Abstract
Genetic information is vital in the cell cycle of DNA-based organisms. DNA polymerases (DNA Pols) are crucial players in transactions dealing with these processes. Therefore, the detailed understanding of the structure, function, and mechanism of these proteins has been the focus of significant effort. Computational simulations have been applied to investigate various facets of DNA polymerase structure and function. These simulations have provided significant insights over the years. This perspective presents the results of various computational studies that have been employed to research different aspects of DNA polymerases including detailed reaction mechanism investigation, mutagenicity of different metal cations, possible factors for fidelity synthesis, and discovery/functional characterization of cancer-related mutations on DNA polymerases.
1. Introduction
The accurate synthesis and maintenance of DNA in cells depend heavily on a complex network of proteins, of which DNA polymerases are crucial players. Each DNA polymerase has a unique task to ensure the successful replication and repair of the genome of a cell. The number of potential errors, mismatches, and damage to DNA is vast, with far-reaching consequences for cell function that can result in disease or, in many cases, death.1−4 These polymerases have complex structures, mechanisms, and dynamics, which in many cases are not fully understood.5−7 Computational investigations of these systems require a variety of techniques and a balance between rigorous theory and computational resources, especially considering the relatively large size of polymerase systems and their multifarious functions.8
DNA polymerases are divided into several families, each of which has similarities in active site structure, processivity, and potential for additional activity (such as exonuclease or lyase capabilities).9−12 In brief, family A polymerases perform excision repair with relatively poor processivity; family B performs DNA repair with the potential to bypass lesions and has variable processivity; family C performs DNA synthesis with a proofreading function and a high level of processivity; family X is involved several types of DNA repair, including base excision repair (BER) and non-homologous end joining (NHEJ); and family Y performs translesion DNA synthesis with a low level of accuracy.8,13−15
Even within families there can be substantial differences in structure and function of the various polymerases. However, overall the polymerase subdomain structures are generally similar and are usually described as a right or left-hand. Within the hand there are the finger, thumb, and palm domains. The palm domain is where the DNA is bound while the polymerase is active, and also where the active site for phosphoryl transfer/nucleotide addition takes place, which is discussed more thoroughly below. The thumb domain is usually thought to be involved in the position and movement of the DNA through the polymerase, and the finger domain is involved with the alignment of the incoming nucleotide with the template strand and, potentially, recognition of the correct incoming nucleotide.16
Computational simulations based on detailed classical and/or quantum analysis have been applied to gain atomic-level insight into this important class of enzymes. Molecular dynamics (MD) simulations allow for the investigation of large and small-scale structural differences over time and can provide better understanding regarding how large, flexible polymerases adapt to relatively small structural changes, such as mutagenic lesions, and how they maintain fidelity.17−21 These simulations can also provide information on large scale motions and structural changes such as switching between polymerase and exonuclease activities.22,23 As a recent example, Kim et al. have provided a novel explanation for how DNA polymerase β (Polβ) becomes inactivated by oxidized guanine by applying targeted MD to obtain the specifics of the transition from the open active state of Polβ to the inactive closed state by disruption of a water network and, eventually, the active site.24 Yang et al. have also used targeted MD to investigate how Polβ changes between correctly paired terminal bases versus incorrectly paired ones, and show aspects of the overall motion of Polβ, and of a rotation of Arg258 in the active site that has been correlated to the shift from the active to inactive state.25 Additionally, molecular mechanics (MM) work by Jia et al. uncovered the N-clasp structural feature of Polκ that allows for nearly error-free bypass of mutagenic lesions.26
Quantum mechanical (QM) and hybrid QM/molecular mechanics (QM/MM) methods have also frequently been used to investigate polymerase function and activity. QM methods alone are highly accurate but are difficult or impossible to apply to large enzymatic systems, and so usually QM/MM is used to obtain information on the complex electronic structure of the metal ions and accurate energetics for reaction mechanisms.7,27−30 QM/MM subdivides the calculated system into a small region of 100 atoms or less that is calculated with QM, usually the active site of the polymerase along with relevant ions and cofactors, and the MM region, containing the rest of the polymerase and solvent. This allows for insight into particulars of the reaction mechanisms and metal catalysis that differ between specific polymerase families and sometimes within polymerases themselves, as discussed further in section 2 and throughout the perspective. These cannot be investigated with MD alone since they require investigation into bond breaking and forming and the movement of electrons. Family X and Y polymerases in particular have been the object of a large number of QM/MM31 simulations.19,32−39 To name a few examples, Hummer et al. have applied combined QM/MM techniques to investigate a variety of aspects about B. halodurans ribonuclease (RNase), including the reactivity of specific metals and the overall mechanism, gaining insights into nucleotide cleavage and transfer reactions.40 Combined QM/MM simulations and structural/biochemical experiments, reported by Perera et al., have given fascinating insight into the role of metal ions in the active site of Polβ, and their dynamics of facilitation and inhibition in relation to pyrophosphorolysis, or the reverse of the usual addition reaction of polymerases.41 QM/MM has also been used to gain mechanistic insight into several Y family polymerases, which are unique in their ability to bypass bulky chemical lesions. Dpo4, which can perform error-prone translesion synthesis, has a particularly solvent-exposed active site; Wang et al. have shown a corresponding mechanism for nucleotidyl-transfer reactions through water-dependent pathways using QM/MM calculations.42 Additionally, QM/MM work by Hoffmann et al. shows a similar water-mediated transfer reaction for Polκ, which has a much higher fidelity as compared to Dpo4.43 Further discussion of mutagenic lesions and Dpo4 is found later in the perspective in section 5.
The use of advanced force fields for classical or QM/MM simulations applied to complex biomolecular systems also provides new insights and results, showing new avenues for exploring the role of electronic polarization and improved treatments of permanent electrostatics.44−48 These advanced methods can improve solvent boundaries for QM/MM49 and the description of metal cations.50−52
We have developed a variety of methods for simulations and analysis, which have been applied to various biochemical systems including DNA polymerases. In this perspective, we discuss our studies on DNA polymerase catalysis in section 2, metal mutagenicity in section 3, fidelity determinants in section 4, mutagenic lesion bypass in section 5, and cancer mutations and their effects on the structure and function of polymerases in section 6.
2. Computational Investigation of DNA Polymerase Reaction Mechanisms
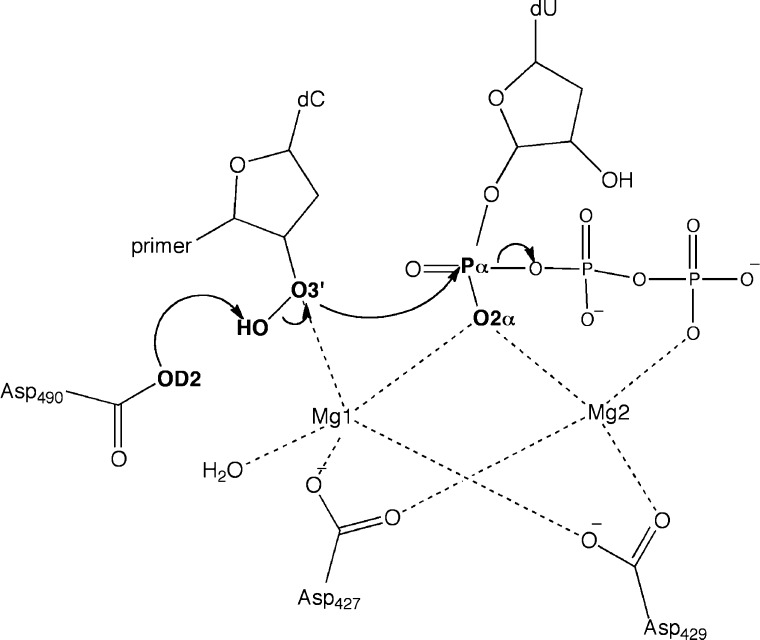
As briefly touched on in the Introduction, DNA polymerases possess a remarkable degree of specificity and variability in function. That said, the reaction mechanism of DNA synthesis is very consistent across polymerases. The reaction mechanism for DNA synthesis by DNA polymerases involves a nucleophilic attack on the alpha phosphate (Pα) of the incoming nucleotide by the O3′ of the primer-terminus nucleotide with the concomitant formation of pyrophosphate (PPi). This reaction results in the formation of an O–P bond and therefore the addition of the incoming nucleotide, as shown schematically in Figure 1. In general, this mechanism is considered to have two metal ions, though that can vary, as will be discussed later in the perspective.53
Figure 1.
Mechanism of polymerase addition reaction for Pol λ. Reproduced from Fang, D., Chaudret, R., Piquemal, J.-P., and Cisneros, G. A. (2013) Toward a Deeper Understanding of Enzyme Reactions Using the Coupled ELF/NCI Analysis: Application to DNA Repair Enzymes. J. Chem. Theo. Comp.9, 2156–2160. Copyright 2013 American Chemical Society.54
Several reaction mechanisms have been investigated to assess the particulars of proton transfer from the O3′ and seem to vary depending on the particular DNA polymerase. The proton transfer from the nucleophilic O may involve a direct proton transfer to an oxygen on the Pα, indirectly through an ordered water, or to one of the conserved aspartate/glutamic acid residues in the active site.36,37,55 We have performed various theoretical studies on the reaction mechanism of DNA polymerase λ (Polλ).56 Polλ is an X family polymerase involved in NHEJ and can fill small DNA gaps (1–2 nucleotides). QM/MM simulations of the reaction mechanism for the three H+ transfer possibilities in Polλ suggest that in this case the proton is transferred to the conserved D490 in the active site.56
DNA polymerases have been shown to be able to perform their catalytic activity using different divalent cations in the active site.4,57 Polλ can employ either Mn2+ or Mg2+ to synthesize DNA.58 We performed QM/MM simulations using both metal cations to determine the differences in the reaction mechanism in Polλ. Our results show that the energy barriers for both Mn2+ and Mg2+ are in good agreement with experimental estimates.56 In both cases, the reaction is calculated to proceed through a two step mechanism involving the initial proton transfer from O3′ to D490, followed by the nucleophilic attack of O3′ on the alpha phosphate of the incoming nucleotide. The energy barrier associated with the reaction is ∼2 kcal/mol smaller for the Mn2+ catalyzed reaction than for Mg2+, in agreement with experiment.
