjects.22 Three CpG islands, two upstream of the transcription start site and one 200 bp downstream of the transcription start site, were hypermethylated in advanced NAFLD subjects.22 These findings in NAFLD subjects are consistent with a report of MAT1A coding region methylation around +88 bp from the transcription start site causing decreased MAT1A transcription in human HCC cells. 21
3.1.2.Inhibition of MATI/III enzymes during liver injury
Oxidative stress caused by alcohol consumption, toxin exposure, septic shock, viral hepatitis, and inflammatory responses mediated by TNF-α and IL-6 inactivated the MATI/III enzyme.18 Increased amounts of reactive oxygen species and NO (caused by high nitric oxide synthase activity) in cirrhotic livers inactivated the MATI/III enzyme.36,37 The loss of MATI/III activity was also observed in experimental models of liver injury such as alcohol feeding in baboons,49 carbon tetrachloride (CCl4) in rats,50 paracetamol in rats,51 and buthionine and sulfoximine intoxication in rats.38
3.1.3.Consequences of MAT1A deficiency
The mechanisms of MAT1A deregulation in liver disease have been extensively studied using the Mat1a-knockout (KO) mouse model (Mat1a-KO).52 Three-month-old Mat1a-KO mice develop hepatic hyperplasia and are susceptible to steatosis in response to a choline-deficient diet. Eight-month-old Mat1a-KO animals spontaneously develop steatohepatitis on a normal diet and by 18 months they develop HCC.52 The livers of Mat1a-deficient mice exhibit increased oxidative stress caused by low glutathione levels and increased activity of cytochrome P450 family 2 subfamily E member 1 (CYP2E1) enzyme,53 which facilitates the release of reactive oxygen species during the metabolism of hepatotoxins such as alcohol, CCl4, and acetaminophen.54 Furthermore, Mat1a-KO mice are highly sensitive to CCl4-mediated liver injury compared to wild type littermates.53 Mitochondrial dysfunction is also evident in Mat1a-deficient mice. Regarding the mechanism involved, Mat1a deficiency leads to a depletion of the mitochondrial chaperone, prohibitin 1 (PHB1), in the liver,55 which is consistent with the finding that PHB1-deficient mouse livers exhibit increased injury, mitochondrial damage, and oxidative stress.56
3.1.4.MAT2A/MAT2B deregulation by factors promoting fibrogenesis
CCl4-induced liver fibrosis leads to the induction of Mat2a mRNA levels.57 Increased levels of MAT2B-encoded MATβ proteins are detected in human cirrhotic liver versus normal controls.58 We and others have shown that rat HSCs, fibrogenic cells of the liver, express Mat2a and Mat2b genes but do not express Mat1a.6,59 Chronic liver injury leads to HSC activation (production of excessive extracellular matrix components), loss of vitamin A, and proliferation.60 In turn, HSC activation in rats induces Mat2a and Mat2b mRNA levels.31,59 Even though the individual MATα2 and MATβ subunits are induced during HSC activation, the MATII enzyme responsible for SAMe production in HSCs exhibits reduced activity. We speculate that this reduction in activity might be caused by an increase in the MATβ:MATα2 ratio, which would lower the Ki of MATII. This is consistent with a decrease in SAMe levels and global DNA hypomethylation during HSC activation.59 Silencing studies have provided mechanistic insights of the role of MAT2A and MAT2B during HSC activation. Silencing MAT2A lowered SAMe levels in activated HSCs decreasing proliferation and increasing apoptosis.59 These findings indicate that MAT2A is required to maintain the intracellular SAMe pool necessary for cell proliferation. Silencing MAT2B in HSCs inhibits activation but in a manner independent of changes in SAMe levels. Interestingly, the MATβ protein promotes HSC activation by inducing two pro-fibrogenic and pro-survival signaling pathways, ERK and phosphatidyl inositol 3 kinase (PI3K). 59 Recently, the regulation of MAT2A and MAT2B genes was studied in primary human HSCs. Unlike rat HSCs, human MAT2A and MAT2B are not controlled transcriptionally but their protein products are subject to post-translational modifications during HSC activation.41 The MATα2 and MATβ subunits are stabilized by phosphorylation during HSC activation, which favors the transition of quiescent HSCs to the activated state. 41
3.2.MATs and liver cancer
3.2.1.MAT1A and liver cancer
The Mat1a-KO mouse model has provided important mechanistic insights of how MAT1A might influence HCC development. Mechanisms of liver cancer development caused by Mat1a deficiency are described below.
3.2.2.Liver cancer stem cells and sustained ERK activation
Mat1a-KO livers contain increased populations of liver cancer stem cells or CD133+/CD49f+ oval cells, which are tumorigenic in nature, and exhibit deregulated ERK activation and increased oncogenic signals (K-ras and survivin).61,62 Sustained ERK activation in these cells allows them to be resistant to the apoptotic effects of transforming growth factor-β (TGF-β), a growth regulator of hepatocytes.62,63 Uncontrolled ERK activation is associated with highly aggressive forms of HCC. 64 In the normal liver, ERK activity is controlled by dual-specificity phosphatase 1 (DUSP1). Transient ERK activation activates DUSP1, which feeds back to regulate ERK and control pro-survival signaling.65 Indeed, low levels of DUSP1 mRNA and protein are one cause of deregulated ERK signaling upon Mat1a deficiency. Exogenous SAMe administration in Mat1a-KO mice normalized DUSP1 levels in the liver.66
3.2.3.Genomic instability
Because MAT1A influences DNA methylation via SAMe levels, a deficiency in MAT1A leading to global DNA hypomethylation is also associated with genomic instability.67 Genomic instability is highly prevalent in HCC and is a well-known prognostic marker.68 In normal cells, genomic instability is controlled by the DNA repair system consisting of the Apurinic/Apyrimidinic Endonuclease 1 (APEX1) protein.69 Mat1a-KO livers and primary de-differentiated mouse hepatocytes with low Mat1a expression and low SAMe levels exhibited a strong downregulation of APEX1.70 SAMe replenishment in culture de-differentiated hepatocytes stabilizes APEX1 proteins. Therefore, Mat1a/SAMe deficiency promotes genomic instability by destabilizing the APEX1 component of the DNA repair system.
