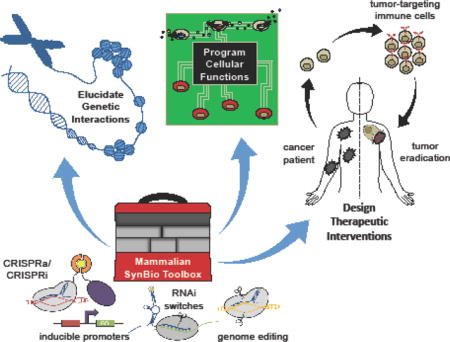
Abstract
The recent expansion of molecular tool kits has propelled synthetic biology toward the design of increasingly sophisticated mammalian systems. Specifically, advances in genome editing, protein engineering, and circuitry design have enabled the programming of cells for diverse applications, including regenerative medicine and cancer immunotherapy. The ease with which molecular and cellular interactions can be harnessed promises to yield novel approaches to elucidate genetic interactions, program cellular functions, and design therapeutic interventions. Here, we review recent advancements in the development of enabling technologies and the practical applications of mammalian synthetic biology.
Graphical abstract
Introduction
Synthetic biology is an interdisciplinary field founded on the application of engineering principles to biology, with the aim of advancing our ability to decode and reprogram living systems with diverse behaviors and functions. While early efforts primarily focused on the development of transcription-based circuits in bacteria [1,2], recent confluence of powerful tools in genome editing, protein engineering, and genetic circuitry design has enabled the engineering of sophisticated mammalian systems and substantive progress toward applications in health and medicine [3,4]. Here, we review the expansion of the mammalian synthetic biology toolbox, as well as how these technologies are being leveraged to yield novel approaches to study cell biology and design personalized therapeutics.
Adapting Genome-Editing Tools for Mammalian Synthetic Biology
Since its inception, synthetic biology has enabled researchers to understand and engineer increasingly complex systems by enhancing our ability to interrogate, modulate, and reprogram biological functions. Increasingly, genome-editing tools have been used to not only modify chromosomal makeup, but also regulate the expression of both endogenous and transgenic genes. Leading technologies for genome editing include zinc-finger nucleases (ZFNs) [5], transcription activator-like effector nucleases (TALENs) [6], and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) [7]. Unlike viral gene-delivery platforms that result in non-site-specific insertion, ZFNs, TALENs, and CRISPR/Cas9 can introduce site-specific gene modifications by cleaving genomic DNA at specific target loci [8]. In mammalian cells, these double-stranded breaks are typically repaired through error-prone non-homologous end joining (NHEJ) to generate frame-shift insertions and deletions (indels), thus disrupting target gene expression [9]. By supplying a homology-directed repair (HDR) template, sequence-defined modifications can also be made with single base-pair resolution [10]. Recent investigations have focused on improving the efficiency of HDR in human cells, particularly through the use of small-molecule inhibitors or RNAi to suppress key enzymes involved in NHEJ-mediated DNA repair [9,11,12].
Among the three main genome-editing tools, CRISPR/Cas9 has become the undisputed favorite in recent years due to its high efficiency and ease of use. Although ZFNs and TALENs were developed earlier than CRISPR/Cas9, both of these methods rely on protein-DNA interactions that require new ZF and TALE proteins to be designed and optimized for each DNA target, thus creating barriers to their widespread use [13]. In contrast, CRISPR/Cas9 complexes are targeted to DNA via Watson-Crick base-pairing between a single guide RNA (sgRNA) and the target DNA sequence [14–16]. The ease with which sgRNAs can be designed and introduced into cells has enabled the engineering of CRISPR/Cas9 as highly predictable and easily multiplexed DNA-binding modules for transcriptional control [17,18] (Figure 1). Numerous web-based sgRNA design algorithms have facilitated the wide adoption of this technology [19,20], and the synthetic biology community and beyond have witnessed an explosion of new applications based on CRISPR/Cas9 [15,16]. For example, a nuclease-null variant of Cas9 (dCas9) has been fused to activator and repressor domains to generate designer transcription factors that can mediate constitutive expression (CRISPRa) or silencing (CRISPRi) of individual endogenous genes in various human cell types (Figure 1A,B) [14–16,21–23•]. In addition, simultaneous co-expression of multiple sgRNAs can direct sustained activation of multiple target loci to trigger genetic scripts, such as the differentiation of induced pluripotent stem cells (iPSCs) or the conversion of fibroblasts into neuronal cells [16,21,22]. To further fine-tune CRISPR/Cas9-based transcription regulation, Zalatan et al. extended conventional sgRNAs with docking sites for RNA-binding proteins to form scaffold RNAs that can mediate differential recruitment of activators and repressors to distinct target loci, further enhancing our ability to perturb and enact genetic programs [23•]. As an alternative to transcriptional control, Moore et al. demonstrated that the intensity and duration of gene expression after transient transfection can be tightly regulated by using a single plasmid to encode for the gene of interest (GOI) as well as CRISPR/Cas9 components that cleave the GOI [24]. This system ensures transience in gene expression and also enables gene-expression calibration by adjusting the protein stability as well as sgRNA/Cas9 complex targeting affinity.