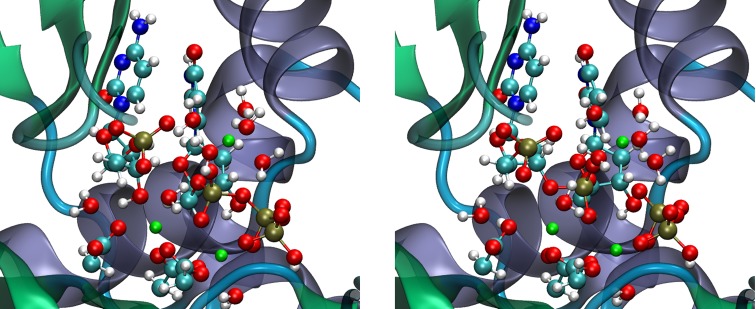
Further insights into the reaction were provided by investigating the impact of individual residues on the critical points along the reaction path. To this end, the non-bonded interaction energy between each residue in the protein and the active site can be analyzed by means of an energy decomposition analysis (EDA). This analysis provides a qualitative assessment of the role of each amino acid on the reaction and can be used to compare to available mutagenesis experiments, or to predict possible mutation sites for subsequent experimental analysis. The residues found to be important for catalysis in Polλ are shown in Figure 2.56 None of the predicted residues had been experimentally investigated in Polλ prior to our simulations. Only three homologous residues had been mutated experimentally in Polβ, and all exhibited impact on catalysis. Following our computational simulations, mutations on Polλ were performed on R38659 and K427,60 which confirmed the role of these residues on activity and stabilization of the DNA substrate. These residues located in the so-called second shell around the active site also gave rise to the analysis of cancer SNPs as described in subsection 6.
Figure 2.
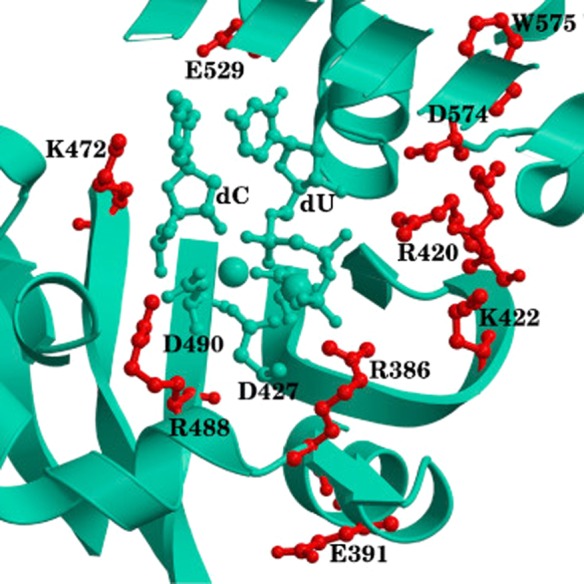
Catalytically relevant residues in Polλ. Residues colored in red have large energetic contributions to transition state stability or are present in all transition states, residues in green correspond to the QM subsystem. Reprinted from Cisneros, G. A., Perera, L., García-Díaz, M., Bebenek, K., Kunkel, T. A., and Pedersen, L. G. (2008) Catalytic mechanism of human DNA polymerase λ with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies. In DNA Repair, Vol. 7, pp 1824–1834, Copyright 2008, with permission from Elsevier.56
E. coli DNA polymerase III (Pol III) is a complex that is relatively large composed of 10 subunits with multiple functions including DNA replication, post-replicative repair, and exonucleolytic proofreading.61,62 We have investigated the ϵ subunit in particular and its function as an exonuclease.63 The ϵ subunit (termed ϵ in the subsequent discussion) preferentially removes incorrectly paired nucleotides and is a crucial component of DNA Pol III’s high rate of fidelity.64,65
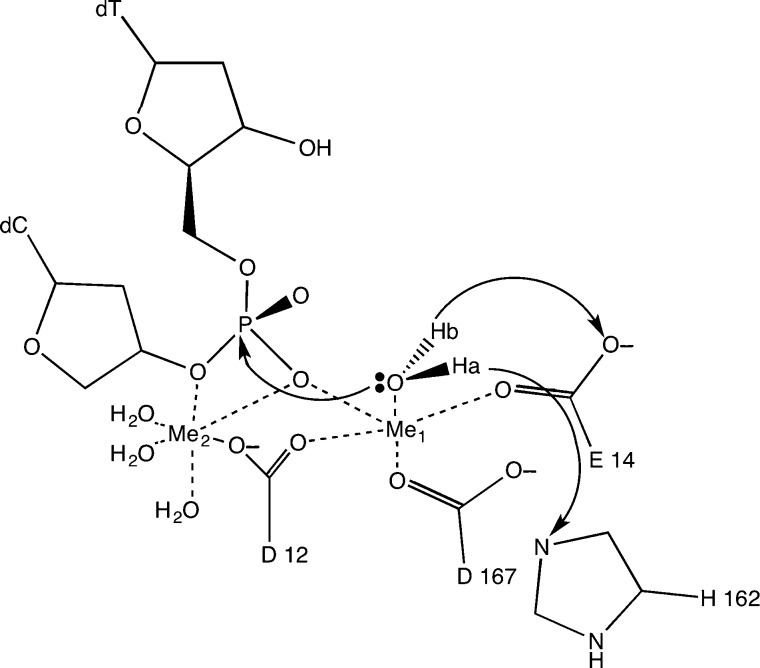
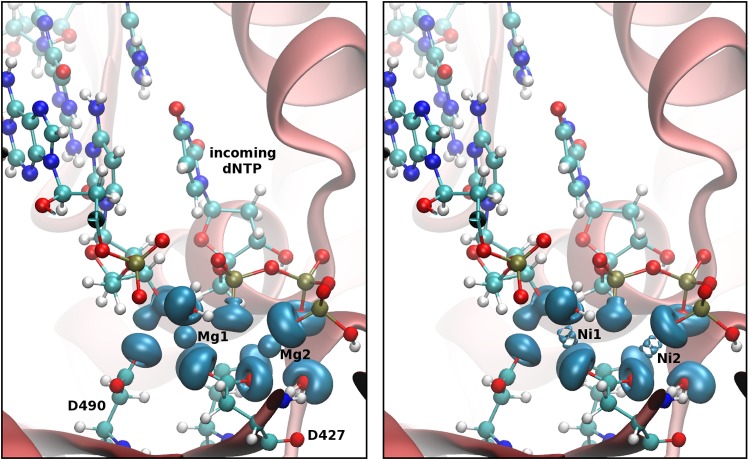
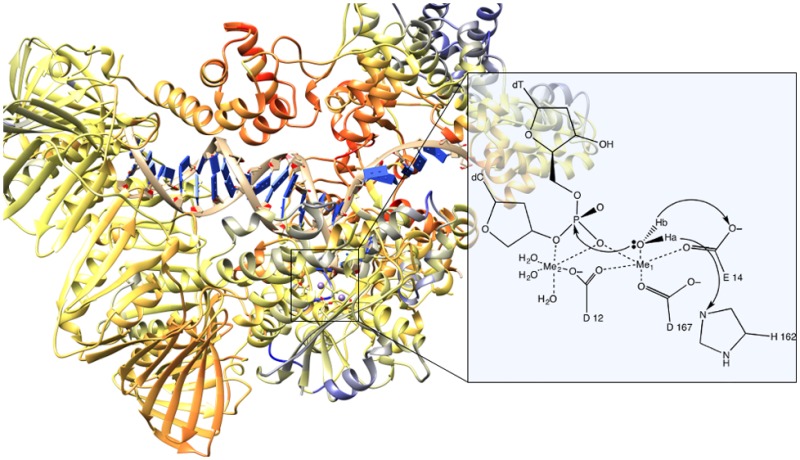
One metal and two metal mechanisms have been proposed66 for the proofreading mechanism; our theoretical work focused on the two-metal mechanism.67 Additionally, the reaction catalyzed by ϵ in vivo has been reported to be faster with Mn2+ than with Mg2+, so the mechanisms for both of these metals were explored.66 In both cases, the two metal mechanism involves a nucleophilic attack on the base to be excised by an attacking hydroxide ion; essentially, the reverse of the standard polymerase addition reaction. The formation of this ion is facilitated by the catalytic metal, Me1, as seen in Figure 3.
Figure 3.
Reaction mechanism for the ϵ subunit of DNA polymerase III holoenzyme.
The ϵ subunit can associate with an additional θ subunit, which has been proposed to promote its activity.68 The θ subunit also has a smaller homologue from bacteriophage P1, HOT, which can also stabilize ϵ and perform a similar function.69 Crystal structures of the ϵ–HOT complex with two metals in the active site were used to explore the mechanism using QM/MM and similar techniques as described for Polλ. Our simulations involved three different systems including the ϵ–HOT complex with Mg2+ or Mn2+, and free–ϵ with Mg2+. The calculated energy barriers suggest that ϵ is slightly more active with Mn2+ than with Mg2+; the ϵ–HOT complex shows a slightly lower activation barrier compared with free-ϵ consistent with experiment.
A particularly interesting aspect of this study involves the number of ligands coordinated to the catalytic metal in the active site. The reported crystal structure for ϵ–HOT, pdbid 2IDO, suggests that the catalytic metal is pentacoordinated.70 This is an unusual coordination number, especially for Mg2+, which is considered to be the natural divalent cation for both DNA polymerases and exonucleases. Moreover, this pentacoordination is preferentially maintained along the reaction path according to our QM/MM simulations.67 The more typical hexacoordination was also investigated but did not result in chemically reasonable structures for the catalytic step. This was observed for both Mn2+, which can maintain a pentacoordinated structure in the first coordination shell, and for Mg2+, for which it is less common but still present in other enzymes. This unusual coordination sphere has been subsequently reported in other enzymes.40,71,72
Additionally, we performed EDA in a similar manner to Polλ to determine residues that either stabilize or destabilize the TS for the proofreading mechanism (Figure 4).67 Several residues near the active site and on HOT were shown to have a significant effects on catalysis. Four of these residues had been studied experimentally previously. All four showed mutator effects proportional to their (de)stabilization energies, with D103 and D129 showing particularly strong mutagenic effects.
Figure 4.