3.2.3.LKB1-AMPK signaling
Mat1a-KO liver also exhibits an induction of the LKB1-AMPK signaling pathway. Liver kinase B1 (LKB1) phosphorylates and activates AMP-activated protein kinase (AMPK) under the conditions of nutrient deprivation and low ATP levels.71 In hepatocytes, AMPK activation increases the cytoplasmic content of HuR, an mRNA stabilizing protein, which in turn stabilizes several cyclin mRNAs leading to cell growth.72 Mat1a deficiency increases both LKB1 and AMPK activity and exogenous SAMe treatment blocks AMPK activation by mediating its dephosphorylation through protein phosphatase 2A.72 Enhanced LKB1 activity was observed in SAMe-deficient cell lines (SAMe-D) derived from Mat1a-KO livers and in human HCC tissues, and is required for the survival of SAMe-D cells.73
3.2.4.Cholangiocarcinoma (CCA) development
MATα1 was recently shown to interact with several transcription factors such as c-Myc, c-MAF, and MAFG to influence liver oncogenesis. C-Myc is a well-known oncogene whose induction is important during cholestatic liver injury and the progression of CCA in a mouse model of cholestasis-associated CCA.74 MAF proteins are activator protein 1 family members that regulate antioxidant stimuli by binding in a homodimeric or heterodimeric state to antioxidant response elements (ARE) on promoters.75 However, their role in CCA progression is unknown. MATα1 and c-MYC/MAFG/c-MAF proteins were recently shown to interact directly with each other at the E-box elements that control their promoters.76 Interestingly, the E-box element in MAT1A is a repressor but it is an enhancer in c-Myc/MAFG/c-MAF promoters. Furthermore, MATα1 suppresses E-box, whereas c-Myc/MAFG/c-MAF activates E-box-driven promoter activity.76 This results in reciprocal regulation between MAT1A and c-Myc/MAFG/c-MAF. MAT1A is expressed at high levels in normal bile duct epithelial cells and is repressed in CCA where MAFG and c-MAF are induced. These changes favor c-MYC induction as well as CCA proliferation because both of these effects are suppressed by the over-expression of MAT1A or by the suppression of MAFG/c-MAF expression (Fig.1). 76
Fig.1.
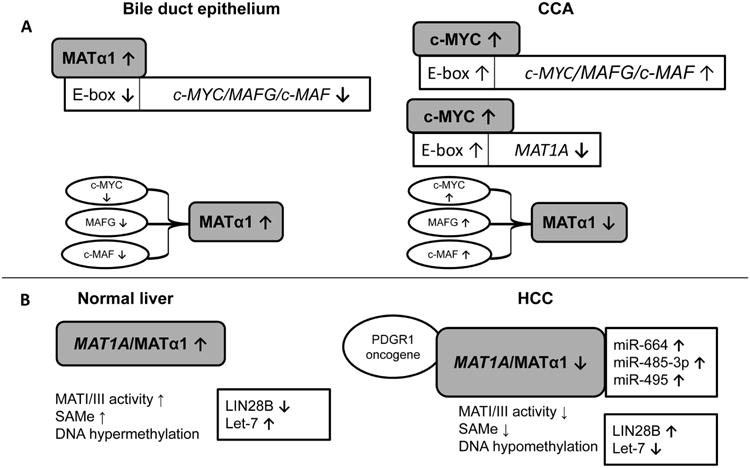
Summary of MAT1A expression and MATα1 interactions in normal and diseased liver. A. MAT1A is highly expressed in normal bile duct epithelial cells but downregulated in CCA. MATα1 interacts with c-MYC, MAFG, and c-MAF, which exhibit the opposite expression pattern to MAT1A. High MATα1 levels in normal bile duct epithelium inhibits E-box-driven c-MYC, MAFG, and c-MAF promoter activity whereas high c-Myc levels in CCA inhibit MAT1A but promote c-MAF and MAFG promoter activities. B. MAT1A expression, regulation, and interaction in normal liver and HCC. High MAT1A expression and MATI/III activity is associated with high SAMe levels, DNA hypermethylation, induction of the tumor suppressor, Let-7 and inhibition of the tumor promoter, LIN28B. During HCC, the microRNA-mediated inhibition of MAT1A expression and loss of MATα1 activity caused by PDGR1 binding leads to a depletion of SAMe, global DNA hypomethylation, induction of LIN28B and inhibition of Let-7.
MAT, methionine adenosyltransferase; c-MYC, myelocytomatosis viral oncogene homolog MAF, musculoaponeurotic fibrosarcoma oncogene; CCA, cholangiocarcinoma; PDGR1, p53 And DNA Damage Regulated 1; LIN28B, lin-28 homolog B [Caenorhabditis elegans]; Let-7, miR-664, miR-485-3p, miR-495 are microRNAs.
3.2.5.Altered MATα1 activity and DNA hypomethylation in liver injury and hepatoma
Recently, the interaction between MATα1 and p53 And DNA Damage Regulated 1 (PDRG1) protein was reported in hepatoma cells, which exhibited decreased MATα1 expression and increased nuclear MATα1 monomers and tetramers.13 PDRG1 is an oncogene that is upregulated in cancers of the bladder, breast, and colon.77 When PDGR1 binds to nuclear MATα1 there was a decrease in MAT activity and DNA was hypomethylated. However, the molecular mechanism of how this interaction affects the downstream functions of MAT has not been further investigated. The associations of MAT1A/MATα1 with liver dysfunction are summarized in Table 2 and Fig.1.