Figure 1.
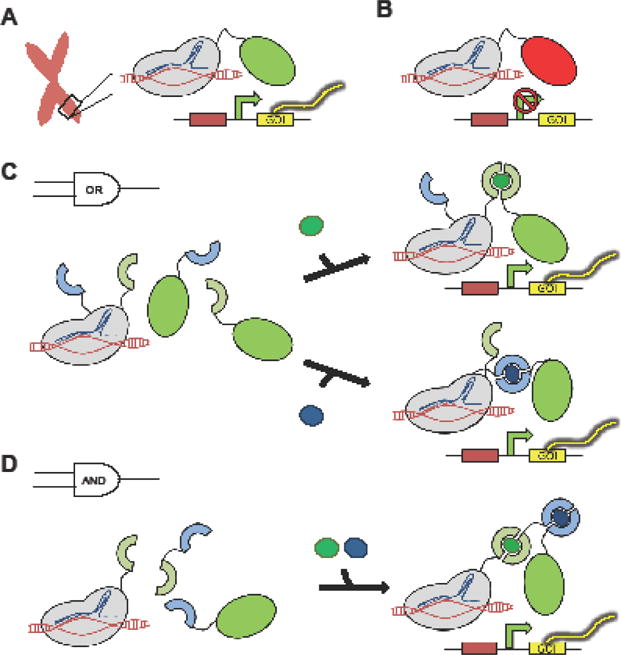
Modular CRISPR/dCas9 transcription factors mediate precise, constitutive or inducible activation or silencing of specific chromosomal loci. (A) Activator or (B) repressor domains fused to dCas9 trigger expression or silencing of targeted endogenous genes, respectively. (C) Heterodimerization domains recruit distinct activator domains in response to different optical or chemical stimuli, yielding OR-gate transcription activation. (D) Recruitment of a transcription activator domain to the target locus is dependent on simultaneous heterodimerization events, generating an AND-gate transcription response.
Additional Tools for Gene-Expression Regulation
Despite the rapid expansion of gene-regulatory devices based on CRISPR/Cas9, inducible promoters and transcription activators remain a dominant tool for mammalian gene- expression regulation [25–28]. Although cataloged inducible promoters are readily available in parts repositories, the vast majority utilizes a very limited set of core promoters, most commonly the minimal cytomegalovirus (minCMV) promoter. To better understand the range of gene expression intensity and inducibility achievable in mammalian cells, our group recently reported the systematic evaluation of a panel of eight mammalian core promoters [25]. By using a synthetic core promoter with significantly reduced basal expression and increased fold-induction compared to minCMV, we were able to engineer human T cells that specifically induce tumor-targeting chimeric antigen receptor expression under hypoxic conditions, which are characteristic of solid tumor microenvironments [25]. Synthetic biologists have long been interested in engineering Boolean-logic operators using inducible promoter systems [1,29], and recent studies have produced increasingly robust gene-expression systems that can demonstrate precise in vivo spatiotemporal control actuated by diverse input signals (e.g., small-molecule ligands such as tetracycline as well as non-molecular cues such as blue light) [26]. Boolean-logic operators can also be programmed by fusing dCas9 and activator or repressor domains to complementary pairs of chemically or optically inducible heterodimerization domains (Figure 1C,D) [27]. In the same vein, a split-intein system was used to inducibly reconstitute transcription factors in response to DNA inputs [28•].