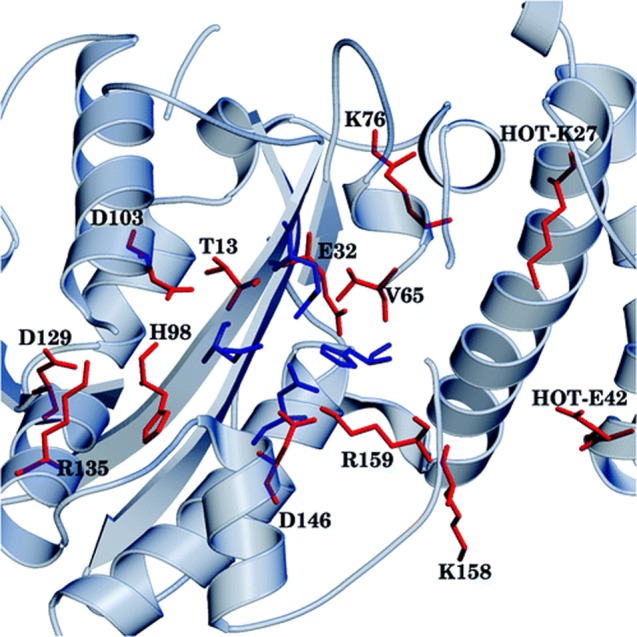
Catalytically important residues in ϵ–HOT. Top: Residues colored in red have large energetic contributions to transition state stability and/or are present in all transition states, residues in blue are active site residues. Reproduced from Cisneros, G. A., Perera, L., Schaaper, R. M., Pedersen, L. C., London, R. E., Pedersen, L. G., and Darden, T. A. (2009) Reaction Mechanism of the ε Subunit of E. coli DNA Polymerase III: Insights into Active Site Metal Coordination and Catalytically Significant Residues. J. Am. Chem. Soc.131, 1550–1556. Copyright 2009 American Chemical Society.67
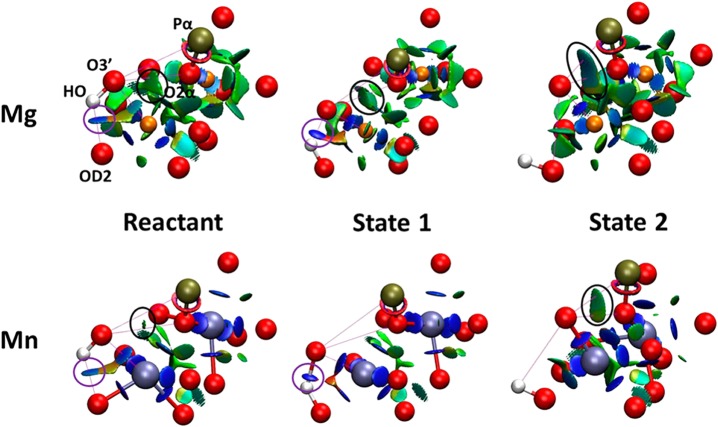
We have also developed several methods to improve simulation accuracy and enable deeper analyses. One of these involves the combination of two quantum interpretative techniques, electron localization function (ELF) and noncovalent interaction (NCI) analysis, to investigate covalent and non-covalent regions. This combination was originally developed to investigate organic reactions in gas phase73 and subsequently extended by us to enzymatic reaction mechanisms.54 We applied ELF/NCI analysis to further examine the role of the electronic structure on the intermolecular interactions and metal coordinations related to the reactions catalyzed by Polλ and ϵ. For Polλ, the ELF/NCI analysis revealed that the interaction between O3′ on the primer and Mg2+ was weaker than for Mn2+, as indicated by the associated NCI surface. Concomitantly, the interaction between O3′ and the phosphate oxygen coordinated to the metal ions was stronger for Mg2+ than for Mn2+ (Figure 5). These results are consistent with our previous QM/MM (and experimental) results where the barrier for proton transfer is higher for Mg2+;63 further details about the ELF results can be seen in Subsection 3.
Figure 5.
NCIplot for Pol λ reactant structures, with strong attractive forces denoted in blue, strong repulsive forces denoted in red, and weak forces denoted in green. Reproduced from Fang, D., Chaudret, R., Piquemal, J.-P., and Cisneros, G. A. (2013) Toward a Deeper Understanding of Enzyme Reactions Using the Coupled ELF/NCI Analysis: Application to DNA Repair Enzymes. J. Chem. Theo. Comp.9, 2156–2160. Copyright 2013 American Chemical Society.54
Similarly, for the ϵ subunit of Pol III, the Mn2+ ion shows electronic density splitting in the surfaces associated with the metal ions. This splitting is not present for Mg2+, indicating that the Mn2+ ion has stronger interactions with pertinent residues in the active site as compared to Mg2+. Interestingly, the catalytic metal exhibits five splitting basins as shown in Figure 6, which is also consistent with the pentacoordination of Me1 as explained above. Further insights into metal mutagenicity using ELF/NCI are discussed below in section 3.
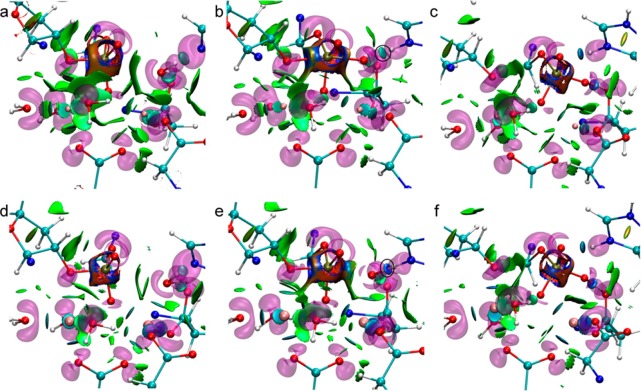
Figure 6.
Combined NCI and ELF surfaces for the ϵ subunit of Pol III. ELF surfaces are shown in transparent purple, and NCI surfaces shown as solid red/green/blue surfaces. Panels a–c show the reactant, transition state and product structures for Mg2+, while panels d–f show the same for Mn2+. Reproduced from Fang, D., Chaudret, R., Piquemal, J.-P., and Cisneros, G. A. (2013) Toward a Deeper Understanding of Enzyme Reactions Using the Coupled ELF/NCI Analysis: Application to DNA Repair Enzymes. J. Chem. Theo. Comp.9, 2156–2160. Copyright 2013 American Chemical Society.54
2.1. Third Metal in Polλ Mechanism
As explained above, the synthesis of DNA by DNA polymerases involves a general two metal ion mechanism. Recently, Nakamura et al. reported the existence of a transient third metal ion in the in crystalo reaction of Polμ based on time-resolved X-ray crystallography.74 Wilson and co-workers have published similar evidence for Polβ.75 On the basis of these results, Perera et al. have performed extensive computational simulations to investigate the role of this third cation in the reaction mechanism of Polβ.27,41 Their results suggest that this third metal does not affect the forward chemical step (DNA synthesis) and significantly impairs the pyrophosphorolysis (backward) reaction.
Polλ is 35% identical to Polβ and thus the question arises of whether this transient third metal is also present in the Polλ reaction. We have explored the possible role of the third metal ion in Polλ using our own QM/MM program, LICHEM.76 As reported in ref (74), the comparison of the calculated energies of Polλ reactant and product structures with a third metal included in the active site results in a stabilization of the product relative to the reactant (Figure 7). This is in contrast to our original reported path for the mechanism of Polλ, where the calculated reaction energy is endoergic by around 4–5 kcal/mol. Thus, our initial calculations are consistent with the work of Perera et al. and Genna et al. in that the third metal stabilizes the product structure and thus reduces the likelihood of the backward reaction.27,77 In addition, the resulting distances for the active site in the optimized structures in Polλ are similar to those reported by Perera et al., with the third metal coordinating to the apical oxygen on Pα with distances around 2.1 Å for reactant and product.
Figure 7.
Close-up of the active site for the optimized reactant (left) and product (right) structures of Polλ with the third metal in the active site.
3. Metal Mutagenicity in DNA Polymerases
The requirement of metal cations to facilitate the reaction in DNA Pols gives rise to the possibility of replacement of the ”natural” cation, Mg2+, by other metals. It is known that several cations are carcinogenic or genotoxic, and these effects are due to the inhibition of DNA repair transactions.78 Indeed, the replacement of Mg2+ ions in the active site of DNA polymerases by metals such as Cr(II), Cd(II), Ni(II), Ca(II), Na(+), or Mn(II) results in either a decrease in synthesis fidelity or outright inhibition.57 Pelletier et al. investigated the effect of different metals in ternary structures of DNA Polβ.79 Their results show that metals other than Mg2+ result in changes in the active site geometry.
On the basis of our previous results on the reaction mechanism of Polλ, we investigated the effect of nine different cations including Mg2+, Na+, Ca2+, Zn2+, Co2+, Cr2+, Cu2+, Mn2+, and Ni2+ using QM/MM methods.80 In addition to the structural effects, we performed electron localization function (ELF) analyses on all the resulting systems to gain insights into the effects of the metals on the electronic structure.
The structures obtained from our simulations indicate that the overall arrangement of the active site is largely unaltered regardless of the cation, with only subtle differences arising such as very slight increases of 0.2–0.4 Å between the metals and the atoms in the first coordination shell. Similarly, the nucleophilic attack distance (O3′ to Pα) increases slightly, ∼0.3 Å, for two of the three inhibitor cations, Na+ and Ca2+.
To further understand the differences between these systems given the very slight changes in geometry, we turned to the analysis of the electronic structure via ELF. The results from the ELF analysis are striking in that out of the nine tested cations, significant differences were observed between the calculated systems with inhibitor and mutagenic cations compared to the Mg2+ system.80 In particular, the ELF analysis indicates that when two Mg2+ occupy the active site, the cations and incoming nucleotide experience a particular electronic polarization. By contrast, the inhibitory (Ca+, Na2+, and Zn2+) and mutagenic (Co2+, Cr2+, Cu2+, Mn2+, and Ni2+ cations exert a different (hyper)polarization on the oxygen atoms on the triphosphate of the incoming nucleotide. In addition, the enzyme environment also significantly affects the metals in that only the Mg2+ shows a single basin around the metals, whereas all other metals are (hyper-)polarized in such a way that the metal exhibits multiple basins (Figure 8).
Figure 8.
