Abstract
Sepsis remains a leading cause of morbidity and mortality among intensive care unit (ICU) patients. For each hour treatment initiation is delayed after diagnosis, sepsis-related mortality increases by approximately eight percent. Therefore, maximizing effective care requires early recognition and initiation of treatment protocols. Antecedent signs and symptoms of sepsis can be subtle and unrecognizable (e.g., loss of autonomic regulation of vital signs), causing treatment delays and harm to the patient. In this work we investigated the utility of high-resolution blood pressure (BP) and heart rate (HR) times series dynamics for the early prediction of sepsis in patients from an urban, academic hospital, meeting the third international consensus definition of sepsis (sepsis-III) during their ICU admission. Using a multivariate modeling approach we found that HR and BP dynamics at multiple time-scales are independent predictors of sepsis, even after adjusting for commonly measured clinical values and patient demographics and comorbidities. Earlier recognition and diagnosis of sepsis has the potential to decrease sepsis-related morbidity and mortality through earlier initiation of treatment protocols.
Introduction
Sepsis is among the leading causes of morbidity and mortality in critically ill patients and is the most expensive condition by healthcare spending [1–2]. The major tenet of sepsis care is prompt recognition and initiation of treatment. Recent studies have shown that survival benefit from early intervention in sepsis is almost entirely dependent on time-to-first antibiotics [3]. However, no clinically validated system exists for accurate, real-time prediction of sepsis onset in the adult intensive care unit (ICU) population. Such a system should provide enough lead time for initiation of administration of pressors, fluids and antibiotics in a proactive and timely manner. Several publications have attempted to predict downstream events related to sepsis (such as organ failure and septic shock [4]), but the key issue (identifying sepsis in a few hours prior to the clinician identifies it) remains unsolved. A major impediment to application of supervised machine learning techniques to the problem of early prediction of sepsis has been the lack of a gold standard for diagnosis of sepsis [5]. More recently, the Third International Consensus Definitions Task Force [6], with the objective of increasing the specificity of diagnosis, defined sepsis as “life-threatening organ dysfunction due to a dysregulated host response to infection.” Subsequent work by Seymour et al. [7] provided a more precise clinical criteria for identifying patients with suspected infection who are at risk of sepsis. They identified an episode of suspected infection as the combination of antibiotics and blood cultures within a specific time epoch, and defined the first of these two events as the “onset” of infection. The onset time of sepsis was then defined as an episode of suspected infection with two points or more change in the Sequential Organ Failure Assessment (SOFA) Score. Using this new definition, Seymour et al. were able to validate the discriminative power of the existing clinical criteria and definitions for sepsis using 1.3 million electronic medical record (EMR) encounters [7].
Much of the existing literature on application of predictive analytics to early prediction of sepsis has focused around the EMR and lab results as features for a supervised machine learning algorithm. However, the resolution of the EMR data is far too coarse to produce individually specific predictions. In fact, often availability of certain lab results in themselves may be indicative of clinical suspicion of sepsis. Conversely, without having a clinician in the loop to recognize early signs of decompensation and order the necessary lab values, an EMR-based predictive analytic algorithm may not have access to the required features to predict sepsis in a timely manner.