3.3.MAT2A/MAT2B and liver cancer
A switch from MAT1A to MAT2A/MAT2B was reported during liver de-differentiation and HCC.45 The expressions of MAT2A and MAT2B are induced in response to the stimulation of liver cancer cells with growth factors such as leptin and hepatocyte growth factor. 78,79 In the last several years, novel interactions of MATα2 and MATβ as independent proteins were reported, indicating they might be potential therapeutic targets. Mechanisms of liver cancer development caused by MAT2A/MAT2B dysregulation are described below.
3.3.1.MAT2A and growth signaling
Silencing MAT2A prevents the pro-survival signaling of leptin by decreasing intracellular SAMe and limiting polyamine biosynthesis78, which is essential for growth.18 Cross talk between MATα2 and polyamine biosynthesis was recently reported in liver cancer cells. The induction of MATα2 enhances polyamine biosynthesis and growth, while increases in polyamines promote MAT2A transcription by a feed-forward mechanism that involves AP-1.80 Recently, MATα2 sumoylation was shown to be a post-translational mechanism that promoted its stability and interaction with the well-known oncoprotein and apoptotic regulator protein, BCL-2, in liver cancer cells (Fig.2). 43 This interaction stabilized BCL-2 proteins. MATα2 also acts as a transcription factor that enhances activity of the BCL-2 promoter.43 SUMO-stabilized MATα2 confers chemoresistance to liver cancer cells by preventing apoptosis mediated by 5-fluorouracil.43 Other post-translational modifications of MATα2 such as acetylation and ubiquitination are important in HCC. In human HCC, the induction of MATα2 is associated with a significant reduction in p300 (E1A binding protein)-mediated K81 acetylation and subsequent ubiquitin protein ligase E3 component n-recognin 4 (UBR4)-mediated ubiquitination/degradation.44 High folate concentrations promoted proliferation in several cancers (ovarian, endometrial, colorectal, kidney, lung, and breast carcinomas).81 Folate deprivation enhances the association of MATα2 with p300 and UBR4, leading to both K81 acetylation and ubiquitination, resulting in its proteasomal degradation. Folate stabilizes MATα2 by dissociating the MATα2-P300-UBR4 complex, and promoting MATα2 interactions with histone deacetylase (HDAC3). Accumulated MATα2 protein facilitates rapid cell proliferation and promotes tumor growth.44 In HCC tissues, a decrease in MATα2 K81 acetylation was associated with an increased expression of HDAC3 compared with normal tissues, thereby proving the relevance of this interaction in liver cancer (Fig.2).44
Fig.2. Summary of MATα2/MATβ interactions in normal and diseased liver.
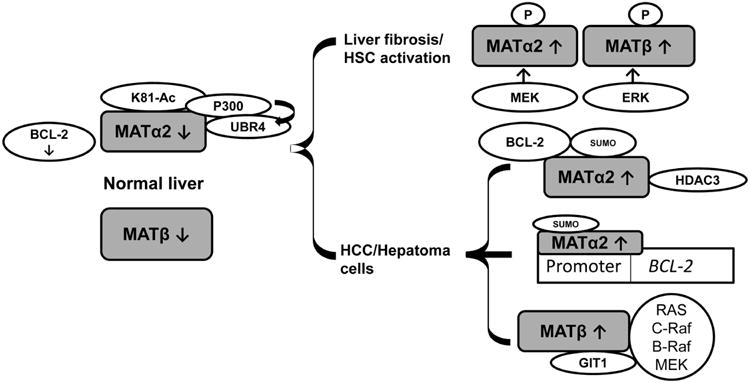
MATα2 and MATβ are expressed at low levels in normal liver. In normal liver, MATα2 acetylation (Ac) at K81 mediated by p300 leads to ubiquitination and degradation by UBR4. In liver disease, two mechanisms of MATα2/MATβ stabilization are associated with highly proliferative phenotypes. In liver fibrosis and HSC activation, MATα2 and MATβ are phosphorylated via MEK/ERK, respectively, leading to their stabilization. In HCC and hepatoma cells, the acetylation of MATα2 at K81 is prevented by HDAC3 binding/deacetylation. MATα2 is sumoylated in HCC, interacts and stabilizes BCL-2 proteins, and stimulates the BCL-2 promoter. MATβ interacts with GIT1 during HCC and recruits RAS/C-Raf/B-Raf and MEK to promote MAPK signaling and growth.
MAT, methionine adenosyltransferase; UBR4, ubiquitin protein ligase E3 component n-recognin 4; MEK, mitogen-activated protein kinase/ERK kinase; ERK, extracellular signal-regulated kinase; HDAC3, histone deacetylase 3; BCL-2, B-Cell CLL/lymphoma 2; GIT1, G-protein-coupled receptor kinase-interacting protein 1; Raf1/C-Raf/B-Raf, Murine Leukemia Viral Oncogene Homolog, protooncogene; MAPK, mitogen-activated protein kinase; SUMO, small ubiquitin modifier; P, phosphorylated protein.
