Abstract
Stochastic optical reconstruction microscopy (STORM) can achieve resolutions of better than 20nm imaging single fluorescently labeled cells. However, when optical aberrations induced by larger biological samples degrade the point spread function (PSF), the localization accuracy and number of localizations are both reduced, destroying the resolution of STORM. Adaptive optics (AO) can be used to correct the wavefront, restoring the high resolution of STORM. A challenge for AO-STORM microscopy is the development of robust optimization algorithms which can efficiently correct the wavefront from stochastic raw STORM images. Here we present the implementation of a particle swarm optimization (PSO) approach with a Fourier metric for real-time correction of wavefront aberrations during STORM acquisition. We apply our approach to imaging boutons 100 μm deep inside the central nervous system (CNS) of Drosophila melanogaster larvae achieving a resolution of 146 nm.
OCIS codes: (220.1080) Active or adaptive optics; (180.2520) Fluorescence microscopy; (010.7350) Wave-front sensing; (110.7348) Aberration compensation; (100.3008) Image recognition, algorithms and filters
1. Introduction
Stochastic Optical Reconstruction Microscopy (STORM), a single molecule localization microscopy technique, was first performed on essentially two-dimensional samples using TIRF Microscopy [1, 2]. STORM Microscopy has reliably been demonstrated on whole cells [3, 4], and has been combined with light-sheet microscopy to facilitate three-dimensional imaging [5–8]. Some of these efforts are still focused on imaging single cells, but in [5], STORM microscopy was combined with light sheet microscopy to image human mammary cell spheroids at a depth of 80 μm with a resolution of ~60nm. In [8], the zebrafish neuromast sensory organ was imaged in a volume ~30µm thick using LLS-PAINT. In both of these efforts, wavefront aberrations were not addressed. Although STORM may be possible as long as the PSF core is intact, precision will be lost due to scattered photons, and in strongly aberrating samples or several mean free paths into a strongly scattering sample, STORM imaging is not possible.
Wavefront aberrations caused by refractive index variations reduce the resolution of STORM by increasing the size of the PSF and reducing the number of photons in the PSF core [9]. The resolution is affected through both reduced localization precision and the reduced number of localizations. Because the resolution of STORM is an order of magnitude better than the resolution of conventional microscopy, even small wavefront aberrations can significantly impact the resolution. Adaptive Optics (AO) can be used to correct wavefront aberrations, restoring the size of the PSF and increasing the number of photons in the PSF core.
AO has been used extensively in biological microscopy [10]. In particular, “sensorless” techniques have been developed in which the wavefront is optimized using a deformable mirror or spatial light modulator without the use of a wavefront sensor [11]. These methods optimize a fitness metric over the wavefront’s degrees of freedom where the fitness metric is a scalar value derived from the acquired image [12]. Wavefront sensorless correction has been applied to widefield [13], confocal [14], multiphoton [15], structured illumination [16, 17], and STORM [9, 18]. The effect of optical aberrations in STORM is characterized in [19]. Model-based algorithms [20] and a variety of optimization algorithms [21] have been applied to sensorless AO. Peak intensity, total image intensity [22], and image sharpness [12] have all been used as fitness metrics. These metrics work well when the intensity of the sample is constant or slowly varying. In STORM imaging, the intensity of each acquired frame of a data set fluctuates strongly because the emission events from individual fluorophores follow an exponential distribution [9].
We have previously reported an approach to AO-STORM that combines a genetic algorithm (GA) with an intensity insensitive Fourier Metric (FM) to permit wavefront correction in real time during imaging, and we demonstrated AO-STORM, imaging neurons in the Drosophila central nervous system (CNS) [9]. The FM applies a high-pass filter to the Fourier Transform of the image and is normalized by the sum of the magnitude of the intensity frequency components. The value of the FM increases as the wavefront improves because the high-frequencies in the OTF are maximized with a flat wavefront, but the FM is relatively insensitive to intensity fluctuations due to the normalization. GAs are modeled on natural selection and generate new wavefronts to apply through mutation and crossover of a population of wavefronts described by a finite set of Zernike modes. Our GA works well but requires a few thousand frames to converge. Because a good STORM image can be acquired in ~10,000 raw frames and the fluorophores have a limited life before photo-bleaching, a faster convergence is desirable. A further problem with the GA is that the random mutation required to sample the search space and avoid local minima results in wasted frames as the GA begins to converge.
Here we present the application of Particle Swarm Optimization (PSO) [23], another machine learning optimization algorithm, to wavefront correction during real-time STORM data acquisition. The PSO algorithm due to its swarm solution finding method has been shown to converge an order of magnitude faster than the GA algorithm which uses an evolutionary scheme to find a solution. We apply this approach to the imaging of boutons in the ventral nerve cord (VNC), 100 µm inside the Drosophila CNS. We optimize the PSO parameters with simulations, and then demonstrate correcting system aberrations and flattening the deformable mirror (DM), imaging a fluorescent microsphere. Finally, we apply the PSO to STORM imaging — First correcting the aberrations induced by the nucleus of a cultured cell and then correcting the sample-induced aberrations in the Drosophila CNS. The algorithm converges in ~500 frames, a significant improvement in convergence time over our previous experiments with the GA.