Post-transcriptional regulatory processes such as alternative splicing and RNA interference (RNAi) also offer unique opportunities for mammalian gene regulation. One example makes use of conditionally active “splice-switching” oligos that either hybridizes with pre-mRNA or becomes cleaved in the presence of light, thereby altering gene expression by masking or revealing splice sites, respectively [30]. Additional examples of RNA-based regulatory schemes include the use of four-way nucleic acid strand exchange to report intracellular mRNA levels [31], and the programming of ribozyme switches to exert cell-cycle control in response to the chemical ligand theophylline [32]. Furthermore, coupling a β-catenin–binding RNA aptamer to microRNA (miRNA) targeted against a green fluorescent protein (GFP) mRNA construct has been shown to enable quantification of nuclear β-catenin concentrations [33]. Interestingly, a similar strategy can be employed in reverse to detect endogenous expression of miRNAs involved in cell development [34,35•]. By expressing an mRNA targeted by miRNA-302a-5p, Parr et al. demonstrated the ability to sort or selectively eliminate undifferentiated human iPSCs from heterogeneous cell populations, on the basis of retained miR-302a-5p expression in undifferentiated cells [35].
Elucidating Genetic Interactions
Along with the diverse tools now available for precise genetic perturbations, next- generation sequencing technologies have empowered the interrogation of gene networks and complex signal-processing relationships in high-throughput fashion. Construction of multiplexed, barcoded CRISPR/Cas9 sgRNA libraries has rapidly evolved to allow combinatorial perturbations and the discovery of cooperative epigenetic modifications that play a role in oncogenesis [36]. Stockman et al. developed a multiplex strategy (MoSAIC) for assessing genetic interactions using CRISPR/Cas9, in which direct barcoding and pairwise assembly of sgRNAs occur in a single cloning step, further improving assembly efficiency and reducing potential library bias associated with multi-step construction methods [37]. Pooled functional and phenotypic screens of RNAi- and CRISPRi-treated cells have revealed the relative impact of individual genes in mediating diverse cell behaviors, including somatic cell reprogramming, the unfolded protein response, and immune cell activation (Figure 2) [38–42]. CRISPR-mediated perturbations can also be carried out in vivo to analyze genetic determinants of physiological immune responses. For example, by evaluating differences in gene expression between activated and dysfunctional tumor-infiltrating T cells, Singer et al. revealed distinct gene modules involved in T-cell activation vs. dysfunction, and further identified GATA-binding protein 3 (GATA3) as an important regulator of T-cell dysfunction [42•]. These findings may prove critical for streamlining therapeutic drug screens, as well as engineering more effective cell- based therapies.
Figure 2.
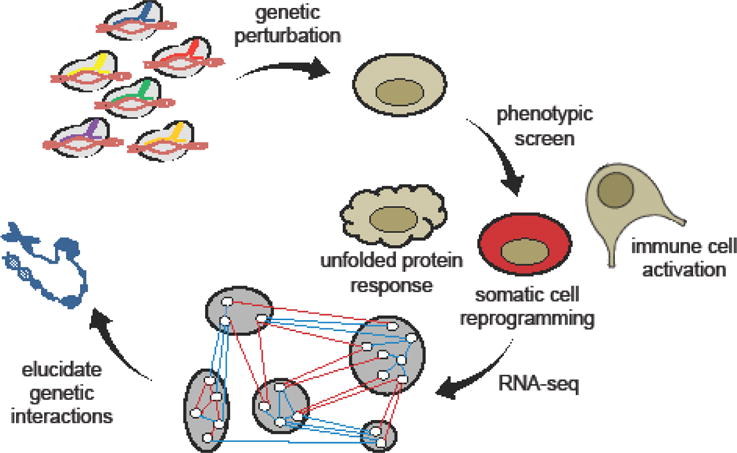
High-throughput mapping of gene networks and epigenetic landscapes. Multiplexed CRISPRi and RNAi libraries can introduce precise genetic perturbations into mammalian cells in high-throughput fashion. Pooled phenotypic and functional screens can subsequently identify key genetic modules and interactions.