The use of continuously measured high-resolution ECG and blood pressure data has provided promising results in the hunt for an accurate predictor. Sepsis is known as a dysregulated immune-mediated host response to infection. According to the anti-inflammatory reflex model [20], action potentials coming from the vagus nerve control lymphocytes to secrete acetylcholine that inhibits the production of pro-inflammatory cytokines through alpha7 nicotinic receptors. In other words, pathogen-induced inflammation increases the activity of vagus nerve, which controls the production of pro-inflammatory cytokines and prevents tissue damage. Although, the relationship amongst inflammation, vagus nerve activity and heart rate variability (HRV) and Baroreflex control of blood pressure (BP) and heart rate (HR) is complex, this model suggests that monitoring indices of heart rate variability and complexity (as markers of vagus nerve activity) may provide a useful tool as a measure of anti-inflammatory reflex in health and disease. Hug et al. [8] demonstrated that transient hypotensive events, identified from the raw blood pressure waveform, which later led to sepsis and a higher mortality, were missed by clinical teams for 4 hours on average. The key to this discovery was the use of signal quality metrics to reprocess the blood pressure waveform and remove untrustworthy data. Changes in blood pressure and heart rate dynamics have been shown to be associated with the onset of decompensation in critically ill patients [9–11]. In particular, Mayaud et al demonstrated that the entropy of heart rate is associated with sepsis in adult critical care subjects [11]. After extracting a cohort of 2155 severe sepsis patients from the MIMIC II database [12] using the Angus criteria [13] they trained a logistic regression algorithm to predict in-hospital mortality. They showed that when using multiscale entropy (MSE) coefficients [14] as inputs, an AUROC of 0.63 ± 0.01 was possible, compared to 0.68% ± 0.01 when using the Acute Physiology Score (APS) with its original coefficients. Combining both gave an AUROC of 0.70 ± 0.01. This motivates the need for high-resolution physiological data on multiple time scales.
In this work, following the latest definition of sepsis [6–7], we sought to predict onset of sepsis four hours ahead of time, using commonly measured vital signs, as well as features derived from ECG and Blood pressure waveforms.
Methods
Description of cohort
Heart rate (from the electrocardiogram) and blood pressure time series at 2 seconds resolution were collected from ICU bedside monitors in an Emory affiliated hospital, using the BedMaster system (Excel Medical Electronics, Jupiter FL, USA), which is a third-party software connected to the hospital’s General Electric (GE) monitors for the purpose of electronic data extraction and storage of high-resolution waveforms. All adult ICU units were included in this study, including Medical and Surgical, Cardiac Care, and Neuro-intensive care units. The bedside monitor data was then matched and time synchronized to each patient’s EMR data. Of the total of 1100 matched patients, 242 (~22%) met the definition of sepsis by Seymour et al. [7] at some time point during their ICU stay. Specifically, all episodes of suspected infection (tsuspicion) were identified as the earlier timestamp of antibiotics and blood cultures within a specific time span; if the antibiotic was given first, the culture sampling must have been obtained within 24 hours. If the culture sampling was first, the antibiotic must have been ordered within 72 hours. The onset time of sepsis (tsepsis) was then defined as an episode of suspected infection with a two or more points change in the SOFA score from up to 48 hours before to up to 24 hours after the tsuspicion. The average length of hospital stay (LOS) among the septic patients was 137.6 [68.2–295.7] hours, and the percentages of in-hospital mortality and in-patient hospice were 15.2% and 13.5%, respectively. The septic patients exhibited a higher average SOFA score compared to non-septic patients (4.8 [3.1–6.8] versus 1.6 [0.6–3.4]).
Features
For every subject, their socio-demographics features (Age, Gender, Weight, Race) were collected. We also included features that were commonly recorded by the bedside nurses including, Mean Arterial Pressure (MAP), Heart Rate (HR), Peripheral capillary Oxygen Saturation (SpO2), Systolic Blood Pressure (SBP), Diastolic Blood Pressure (DBP), Respiration Rate (Resp), Glasgow Coma Score (GCS), and Temperature (Temp). Each of the above mentioned features were discretized into 8 bins. We also extracted a few features that capture history, comorbidity, and the clinical context of the patient, including Charlson Comorbidity Index, Mechanical Ventilation, Unit Information (Surgical, Cardiac Care, or Neuro-intensive care), as well as Surgical Specialty (Cardiovascular, Neuro, Ortho-Spine, Oncology, Urology, etc.) and Wound Type (clean, contaminated, dirty, or infected) if the patient had a surgery in past 72 hours.