2. Materials and methods
2.1. Particle swarm optimization algorithm
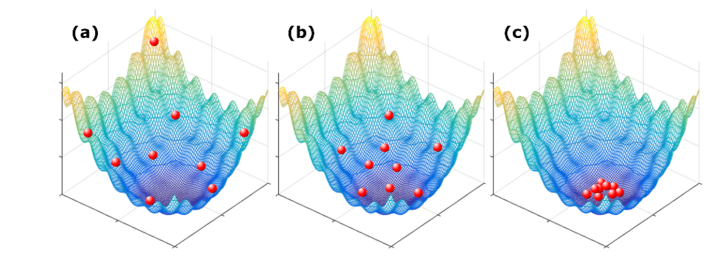
The PSO algorithm is modeled on the collective behavior of swarming insects searching for a food source. Figure 1 shows a demonstration of how particles in a swarm are searching for a solution in a 2-dimensional space. Each individual in the swarm searches the 2-dimensional space, remembers the best position it found and also has access to the swarm’s memory of the best location. The algorithm updates the velocity and position of each individual for the next iteration. Different weights are given to the particle’s personal best location and the swarm’s best location in order to lead the individuals to the best location while avoiding an excessive number of iterations but with enough variation among the individuals to effectively search the parameter space. First, the positions of the individuals in the population are generated from a uniform random distribution to uniformly cover the search space. The individuals are then evaluated by applying the wavefront and measuring the fitness value. The velocity and position of each individual are then updated based on the measured fitness values. The updated values of position and velocity at iteration, t + 1, are given by
| (1) |
| (2) |
whereandare the position and velocity of the particle i at the t iteration. is the ith particles best location and is the collective best position of all particles over all iterations,. andare random weights given to the individual and global best positions in determining the velocity of the particle. In each iteration, these values are updated through a random process with uniform distribution between zero and the given maximum value. The randomness of the weights improves the ability of the algorithm to work in a dynamic search space. The search is terminated and the global best position is selected when either the metric reaches a pre-determined value or a maximum number of iterations is reached. In our application, each wavefront (particle) searches the n-dimensional space of Zernike modes (n being the number of Zernike modes) to find the position that yields the best value of the FM.
Fig. 1.
A demonstration of Particle Swarm Optimization in a 2 dimensional space. The red particles are searching for the minimum of the surface. (a-c) show early, mid, and late stages of the convergence.
We performed simulations of STORM imaging to evaluate the PSO algorithm and determine the best parameters. As described previously [9, 24], the simulated object consists of 1000 fluorophores placed in a circle of radius 250nm. The wavefront is described by the first 15 Zernike modes. This number is chosen for our simulation to be representative of our optical setup, because our deformable mirror can most efficiently correct for these modes. For each raw frame of the STORM data set, a small subset of fluorophores described by a Poisson process with mean value µ = 3 is chosen to emit. The image is then generated by multiplying the wavefront by a phase shift corresponding to the fluorophore position and adding the magnitude squared of the Fourier Transform to the image for each fluorophore. Finally, Poisson noise is added to the image.
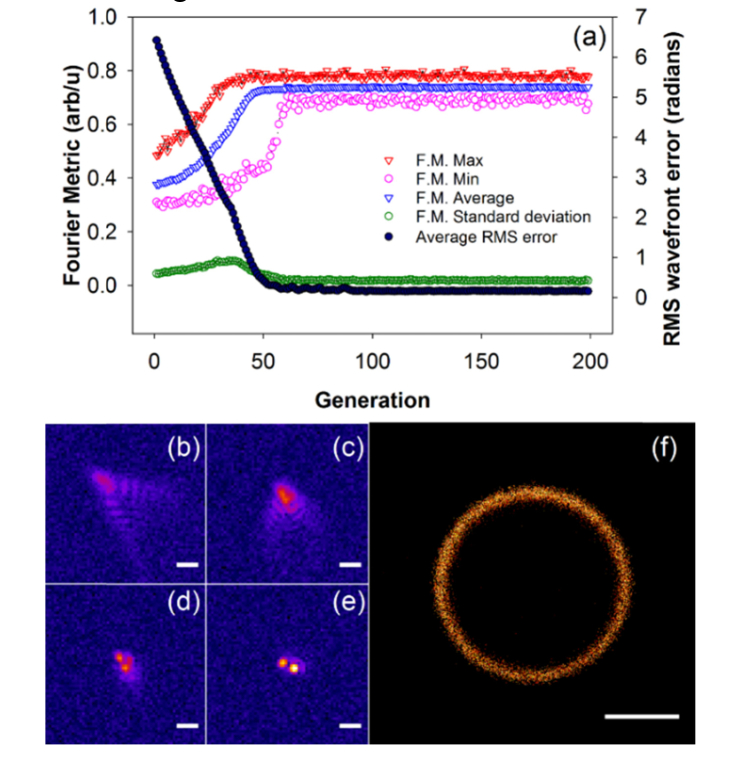
We then optimized the parameters and, and the population size. Population sizes between 20 and 50 are optimum. If the population is too small, there are not enough particles to explore the space adequately and, if it is too large, the convergence is slow. Optimal ranges for and are between 0.5 and 2.0 with higher weight given to the global position,. With lower weights the algorithm converges slower, and with higher values the particles move too fast and jump over the solution without effectively evaluating the local neighborhood, which again slows down the convergence. Initial velocities uniformly distributed between 0.1 and 0.5 radians/frame were found to be optimal. We used the optimized parameters (velocity: −0.1 ~0.1 radians/frame,and: 1.0, population size: 25 wavefronts, and position range: −5 ~5 radians), generated a target aberrated wavefront with RMS error of 0.33 radians, and ran a simulation for 200 generations; the results are shown in Fig. 2, which shows convergence within 50 generations or 1250 frames. The implementation of the particle swarm optimization algorithm we use is the open-source Python package, DEAP [25].