3.3.2.MAT2B and growth signaling
The MATβ protein was originally thought to be a regulatory subunit of MATII that lowered the Ki for the product, SAMe, and the Km for the substrate, methionine.9 However, in recent years, several interactions of MATβ were reported to be independent of its association with MATα2. The MATβ protein provides a proliferative advantage to liver cancer cells because knockdown of the MAT2B gene inhibited DNA synthesis and growth in HCC cell lines.58,78 The mechanism by which MATβ induces growth involves the induction of pro-survival signaling in liver cancer cells. Indeed, silencing MAT2B inhibited activation of the mitogen-activated protein kinase (MAPK) pathway/ERK phosphorylation induced by the growth factor leptin, in HCC cell lines.78 MATβ also induced activation of the PI3K/AKT phosphorylation pathway in response to leptin stimulation.78 Because the ERK/MATβ interaction appears to favor cell proliferation, further studies to provide mechanistic insights into how this system functions in liver cancer cells have been undertaken. Recent work from Peng et al. has shown that the MATβ protein is a scaffolding partner of G-protein-coupled receptor kinase-interacting protein 1 (GIT1) (Fig.2).82 The MATβ-GIT1 complex efficiently binds to the MAPK components, mitogen-activated protein kinase/ERK kinase (MEK) and ERK and causes their activation. In the classical pathway of MAPK activation, signal transduction from Raf kinases (C-Raf, B-Raf or A-Raf) to MEK induces the phosphorylation and activation of MEKs.83 MATβ-GIT1 activates and recruits C-Raf and B-Raf to MEK in liver cancer cell lines to promote MEK activation.84 In addition, B-Raf and C-Raf form homodimers and heterodimers with enhanced activity compared to monomeric states.85 The MATβ-GIT1 complex also induces heterodimerization between B-Raf and C-Raf,84 further amplifying the MEK signal. The MATβ-GIT1 interaction has a marked effect on cell survival signaling in HCC. Furthermore, when a human hepatoma cell line (Huh7) overexpressing MAT2B or GIT1 was injected into a mouse orthotopic HCC model, it enhanced tumor growth compared with the empty vector control. However, combining MAT2B with GIT1 dramatically induced growth and distant metastasis compared with individual genes alone.82 In this orthotopic model, the enhanced recruitment of C-Raf and B-Raf to MEK1 was observed. Therefore, MATβ-GIT1 promotes tumor survival and metastasis with a positive effect on MAPK signaling.84 The associations of MATα2/MATβ with liver dysfunction are summarized in Table 2 and Fig.2.
4.MATs As Therapeutic Targets
4.1.Repletion of SAMe reserves
Deregulated methionine metabolism as observed in the Mat1a-KO mouse model leads to SAMe deficiency.52 MAT1A silencing is also observed in human liver cirrhosis,48 advanced NAFLD,22 alcoholic hepatitis,86 and HCC.45 Therefore, the repletion of SAMe content appears might be an obvious therapy to overcome the loss of SAMe caused by MAT deficiency. Indeed, clinical studies have shown that long-term SAMe therapy improved the survival or delayed liver transplantation in patients with alcoholic liver cirrhosis, especially in those with less advanced liver disease.87 However, the beneficial effect of SAMe therapy in human liver cirrhosis was not confirmed in another large randomized placebo-controlled trial. 45 In a CCl4 model of liver injury, SAMe prevented the activation of a transgenic collagen promoter in HSCs.88 In primary HSCs, SAMe down-regulated basal and TGF-β-induced extracellular matrix protein, α-smooth muscle actin (α-SMA). 88 SAMe also inhibited the growth of liver cancer cells when administered at a pharmacological dosage and was anti-apoptotic in normal hepatocytes but pro-apoptotic in cancerous hepatocytes.18,78 Even though SAMe is chemopreventive before HCC has become established, it was ineffective at treating established tumors in an orthotopic HCC model.89 This is because SAMe buildup in the liver was prevented by the compensatory induction of enzymes such as glycine-N methyl transferase (GNMT) that catabolize SAMe.89 Normal hepatic SAMe levels were insufficient to mediate a pro-apoptotic effect on liver cancer cells. However, GNMT expression is often downregulated in human cirrhosis and HCC,45 so the efficacy of SAMe for HCC treatment in humans remains to be studied. In addition to SAMe, approaches that target MAT enzymes or their interactions and post-translational modifications need to be considered. Some of these approaches are described below.
4.2.Restoration of endogenous MAT1A expression
The forced expression of MAT1A in Huh7 cells resulted in a stable increase in SAMe levels in vitro and in vivo in a xenograft model, the induction of tumor suppressor genes, apoptosis, the downregulation of angiogenesis genes, and reduced cell growth compared to control tumors.90 However, this is impractical for therapy; therefore, a better approach might be to enhance MAT1A expression in HCC by targeting microRNAs that regulate MAT1A. MiR-664, miR-485-3p, and miR-495 target MAT1A in HCC. Inhibition of these miRs may provide an effective therapeutic strategy for HCC.25 Restoring MAT1A expression by the above means might restore SAMe levels in the liver. Physiological SAMe levels maintain MAT1A expression but inhibit the expression of MAT2A/MAT2B that induces growth in HCC cell lines.70,78 Because a reduction in MAT1A/SAMe levels is associated with a concomitant induction of MAT2A/MAT2B, this is a reasonable approach to target proliferative responses in HCC.