ELF analysis for the active site of Polλ with Mg2+ (left) and Ni2+ (right) occupying the catalytic and nucleotide binding metal sites. Reproduced from Chaudret, R., Piquemal, J.-P., and Cisneros, G. A. (2011) Correlation between electron localization and metal ion mutagenicity in DNA synthesis from QM/MM calculations. Phys. Chem. Chem. Phys.13, 11239–11247, with permission from the PCCP Owner Societies.80
Thus, the detailed analysis of the electronic structure of Polλ suggests that the replacement of the natural metals by other cations results in a change in polarization that may be responsible for the observed inhibitory or mutagenic effects of the replacing cations depending on their identity.80
4. DNA Synthesis Fidelity Checking
The accurate replication of the genomic data is crucial for organismal survival. DNA polymerases are arguably some of the most important enzymes in the transactions involved in replication. For example, the replication error rates in E. coli are around 1 in 10–10.64 This very low error rate (high fidelity) is due to at least three steps that are involved in the replication of DNA: (1) base selection, (2) proofreading, and (3) mismatch repair. DNA Pol I is a key player in the achievement of the low error rates due to its participation in two of the three steps via three enzymatic activities: DNA polymerase, 5′ →3′ exonuclease, and 3′ →5′ exonuclease.81
The Klenow fragment (KF) is a functional fragment of the POLA gene, which codes for PolI, and contains the polymerase and 3′ →5′ exonuclease activities.82−91 The KF has provided a very useful system to learn a great deal about DNA polymerases including reasons for nucleotide misinsertion discrimination and decrease in synthesis fidelity rates, among other insights.84,91 The KF is a member of the A family of polymerases, which includes various other highly studied polymerases such as DNA PolI from Thermus aquaticus, Klentaq fragment, and Bacillus stearothermophilus [Bacillus fragment (BF)].
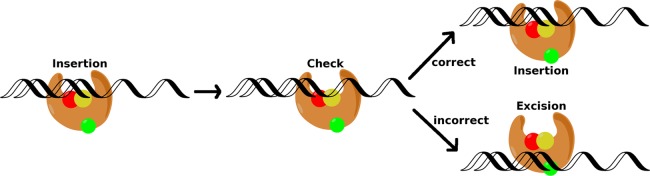
The discrimination of correct versus incorrect nucleotide insertion in KF has been investigated with single-molecule Förster resonance energy transfer (FRET) by Christian et al.92 The single-molecule results indicated a movement along the DNA following synthesis that had not been previously observed. The authors proposed that KF translocates two bases downstream along the DNA strand following a nucleotide incorporation before backtracking for the next round of incorporation (Figure 9).92 On the basis of these results, Christian et al. proposed a possible post-instertion fidelity checking site to check that the newly incorporated base is not mismatched with the templating base. The checking site is assumed to be located immediately behind the pre-insertion site (n – 1 position along the DNA)
Figure 9.
Schematic diagram of the proposed fidelity checking process in DNA polymerase I. Following nucleotide insertion, the DNA is translocated to the checking site. If the inserted base pair is a correct pair, the polymerase continues. If not, the DNA is shifted to the exonuclease domain and the incorrect base pair is excised. Reproduced from Graham, S.E., Syeda, F., and Cisneros, G. A. (2012) Computational Prediction of Residues Involved in Fidelity Checking for DNA Synthesis in DNA Polymerase I. Biochemistry51, 2569–2578. Copyright 2012 American Chemical Society.93
We employed various computational techniques to investigate the location of potential residues that could be involved in the putative checking site on KF, BF, and Klentaq.93,94 MD simulations were performed on all three polymerases containing correctly or incorrectly paired bases with either a blunt end (KF, BF)93 or a two-base overhang template (Klentaq).94 The resulting ensembles were subjected to various analyses including energy decomposition (EDA), electrostatic free energy response (EFER),95,96 and non-covalent interaction (NCI)97,98 to investigate whether/if any residues show changes in interaction with correctly or incorrectly paired bases.
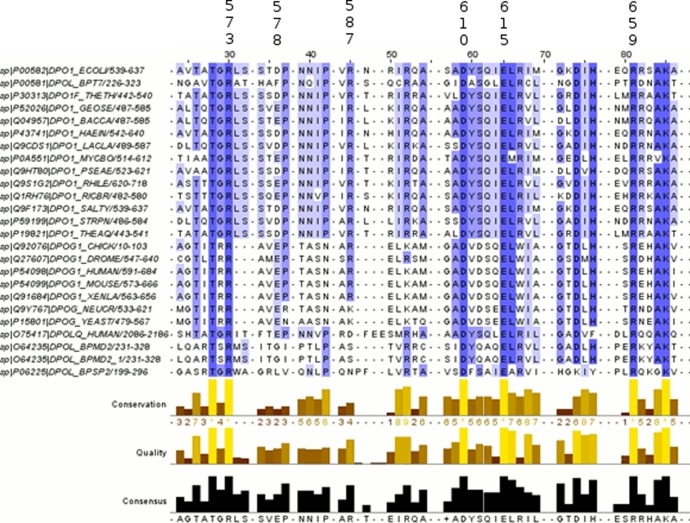
The interaction analyses revealed that there are a number of residues around the putative checking site (Figure 10). Correlation of EDA, EFER, and NCI analyses for all three systems (KF, BF, and Klentaq) suggests that there are six residues with altered interactions with mismatched bases compared to correctly paired bases in the putative checking site. Figure 11 shows a condensed sequence alignment for A family polymerases that indicates the six residues with altered interactions.
Figure 10.
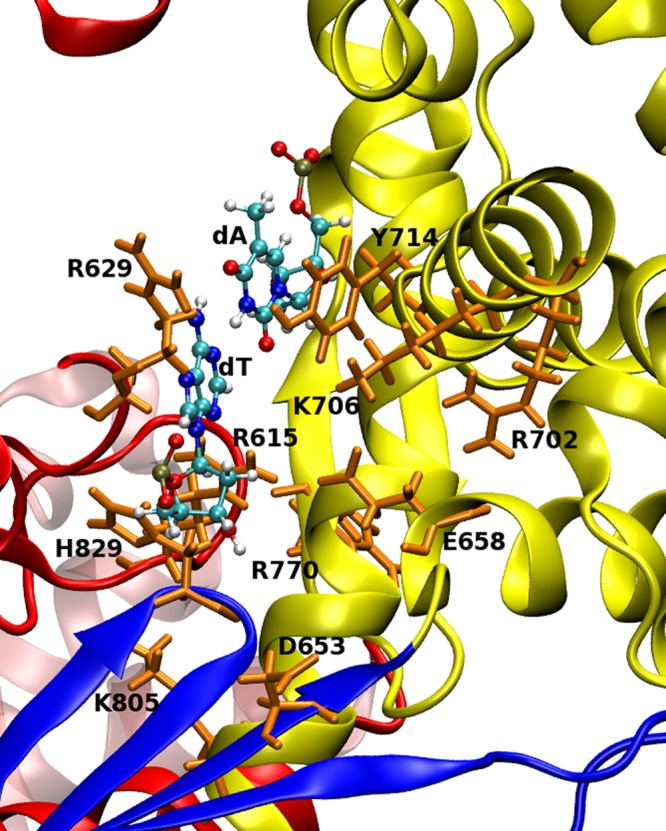
Positions of residues with altered interactions in KF and BF (orange sticks). The structure and numbering correspond to BF. Polymerase subdomains are colored red, blue, yellow, and mauve for the palm, fingers, thumb, and exonuclease, respectively. DNA bases (T:A) in the preinsertion site are shown as balls and sticks. Reproduced from Graham, S.E., Syeda, F., and Cisneros, G. A. (2012) Computational Prediction of Residues Involved in Fidelity Checking for DNA Synthesis in DNA Polymerase I. Biochemistry51, 2569–2578. Copyright 2012 American Chemical Society.93
Figure 11.
Condensed alignment for family A DNA polymerases with the common residues with altered interactions in the putative (n – 1) checking site indicated. The numbering corresponds to Klentaq. Reproduced from Elias, A. A., and Cisneros, G. A. (2014) Computational Study of Putative Residues Involved in DNA Synthesis Fidelity Checking in Thermus aquaticus DNA Polymerase I. In Adv. Protein. Chem. Struct. Biol.96, 39–75, Copyright 2014, with permission from Elsevier.94
Various residues that show altered interactions from our computational analysis have been reported to result in changes to DNA fidelity synthesis in KF and Klentaq. In particular, residues R668 and R682 in KF (R615, R629 in BF and R573, R587 in Klentaq) have been reported to be important in the partitioning of the DNA primer terminus between the polymerase and exonuclease active sites as well as mispair discrimination for synthesis fidelity.88,89 The correlation between experimental mutagenesis studies indicating the role of these residues in fidelity and the possible involvement of these residues in the fidelity-checking step provide possible targets for further investigation of this intriguing hypothesis.
5. Effects of Mutagenic Lesions on Structure and Function of DNA Polymerase IV
Y-family polymerases, such as DNA polymerase IV (Dpo4), have large, flexible, and solvent exposed active sites that can accommodate bulky lesions, which is likely crucial to their ability to perform translesion synthesis (TLS). DNA can be damaged in a variety of ways, including the incorporation of endogenously produced molecules such as polycyclic aromatic hydrocarbons (PAHs).99 Benzo[a]pyrene (B[a]P)is a particularly bulky adduct that can add to DNA in several conformations with different effects on DNA replication.5,100
We have recently carried out computational simulations to aid in the understanding of biochemical and single molecule FRET experiments for Dpo4 with B[a]P damage. The experimental results indicate that the cis conformation of B[a]P adducted to guanine exhibits two different behaviors depending on the solvent environment; in a pure water system, the adduct takes the place of the guanine residue, intercalated within the DNA helix, and replication does not proceed past that point.101 In a 10% dimethyl sulfoxide (DMSO)/90% water mixture, two different FRET states are observed and Dpo4 is able to continue the DNA synthesis (TLS). Additionally, a G-G mismatch is more stable than the correct G-C base pairing, resulting in increased errors in TLS.
We performed classical MD simulations on Dpo4 with B[a]P incorporated to the DNA in pure water and in the water/DMSO mixture.101 Our results indicate that the B[a]P indeed intercalates with the DNA and does not allow the adduct to be everted from the double strand when in pure water. Conversely, in the water/DMSO mixture, the B[a]P adduct flips out into an open space next to the active site underneath the finger domain in a solvent exposed conformation, allowing replication to continue (Figure 12). This is enabled by DMSO reducing the dielectric constant of the solvent around the B[a]P adduct (effectively microsolvating the B[a]P) and allowing it to be everted from the double helix and thus avoiding the arrest of the polymerase.