Sensing, Recording, and Reprogramming Cellular Functions
In addition to performing genetic perturbations, the CRISPR/Cas9 system has also been repurposed to interrogate environmental cues and their interactions with mammalian cells. Perli et al. recently described mSCRIBE, a device that utilizes an adaptive, self-targeting sgRNA to convert molecular exposure to analog memory in the form of mutational load [43••]. By triggering expression of mSCRIBE in response to NF-κB activation, the researchers were able to record the duration of tumor necrosis factor alpha (TNF-α) exposure in vitro and lipopolysaccharide-induced inflammation in vivo [43••]. Utilizing a similar strategy, Kalhor et al. traced cell lineages by barcoding cells with arrays of self-targeting sgRNAs that evolve into unique signatures that record the cumulative mutational history from prior cell generations [44]. More generally, greater understanding of programmed cellular responses to environmental cues is essential for engineering systems that interface robustly with physiological stimuli. For example, cell development and lineage commitment are heavily influenced by exposure to competing physicochemical signals from the local microenvironment. To study such signals, Sokolik et al. introduced a synthetic circuit encoding light-inducible expression of the neural differentiation factor Brain 2 (Brn2) into mouse embryonic stem cells to quantify the impact of fluctuations in environmental cues during cellular signal processing [45]. The cells demonstrated remarkable ability to separate signals from noise, requiring a threshold signaling amplitude and duration in order to trigger differentiation into a neuronal phenotype [45]. By controlled manipulation or silencing of specific signaling pathways, iPSCs, progenitors, and fibroblasts can be reprogrammed to provide renewable sources of inflammation-resistant cartilage, insulin- secreting beta-like cells, and muscle cells for regenerative medicine [46–48].
The ability to reprogram individual cell behaviors can be further leveraged to engineer multicellular interactions and devices with greater computational power. Modular cell-surface receptor technologies enable cells to sense extracellular inputs and communicate with novel outputs in autocrine or paracrine fashion [49–51]. For example, implementation of a modular extracellular sensor architecture rewired human T cells to secrete the immunostimulatory cytokine interleukin-2 in response to vascular endothelial growth factor, an extracellular protein with potent immunosuppressive effects in tumor microenvironments [51]. Naturally occurring intercellular communication modalities such as exosomes can also be repackaged and readdressed to exchange messages between cell populations [52]. Distinct responsibilities can also be assigned to different cells to enhance the signal processing speed of entire cell consortia. For example, using mammalian olfaction as a blueprint, Müller et al. developed a cell consortium that converts analog fragrance sensing to digitized reporter expression [53••]. These examples illustrate the vast potential of mammalian cells to process complex stimuli and instruct useful functions.
Designing Therapeutic Interventions
With the development of increasingly robust tools to detect and respond to physiological stimuli, the promise of mammalian synthetic biology is now being realized in personalized medicine. Many molecular determinants of disease can now be rapidly identified and targeted with the help of synthetic biological circuitry [54,55]. Furthermore, disease treatment using engineered cells has emerged as a particularly exciting application area in recent years [49,56,57]. In contrast to biochemical drugs, therapeutic cells are living agents, with the capacity to proliferate, adapt, and mediate sustained therapeutic benefit. Cell-based regulatory devices have been successfully engineered to act as closed-loop controllers for a wide array of therapeutic applications in vivo, including regulation of hepatocyte growth factor expression in response to bile-acid accumulation caused by drug-induced liver failure [54], insulin secretion for glycemic control [58], and suppression of thyroid-stimulating hormone receptor in Graves’ disease [59]. Although each of these systems functions in different disease states, they share similar design structures, highlighting the modularity of synthetic biology approaches to biological design.