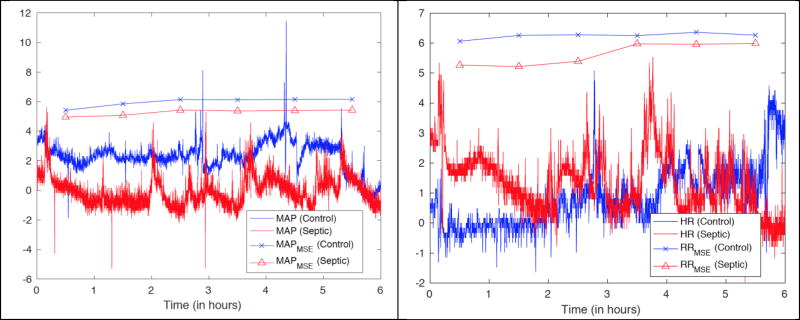
We also calculated the following features from the HR and MAP time series (2 sec resolution) derived from the bedside monitor’s proprietary software from the ECG and BP waveforms: standard Deviation of HR (HRSTD), Standard Deviation of MAP (MAPSTD), Multiscale Entropy and Conditional Multiscale Entropy of (60/HR or RR intervals) and MAP (Over 17 Scales; RRMSE and RRMSCE, and MAPMSE and MAPMSCE, respectively). The time series-related features were calculated using 6 hours time windows, with one hour strides. For each window (6 hour × 60 min/hour × 30 samples / min = 10800 samples) we considered 17 different scales ( scales 1, 4, 7, …, 49), over which sample entropy [14] and conditional entropy [15] were calculated. A comparison between the Multiscale entropy features of a septic and a control subject is shown in Fig. 1.
Figure 1.
(Left panel) Plot of MAP time series for a septic and a control subject, and their corresponding multiscale entropy at Scale 4. (Right panel) Plot of HR time series for a septic and a control subject, and their corresponding multiscale entropy at Scale 4. The HR and MAP time series have been normalized by their standard deviations for illustration purposes
All clinical features with a higher sampling frequency than 1/hour were further uniformly resampled into one hour time bins, by taking the median values if multiple measurement were available, updated hourly if/when new data became available, and otherwise the old values were kept (sample-and-hold interpolation). Mean imputation was used to replace all remaining missing values (mainly at the start of each record).
Algorithm
As noted above, to predict sepsis or suspicion of sepsis 4 hours ahead of time, we considered three separate models, based on, 1) entropy features derived from the bedside monitor data, 2) combining the EMR features, and socio-demographic-patient history features, and 3) combining features from models 1 and 2. An Elastic Net logistic classifier was trained for all the three models, where an internal 10 fold cross validation was performed to determine the optimal regularization parameter (λ).
Statistical Methods
For all continuous variables we report medians ([25-percentile, 75-percentile]) and utilize a two-sided Wilcoxon rank-sum test while comparing two populations. All classification results are based on a 10 fold cross-validation (80% training 20% testing), and the area under receiver operating characteristic (AUROC) curves is reported, as well as specificity and accuracy at a fixed 85% sensitivity level. We combined all the predictions (probabilities of being septic) across all the ten folds to report a single pooled AUROC [19].
Results
We evaluated a total of 242 subjects for this study. The median [25-percentile, 75-percentile] age of the cohort was 59 [46, 69] years, and approximately 47% were female. The baseline characteristics of the entire patient population have been tabulated in Table 1. Notably, onset of sepsis is associated with a drop in MAP (76 [70 87] vs. 80.3 [70.0 94.0]) as well as SBP and DBP, and an increase in HR.
Table 1.