Figure 12.
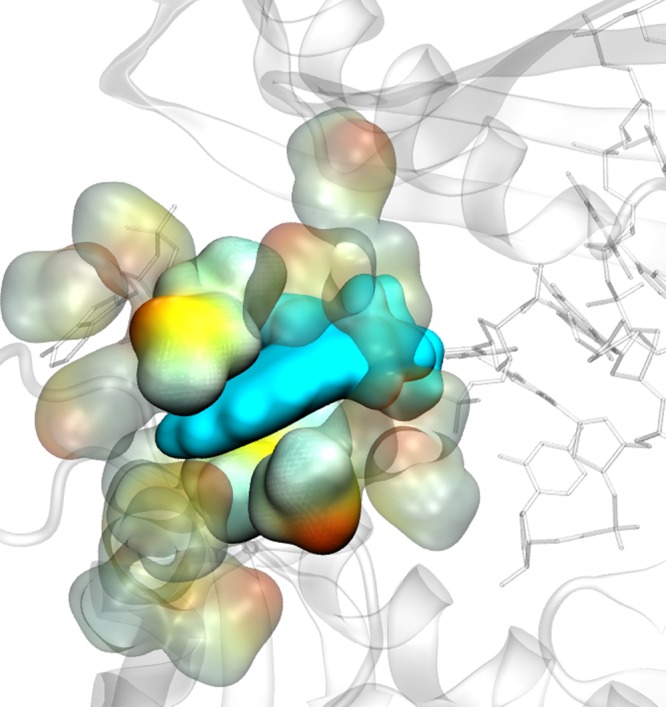
Microsolvation of the flipped out conformation of cis-benzo[a]pyrene (blue surface) with dimethyl sulfoxide (yellow and green surfaces).
6. Discovery and Characterization of Cancer Mutants on DNA Pols
The discovery of various residues involved in catalyis in the “2nd-shell” of Polλ (Figure 2) raised a question about how many of these residues are conserved among all human polymerases. A detailed sequence–structure alignment reveals that five of the nine catalytically relevant second-shell residues are at least partially conserved (Figure 13). This points to the evolutionary importance of these residues for polymerase activity and/or function.
Figure 13.
Second-shell residues conserved among 14 different human polymerases.
On the basis of these results, we wondered if there are any natural variants associated with a disease state that result in mutations of these second-shell residues and how these may affect the structure and function. Several polymerases have been linked specifically to cancer, such as Polη, which has a well-documented relationship with skin cancer.3,102,103 To this end, we developed a methodology that combines a targeted search for single nucleotide polymorphisms (SNPs) on selected genes, statistical validation of the uncovered SNPs, and computational simulations of the wild-type and SNP variants to determine if/whether the disease variant shows a difference in its structure or function. The search algorithm was implemented in a computer program called Hypothesis Driven–SNP–Search, or HyDn–SNP–S.104
The first application of HyDn–SNP–S was performed to uncover SNPs on all DNA polymerase genes for four different genome-wide association studies comprising melanoma, lung, prostate, and breast cancer (Figure 14). Following logistical regression, 79 exonic non-synonymous SNPs were found to be statistically significantly associated with disease status.104 Further analysis to determine derived haplotypes of the uncovered SNPs with the selected cancer phenotypes resulted in two new haplotypes for breast and prostate cancer. The first haplotype is composed of two SNPs on POLL, rs3730477 and rs3730463, and results in a three-fold increase in risk of breast cancer. The second haplotype is constructed from three SNPs on POLG, rs3087374, rs2351000, and rs2247233. The resulting haplotype indicates having two copies of the G–C–A haplotype (p = 0.008) results in a 9.64-fold increase in prostate cancer disease status.104
Figure 14.
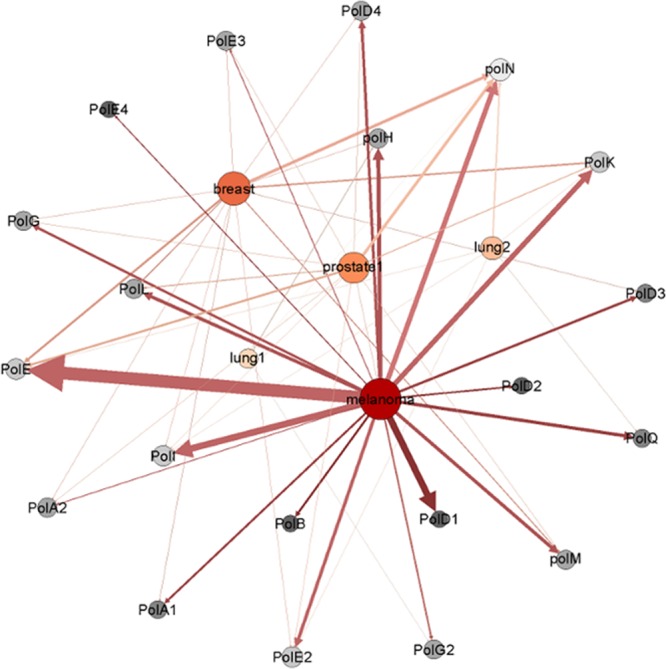
Edge–node network of the HyDn–SNPs results for DNA polymerases. Phenotypes and polymerases are shown as nodes, edges are weighted by total number of SNPs connecting each phenotype to each polymerase. Reproduced from Swett, R. J., Elias, A., Miller, J. A., Dyson, G. E., and Cisneros, G. A. (2013) Hypothesis driven single nucleotide polymorphism search (HyDnSNP-S). In DNA Repair12, 733–740, Copyright 2013, with permission from Elsevier.104
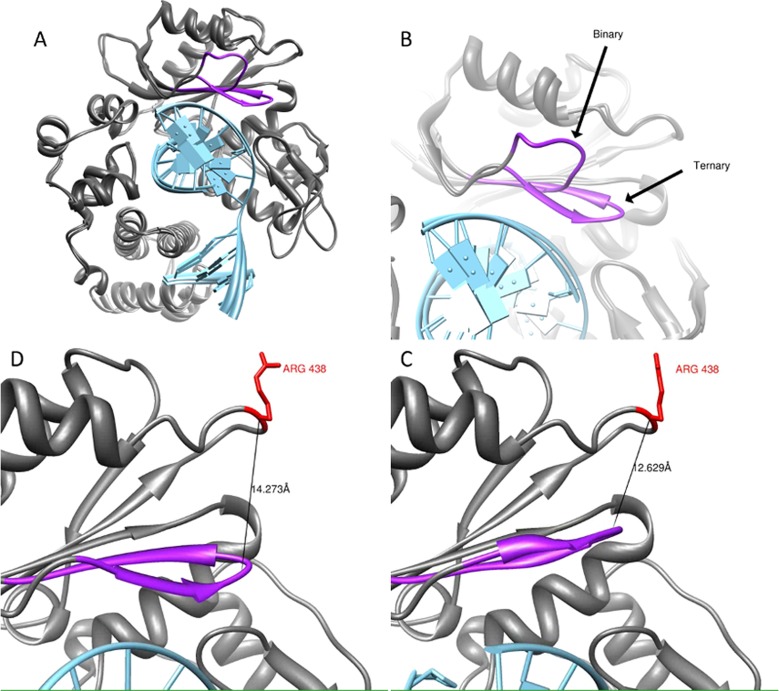
In addition to uncovering a large number of previously unknown cancer missense SNPs on DNA polymerases, we performed computational simulations on one particular SNP. One of the SNPs on Polλ, rs3730477, is associated with breast cancer status and results in the R438W variant. R438 is located around 12 Å away from the active site (Figure 15). This residue sits at the end of loop1, which is crucial for Polλ fidelity, and the SNP variant has been shown to give rise to chromosomal abnormalities.105,106 We performed MD simulations on the binary and ternary structures of wild-type and the R438W variant to determine the possible impact of the breast-cancer mutation on the structure or function of Polλ. Our MD simulations indicate that the mutation of R by W at position 438 results in a change in the dynamics of loop 1, without affecting the overall motions of the rest of the protein. Our results suggest that the alteration of the motion of loop 1, which is experimentally known to be crucial for fidelity, could affect the function of this critical structural feature and help explain the observed decrease in fidelity.104 Recently, on the basis of our work, Sweasy and co-workers have confirmed that rs3730477 can be used as a biomarker for breast cancer, and, more importantly for individuals with this particular mutation, estrogen treatment may increase the risk of breast cancer status.107
Figure 15.
(A) Overlay of Polλ in the binary and ternary conformations. DNA is shown in light blue, and Loop 1 is shown in purple. (B) Differences in Loop 1 orientation between the two conformations. Distance between position 438 and Loop 1 following an interpolation between the two structures at its (D) furthest and (C) closest approaches. Reproduced from Swett, R. J., Elias, A., Miller, J. A., Dyson, G. E., and Cisneros, G. A. (2013) Hypothesis driven single nucleotide polymorphism search (HyDnSNP-S). In DNA Repair12, 733–740, Copyright 2013, with permission from Elsevier.104
7. Summary and Perspective
Computational simulations of DNA polymerases using classical, quantum mechanical, and hybrid QM/MM methods have enabled a deeper understanding of a variety of facets related to their structure, function, and mechanism. Detailed analysis at the atomic and electronic structure level enabled by these methods has been shown to be useful to gain insights into factors of metal mutagenicity, nucleotide discrimination, and impact of protein residues on catalytic activity. The application of novel bioinformatic techniques to uncover cancer-associated SNPs in DNA polymerases has resulted in a large number of statistically significantly associated biomarkers on DNA polymerases for four distinct cancer phenotypes. The use of computational simulations on these cancer mutants has provided possible explanations for the impact of these cancer mutations on the function of the proteins.
In the future, it is expected that the improvement of simulation methods, including more accurate classical potentials, coupled with better levels of QM theory, enabled by larger computing capacity will provide more opportunities for deeper insights into these systems. For example, longer simulation times will enable the possibility to study the transfer of ssDNA from the polymerase to the exonuclease to investigate steps required for proofreading. Continuing the use of computational simulations to investigate the impact of cancer-associated SNPs on DNA polymerase structure, function, and dynamics will provide the possibility to investigate the effect of these cancer mutations at an atomic level. It is clear that the use of computational simulations of DNA polymerases is a robust and well-established field that has provided important contributions and will continue to aid in the detailed understanding of these important enzymes.