Modular design principles have also contributed to the rapid rise of cell-based immunotherapy, particularly the use of T cells expressing chimeric antigen receptors (CARs). While CD19 CAR-T cell therapy has demonstrated curative potential in human patients with relapsed B-cell malignancies, challenges such as a lack of tumor-exclusive antigens and the possibility of tumor escape via loss of the targeted antigen currently limit the application of adoptive T-cell therapy as a front-line cancer-treatment option [55]. CARs traditionally utilize an antibody-derived single-chain variable fragment (scFv) to redirect T cells against target cells that express a cognate antigen. Several studies have demonstrated the ability to fine-tune T-cell activation by incorporating additional scFv domains to program Boolean logic (Figure 3) [56,60,61]. As one example, our group developed an OR-gate CAR that prevents the mutational escape of malignant B cells in vivo by enabling T cells to recognize two pan—B-cell markers, CD19 and CD20 (Figure 3A,B) [56]. As an approach to increase tumor-targeting specificity, Roybal et al. described the design of synthetic Notch receptors that sequester transcription factors at the cell membrane until they encounter a cognate antigen. Antigen-binding triggers transcription-factor release and the expression of a CAR, which can subsequently trigger T-cell activation upon an encounter with a second antigen (Figure 3C–E) [57••]. In this manner, T-cell activation and cytotoxic outputs are subject to AND-gate logic and restricted to tissues that express both antigens, thus reducing off-tumor toxicity [57••].
Figure 3.
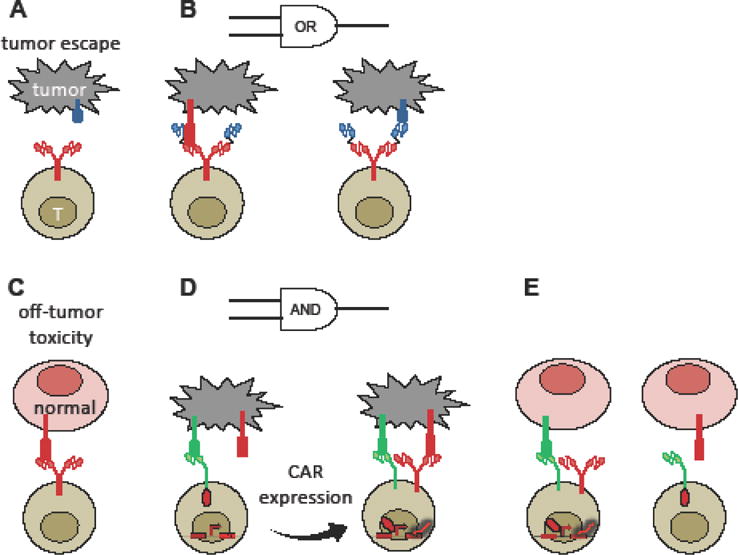
Synthetic receptor systems enable T cells to interrogate target cells with Boolean logic, thus preventing tumor escape and reducing off-tumor toxicity. (A) Tumor heterogeneity and selective loss of target antigen can lead to tumor cells that escape detection by single-input CAR-T cells. (B) A bi-specific, OR-gate CAR construct containing two scFvs enables T cells to target tumor cells that retain either of two target antigens, thus lowering the probability of tumor escape. (C) Single-input CAR-T cells may trigger off-tumor toxicity against healthy tissues that express the target antigen at low basal levels. (D) A synthetic Notch receptor releases a transcription factor that drives CAR expression in response to target-antigen encounter. CAR activation then triggers the T cell to lyse tumor cells that also express a second target antigen. (E) Since the synthetic Notch receptor cannot trigger T-cell activation or cytotoxicity on its own, normal cells that express either, but not both, of the target antigens are protected from CAR-T cell-mediated toxicity.
Precision genome editing technologies are also being explored to treat debilitating and fatal genetic diseases including sickle-cell anemia and Duchenne muscular dystrophy [10,62]. Notably, CRISPR/Cas9-mediated gene correction was able to partially rescue dystrophin expression in a postnatal DMD mouse model [10]. While concerns over potential off-target genome-editing effects are still under evaluation, the progression of genome-editing technologies presents renewed promise for gene therapy.