Baseline characteristics of the patient population
| Clinical Variables | Baseline | Septic | p-value |
|---|---|---|---|
| MAP | 80.3 [70.0 94.0] | 76 [70 87] | 0.02 |
| HR | 90.0 [75.0 104.5] | 92.0 [78.0 110.0] | <0.01 |
| SpO2 | 98.0 [96.0 100.0] | 98.0 [96.0 100.0] | 0.15 |
| SBP | 119.0 [105.0 140.0] | 116.0 [102.0 136.0] | <0.01 |
| DBP | 62.0 [53.4 73.2] | 58.0 [52.0 66.0] | <0.01 |
| Respiration Rate | 18.0 [12.5 22.0] | 17.0 [10.0 22.0] | 0.29 |
| GCS | 10.0 [6.0 15.0] | 9.0 [5.0 14.0] | 0.13 |
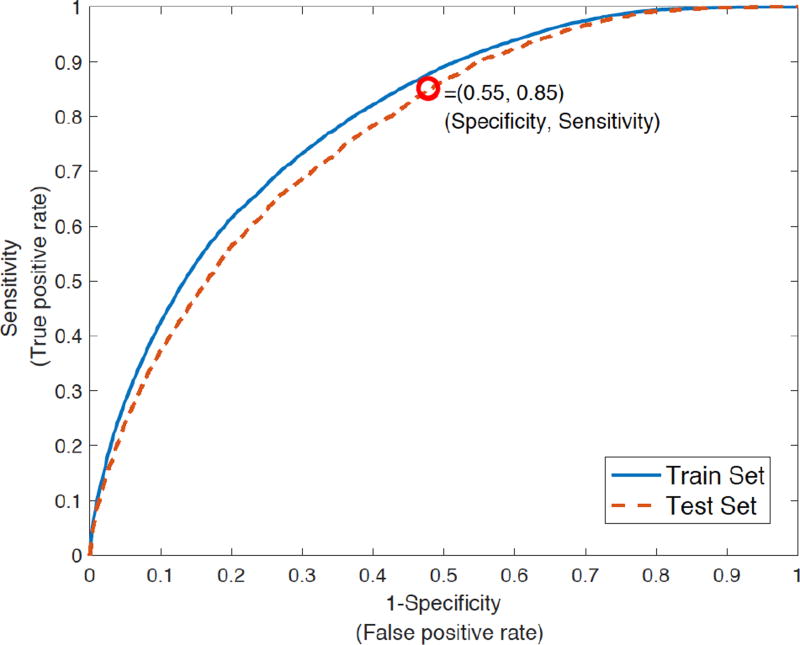
Table 2 provides a summary of the performance of all the three models. The Entropy features alone (Model 1) achieved a pooled AUROC of 0.67, with an overall accuracy of 47% on the testing set. A statistical significance test was performed to determine the importance of the entropy features, and 14 out of the 68 entropy features had p-values less than α= 0.05 (including scales 4, 7, and 10). Next, the model based on combining the EMR features and socio-demographic-patient history features (Model 2) achieved a pooled AUROC of 0.70, with an overall accuracy of 50%. Finally, by combining all the features (Model 3), the classifier achieved a pooled AUROC of 0.78, with an overall accuracy of 61% (See Fig. 2).
Table 2.
Performance summary of all three models.
| Model | AUC ROC Test (Train) |
Specificity* Test (Train) |
Accuracy* Test (Train) |
|---|---|---|---|
| Entropy features | 0.67 (0.68) | 0.41 (0.42) | 0.47 (0.49) |
| EMR + Socio-demographic patient history features | 0.70 (0.72) | 0.45 (0.46) | 0.50 (0.52) |
| Entropy + EMR + Socio-demographic patient history features | 0.78 (0.80) | 0.55 (0.57) | 0.61 (0.62) |
Model sensitivity was fixed at 0.85
Figure 2.
Area under the receiver-operating characteristic (ROC) curve of Model 3 (Combining Entropy, EMR, and socio-demographic-patient history features). The AUROC on test set and training set was 0.78 and 0.80 respectively. The model 3 achieved a specificity of 0.55 at 0.85 sensitivity level (for the test set).