4.3.Therapeutic intervention by targeting MAT2A and MAT2B
Therapeutic approaches targeting MAT2A and MAT2B may also be effective in liver disease. Molecules such as the fluorinated N,N-dialkylaminostilbenes (FIDAS reagents) bind to the active site of MATα2 and prevent its induction in colon cancer.91 Similar molecules that inhibit MATα2 and MATβ expression, post-translational modifications, stability, interactions, and activity could be used to target liver fibrosis and HCC. A thorough structural analysis of these proteins for the design of such drugs is required. Inhibiting the expression of MATα2 and MATβ suppressed HSC activation and might be a target for liver fibrosis.59 The over-expression of miR-21-3p targeted MAT2A and MAT2B and might also be used to target HCC.35 Targeting MAT post-translational modifications might also be a reasonable way to control liver disease. Recent work showed that the sumoylation of MATα2 stabilizes it, facilitated BCL-2 induction during HCC, and induced chemoresistance in HCC cells.43 Therefore, blocking the sumoylation of MATα2 at specific residues using small molecules might provide a therapeutic advantage for HCC treatment. Similarly, the phosphorylation of MATα2 and MATβ at specific residues stabilized these proteins and facilitated HSC activation.41 Because HSC activation is an important mediator of liver fibrosis, blocking the site-specific phosphorylation of these proteins might be an effective liver fibrosis treatment. MATβ has been studied for its interaction with the ERK/MEK MAPK pathway and its association with cell survival in HCC.78, 82 The novel interaction of MATβ and GIT1 was shown to amplify Raf-mediated MAPK activation and therefore might be targeted to inhibit MAPK signaling and cell proliferation during HCC. 84
5.Conclusion
MAT is an enzyme that produces SAMe, which is essential for all methylation reactions. MAT genes and isoenzymes are dysregulated in liver injury and cancer: MAT1A expression is reduced and its encoded isoenzymes MATI/III are inactivated. In contrast, MAT2A and MAT2B are induced in liver cancer. Furthermore, MATα1, MATα2, and MATβ subunits exhibit distinct interactions and post-translational modifications that have both SAMe-dependent and independent effects. A summary of the key findings are provided below:
Liver pathologies (alcoholic hepatitis, non-alcoholic steatohepatitis, liver cirrhosis, HCC and CCA) are associated with reduced MAT1A expression.
Reduced MAT1A expression is associated with SAMe deficiency in the liver.
Liver fibrosis and HCC are associated with increased MAT2A and MAT2B expression.
MAT2A and MAT2B exhibit a strong association with proliferative phenotype in the liver.
The future direction of this research will be to develop molecular approaches that stabilize MATα1 and destabilize MATα2/MATβ to provide effective strategies for the treatment of liver fibrosis, cirrhosis, HCC, and CCA.
Acknowledgments
This work was supported by USA National Institutes of Health(NIH) grants R01DK51719 (SC Lu) and R01DK107288 (SC Lu and K Ramani).
Footnotes
Conflicts of Interest: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Detich N, Hamm S, Just G, Knox JD, Szyf M. The methyl donor S-Adenosylmethionine inhibits active demethylation of DNA: a candidate novel mechanism for the pharmacological effects of S-Adenosylmethionine. J Biol Chem. 2003;278:20812–20820. doi: 10.1074/jbc.M211813200. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlin ME, Ubagai T, Mudd SH, et al. Methionine adenosyltransferase I/III deficiency: novel mutations and clinical variations. Am J Hum Genet. 2000;66:347–355. doi: 10.1086/302752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotb M, Mudd SH, Mato JM, et al. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet. 1997;13:51–52. doi: 10.1016/s0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 4.Lu SC, Gukovsky I, Lugea A, et al. Role of S-adenosylmethionine in two experimental models of pancreatitis. FASEB J. 2003;17:56–58. doi: 10.1096/fj.01-0752fje. [DOI] [PubMed] [Google Scholar]
- 5.Horikawa S, Tsukada K. Molecular cloning and developmental expression of a human kidney S-adenosylmethionine synthetase. FEBS Lett. 1992;312:37–41. doi: 10.1016/0014-5793(92)81405-b. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu-Saito K, Horikawa S, Kojima N, Shiga J, Senoo H, Tsukada K. Differential expression of S-adenosylmethionine synthetase isozymes in different cell types of rat liver. Hepatology. 1997;26:424–431. doi: 10.1002/hep.510260224. [DOI] [PubMed] [Google Scholar]
- 7.González B, Pajares MA, Hermoso JA, Guillerm D, Guillerm G, Sanz-Aparicio J. Crystal structures of methionine adenosyltransferase complexed with substrates and products reveal the methionine-ATP recognition and give insights into the catalytic mechanism. J Mol Biol. 2003;331:407–416. doi: 10.1016/s0022-2836(03)00728-9. [DOI] [PubMed] [Google Scholar]
- 8.Murray B, Antonyuk SV, Marina A, et al. Structure and function study of the complex that synthesizes S-adenosylmethionine. IUCrJ. 2014;1:240–249. doi: 10.1107/S2052252514012585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halim AB, LeGros L, Geller A, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J Biol Chem. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Ara AI, Magilnick N, et al. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134:281–291. doi: 10.1053/j.gastro.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pajares MA, Alvarez L, Pérez-Sala D. How are mammalian methionine adenosyltransferases regulated in the liver? A focus on redox stress. FEBS Lett. 2013;587:1711–1716. doi: 10.1016/j.febslet.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 12.Reytor E, Pérez-Miguelsanz J, Alvarez L, Pérez-Sala D, Pajares MA. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 2009;23:3347–3360. doi: 10.1096/fj.09-130187. [DOI] [PubMed] [Google Scholar]