Conclusion
The rapid progression of mammalian synthetic biology can be traced to the development of enabling technologies that can probe, disrupt, and regulate gene expression with unprecedented scalability. In conjunction with advances in next-generation sequencing technologies, CRISPR/Cas9 has been a dominant driver in recent developments of new tools used to elucidate gene networks and pinpoint molecular determinants of cell physiology. New methodologies and tools may soon push the boundaries of high-throughput screening even further. For example, the Type V CRISPR-Cas Cpf1 system utilizes guide RNAs that are just 42 nucleotides long, a length that can be readily synthesized commercially [63]. Cpf1 also generates double-stranded breaks with 5’-overhangs, a feature that may enhance HDR efficiencies. Similarly, more efficient genome-editing strategies can improve cell-device manufacturing processes and potentially reduce concerns around off-target genome modifications. The continued development of multicellular systems will further enable us to recapitulate and employ the computational power of cells to design living systems with diverse functional outputs. As mammalian synthetic biology comes of age, the increasing ease with which molecular and cellular interactions can be harnessed promises to drive our pursuit of biological understanding and therapeutic application to the next frontier.
Highlights.
Mammalian synbio toolkit spans gene editing, protein engineering, and genetic circuitry
design
Genetic interactions can be interrogated in detail with CRISPR-based devices
Novel synthetic proteins and genetic circuits can record, perturb, and reprogram cell
functions
Mammalian synbio is contributing to the emergence of cell-based therapy as a new pillar in medicine
Acknowledgments
We acknowledge all the works that could not be cited due to space constraints. This material is based in part upon work supported by the National Institutes of Health (DP5OD012133-01 and P50CA092131) and the National Science Foundation (CBET 1533767), P.H. is supported by the NIH Biotechnology Training in Biomedical Sciences and Engineering Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31:448–52. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 3.Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat Rev Mol Cell Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojima R, Aubel D, Fussenegger M. Toward a world of theranostic medication: Programming biological sentinel systems for therapeutic intervention. Adv Drug Deliv Rev. 2016;105:66–76. doi: 10.1016/j.addr.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Gaj T, Wallen MC, Barbas CF. Improved Cell-Penetrating Zinc-Finger Nuclease Proteins for Precision Genome Engineering. Mol Ther Acids. 2015;4:e232. doi: 10.1038/mtna.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mussolino C, Alzubi J, Fine EJ, Morbitzer R, Cradick TJ, Lahaye T, Bao G, Cathomen T. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014;42:6762–6773. doi: 10.1093/nar/gku305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–91. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Gaj T, Yang Y, Wang N, Shui S, Kim S, Kanchiswamy CN, Kim J-S, Barbas CF. Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat Protoc. 2015;10:1842–59. doi: 10.1038/nprot.2015.117. [DOI] [PubMed] [Google Scholar]
- 9.Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, et al. Small Molecules Enhance CRISPR Genome Editing in Pluripotent Stem Cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 12.Pinder J, Salsman J, Dellaire G. Nuclear domain “knock-in” screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–92. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez F, Zhu Z, Shi Z-D, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–26. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black JB, Adler AF, Wang H-G, D’Ippolito AM, Hutchinson HA, Reddy TE, Pitt GS, Leong KW, Gersbach CA. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell. 2016;19:406–414. doi: 10.1016/j.stem.