Fig. 2.
Simulation of AO-STORM using PSO for correction of random wavefront aberrations. (a) shows a graph of the metric values and rms wavefront error plotted against the generation. (b-e) show individual frames at generations 0, 13, 41, and 51 respectively. (f) is the reconstructed image from 2000 frames after convergence of the algorithm. Scale bars for are 1 μm for (b-e) and 200 nm for (f).
2.2. Optical setup
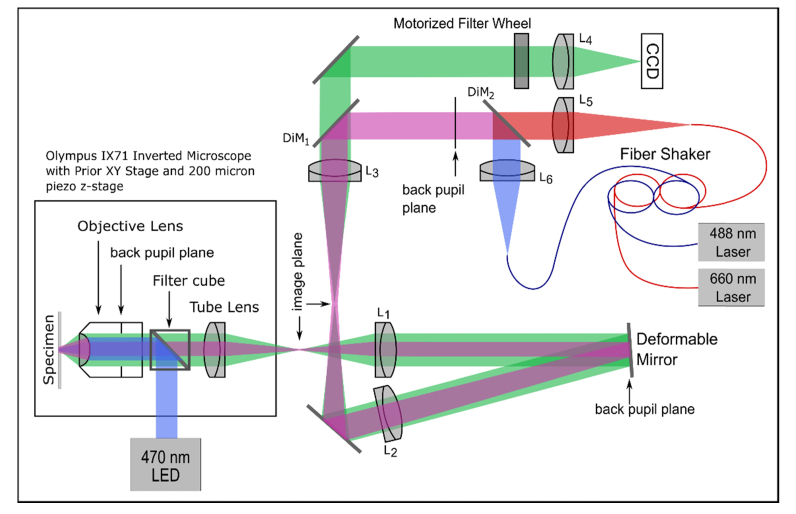
Data acquisition was performed on our optical setup consisting of an Olympus IX71 inverted microscope with external optics to re-image the back pupil plane of the 60x oil objective lens (Plan Apo N) onto a deformable mirror (Mirao 52E, Imagine Optic), Fig. 3. The diameter of the deformable mirror limits the system NA to 1.28. After the deformable mirror, a further image plane is generated which is then relayed to an Andor EMCCD camera (DV887DCS-BV with 14bit ADC). The image is magnified 3x so that the image is sampled at the Nyquist frequency by the CCD. A CCD pixel corresponds to 89nm at the sample plane. For imaging of fluorescent microspheres, a 470nm LED (Thorlabs M470L2) is fitted through the back port of the IX71 microscope. The LED light is reflected to the objective through a filter cube (Emission filter: 535/50 nm, Dichroic mirror: 505 nm longpass, and Excitation filter: 480/40 nm, Olympus) and an additional filter at 520nm (Semrock FF01-520/35-25) right before the camera. The filter cube was removed for laser illumination and STORM imaging. For QD 565 nm imaging a 504 nm filter (Semrock FF01-504/12-25) was placed in the imaging path. For Alexa 647 imaging a 700 nm filter (Semrock FF01-515/588/700-25) was used. The QDs are excited with a 488nm laser (Cyan 488, Newport) which is coupled into a 100µm core diameter fiber. The fiber is shaken using a fiber shaker to remove speckle [26]. To excite Alexa Fluor 647, a 660nm laser (Obis 660LX, Coherent) is coupled into a separate fiber attached to the shaker. For laser excitation, a multiband dichroic (XF2054, Omega Optical) is used (DiM1). The lasers are collimated using 7.86 mm fiber collimators (Thorlabs F240SMA), and combined with a 550 nm longpass dichroic (Thorlabs DMLP550T). The microscope is equipped with a PriorProScan xy stage and a Prior NanoScanZ piezo stage for focusing. For filtering and other image processing methods Python(x,y) version 2.7.6.1 was used. For reconstruction of all STORM images in this paper we used rapidSTORM versions 2.21 and 3.2 [27].
Fig. 3.
Diagram of the microscope. L1: 350mm efl achromat. L2: 350mm efl achromat. L3: 100mm efl achromat. L4: 300mm efl achromat. L5 and L6: 7.86 mm. DiM1: multiband dichroic mirror. DiM2: 550 nm long pass.