- 13.Pérez C, Pérez-Zúñiga FJ, Garrido F, Reytor E, Portillo F, Pajares MA. The Oncogene PDRG1 Is an Interaction Target of Methionine Adenosyltransferases. PLoS One. 2016;11:e0161672. doi: 10.1371/journal.pone.0161672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh Y, Ikura T, Hoshikawa Y, et al. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011;41:554–566. doi: 10.1016/j.molcel.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 15.Ramani K, Lu SC. Methionine adenosyltransferase genes in liver health and disease. In: Tirosh O, editor. Liver Metabolism and Fatty Liver Disease. Taylor & Francis Group/CRC Press; 2014. pp. 103–124. [Google Scholar]
- 16.Zeng Z, Huang ZZ, Chen C, Yang H, Mao Z, Lu SC. Cloning and functional characterization of the 5′-flanking region of human methionine adenosyltransferase 1A gene. Biochem J. 2000;346:475–482. [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda R, Nishida T, Watanabe F, et al. Involvement of CCAAT/enhancer binding protein-beta (C/EBPbeta) in epigenetic regulation of mouse methionine adenosyltransferase 1A gene expression. Int J Biochem Cell Biol. 2008;40:1956–1969. doi: 10.1016/j.biocel.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Mato JM, Corrales FJ, Lu SC, Avila MA. S-adenosylmethionine: a control switch that regulates liver function. FASEB J. 2002;16:15–26. doi: 10.1096/fj.01-0401rev. [DOI] [PubMed] [Google Scholar]
- 19.Torres L, Avila MA, Carretero MV, et al. Liver-specific methionine adenosyltransferase MAT1A gene expression is associated with a specific pattern of promoter methylation and histone acetylation: implications for MAT1A silencing during transformation. FASEB J. 2000;14:95–102. doi: 10.1096/fasebj.14.1.95. [DOI] [PubMed] [Google Scholar]
- 20.Frau M, Tomasi ML, Simile MM, et al. Role of transcriptional and posttranscriptional regulation of methionine adenosyltransferases in liver cancer progression. Hepatology. 2012;56:165–175. doi: 10.1002/hep.25643. [DOI] [PubMed] [Google Scholar]
- 21.Tomasi ML, Li TW, Li M, Mato JM, Lu SC. Inhibition of human methionine adenosyltransferase 1A transcription by coding region methylation. J Cell Physiol. 2012;227:1583–1591. doi: 10.1002/jcp.22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vázquez-Chantada M, Fernández-Ramos D, Embade N, et al. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koturbash I, Melnyk S, James SJ, Beland FA, Pogribny IP. Role of epigenetic and miR-22 and miR-29b alterations in the downregulation of Mat1a and Mthfr genes in early preneoplastic livers in rats induced by 2-acetylaminofluorene. Mol Carcinog. 2013;52:318–327. doi: 10.1002/mc.21861. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Cho ME, Li TW, et al. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123:285–298. doi: 10.1172/JCI63861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Huang ZZ, Wang J, Lu SC. The role of c-Myb and Sp1 in the upregulation of methionine adenosyltransferase 2A gene expression in human hepatocellular carcinoma. FASEB J. 2001;15:1507–1516. doi: 10.1096/fj.01-0040com. [DOI] [PubMed] [Google Scholar]
- 27.Yang H, Huang ZZ, Zeng Z, Chen C, Selby RR, Lu SC. Role of promoter methylation in increased methionine adenosyltransferase 2A expression in human liver cancer. Am J Physiol Gastrointest Liver Physiol. 2001;280:G184–G190. doi: 10.1152/ajpgi.2001.280.2.G184. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez JL, Boukaba A, Sandoval J, et al. Transcription of the MAT2A gene, coding for methionine adenosyltransferase, is upregulated by E2F and Sp1 at a chromatin level during proliferation of liver cells. Int J Biochem Cell Biol. 2007;39:842–850. doi: 10.1016/j.biocel.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Yang H, Sadda MR, Yu V, et al. Induction of human methionine adenosyltransferase 2A expression by tumor necrosis factor alpha. Role of NF-κB and AP-1. J Biol Chem. 2003;278:50887–50896. doi: 10.1074/jbc.M307600200. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Liu L, Zhao Y, et al. Hypoxia induces genomic DNA demethylation through the activation of HIF-1α and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10:1113–1123. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- 31.Ramani K, Tomasi ML. Transcriptional regulation of methionine adenosyltransferase 2A by peroxisome proliferator-activated receptors in rat hepatic stellate cells. Hepatology. 2012;55:1942–1953. doi: 10.1002/hep.25594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo YT, Leng XS, Li T, et al. Effect of ligand of peroxisome proliferator-activated receptor gamma on the biological characters of hepatic stellate cells. World J Gastroenterol. 2005;11:4735–4739. doi: 10.3748/wjg.v11.i30.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hellemans K, Michalik L, Dittie A, et al. Peroxisome proliferator-activated receptor-beta signaling contributes to enhanced proliferation of hepatic stellate cells. Gastroenterology. 2003;124:184–201. doi: 10.1053/gast.2003.50015. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Zheng Y, Li TW, et al. Methionine adenosyltransferase 2B, HuR, and sirtuin 1 protein cross-talk impacts on the effect of resveratrol on apoptosis and growth in liver cancer cells. J Biol Chem. 2013;288:23161–23170. doi: 10.1074/jbc.M113.487157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo TF, Tsai WC, Chen ST. MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One. 2013;8:e75628. doi: 10.1371/journal.pone.0075628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avila MA, Mingorance J, Martínez-Chantar ML, et al. Regulation of rat liver S-adenosylmethionine synthetase during septic shock: role of nitric oxide. Hepatology. 1997;25:391–396. doi: 10.1002/hep.510250222. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Mato I, Castro C, Ruiz FA, Corrales FJ, Mato JM. Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J Biol Chem. 1999;274:17075–17079. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 38.Corrales F, Ochoa P, Rivas C, Martin-Lomas M, Mato JM, Pajares MA. Inhibition of glutathione synthesis in the liver leads to S-adenosyl-L-methionine synthetase reduction. Hepatology. 1991;14:528–533. [PubMed] [Google Scholar]