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore R, Spinhirne A, Lai MJ, Preisser S, Li Y, Kang T, Bleris L. CRISPR-based selfcleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res. 2015;43:1297–1303. doi: 10.1093/nar/gku1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ede C, Chen X, Lin M-Y, Chen YY. Quantitative Analyses of Core Promoters Enable Precise Engineering of Regulated Gene Expression in Mammalian Cells. ACS Synth Biol. 2016;5:395–404. doi: 10.1021/acssynbio.5b00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Li T, Wang X, Du Z, Liu R, Yang Y. Synthetic dual-input mammalian genetic circuits enable tunable and stringent transcription control by chemical and light. Nucleic Acids Res. 2016;44:2677–2690. doi: 10.1093/nar/gkv1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Xiong X, Wong S, Charles EJ, Lim WA, Qi LS. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods. 2016;13:1043–1049. doi: 10.1038/nmeth.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Slomovic S, Collins JJ. DNA sense-and-respond protein modules for mammalian cells. Nat Methods. 2015;12:1085–1090. doi: 10.1038/nmeth.3585. [DOI] [PubMed] [Google Scholar]
- 29.Gaber R, Lebar T, Majerle A, Ŝter B, Dobnikar A, Bencina M, Jerala R. Designable DNA- binding domains enable construction of logic circuits in mammalian cells. Nat Chem Biol. 2014;10:203–8. doi: 10.1038/nchembio.1433. [DOI] [PubMed] [Google Scholar]
- 30.Hemphill J, Liu Q, Uprety R, Samanta S, Tsang M, Juliano RL, Deiters A. Conditional Control of Alternative Splicing through Light-Triggered Splice-Switching Oligonucleotides. J Am Chem Soc. 2015;137:3656–3662. doi: 10.1021/jacs.5b00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groves B, Chen Y-J, Zurla C, Pochekailov S, Kirschman JL, Santangelo PJ, Seelig G. Computing in mammalian cells with nucleic acid strand exchange. Nat Nanotechnol. 2015;11:287–294. doi: 10.1038/nnano.2015.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei KY, Smolke CD. Engineering dynamic cell cycle control with synthetic small molecule-responsive RNA devices. J Biol Eng. 2015;9:21. doi: 10.1186/s13036-015-0019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloom RJ, Winkler SM, Smolke CD. A quantitative framework for the forward design of synthetic miRNA circuits. Nat Methods. 2014;11:1147–1153. doi: 10.1038/nmeth.3100. [DOI] [PubMed] [Google Scholar]
- 34.Miki K, Endo K, Takahashi S, Funakoshi S, Takei I, Katayama S, Toyoda T, Kotaka M, Takaki T, Umeda M, et al. Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell. 2015;16:699–711. doi: 10.1016/j.stem.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 35••.Parr CJC, Katayama S, Miki K, Kuang Y, Yoshida Y, Morizane A, Takahashi J, Yamanaka S, Saito H. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci Rep. 2016;6:32532. doi: 10.1038/srep32532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong ASL, Choi GCG, Cui CH, Pregernig G, Milani P, Adam M, Perli SD, Kazer SW, Gaillard A, Hermann M, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci U S A. 2016;113:2544–9. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stockman VB, Ghamsari L, Lasso G, Honig B, Shapira SD, Wang HH, Boone C, Bussey H, Andrews B, Tong A, et al. A High-Throughput Strategy for Dissecting Mammalian Genetic Interactions. PLoS One. 2016;11:e0167617. doi: 10.1371/journal.pone.0167617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toh C-XD, Chan J-W, Chong Z-S, Wang HF, Guo HC, Satapathy S, Ma D, Goh GYL, Khattar E, Yang L, et al. RNAi Reveals Phase-Specific Global Regulators of Human Somatic Cell Reprogramming. Cell Rep. 2016;15:2597–2607. doi: 10.1016/j.celrep.2016.05.049. [DOI] [PubMed] [Google Scholar]
- 39.Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882. e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866. e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896. e15. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 42•.Singer M, Wang C, Kuchroo VK, Regev A, Anderson AC, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell. 2016;166:1500–1511. e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Perli SD, Cui CH, Lu TK. Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science. 2016;353:aag0511–aag0511. doi: 10.1126/science.aag0511. [DOI] [PubMed] [Google Scholar]