2.3. Fluorescent microsphere and C.elegans sample preparation
200nm Yellow-Green Fluorescent microspheres were purchased from Life technologies. For fluorescent microsphere slides, we first dried 200nm fluorescent microspheres on number 1.5 coverslips. The coverslips were sonicated in Sodium hydroxide (J.T. Baker) for 30 minutes followed by 30 minutes sonication in 98% Ethanol (J.T. Baker) after washing 3 times in deionized water. The coverslips were mounted on slides using glycerol.
For C. elegans samples, we dried 15µl of a solution containing 200nm yellow-green fluorescent microspheres - diluted by a factor of 103 in deionized water - on an electrostatically charged glass slide (Shandon Colorfrost from Thermo Scientific). 5µl of a 50mM solution of tetramisole (L9756, Sigma) was applied to the slide, and C. elegans roundworms were then transferred to the slide on the dried layer of microspheres. After allowing most of the tetramisole solution to dry, a drop of glycerol was applied and a no. 1.5 coverslip was fixed on top with nail polish.
2.4. Preparation of cultured cells
Hela cells were cultured in Dulbecco’s Modified Eagle’s Medium with 10% fetal bovine serum (FBS), Glutamine, and Penicillin/streptomycin. For imaging purposes, the cells were plated onto a cover-glass-bottom (#1.5) 35mm petry dish at an initial confluency of about 50% and cultured for one day to let the cells attach to the dish. For immunostaining, the cells were washed with Phosphate-Buffered Saline (PBS) once and fixed in a 4% Paraformaldehyde in PBS solution for 10 min. After 3 washes with PBS, the cells were blocked by incubation with blocking buffer (3% Bovine Serum Albumin (BSA), 10% normal serum in PBS), for 2 hours. Then the blocking buffer was aspirated, and the cells were incubated with Anti α-Tubulin Rabbit primary antibody (abcam) diluted in 3% BSA at 4°C overnight. The cells were washed 3 times with the washing buffer (0.2% BSA and 0.5% Triton X-100 in PBS) for 10 minutes per wash. Qdot 565 goat F(ab')2 anti-rabbit IgG conjugate (H + L) (Life Technologies) secondary antibody was added to the sample in 6% BSA and incubated for 2 hours, protected from light. The cells were washed again 3 times with washing buffer and once with PBS for 10 minutes per wash and stored in PBS before imaging. Immediately before imaging, the buffer was switched to a solution containing 20% glycerol (v/v) for preserving QDs from fast oxidation [28].
2.5. Preparation of the Drosophila central nervous system (CNS)
Intact CNS tissues were freshly dissected from 74 AEL Drosophila larvae and immediately fixed in 4% paraformaldehyde for 1-1.5 hours. The tissues were then washed 6 times, 20 minutes each time, in PBS solution containing 0.1% TritonX-100 (PBT). About 20 tissues were incubated in 1ml rabbit anti-NPF primary antibody overnight. The primary antibody was pre-incubated with 50μg/ml C8 peptide at 4°C for 12 hours to block non-specific binding [29]. After additional 6 PBT washes, the tissues were incubated in Alexa Fluor 647 F(ab')2 Fragment of Goat Anti-Rabbit IgG (H + L) secondary antibody purchased from Life Technologies diluted 1:500 from the original concentration overnight. The tissues were washed with PBT six times after staining. The tissues were placed in the STORM imaging buffer with MEA [30], 20 minutes prior to imaging. The STORM imaging buffer with MEA buffer for Alexa 647 was prepared according to the Nikon N-STORM protocol, by mixing 310µl Buffer B, 35µl MEA buffer, and 3.5µl GLOX solution. Buffer B was made by combining 50 mM Tris pH 8.0, 10 mM NaCl, and 10% w/v glucose. 1M MEA buffer was made by mixing 77mg of Cysteamine (Sigma-Aldrich #30070) in 1ml of 0.25N HCl. GLOX solution was made by mixing 0.56 mg/mL Glucose Oxidase (Sigma-Aldrich #G2133), 0.17 mg/mL Catalase (Sigma-Aldrich #C40), in 200µl of a buffer containing 10mM Tris ph8.0 and 50mM NaCl. The solution was vortexed, spinned down at 14,000 rpm, and only the supernatant was used.