Acknowledgments
Resources from the National Institutes of Health–NIGMS, the NVIDIA Foundation Compute the Cure program, and computing time from Wayne State’s C&IT and UNT’s CASCaM are gratefully acknowledged.
Glossary
Abbreviations
- Pol
polymerase
- BER
base excision repair
- NHEJ
nonhomologous end joining
- MD
molecular dynamics
- QM
quantum mechanics
- QM/MM
quantum mechanics/molecular mechanics
- Pα
alpha phosphate
- PPi
pyrophosphate
- EDA
energy decomposition analysis
- SNP
single nucleotide polymorphism
- HOT
homologue of theta
- TS
transition state
- ELF
electron localization function
- NCI
noncovalent interaction
- KF
Klenow fragment
- FRET
Förster resonance energy transfer
- EFER
electrostatic free energy response
- Dpo4
DNA polymerase IV
- TLS
translesion synthesis
- PAH
polycyclic aromatic hydrocarbon
- DMSO
dimethyl sulfoxide
- B[a]P
benzo[a]pyrene
Biographies
Alice R. Walker obtained her B.Sc. in Chemistry from the University of Michigan-Dearborn and is currently a Ph.D. candidate at the University of North Texas under Dr. G. Andrés Cisneros. Her research interests are centered around various aspects of computational biochemistry, particularly modifying and applying quantum mechanical/molecular mechanics to biochemical systems associated with cancer and disease.
G. Andrés Cisneros received a B.Sc. in Chemistry from the National Autonomous University of Mexico (UNAM) and a Ph.D. in Chemistry from Duke University. Afterward, he worked as an intramural postdoctoral fellow in the National Institute of Environmental Health Sciences/NIH. He is currently an Associate Professor in the Department of Chemistry at the University of North Texas and a member of CASCaM at UNT. His research interests include the study of DNA replication/repair systems, development, and application of accurate methods for chemical/biochemical simulations with emphasis on force field and QM/MM development and simulation of ionic liquid systems.
This work was supported by the National Institutes of Health–NIGMS, Grant Nos. R01GM108583, R01GM118501 to G.A.C., and by the NVIDIA Foundation Compute the Cure program.
The authors declare no competing financial interest.
References
- Starcevic D.; Dalal S.; Sweasy J. B. (2004) Is there a link between DNA polymerase β and cancer?. Chem. Commun. 3, 98–1001. [PubMed] [Google Scholar]
- Iwanaga A.; Ouchida M.; Miyazaki K.; Hori K.; Mukai T. (1999) Functional mutation of DNA polymerase β found in human gastric cancer - inability of the base excision repair in vivos. Mutat. Res., DNA Repair 435, 121–128. 10.1016/S0921-8777(99)00036-1. [DOI] [PubMed] [Google Scholar]
- Lange S. S.; Takata K.; Wood R. D. (2011) DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110. 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A.; Monnat R. J. (2008) DNA polymerases and human disease. Nat. Rev. Genet. 9, 594–604. 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- Broyde S.; Wang L.; Rechkoblit O.; Geacintov N. E.; Patel D. J. (2008) Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 33, 209–219. 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland A. J.; Roitberg A. E.; Tuñón I. (2012) Enzyme dynamics and catalysis in the mechanism of DNA polymerase. Theor. Chem. Acc. 131, 1286. 10.1007/s00214-012-1286-8. [DOI] [Google Scholar]
- Johnson K. A. (2010) The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta, Proteins Proteomics 1804, 1041–8. 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. (2003) Considering the cancer consequences of altered DNA polymerase function. Cancer Cell 3, 105–110. 10.1016/S1535-6108(03)00027-8. [DOI] [PubMed] [Google Scholar]
- Aravind L.; Koonin E. V. (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 26, 3746–52. 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouge J.; Ralec C.; Henneke G.; Delarue M. (2012) Molecular recognition of canonical and deaminated bases by P. abyssi family B DNA polymerase. J. Mol. Biol. 423, 315–36. 10.1016/j.jmb.2012.07.025. [DOI] [PubMed] [Google Scholar]
- Jozwiakowski S. K.; Keith B. J.; Gilroy L.; Doherty A. J.; Connolly B. A. (2014) An archaeal family-B DNA polymerase variant able to replicate past DNA damage: occurrence of replicative and translesion synthesis polymerases within the B family. Nucleic Acids Res. 42, 9949–9963. 10.1093/nar/gku683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare V.; Eckert K. A. (2002) The proofreading 3′->5′ exonuclease activity of DNA polymerases: a kinetic barrier to translesion DNA synthesis. Mutat. Res., Fundam. Mol. Mech. Mutagen. 510, 45–54. 10.1016/S0027-5107(02)00251-8. [DOI] [PubMed] [Google Scholar]
- Yang W. (2014) An Overview of Y-Family DNA Polymerases and a Case Study of Human DNA Polymerase β. Biochemistry 53, 2793–2803. 10.1021/bi500019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K.; Kunkel T. A. (2004) Functions of DNA polymerases. Adv. Protein Chem. 69, 137–165. 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- Moon A. F.; García-Díaz M.; Batra V. K.; Beard W. A.; Bebenek K.; Kunkel T. A.; Wilson S. H.; Pedersen L. C. (2007) The X family portrait: Structural insights into biological functions of X family polymerases. DNA Repair 6, 1709–1725. 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federley R. G.; Romano L. J. (2010) DNA polymerase: structural homology, conformational dynamics, and the effects of carcinogenic DNA adducts. J. Nucleic Acids 2010, 1. 10.4061/2010/457176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker R.; Oelschlaeger P.; Warshel A. (2009) A binding free energy decomposition approach for accurate calculations of the fidelity of DNA polymerases. Proteins: Struct., Funct., Genet. 78, 671–680. 10.1002/prot.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.; Li G. (2014) Bridging the Missing Link between Structure and Fidelity of the RNA-Dependent RNA Polymerase from Poliovirus through Free Energy Simulations. J. Chem. Theory Comput. 10, 5195–5205. 10.1021/ct5006449. [DOI] [PubMed] [Google Scholar]
- Florián J.; Goodman M. F.; Warshel A. (2003) Computer simulation studies of the fidelity of DNA polymerases. Biopolymers 68, 286–299. 10.1002/bip.10244. [DOI] [PubMed] [Google Scholar]
- Meli M.; Sustarsic M.; Craggs T. D.; Kapanidis A. N.; Colombo G. (2016) DNA Polymerase Conformational Dynamics and the Role of Fidelity-Conferring Residues: Insights from Computational Simulations. Front. Mol. Biosci. 3, 20. 10.3389/fmolb.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa I. M.; Shen H.; Morton B.; Colina C. M.; Cameron C. E. (2011) Molecular Dynamics Simulations of Viral RNA Polymerases Link Conserved and Correlated Motions of Functional Elements to Fidelity. J. Mol. Biol. 410, 159–181. 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euro L.; Haapanen O.; Róg T.; Vattulainen I.; Suomalainen A.; Sharma V. (2017) Atomistic Molecular Dynamics Simulations of Mitochondrial DNA Polymerase γ: Novel Mechanisms of Function and Pathogenesis. Biochemistry 56, 1227–1238. 10.1021/acs.biochem.6b00934. [DOI] [PubMed] [Google Scholar]
- Miller B. R.; Parish C. A.; Wu E. Y. (2014) Molecular Dynamics Study of the Opening Mechanism for DNA Polymerase I. PLoS Comput. Biol. 10, e1003961. 10.1371/journal.pcbi.1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.; Freudenthal B. D.; Beard W. A.; Wilson S. H.; Schlick T. (2016) Insertion of oxidized nucleotide triggers rapid DNA polymerase opening. Nucleic Acids Res. 44, 4409–24. 10.1093/nar/gkw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Beard W. A.; Wilson S. H.; Broyde S.; Schlick T. (2002) Polymerase β simulations suggest that Arg258 rotation is a slow step rather than large subdomain motions per se. J. Mol. Biol. 317, 651–671. 10.1006/jmbi.2002.5450. [DOI] [PubMed] [Google Scholar]
- Jia L.; Geacintov N. E.; Broyde S. (2008) The N-clasp of human DNA polymerase kappa promotes blockage or error-free bypass of adenine- or guanine-benzo[a]pyrenyl lesions. Nucleic Acids Res. 36, 6571–84. 10.1093/nar/gkn719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera L.; Freudenthal B. D.; Beard W. A.; Pedersen L. G.; Wilson S. H. (2017) Revealing the role of the product metal in DNA polymerase β catalysis. Nucleic Acids Res. gkw1363. 10.1093/nar/gkw1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik M. M.; Bhattacharjee N.; Feliks M.; Ng K. K.-S.; Field M. J. (2017) Norovirus RNA-dependent RNA polymerase: A computational study of metal-binding preferences. Proteins: Struct., Funct., Genet. 85, 1435. 10.1002/prot.25304. [DOI] [PubMed] [Google Scholar]
- Parks J. M.; Kondru R. K.; Hu H.; Beratan D. N.; Yang W. (2008) Hepatitis c virus ns5b polymerase: Qm/mm calculations show the important role of the internal energy in ligand binding. J. Phys. Chem. B 112, 3168–3176. 10.1021/jp076885j. [DOI] [PubMed] [Google Scholar]
- Carvalho A. T. P.; Fernandes P. A.; Ramos M. J. (2011) The Catalytic Mechanism of RNA Polymerase II. J. Chem. Theory Comput. 7, 1177–1188. 10.1021/ct100579w. [DOI] [PubMed] [Google Scholar]
- Warshel A.; Levitt M. (1976) Theoretical studies of enzymatic reactions: dielectric electrostatic and steric stabilization of the carbonium ion in the reaction of lyzozyme. J. Mol. Biol. 103, 227–249. 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- Florián J.; Goodman M. F.; Warshel A. (2002) Theoretical Investigation of the Binding Free Energies and Key Substrate-Recognition Components of the Replication Fidelity of Human DNA Polymerase β. J. Phys. Chem. B 106, 5739–5753. 10.1021/jp020790u. [DOI] [Google Scholar]