- 44.Kalhor R, Mali P, Church GM. Rapidly evolving homing CRISPR barcodes. Nat Methods. 2017;14:195–200. doi: 10.1038/nmeth.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokolik C, Liu Y, Bauer D, McPherson J, Broeker M, Heimberg G, Qi LS, Sivak DA, Thomson M. Transcription Factor Competition Allows Embryonic Stem Cells to Distinguish Authentic Signals from Noise. Cell Syst. 2015;1:117–129. doi: 10.1016/j.cels.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunger JM, Zutshi A, Willard VP, Gersbach CA, Guilak F. CRISPR/Cas9 editing of induced pluripotent stem cells for engineering inflammation-resistant tissues. Arthritis Rheumatol. 2016 doi: 10.1002/art.39982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saxena P, Heng BC, Bai P, Folcher M, Zulewski H, Fussenegger M. A programmable synthetic lineage-control network that differentiates human IPSCs into glucose- sensitive insulin-secreting beta-like cells. Nat Commun. 2016;7:11247. doi: 10.1038/ncomms11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Xu L, Gibson TM, Gersbach CA, Sullenger BA. Differential effects of toll-like receptor stimulation on mRNA-driven myogenic conversion of human and mouse fibroblasts. Biochem Biophys Res Commun. 2016;478:1484–90. doi: 10.1016/j.bbrc.2016.08.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, Lim WA. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schukur L, Geering B, Charpin-El Hamri G, Fussenegger M. Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Sci Transl Med. 2015;7:318r–a201. doi: 10.1126/scitranslmed.aac4964. [DOI] [PubMed] [Google Scholar]
- 51.Schwarz KA, Daringer NM, Dolberg TB, Leonard JN. Rewiring human cellular input- output using modular extracellular sensors. Nat Chem Biol. 2016;13:202–209. doi: 10.1038/nchembio.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung ME, Leonard JN. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J Extracell vesicles. 2016;5:31027. doi: 10.3402/jev.v5.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Müller M, Ausländer S, Spinnler A, Auslander D, Sikorski J, Folcher M, Fussenegger M. Designed cell consortia as fragrance-programmable analog-to-digital converters. Nat Chem Biol. 2017;13:309–316. doi: 10.1038/nchembio.2281. [DOI] [PubMed] [Google Scholar]
- 54.Bai P, Ye H, Xie M, Saxena P, Zulewski H, Charpin-El Hamri G, Djonov V, Fussenegger M. A synthetic biology-based device prevents liver injury in mice. J Hepatol. 2016;65:84–94. doi: 10.1016/j.jhep.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakravarti D, Cho JH, Weinberg BH, Wong NM, Wong WW. Synthetic biology approaches in cancer immunotherapy, genetic network engineering, and genome editing. Integr Biol. 2016;8:504–517. doi: 10.1039/c5ib00325c. [DOI] [PubMed] [Google Scholar]
- 56.Zah E, Lin M-Y, Silva-Benedict A, Jensen MC, Chen YY. T cells expressing CD19/CD20 bi-specific chimeric antigen receptors prevent antigen escape by malignant B cells. Cancer Immunol Res. 2016;4:498–508. doi: 10.1158/2326-6066.CIR-15-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Roybal KT, Rupp LJ, Morsut L, Walker WJ, McNally KA, Park JS, Lim WA. Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie M, Ye H, Wang H, Charpin-El Hamri G, Lormeau C, Saxena P, Stelling J, Fussenegger M. β-cell-mimetic designer cells provide closed-loop glycemic control. Science (80-) 2016;354:1296–1301. doi: 10.1126/science.aaf4006. [DOI] [PubMed] [Google Scholar]
- 59.Saxena P, Charpin-El Hamri G, Folcher M, Zulewski H, Fussenegger M. Synthetic gene network restoring endogenous pituitary-thyroid feedback control in experimental Graves’ disease. Proc Natl Acad Sci. 2016;113:1244–1249. doi: 10.1073/pnas.1514383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grada Z, Hegde M, Byrd T, Shaffer DR, Ghazi A, Brawley VS, Corder A, Schönfeld K, Koch J, Dotti G, et al. TanCAR: A Novel Bispecific Chimeric Antigen Receptor for Cancer Immunotherapy. Mol Ther Nucleic Acids. 2013;2:e105. doi: 10.1038/mtna.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hegde M, Mukherjee M, Grada Z, Pignata A, Landi D, Navai SA, Wakefield A, Fousek K, Bielamowicz K, Chow KKH, et al. Tandem CAR T cells targeting HER2 and IL13Ra2 mitigate tumor antigen escape. J Clin Invest. 2016;126:3036–3052. doi: 10.1172/JCI83416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, Heo S-J, Mitros T, Munoz DP, Boffelli D, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360r–a134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]