- 39.Pajares MA, Durán C, Corrales F, Mato JM. Protein kinase C phosphorylation of rat liver S-adenosylmethionine synthetase: dissociation and production of an active monomer. Biochem J. 1994;303:949–955. doi: 10.1042/bj3030949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyahara T, Schrum L, Rippe R, et al. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J Biol Chem. 2000;275:35715–35722. doi: 10.1074/jbc.M006577200. [DOI] [PubMed] [Google Scholar]
- 41.Ramani K, Donoyan S, Tomasi ML, Park S. Role of methionine adenosyltransferase α2 and β phosphorylation and stabilization in human hepatic stellate cell trans-differentiation. J Cell Physiol. 2015;230:1075–1085. doi: 10.1002/jcp.24839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghioni P, D'Alessandra Y, Mansueto G, et al. The protein stability and transcriptional activity of p63alpha are regulated by SUMO-1 conjugation. Cell Cycle. 2005;4:183–190. doi: 10.4161/cc.4.1.1359. [DOI] [PubMed] [Google Scholar]
- 43.Tomasi ML, Ryoo M, Ramani K, et al. Methionine adenosyltransferase α2 sumoylation positively regulate Bcl-2 expression in human colon and liver cancer cells. Oncotarget. 2015;6:37706–37723. doi: 10.18632/oncotarget.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang HB, Xu YY, Zhao XN, Zou SW, Zhang Y, Zhang M, et al. Acetylation of MAT IIα represses tumour cell growth and is decreased in human hepatocellular cancer. Nat Commun. 2015;6:6973. doi: 10.1038/ncomms7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan DM, Hoffman JL. Fractionation and kinetic properties of rat liver and kidney methionine adenosyltransferase isozymes. Biochemistry. 1983;22:1636–1641. doi: 10.1021/bi00276a017. [DOI] [PubMed] [Google Scholar]
- 47.Kinsell LW, Harper HA, Barton HC, Hutchin ME, Hess JR. Studies in methionine and sulfur metabolism. I. The fate of intravenously administered methionine, in Normal individuals and in patients with liver damage. J Clin Invest. 1948;27:677–688. doi: 10.1172/JCI102016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avila MA, Berasain C, Torres L, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 49.Lieber CS, Casini A, DeCarli LM, et al. S-adenosyl-L-methionine attenuates alcohol-induced liver injury in the baboon. Hepatology. 1990;11:165–172. doi: 10.1002/hep.1840110203. [DOI] [PubMed] [Google Scholar]
- 50.Corrales F, Giménez A, Alvarez L, et al. S-Adenosylmethionine treatment prevents carbon tetrachloride-induced S-adenosylmethionine synthetase inactivation and attenuates liver injury. Hepatology. 1992;16:1022–1027. doi: 10.1002/hep.1840160427. [DOI] [PubMed] [Google Scholar]
- 51.Stramentinoli G, Pezzoli C, Galli-Kienle M. Protective role of S-adenosyl-l-methionine against acetaminophen induced mortality and hepatotoxicity in mice. Biochem Pharmacol. 1979;28:3567–3571. doi: 10.1016/0006-2952(79)90401-5. [DOI] [PubMed] [Google Scholar]
- 52.Lu SC, Alvarez L, Huang ZZ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 54.Wu D, Cederbaum AI. Opposite action of S-adenosyl methionine and its metabolites on CYP2E1-mediated toxicity in pyrazole-induced rat hepatocytes and HepG2 E47 cells. Am J Physiol Gastrointest Liver Physiol. 2006;290:G674–G684. doi: 10.1152/ajpgi.00406.2005. [DOI] [PubMed] [Google Scholar]
- 55.Santamaria E, Avila MA, Latasa MU, et al. Functional proteomics of non-alcoholic steatohepatitis: mitochondrial proteins as targets of S-adenosylmethionine. Proc Natl Acad Sci USA. 2003;100:3065–3070. doi: 10.1073/pnas.0536625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ko KS, Tomasi ML, Iglesias-Ara A, et al. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology. 2010;52:2096–2108. doi: 10.1002/hep.23919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang HL, Lai JJ, Lin WL, Lin WC. A fermented substance from Aspergillus phoenicis reduces liver fibrosis induced by carbon tetrachloride in rats. Biosci Biotechnol Biochem. 2007;71:1154–1161. doi: 10.1271/bbb.60604. [DOI] [PubMed] [Google Scholar]
- 58.Martínez-Chantar ML, García-Trevijano ER, Latasa MU, et al. Methionine adenosyltransferase II beta subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940–948. doi: 10.1053/gast.2003.50151. [DOI] [PubMed] [Google Scholar]
- 59.Ramani K, Yang H, Kuhlenkamp J, et al. Changes in the expression of methionine adenosyltransferase genes and S-adenosylmethionine Homeostasis during hepatic stellate cell activation. Hepatology. 2010;51:986–995. doi: 10.1002/hep.23411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maher JJ, McGuire RF. Extracellular matrix gene expression increases preferentially in rat lipocytes and sinusoidal endothelial cells during hepatic fibrosis in vivo. J Clin Invest. 1990;86:1641–1648. doi: 10.1172/JCI114886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rountree CB, Senadheera S, Mato JM, Crooks GM, Lu SC. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology. 2008;47:1288–1297. doi: 10.1002/hep.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding W, Mouzaki M, You H, et al. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49:1277–1286. doi: 10.1002/hep.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen LN, Furuya MH, Wolfraim LA, et al. Transforming growth factor-beta differentially regulates oval cell and hepatocyte proliferation. Hepatology. 2007;45:31–41. doi: 10.1002/hep.21466. [DOI] [PubMed] [Google Scholar]