3. Results
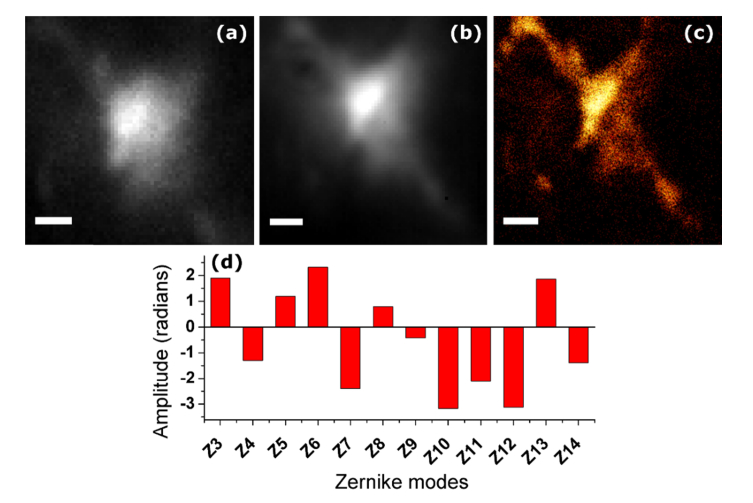
Using optimized parameters determined from the simulations, we corrected the system aberrations, including flattening the deformable mirror (DM). We measured a total RMS wavefront error of 5.7 radians using phase retrieval [31], when all DM actuators were set to 0V. Figure 4 shows the results of correcting system aberrations by imaging a 200nm yellow-green (YG) fluorescent microsphere. Since we are only correcting the system aberrations and deformation of the DM which are low-order, the wavefronts are described by 8 Zernike modes: defocus, astigmatism (90° and 45°), coma (horizontal and vertical), trefoil (2 orders), and spherical aberration. We use the root mean square normalized Zernike modes with ordering as in [32] except where noted. The algorithm started with 0V on all DM actuators. Figure 4(a) shows the PSF before correction. Figure 4(b) is the image after the correction is completed. The algorithm ran for 50 generations with a population of 50 and reached an optimum value after 10 generations (less than 500 frames). The values of and are both 2.0. The speeds along each mode for each particle were chosen from a uniform distribution with range −0.5 to 0.5 radians and the position range was −5 to 5 radians. The removed wavefront error was 4.7 radians RMS (excluding defocus), and the peak intensity of the 3D PSF was increased by a factor of 3.2. A phase retrieval calculation on the corrected PSF yielded a Strehl ratio of 42% (0.95 radians residual RMS wavefront error) [33]. In comparison to the GA, PSO requires approximately 75% fewer iterations for convergence [9]. Supplemental Visualization 1 (22.6MB, avi) shows the convergence of the algorithm for the bead experiment.
Fig. 4.
PSO-AO correction on a single fluorescent microsphere. (a) PSF before correction. (b) The final frame of the optimization showing the corrected PSF. (c) Zernike coefficients of the correction (Noll ordering). The experiment was performed with particles placed in the position range from −5 to 5 radians. The velocity range was −0.5 to 0.5. The population size was 50. Values ofand were both 2.0. Sigma for the metric was 0.1 μm−1. The exposure time was 50 ms. A 470 nm LED was used for illumination. Scale bars are 1 µm. A video of this process is shown in supplemental Visualization 1 (22.6MB, avi) .
Our first test of aberrating tissue is the C. elegans roundworm. In Fig. 5(a) we show 200nm YG microspheres placed under the worm. The images of beads (essentially, the PSF) under the worm show strong optical distortions compared to images of beads not under the worm, at the top and bottom of the image. We applied PSO-AO to correct the aberrations due to the roundworm, as shown in Fig. 5(b). Because older C. elegans can have a non-homogeneous internal structure we corrected 12 modes corresponding to Zernike orders 2, 3, and 4 which include defocus, astigmatism, coma, trefoil and spherical aberration. The PSO parameters were the same as above. The algorithm started with a DM setting that corrected system aberrations so that a 200nm fluorescent bead on a coverslip was well corrected. The total RMS wavefront error removed was 5.4 radians. We estimated the Strehl ratio outside of the worm to be 70% from phase retrieval measurements on individual microspheres. By comparing the intensity of beads under the worm to the intensity of beads outside the worm, we estimate the Strehl ratio under the worm after correction to be ~40%.
Fig. 5.
200nm YG beads under a C. elegans roundworm. The ROI used for correction is shown as a box in (a) and (b). The aberrated image is shown in (a). The bead images under the worm are distorted. After correction, the bead images under the worm are corrected as shown in (b). (a) and (b) are shown using the same intensity scale. We can see the increase in intensity under the center of the worm in (b). (c) shows the coefficients of the Zernike modes applied to the wavefront by the PSO algorithm (Noll ordering). Scale bars are 5 µm.