Discussion
Our results demonstrate that the dynamics of the HR and BP time series (at varying scales) provide useful predictors of sepsis in the critical care setting four hours before suspicion of sepsis. Inclusion of patients’ demographics, comorbidities, and history of surgery provided a boost in AUROC from 0.67 to 0.78 (p<0.01) and similar improvement in specificity (41% versus 55%) at 85% sensitivity level.
Many predictions focus on death, an outcome for which there is little that can be directly modified in management [16–17]. Earlier recognition and diagnosis of adverse events such as sepsis has the potential to decrease morbidity and mortality through earlier initiation of treatment protocols. However, even when an outcome can be modified, it may be too late to have significant impact. Detection or classification of sepsis or associated sequelae, such as death, has received significant attention in the literature (clinical, engineering and statistical/machine learning). However, prediction has almost exclusively focused on the downstream events to onset of sepsis (such as the septic shock) [4] when it may be too late to effectively intervene in many cases.
This paper is among the first of its kind to look at combining different “granularities” of data to evaluate their combined effect. We note that the InSight algorithm [18] utilized a minimal set of commonly available EHR variables (including blood pressure, heart rate, respiration rate, temperature, SpO2, and GCS) to achieve a slightly lower AUC of 0.74 (± 0.01) for 4 hours ahead prediction of sepsis. However, the authors followed the sepsis III definition, they opted for predicting the time of two or more points change in the SOFA score (tsofa), which is arguably easier to detect than tsepsis given the lab data. In our cohort, roughly 20% of the time tsofa occurs after tsepsis, which may be a consequence of the fact that calculation of an updated SOFA score requires ordering of labs, which is more likely to occur if there is already a clinical suspicion of sepsis. In general, comparison of various algorithms within the literature has been made complicated due to scarcity of standardized publicly available ICU datasets, in addition to differences in definitions of sepsis and data elements collected across institutions. This may include utilization of acausal measurements, which occurs when the order-time of labs are used in prediction models [4, 18] rather than the time when laboratory data become available to the bedside clinicians. Therefore, validation and prospective studies are needed to shed light on the strengths and weaknesses of various algorithms.
Some of the limitations of the current study include the relatively small sample size and the utilization of a single clinical cohort from an urban, academic hospital. Our final model achieved an AUROC of 0.78 which is slightly higher than the existing works [18]. We anticipate that use of high resolution BP, HR and Resp dynamics, including waveform morphology and coupling features, will add further predictive power to our and other existing models and provide a more objective metric of early identification of sepsis and other types of decompensation in critically ill patients.
Conclusion
Multiscale entropy-based measures of HR and BP dynamics can independently predict sepsis four hours prior to its onset. Predictive performance is greatly improved by creating a model that includes other relevant clinical data available from the EMR in real time. Future studies will validate these findings in a larger cohort, and add respiratory dynamics and more advanced derivatives of the ECG and BP waveform to test for further improvement in sepsis prediction.
Acknowledgments
SN is funded by the National Institutes of Health, award # K01ES025445. QL is partially supported by the Surgical Critical Care Initiative (SC2i), funded by the Department of Defense’s Defense Health Program – Joint Program Committee 6 / Combat Casualty Care (USUHS HT9404-13-1-0032 and HU0001-15-2-0001). The opinions or assertions contained herein are the private ones of the author/speaker and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences or any other agency of the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, Rochwerg B. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Medicine. 2017 Mar 1;43(3):304–77. doi: 10.1007/s00134-017-4683-6. [DOI] [PubMed] [Google Scholar]