- 64.Schmitz KJ, Wohlschlaeger J, Lang H, et al. Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J Hepatol. 2008;48:83–90. doi: 10.1016/j.jhep.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 65.Calvisi DF, Pinna F, Meloni F, et al. Dual-specificity phosphatase 1 ubiquitination in extracellular signal-regulated kinase-mediated control of growth in human hepatocellular carcinoma. Cancer Res. 2008;68:4192–4200. doi: 10.1158/0008-5472.CAN-07-6157. [DOI] [PubMed] [Google Scholar]
- 66.Tomasi ML, Ramani K, Lopitz-Otsoa F, et al. S-adenosylmethionine regulates dual-specificity mitogen-activated protein kinase phosphatase expression in mouse and human hepatocytes. Hepatology. 2010;51:2152–2161. doi: 10.1002/hep.23530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calvisi DF, Simile MM, Ladu S, et al. Altered methionine metabolism and global DNA methylation in liver cancer: relationship with genomic instability and prognosis. Int J Cancer. 2007;121:2410–2420. doi: 10.1002/ijc.22940. [DOI] [PubMed] [Google Scholar]
- 68.Mann CD, Neal CP, Garcea G, Manson MM, Dennison AR, Berry DP. Prognostic molecular markers in hepatocellular carcinoma: a systematic review. Eur J Cancer. 2007;43:979–992. doi: 10.1016/j.ejca.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 70.Tomasi ML, Iglesias-Ara A, Yang H, et al. S-adenosylmethionine regulates apurinic/apyrimidinic endonuclease 1 stability: implication in hepatocarcinogenesis. Gastroenterology. 2009;136:1025–1036. doi: 10.1053/j.gastro.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martínez-Chantar ML, Vázquez-Chantada M, Garnacho M, et al. S-adenosylmethionine regulates cytoplasmic HuR via AMP-activated kinase. Gastroenterology. 2006;131:223–232. doi: 10.1053/j.gastro.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 73.Martínez-López N, Varela-Rey M, Fernández-Ramos D, et al. Activation of LKB1-Akt pathway independent of phosphoinositide 3-kinase plays a critical role in the proliferation of hepatocellular carcinoma from nonalcoholic steatohepatitis. Hepatology. 2010;52:1621–1631. doi: 10.1002/hep.23860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang H, Li TW, Peng J, et al. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology. 2011;141:378–388. doi: 10.1053/j.gastro.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu SC. Glutathione synthesis. Biochim Biophys Acta. 2013;1830:3143–3153. doi: 10.1016/j.bbagen.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang H, Liu T, Wang J, et al. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans(‡) Hepatology. 2016;64:439–455. doi: 10.1002/hep.28541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Zhang X, Wang L, et al. MicroRNA-214 suppresses oncogenesis and exerts impact on prognosis by targeting PDRG1 in bladder cancer. PLoS One. 2015;10:e0118086. doi: 10.1371/journal.pone.0118086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Ramani K, Yang H, Xia M, Ara AI, Mato JM, Lu SC. Leptin's mitogenic effect in human liver cancer cells requires induction of both methionine adenosyltransferase 2A and 2beta. Hepatology. 2008;47:521–531. doi: 10.1002/hep.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang H, Magilnick N, Noureddin M, Mato JM, Lu SC. Effect of hepatocyte growth factor on methionine adenosyltransferase genes and growth is cell density-dependent in HepG2 cells. J Cell Physiol. 2007;210:766–773. doi: 10.1002/jcp.20891. [DOI] [PubMed] [Google Scholar]
- 80.Tomasi ML, Ryoo M, Skay A, et al. Polyamine and methionine adenosyltransferase 2A crosstalk in human colon and liver cancer. Exp Cell Res. 2013;319:1902–1911. doi: 10.1016/j.yexcr.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hartmann LC, Keeney GL, Lingle WL, et al. Folate receptor overexpression is associated with poor outcome in breast cancer. Int J Cancer. 2007;121:938–942. doi: 10.1002/ijc.22811. [DOI] [PubMed] [Google Scholar]
- 82.Peng H, Dara L, Li TW, et al. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology. 2013;57:2299–2313. doi: 10.1002/hep.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marais R, Light Y, Paterson HF, Mason CS, Marshall CJ. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem. 1997;272:4378–4383. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 84.Peng H, Li TW, Yang H, Moyer MP, Mato JM, Lu SC. Methionine adenosyltransferase 2B-GIT1 complex serves as a scaffold to regulate Ras/Raf/MEK1/2 activity in human liver and colon cancer cells. Am J Pathol. 2015;185:1135–1144. doi: 10.1016/j.ajpath.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matallanas D, Birtwistle M, Romano D, et al. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee TD, Sadda MR, Mendler MH, et al. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin Exp Res. 2004;28:173–181. doi: 10.1097/01.ALC.0000108654.77178.03. [DOI] [PubMed] [Google Scholar]
- 87.Mato JM, Cámara J, Fernández de Paz J, et al. S-adenosylmethionine in alcoholic liver cirrhosis: a randomized, placebo-controlled, double-blind, multicenter clinical trial. J Hepatol. 1999;30:1081–1089. doi: 10.1016/s0168-8278(99)80263-3. [DOI] [PubMed] [Google Scholar]
- 88.Nieto N, Cederbaum AI. S-adenosylmethionine blocks collagen I production by preventing transforming growth factor-beta induction of the COL1A2 promoter. J Biol Chem. 2005;280:30963–30974. doi: 10.1074/jbc.M503569200. [DOI] [PubMed] [Google Scholar]
- 89.Lu SC, Ramani K, Ou X, et al. S-adenosylmethionine in the chemoprevention and treatment of hepatocellular carcinoma in a rat model. Hepatology. 2009;50:462–471. doi: 10.1002/hep.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li J, Ramani K, Sun Z, et al. Forced expression of methionine adenosyltransferase 1A in human hepatoma cells suppresses in vivo tumorigenicity in mice. Am J Pathol. 2010;176:2456–2466. doi: 10.2353/ajpath.2010.090810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang W, Sviripa V, Chen X, et al. Fluorinated N, N-dialkylaminostilbenes repress colon cancer by targeting methionine S-adenosyltransferase 2A. ACS Chem Biol. 2013;8:796–803. doi: 10.1021/cb3005353. [DOI] [PMC free article] [PubMed] [Google Scholar]