- Florián J.; Warshel A.; Goodman M. F. (2002) Molecular Dynamics Free-Energy Simulations of the Binding Contribution to the Fidelity of T7 DNA Polymerase. J. Phys. Chem. B 106, 5754–5760. 10.1021/jp020791m. [DOI] [Google Scholar]
- Florian J.; Goodman M. F.; Warshel A. (2005) Computer simulations of protein functions: Searching for the molecular origin of the replication fidelity of DNA polymerases. Proc. Natl. Acad. Sci. U. S. A. 102, 6819–6824. 10.1073/pnas.0408173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R.; Schlick T. (2006) Correct and incorrect nucleotide incorporation pathways in DNA polymerase β. Biochem. Biophys. Res. Commun. 350, 521–529. 10.1016/j.bbrc.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojin M. D.; Schlick T. (2007) A quantum mechanical investigation of possible mechanisms for the nucleotidyl transfer reaction catalyzed by DNA polymerase β. J. Phys. Chem. B 111, 11244–11252. 10.1021/jp071838c. [DOI] [PubMed] [Google Scholar]
- Wang L.; Yu X.; Hu P.; Broyde S.; Zhang Y. (2007) A water-mediated and substrate-assisted catalytic mechanism for Sulfobolus solfataricus DNA polymerase IV. J. Am. Chem. Soc. 129, 4731–4737. 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick T.; Arora K.; Beard W. A.; Wilson S. H. (2012) Perspective: pre-chemistry conformational changes in DNA polymerase mechanisms. Theor. Chem. Acc. 131, 1287. 10.1007/s00214-012-1287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerlin S. C. L.; Sharma P. K.; Prasad R. B.; Warshel A. (2013) Why nature really chose phosphate. Q. Rev. Biophys. 46, 1–132. 10.1017/S0033583512000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosta E.; Yang W.; Hummer G. (2014) Calcium Inhibition of Ribonuclease H1 Two-Metal Ion Catalysis. J. Am. Chem. Soc. 136, 3137–3144. 10.1021/ja411408x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera L.; Freudenthal B. D.; Beard W. A.; Shock D. D.; Pedersen L. G.; Wilson S. H. (2015) Requirement for transient metal ions revealed through computational analysis for DNA polymerase going in reverse. Proc. Natl. Acad. Sci. U. S. A. 112, E5228–E5236. 10.1073/pnas.1511207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Schlick T. (2008) Quantum Mechanics/Molecular Mechanics Investigation of the Chemical Reaction in Dpo4 Reveals Water-Dependent Pathways and Requirements for Active Site Reorganization. J. Am. Chem. Soc. 130, 13240–13250. 10.1021/ja802215c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior-Hoffmann L.; Wang L.; Wang S.; Geacintov N. E.; Broyde S.; Zhang Y. (2012) Preferred WMSA catalytic mechanism of the nucleotidyl transfer reaction in human DNA polymerase κ elucidates error-free bypass of a bulky DNA lesion. Nucleic Acids Res. 40, 9193–205. 10.1093/nar/gks653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros G. A. (2012) Application of Gaussian Electrostatic Model (GEM) Distributed Multipoles in the Force Field. J. Chem. Theory Comput. 12, 5072–5080. 10.1021/ct300630u. [DOI] [PubMed] [Google Scholar]
- Starovoytov O. N.; Torabifard H.; Cisneros G. A. (2014) Development of AMOEBA Force Field for 1, 3-Dimethylimidazolium Based Ionic Liquids. J. Phys. Chem. B 118, 7156–7166. 10.1021/jp503347f. [DOI] [PubMed] [Google Scholar]
- Torabifard H.; Starovoytov O. N.; Ren P.; Cisneros G. A. (2015) Development of an AMOEBA water model using GEM distributed multipoles. Theor. Chem. Acc. 134, 1–10. 10.1007/s00214-015-1702-y. [DOI] [Google Scholar]
- Karttunen M., Rottler J., Vattulainen I., and Sagui C. (2008) Computational Modeling of Membrane Bilayers. Current Topics in Membranes, Vol. 60, pp 49–89, Elsevier Inc. [Google Scholar]
- Warshel A.; Kato M.; Pisliakov A. V. (2007) Polarizable Force Fields: History, Test Cases, and Prospects. J. Chem. Theory Comput. 3, 2034–2045. 10.1021/ct700127w. [DOI] [PubMed] [Google Scholar]
- Boulanger E.; Thiel W. (2012) Solvent Boundary Potentials for Hybrid QM/MM Computations Using Classical Drude Oscillators: A Fully Polarizable Model. J. Chem. Theory Comput. 8, 4527–4538. 10.1021/ct300722e. [DOI] [PubMed] [Google Scholar]
- Chaudret R.; Gresh N.; Narth C.; Lagardere L.; Darden T. A.; Cisneros G. A.; Piquemal J.-P. (2014) S/G-1: An ab Initio Force-Field Blending Frozen Hermite Gaussian Densities and Distributed Multipoles. Proof of Concept and First Applications to Metal Cations. J. Phys. Chem. A 118, 7598–7612. 10.1021/jp5051657. [DOI] [PubMed] [Google Scholar]
- Chaudret R.; Ulmer S.; van Severen M.-C.; Gresh N.; Parisel O.; Cisneros G. A.; Darden T. A.; Piquemal J.-P.; et al. (2008) Progress Towards Accurate Molecular Modeling of Metal Complexes Using Polarizable Force Fields. AIP Conf. Proc. 1102, 185–192. 10.1063/1.3108373. [DOI] [Google Scholar]
- Lamoureux G.; Harder E.; Vorobyov I.; Roux B.; MacKerell A. (2006) A polarizable model of water for molecular dynamics simulations of biomolecules. Chem. Phys. Lett. 418, 245–249. 10.1016/j.cplett.2005.10.135. [DOI] [Google Scholar]
- Beese L. S.; Steitz T. A. (1991) Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA poymerase I: a two metal ion mechanism. EMBO J. 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D.; Chaudret R.; Piquemal J.-P.; Cisneros G. A. (2013) Toward a Deeper Understanding of Enzyme Reactions Using the Coupled ELF/NCI Analysis: Application to DNA Repair Enzymes. J. Chem. Theory Comput. 9, 2156–2160. 10.1021/ct400130b. [DOI] [PubMed] [Google Scholar]
- Lin P.; Pedersen L. C.; Batra V. K.; Beard W. A.; Wilson S. H.; Pedersen L. G. (2006) Energy analysis of chemistry for correct insertion by DNA polymerase β. Proc. Natl. Acad. Sci. U. S. A. 103, 13294–13299. 10.1073/pnas.0606006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros G. A.; Perera L.; García-Díaz M.; Bebenek K.; Kunkel T. A.; Pedersen L. G. (2008) Catalytic mechanism of human DNA polymerase λ with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies. DNA Repair 7, 1824–1834. 10.1016/j.dnarep.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirover M. A.; Loeb L. A. (1976) Infidelity of DNA synthesis in vitro: Screening for potential metal mutagens or carcinogens. Science 194, 1434–1436. 10.1126/science.1006310. [DOI] [PubMed] [Google Scholar]
- Blanca G.; Shevelev I.; Ramadan K.; Villani G.; Spadari S.; Hübscher U.; Maga G. (2003) Human DNA polymerase λ diverged in evolution from DNA polymerase β toward specific Mn+2 dependence: a kinetic and thermodynamic study. Biochemistry 42, 7467–7476. 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- Fowler J. D.; Brown J. A.; Kvaratskhelia M.; Suo Z. (2009) Probing conformational changes of human DNA polymerase lambda using mass spectrometry-based protein footprinting. J. Mol. Biol. 390, 368–79. 10.1016/j.jmb.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K.; Garcia-Diaz M.; Zhou R.-Z.; Povirk L. F.; Kunkel T. A. (2010) Loop 1 modulates the fidelity of DNA polymerase lambda. Nucleic Acids Res. 38, 5419–31. 10.1093/nar/gkq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz S.; Kwack S.; Goodman M. F.; O’Donnel M.; Echols H. (1991) Specificity and enzymatic mechanism of the editing exonuclease of Escherichia coli DNA polymerase III. J. Biol. Chem. 266, 7888–7892. [PubMed] [Google Scholar]
- McHenry C. S. (1991) DNA polymerase III holoenzyme. J. Biol. Chem. 266, 19127–19130. [PubMed] [Google Scholar]
- Cisneros G. A.; Perera L.; Schaaper R. M.; Pedersen L. C.; London R. E.; Pedersen L. G.; Darden T. A. (2009) Reaction mechanism of the ε subunit of E. coli DNA polymerase III: insights into active site metal coordination and catalytically significant residues. J. Am. Chem. Soc. 131, 1550–1556. 10.1021/ja8082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M. (1993) Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268, 23762–23765. [PubMed] [Google Scholar]
- Derbyshire V.; Pinsonneault J. K.; Joyce C. M. (1995) Structure-function analysis of 3′-5′ exonuclease of DNA polymerases. Methods Enzymol. 262, 363–385. 10.1016/0076-6879(95)62030-3. [DOI] [PubMed] [Google Scholar]
- Hamdan S.; Bulloch E. M.; Thompson P. R.; Beck J. L.; Yang J. Y.; Crowther J. A.; Lilley P. E.; Carr P. D.; Ollis D. L.; Brown S. E.; Dixon N. E. (2002) Hydrolysis of the 5′-p-nitrophenyl ester of TMP by the proofreading exonuclease (ϵ) subunit of Escherichia coli DNA polymerase III. Biochemistry 41, 5266–5275. 10.1021/bi0159480. [DOI] [PubMed] [Google Scholar]