- 2.Soong J, Soni N. Sepsis: recognition and treatment. Clinical Medicine. 2012 Jun 1;12(3):276–80. doi: 10.7861/clinmedicine.12-3-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frost R, Newsham H, Parmar S, Gonzalez-Ruiz A. Impact of delayed antimicrobial therapy in septic ITU patients. Critical Care. 2010 Apr 1;14(S2):P20. [Google Scholar]
- 4.Saria S, Rajani AK, Gould J, Koller D, Penn A. Integration of early physiological responses predicts later illness severity in preterm infants. Science Translational Medicine. 2010;2(48):48–65. doi: 10.1126/scitranslmed.3001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obermeyer Z, Emanuel EJ. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. The New England Journal of Medicine. 2016 Sep 29;375(13):1216. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS. The third international consensus definitions for sepsis and septic shock (sepsis-3) Journal of the American Medical Association (JAMA) 2016 Feb 23;315(8):801–10. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, Rubenfeld G, Kahn JM, Shankar-Hari M, Singer M, Deutschman CS. Assessment of clinical criteria for sepsis: for the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) Jama. 2016 Feb 23;315(8):762–74. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hug CW, Clifford GD, Reisner AT. Clinician blood pressure documentation of stable intensive care patients: an intelligent archiving agent has a higher association with future hypotension. Critical Care Medicine. 2011 May;39(5):1006. doi: 10.1097/CCM.0b013e31820eab8e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchman TG. Nonlinear dynamics, complex systems, and the patho-biology of critical illness. Current Opinion Crit. Care. 2004;10(5):378–382. doi: 10.1097/01.ccx.0000139369.65817.b6. [DOI] [PubMed] [Google Scholar]
- 10.Lehman Li-wei, Mark Roger, Nemati Shamim. A Model-based Machine Learning Approach to Probing Autonomic Regulation from Nonstationary Vital-Signs Time Series. IEEE Journal of Biomedical and Health Informatics. 2016;(99):2016. doi: 10.1109/JBHI.2016.2636808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayaud L, Tarassenko L, Annane D, Clifford G. Predictive power of heart rate complexity to estimate severity in severe sepsis patients. Journal of Critical Care. 2013 Dec 1;28(6):e37. [Google Scholar]
- 12.Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Scientific Data. 2016;3 doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Critical care medicine. 2001 Jul 1;29(7):1303–10. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Costa M, Goldberger AL, Peng CK. Multiscale entropy analysis of complex physiologic time series. Physical review letters. 2002 Jul 19;89(6):068102. doi: 10.1103/PhysRevLett.89.068102. [DOI] [PubMed] [Google Scholar]
- 15.Nemati S, Edwards BA, Lee J, Pittman-Polletta B, Butler JP, Malhotra A. Respiration and heart rate complexity: effects of age and gender assessed by band-limited transfer entropy. Respiratory physiology & neurobiology. 2013 Oct 1;189(1):27–33. doi: 10.1016/j.resp.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor RA, Pare JR, Venkatesh AK, Mowafi H, Melnick ER, Fleischman W, Hall MK. Prediction of In-hospital Mortality in Emergency Department Patients With Sepsis: A Local Big Data–Driven, Machine Learning Approach. Academic Emergency Medicine. 2016 Feb 1; doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gultepe E, Green JP, Nguyen H, Adams J, Albertson T, Tagkopoulos I. From vital signs to clinical outcomes for patients with sepsis: a machine learning basis for a clinical decision support system. Journal of the American Medical Informatics Association. 2014 Mar 1;21(2):315–25. doi: 10.1136/amiajnl-2013-001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desautels T, Calvert J, Hoffman J, Jay M, Kerem Y, Shieh L, Shimabukuro D, Chettipally U, Feldman MD, Barton C, Wales DJ. Prediction of Sepsis in the Intensive Care Unit With Minimal Electronic Health Record Data: A Machine Learning Approach. JMIR medical informatics. 2016 Jul;4(3) doi: 10.2196/medinform.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Airola A, Pahikkala T, Waegeman W, De Baets B, Salakoski T. A comparison of AUC estimators in small-sample studies. Machine Learning in Systems Biology. 2009 Mar 2;:3–13. [Google Scholar]
- 20.Huston JM, Tracey KJ. The pulse of inflammation: heart rate variability, the cholinergic anti-inflammatory pathway and implications for therapy. Journal of Internal medicine. 2011 Jan 1;269(1):45–53. doi: 10.1111/j.1365-2796.2010.02321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]