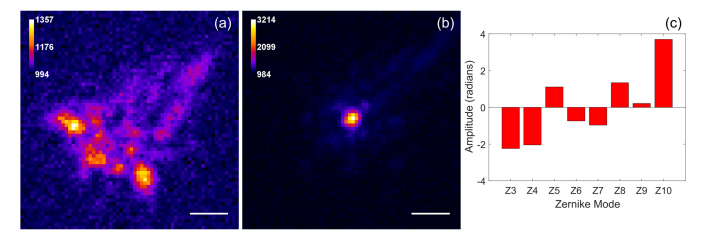
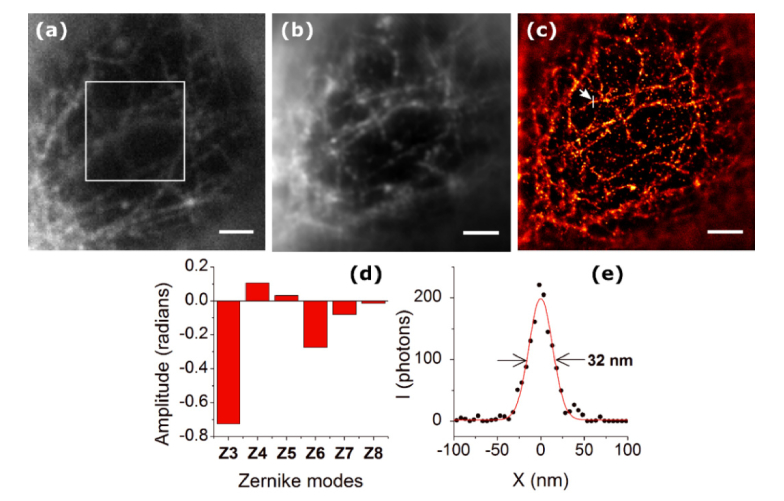
Next, we applied the PSO-AO algorithm to STORM imaging. We imaged microtubules stained with 565nm Quantum Dots (QDs) in a HeLa cell. We applied PSO-AO-STORM to imaging microtubules through the nucleus. We focused on the microtubules beneath the nucleus farther from objective lens. We anticipated the aberrations due to the shape of the nucleus to be mainly low order; therefore, we corrected defocus, astigmatism, coma, and spherical aberration. The population size was 25; and were 1.0 and 2.0 respectively. The cutoff value (sigma) of the high frequency filter in the Fourier metric was 0.74 µm−1. The starting wavefront was already corrected for system aberrations, and the amount of wavefront error removed was 0.78 radians. Figure 6(a) shows a widefield image before correction. The boxed area in Fig. 6(a) shows the input to the metric function. PSO-AO was run for 100 generations, and the metric increased by ~60%. After correction, we obtained 10,000 frames with the best wavefront. Figure 6(b) shows an average of the frames (widefield after correction image), and the reconstructed super-resolution image is shown in Fig. 6(c). Although careful consideration was taken to be as close as possible to the imaging plane, the algorithm still has corrected some defocus, Fig. 6(d). We measured a full width at half-maximum of 32 nm for the microtubule indicated in Fig. 6(c).
Fig. 6.
Correction of aberrations caused by the nucleus of a Hela cell. (a) shows a widefield image of microtubules labeled with 565nm Quantum Dots before correction. The correction ROI is shown in the white box. In (b), an average of all frames from the STORM data set after AO correction is shown. (c) shows the final STORM image, reconstructed using the frames after correction. (d) shows the coefficients of the Zernike modes applied to the wavefront. (e) The cross-section of a microtubule specified with an arrow in (c), shows a FWHM of 32 nm. Scale bars are 2 µm.
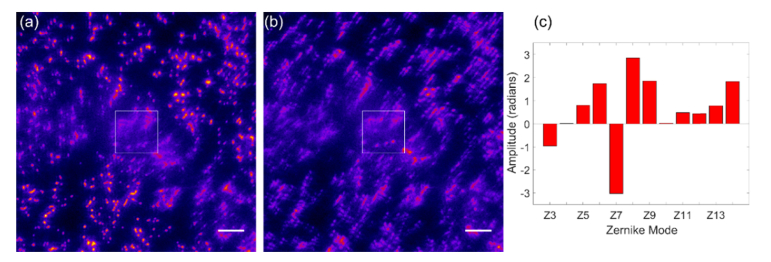
We next tested PSO-AO on Neuropeptide F (NPF) positive boutons labeled with Alexa647 at the depth of about 100 μm in the VNC region of Drosophila CNS, as shown in Fig. 7. A widefield image with only system aberration correction is shown in Fig. 7(a). The population size was 25. Both andwere 1.0. Particle positions ranged from −5 to 5 radians, and the velocity ranged from 0.1 to 0.1 radians. The sigma of the merit function was 0.4 µm−1. In this experiment, we corrected 12 Zernike modes, because the CNS has a non-homogenous shape. The algorithm started from a baseline of 0V on all actuators, and removed a total RMS wavefront error of 6.94 radians, which translates to roughly 4 radians of error induced by the sample. The wavefront converged in 25 generations, and the corrected wavefront was used to acquire 10,000 images to reconstruct a super-resolution image. An after correction widefield image was generated by averaging the acquired frames, Fig. 7(b). The resulting STORM image is shown in Fig. 7(c). We measured a resolution of 146 nm estimated using Fourier Ring Correlation (FRC) analysis [34]. The distribution of Zernike mode coefficients confirms the sample required correction of all the modes used in this experiment.
Fig. 7.
PSO-AO-STORM microscopy applied to a Bouton in the ventral nerve cord of the Drosophila larva. A system-corrected widefield image before PSO-AO is shown in (a). After correction widefield image (b) is generated by averaging all the frames in the STORM data set. (c) shows the PSO-AO- STORM image reconstructed using 10,000 frames after convergence of the algorithm. (d) shows the coefficients of the Zernike modes. Scale bars are 2 µm.