- Cisneros G. A.; Perera L.; Schaaper R. M.; Pedersen L. C.; London R. E.; Pedersen L. G.; Darden T. A. (2009) Reaction Mechanism of the ϵ Subunit of E. coli DNA Polymerase III: Insights into Active Site Metal Coordination and Catalytically Significant Residues. J. Am. Chem. Soc. 131, 1550–1556. 10.1021/ja8082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft-Benz S. A.; Schaaper R. M. (2004) The θ subunit of Escherichia coli DNA polymerase III: a role in sabilizing the ϵ proofreading subunit. J. Bacteriol. 186, 2774–2780. 10.1128/JB.186.9.2774-2780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikova A. K.; Schaaper R. M. (2005) The bacteriophage P1 hot gene product can substitute for the Escherichia coli DNA polymerase III θ subunit. J. Bacteriol. 187, 5528–5536. 10.1128/JB.187.16.5528-5536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby T. W.; Harvey S.; DeRose E. F.; Chalov S.; Chikova A. K.; Perrino F. W.; Schaaper R. M.; London R. E.; Pedersen L. C. (2006) Structure of the E. coli DNA polymerase III ϵ-HOT proofreading complex. J. Biol. Chem. 281, 38466–38471. 10.1074/jbc.M606917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.; Lee J. Y.; Nowotny M. (2006) Making and breaking nucleic acids: Two-Mg2+-Ion catalysis and substrate specificity. Mol. Cell 22, 5–13. 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Rosta E.; Nowotny M.; Yang W.; Hummer G. (2011) Catalytic Mechanism of RNA Backbone Cleavage by Ribonuclease H from Quantum Mechanics/Molecular Mechanics Simulations. J. Am. Chem. Soc. 133, 8934–8941. 10.1021/ja200173a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet N.; Chaudret R.; Contreras-García J.; Yang W.; Silvi B.; Piquemal J.-P. (2012) Coupling Quantum Interpretative Techniques: Another Look at Chemical Mechanisms in Organic Reactions. J. Chem. Theory Comput. 8, 3993–3997. 10.1021/ct300234g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T.; Zhao Y.; Yamagata Y.; Hua Y.-J.; Yang W. (2012) Watching {DNA} Polymerase ν make a phosphodiester bond. Nature 487, 196–201. 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenthal B.; Beard W.; Shock D.; Wilson S. (2013) Observing a {DNA} Polymerase Choose Right from Wrong. Cell 154, 157–168. 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz E. G.; Walker A. R.; Lagardère L.; Lipparini F.; Piquemal J.-P.; Andres Cisneros G. (2016) LICHEM: A QM/MM program for simulations with multipolar and polarizable force fields. J. Comput. Chem. 37, 1019–1029. 10.1002/jcc.24295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genna V.; Vidossich P.; Ippoliti E.; Carloni P.; De Vivo M. (2016) A Self-Activated Mechanism for Nucleic Acid Polymerization Catalyzed by DNA/RNA Polymerases. J. Am. Chem. Soc. 138, 14592–14598. 10.1021/jacs.6b05475. [DOI] [PubMed] [Google Scholar]
- Hartwig A. (1998) Carcinogenicity of metal compounds: possible role of DNA repair inhibition. Toxicol. Lett. 102–103, 235–239. 10.1016/S0378-4274(98)00312-9. [DOI] [PubMed] [Google Scholar]
- Pelletier H.; Sawaya M. R.; Wolfle W.; Wilson S. H.; Kraut J. (1996) A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry 35, 12762–12777. 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- Chaudret R.; Piquemal J.-P.; Andres Cisneros G. (2011) Correlation between electron localization and metal ion mutagenicity in DNA synthesis from QM/MM calculations. Phys. Chem. Chem. Phys. 13, 11239–11247. 10.1039/c0cp02550j. [DOI] [PubMed] [Google Scholar]
- Joyce C. M.; Grindley N. D. (1984) Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J. Bacteriol. 158, 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermek O.; Grindley N. D. F.; Joyce C. M. (2010) Distinct roles of the active site Mg2+ ligands, D882 and L705 of DNA polymerase I (Klenow Fragment) during the prechemistry conformational transitions. J. Biol. Chem. 286, 3755. 10.1074/jbc.M110.167593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K.; Modak M. J. (2003) Presence of 18-Å long hydrogen bond track in the active site of Escherichia coli DNA polymerase I (Klenow Fragment): Its requirement in the stabilization of enzyme-template-primer complex. J. Biol. Chem. 278, 11289–11302. 10.1074/jbc.M211496200. [DOI] [PubMed] [Google Scholar]
- Kuchta R. D.; Benkovic P.; Benkovic S. J. (1988) Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry 27, 6716–6725. 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- Beese L. S.; Friedman J. M.; Steitz T. A. (1993) Crystal structures of the Klenow fragment of DNA polymerase I complexed with deoxynucleoside triphosphate and pyrophosphate. Biochemistry 32, 14095–14101. 10.1021/bi00214a004. [DOI] [PubMed] [Google Scholar]
- Freemont P. S.; Friedman J. M.; Beese L. S.; Sanderson M. R.; Steitz T. A. (1988) Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc. Natl. Acad. Sci. U. S. A. 85, 8924–8928. 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K.; Modak M. J. (2005) Contribution of Polar Residues of the J-Helix in the 3′-5′ exonuclease activity of Escherichia coli DNA poymerase I (Klenow fragment): Q677 regulates the removal of terminal mismatch. Biochemistry 44, 8101–8110. 10.1021/bi050140r. [DOI] [PubMed] [Google Scholar]
- Thompson E. H. Z.; Bailey M. F.; van der Schans E. J. C.; Joyce C. M.; Millar D. P. (2002) Determinants of DNA Mismatch Recognition within the Polymerase Domain of the Klenow Fragment. Biochemistry 41, 713–722. 10.1021/bi0114271. [DOI] [PubMed] [Google Scholar]
- Minnick D. T.; Bebenek K.; Osheroff W. P.; Turner R. M.; Astatke M.; Liu L.; Kunkel T. A.; Joyce C. M. (1999) Side Chains That Influence Fidelity at the Polymerase Active Site of Escherichia coli DNA Polymerase I (Klenow Fragment). J. Biol. Chem. 274, 3067–3075. 10.1074/jbc.274.5.3067. [DOI] [PubMed] [Google Scholar]
- Bebenek K.; Joyce C. M.; Fitzgerald M.; Kunkel T. A. (1990) The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 265, 13878–13887. [PubMed] [Google Scholar]
- Loh E.; Loeb L. E. (2005) Mutability of DNA polymerase I: Implications for the creation of mutant DNA polymerases. DNA Repair 4, 1390–1398. 10.1016/j.dnarep.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Christian T.; Romano L.; Rueda D. (2009) Single-molecule measurements of synthesis by DNA polymerase with base-pair resolution. Proc. Natl. Acad. Sci. U. S. A. 106, 21109–21114. 10.1073/pnas.0908640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham S. E.; Syeda F.; Cisneros G. A. (2012) Computational Prediction of Residues Involved in Fidelity Checking for DNA Synthesis in DNA Polymerase I. Biochemistry 51, 2569–2578. 10.1021/bi201856m. [DOI] [PubMed] [Google Scholar]
- Elias A. A.; Cisneros G. A. (2014) Chapter Two-Computational Study of Putative Residues Involved in DNA Synthesis Fidelity Checking in Thermus aquaticus DNA Polymerase I. Adv. Protein Chem. Struct. Biol. 96, 39–75. 10.1016/bs.apcsb.2014.06.003. [DOI] [PubMed] [Google Scholar]
- Florián J.; Goodman M. F.; Warshel A. (2003) Computer simulation of the chemical catalysis of DNA polymerases: Discriminating between alternative nucleotide insertion mechanisms for T7 DNA polymerase. J. Am. Chem. Soc. 125, 8163–8177. 10.1021/ja028997o. [DOI] [PubMed] [Google Scholar]
- Florián J.; Goodman M. F.; Warshel A. (2005) Computer simulations of protein functions: Searching for the molecular origin of the replication fidelity of DNA polymerases. Proc. Natl. Acad. Sci. U. S. A. 102, 6819–6824. 10.1073/pnas.0408173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. R.; Keinan S.; Mori-Sanchez P.; Contreras-Garcia J.; Cohen A. J.; Yang W. (2010) Revealing Noncovalent Interactions. J. Am. Chem. Soc. 132, 6498–6506. 10.1021/ja100936w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-García J.; Johnson E. R.; Keinan S.; Chaudret R.; Piquemal J.-P.; Beratan D. N.; Yang W. (2011) NCIPLOT: A Program for Plotting Noncovalent Interaction Regions. J. Chem. Theory Comput. 7, 625. 10.1021/ct100641a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelboin H. V. (1980) Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol. Rev. 60, 1107–66. [DOI] [PubMed] [Google Scholar]
- Ling H.; Sayer J. M.; Plosky B. S.; Yagi H.; Boudsocq F.; Woodgate R.; Jerina D. M.; Yang W. (2004) Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 101, 2265–2269. 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage P. S., Walker A. R., Brenlla A., Cisneros G. A., Romano L. J., and Rueda D. (2017) Bulky Lesion Bypass Requires Dpo4 Binding in Distinct Conformations (in preparation). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makridakis N. M.; Reichardt J. K. V. (2012) Translesion DNA polymerases and cancer. Front. Genet. 3, 174. 10.3389/fgene.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick E.; White L. M.; Elliott N. A.; Berg D.; Kiviat N. B.; Loeb L. A. (2006) Mutations in DNA polymerase η are not detected in squamous cell carcinoma of the skin. Int. J. Cancer 119, 2225–2227. 10.1002/ijc.22099. [DOI] [PubMed] [Google Scholar]
- Swett R. J.; Elias A.; Miller J. A.; Dyson G. E.; Andres Cisneros G. (2013) Hypothesis driven single nucleotide polymorphism search (HyDn-SNP-S). DNA Repair 12, 733–740. 10.1016/j.dnarep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K.; Garcia-Diaz M.; Zhou R.-Z.; Povirk L. F.; Kunkel T. A. (2010) Loop 1 modulates the fidelity of DNA polymerase λ. Nucleic Acids Res. 38, 5419–5431. 10.1093/nar/gkq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrados G.; Capp J.-P.; Canitrot Y.; García-Díaz M.; Bebenek K.; Kirchhoff T.; Villanueva A.; Boudsocq F.; Bergoglio V.; Cazaux C.; Kunkel T. A.; Hoffmann J.-S.; Blanco L. (2009) Characterization of a natural mutator variant of human DNA polymerase λ which promotes chromosomal instability by compromising NHEJ. PLoS One 4, e7290. 10.1371/journal.pone.0007290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec A. A.; Bush K. B.; Towle-Weicksel J. B.; Taylor B. F.; Schulz V.; Weidhaas J. B.; Tuck D. P.; Sweasy J. B. (2016) Estrogen Drives Cellular Transformation and Mutagenesis in Cells Expressing the Breast Cancer–Associated R438W DNA Polymerase Lambda Protein. Mol. Cancer Res. 14, 1068–1077. 10.1158/1541-7786.MCR-16-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]