4. Discussion
In thick samples, the resolution of STORM microscopy is degraded by aberrations and scattering. The aberrations increase the size of the PSF and reduce the number of photons in the PSF core both of which serve to reduce the localization precision. The resolution is further reduced because fewer localizations are detected. Scattering increases the background and reduces the number of photons in each PSF further degrading the localization precision. Correcting wavefront aberrations partly compensates for the degradation in imaging in thick samples. While the resolution achieved in the final image is low compared to what can be achieved by STORM in thin samples, it represents a significant improvement over what is possible without wavefront correction. To estimate the resolution improvement from wavefront correction, we have developed an expression for the localization precision for imaging with aberrations
| (3) |
where s is the PSF width, p is the ratio of the reduction of the Strehl ratio, dx is the pixel size, and A is the area over which the scattered photons are distributed, due to aberrations. The 4 radian wavefront error induced by the Drosophila CNS reduces the localization precision by a factor of 45%. Imaging NPF boutons, PSO-AO converged in roughly 25 generations or 625 raw images. Whereas GA-AO required roughly 5000 raw frames. The faster convergence time of PSO compared to GA optimization has been established in the literature [35–37].
In conclusion, we have presented a new machine learning approach for wavefront optimization for STORM imaging using Particle Swarm Optimization. We showed that the combination of the PSO algorithm and an intensity insensitive Fourier metric can effectively correct wavefront aberrations during the acquisition of images of stochastically blinking fluorophores. We demonstrated the PSO-AO-STORM algorithm imaging Neuropeptide F in the Drosophila CNS at a depth of about 100 μm. We demonstrated that PSO-AO-STORM converges a factor of 10 faster than the Genetic Algorithm. In this work and our previous implementation of GAO-AO-STORM, we have achieved imaging in the range of 100-150 μm inside a biological tissue with both refractive and scattering characteristics. STORM deeper inside tissue samples remains a challenging task. With improvements in AO elements (speed, number of actuators) and computational machines, and with the help of techniques such as multi-conjugate AO and woofer-tweeter AO systems [38], many aberrations can be corrected. Scattering presents different challenges and will be harder to correct. Since scattering tissues consist of several layers with independent optical characteristics, future implementations of AO-STORM must take advantage of methods such as multi-conjugate AO [39], or scattering correction [40]. Another challenge is imaging larger fields of view, which could be attained by segmenting of the pupil [41] and correcting for smaller parts of the field of view independently.
Acknowledgments
We thank Dr. James D. Lauderdale for providing facilities for cell culture, and Ms. Snehal Chaudhari and Dr. Edward Kipreos for the roundworms.
Funding
National Science Foundation (NSF) (1350654, 1555576).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References and links
- 1.Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F., “Imaging intracellular fluorescent proteins at nanometer resolution,” Science 313(5793), 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Rust M. J., Bates M., Zhuang X., “Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM),” Nat. Methods 3(10), 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.York A. G., Ghitani A., Vaziri A., Davidson M. W., Shroff H., “Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes,” Nat. Methods 8(4), 327–333 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang B., Jones S. A., Brandenburg B., Zhuang X., “Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution,” Nat. Methods 5(12), 1047–1052 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cella Zanacchi F., Lavagnino Z., Perrone Donnorso M., Del Bue A., Furia L., Faretta M., Diaspro A., “Live-cell 3D super-resolution imaging in thick biological samples,” Nat. Methods 8(12), 1047–1049 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Cella Zanacchi F., Lavagnino Z., Faretta M., Furia L., Diaspro A., “Light-Sheet Confined Super-Resolution Using Two-Photon Photoactivation,” PLoS One 8(7), e67667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meddens M. B. M., Liu S., Finnegan P. S., Edwards T. L., James C. D., Lidke K. A., “Single objective light-sheet microscopy for high-speed whole-cell 3D super-resolution,” Biomed. Opt. Express 7(6), 2219–2236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legant W. R., Shao L., Grimm J. B., Brown T. A., Milkie D. E., Avants B. B., Lavis L. D., Betzig E., “High-density three-dimensional localization microscopy across large volumes,” Nat. Methods 13(4), 359–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tehrani K. F., Xu J., Zhang Y., Shen P., Kner P., “Adaptive optics stochastic optical reconstruction microscopy (AO-STORM) using a genetic algorithm,” Opt. Express 23(10), 13677–13692 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Kubby J. A., ed., Adaptive Optics for Biological Imaging (CRC Press, New York, NY, 2013). [Google Scholar]
- 11.Booth M. J., “Wavefront sensorless adaptive optics for large aberrations,” Opt. Lett. 32(1), 5–7 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Debarre D., Booth M. J., Wilson T., “Image based adaptive optics through optimisation of low spatial frequencies,” Opt. Express 15(13), 8176–8190 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Kner P., Sedat J. W., Agard D. A., Kam Z., “High-resolution wide-field microscopy with adaptive optics for spherical aberration correction and motionless focusing,” J. Microsc. 237(2), 136–147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booth M. J., Neil M. A., Juskaitis R., Wilson T., “Adaptive aberration correction in a confocal microscope,” Proc. Natl. Acad. Sci. U.S.A. 99(9), 5788–5792 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Débarre D., Botcherby E. J., Watanabe T., Srinivas S., Booth M. J., Wilson T., “Image-based adaptive optics for two-photon microscopy,” Opt. Lett. 34(16), 2495–2497 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Débarre D., Botcherby E. J., Booth M. J., Wilson T., “Adaptive optics for structured illumination microscopy,” Opt. Express 16(13), 9290–9305 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Thomas B., Wolstenholme A., Chaudhari S. N., Kipreos E. T., Kner P., “Enhanced resolution through thick tissue with structured illumination and adaptive optics,” J. Biomed. Opt. 20(2), 26006 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Burke D., Patton B., Huang F., Bewersdorf J., Booth M. J., “Adaptive optics correction of specimen-induced aberrations in single-molecule switching microscopy,” Optica 2, 177–185 (2015). [Google Scholar]
- 19.Coles B. C., Webb S. E. D., Schwartz N., Rolfe D. J., Martin-Fernandez M., Lo Schiavo V., “Characterisation of the effects of optical aberrations in single molecule techniques,” Biomed. Opt. Express 7(5), 1755–1767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booth M., “Wave front sensor-less adaptive optics: a model-based approach using sphere packings,” Opt. Express 14(4), 1339–1352 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Wright A. J., Burns D., Patterson B. A., Poland S. P., Valentine G. J., Girkin J. M., “Exploration of the optimisation algorithms used in the implementation of adaptive optics in confocal and multiphoton microscopy,” Microsc. Res. Tech. 67(1), 36–44 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Albert O., Sherman L., Mourou G., Norris T. B., Vdovin G., “Smart microscope: an adaptive optics learning system for aberration correction in multiphoton confocal microscopy,” Opt. Lett. 25(1), 52–54 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Eberhart R., Kennedy J., “A new optimizer using particle swarm theory,” Proceedings of the Sixth International Symposium on Micro Machine and Human Science 6, 39–43 (1995). [Google Scholar]
- 24.Forouhesh Tehrani K., Kner P., “Point spread function optimization for STORM using adaptive optics,” Proc. SPIE 8978, 01–10 (2014). [Google Scholar]
- 25.Fortin F.-A., Rainville F.-M. D., Gardner M.-A., Parizeau M., Gagné C., “DEAP: Evolutionary Algorithms Made Easy,” J. Mach. Learn. Res. 13, 2171–2175 (2012). [Google Scholar]
- 26.Shao L., Kner P., Rego E. H., Gustafsson M. G., “Super-resolution 3D microscopy of live whole cells using structured illumination,” Nat. Methods 8(12), 1044–1046 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Wolter S., Schüttpelz M., Tscherepanow M., VAN DE Linde S., Heilemann M., Sauer M., “Real-time computation of subdiffraction-resolution fluorescence images,” J. Microsc. 237(1), 12–22 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Xu J., Tehrani K. F., Kner P., “Multicolor 3D Super-resolution Imaging by Quantum Dot Stochastic Optical Reconstruction Microscopy,” ACS Nano 9(3), 2917–2925 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Shen P., Cai H. N., “Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food,” J. Neurobiol. 47(1), 16–25 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Dempsey G. T., Vaughan J. C., Chen K. H., Bates M., Zhuang X., “Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging,” Nat. Methods 8(12), 1027–1036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deming R. W., “Phase retrieval from intensity-only data by relative entropy minimization,” J. Opt. Soc. Am. A 24(11), 3666–3679 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Wyant J. C., “Basic Wavefront Aberration Theory for Optical Metrology,” in Applied Optics and Optical Engineering, Shannon R. R., Wyant J. C., eds. (Academic Press, 1992), pp. 28–39. [Google Scholar]
- 33.Kner P., Winoto L., Agard D. A., Sedat J. W., “Closed loop adaptive optics for microscopy without a wavefront sensor,” Proc SPIE Int Soc Opt Eng 7570, 6–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banterle N., Bui K. H., Lemke E. A., Beck M., “Fourier ring correlation as a resolution criterion for super-resolution microscopy,” J. Struct. Biol. 183(3), 363–367 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Hassan R., Cohanim B., Weck O. d., Venter G., “A Comparison of Particle Swarm Optimization and the Genetic Algorithm,” 46th AIAA/ASME/ASCE/AHS/ASC Structures, Structural Dynamics and Materials Conference (2005). [Google Scholar]
- 36.Panda S., Padhy N. P., “Comparison of particle swarm optimization and genetic algorithm for FACTS-based controller design,” Appl. Soft Comput. 8, 1418–1427 (2008). [Google Scholar]
- 37.Duan Y., Harley R. G., Habetler T. G., “Comparison of Particle Swarm Optimization and Genetic Algorithm in the design of permanent magnet motors,” IEEE Power Electronics and Motion Control Conference 6, 822–825 (2009). [Google Scholar]
- 38.Li Q., Reinig M., Kamiyama D., Huang B., Tao X., Bardales A., Kubby J., “Woofer-tweeter adaptive optical structured illumination microscopy,” Photonics Research 5, 329–334 (2017). [Google Scholar]
- 39.Wu T. W., Cui M., “Numerical study of multi-conjugate large area wavefront correction for deep tissue microscopy,” Opt. Express 23(6), 7463–7470 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Zhang X., Kner P., “Binary wavefront optimization using a genetic algorithm,” J. Opt. 16, 125704 (2014). [Google Scholar]
- 41.Park J.-H., Kong L., Zhou Y., Cui M., “Large-field-of-view imaging by multi-pupil adaptive optics,” Nat. Methods 14(6), 581–583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]