Abstract
A genome-wide association study (GWAS) correlates marker and trait variation in a study sample. Each subject is genotyped at a multitude of SNPs (single nucleotide polymorphisms) spanning the genome. Here we assume that subjects are randomly collected unrelateds and that trait values are normally distributed or can be transformed to normality. Over the past decade, geneticists have been remarkably successful in applying GWAS analysis to hundreds of traits. The massive amount of data produced in these studies presents unique computational challenges. Penalized regression with LASSO or MCP penalties is capable of selecting a handful of associated SNPs from millions of potential SNPs. Unfortunately, model selection can be corrupted by false positives and false negatives, obscuring the genetic underpinning of a trait. Here we compare LASSO and MCP penalized regression to iterative hard thresholding (IHT). On GWAS regression data, IHT is better at model selection and comparable in speed to both methods of penalized regression. This conclusion holds for both simulated and real GWAS data. IHT fosters parallelization and scales well in problems with large numbers of causal markers. Our parallel implementation of IHT accommodates SNP genotype compression and exploits multiple CPU cores and graphics processing units (GPUs). This allows statistical geneticists to leverage commodity desktop computers in GWAS analysis and to avoid supercomputing.
Keywords: genetic association studies, greedy algorithm, parallel computing, sparse regression
1 Introduction
Over the past decade, genome-wide association studies (GWASs) have benefitted from technological advances in dense genotyping arrays, high-throughput sequencing, and more powerful computing resources. Yet researchers still struggle to find the genetic variants that account for the missing heritability of many traits. It is now common for consortia studying a complex trait such as height to pool results across multiple sites and countries. Meta-analyses have discovered hundreds of statistically significant SNPs, each of which explains a small fraction of the total heritability. A drawback of GWAS meta-analysis is that it relies on univariate regression rather than on more informative multivariate regression (J. Yang et al., 2010). Because the number of SNPs (predictors) in a GWAS vastly exceeds the number of study subjects (observations), statistical geneticists have resorted to machine learning techniques such as penalized regression (Lange, Papp, Sinsheimer, & Sobel, 2014) for model selection.
In the statistical setting of n subjects and p predictors with n ≪ p, penalized regression estimates a sparse parameter vector β ∈ Rp by minimizing an appropriate objective function f(β) + λp(β), where f(β) is a convex loss, p(β) is a suitable penalty, and λ is a tuning constant controlling the sparsity of β. The most popular and mature sparse regression tool is LASSO (ℓ1) regression (S. Chen & Donoho, 1994; Tibshirani, 1996). Unfortunately, LASSO parameter estimates are biased towards zero (Hastie, Friedman, & Tibshirani, 2009), usually severely so. As a consequence of shrinkage, LASSO regression lets too many false positives enter a model. Since GWAS is often followed by expensive biological validation studies, there is value in reducing false positive rates. To counteract the side effects of shrinkage, Zhang (Zhang, 2010) recommends the minimax concave penalty (MCP) as an alternative to the ℓ1 penalty. Other nonconvex penalties exist, but MCP is probably the simplest to implement. MCP also has provable convergence guarantees. In contrast to the LASSO, which admits too many false positives, MCP tends to allow too few predictors to enter a model. Thus, its false negative rate is too high. Our subsequent numerical examples confirm these tendencies.
Surprisingly few software packages implement efficient penalized regression algorithms for GWAS. The R packages glmnet and ncvreg are ideal candidates, given their ease of use, maturity of development, and wide acceptance. The former implements LASSO-penalized regression (Friedman, Hastie, & Tibshirani, 2010; Lange, 2010; Tibshirani, 1996), while the latter implements both LASSO- and MCP-penalized regression (Breheny & Huang, 2011; Zhang, 2010). Both packages provide excellent functionality for moderately sized problems. However, R’s poor memory management hinders the scalability of both algorithms. In fact, analysis on a typical workstation is limited to at most a handful of chromosomes at a time. Larger problems must appeal to cluster or cloud computing. Neither glmnet nor ncvreg natively support the compressed PLINK binary genotype file (BED file) format so effective in storing and distributing GWAS data (Purcell et al., 2007). Scalable implementations of LASSO for GWAS with PLINK files appear in the packages Mendel, gpu-lasso, SparSNP, and the beta version of PLINK 1.9 (Abraham, Kowalczyk, Zobel, & Inouye, 2012; Chang et al., 2015; G. K. Chen, 2012; Lange et al., 2013; Wu & Lange, 2008). All of these packages include parallel computing capabilities for large GWAS datasets. Mendel, gpu-lasso, and PLINK 1.9 beta have multicore acceleration capabilities, while SparSNP can be run on compute clusters. To our knowledge, only Mendel supports MCP regression with PLINK files.
As an alternative to penalized regression, one can tackle sparsity directly through greedy algorithms for sparse reconstruction. The matching pursuit (MP) (Mallat & Zhang, 1993) algorithm from the signals processing literature reconstructs a signal by adding predictors piecemeal, eventually yielding a sparse representation of the signal. This is a generalization of the older statistical procedure of forward stagewise regression (Donoho, Tsaig, Drori, & Starck, 2012). Similar algorithms treated in the signal processing literature include hard thresholding pursuit (HTP) (Bahmani, Raj, & Boufounos, 2013; Foucart, 2011; Yuan, Li, & Zhang, 2014), orthogonal matching pursuit (OMP) (Pati, Rezaiifar, & Krishnaprasad, 1993; Tropp & Gilbert, 2007), compresive sample matching pursuit (CoSaMP) (Needell & Tropp, 2009) and subspace pursuit (SP) (Dai & Milenkovic, 2009).
An alternative approach is to handle sparsity through projection onto sparsity sets (Blumensath, 2012; Blumensath & Davies, 2008, 2009, 2010). Iterative hard thresholding (IHT) minimizes a loss function f(β) subject to the sparsity constraint ||β||0 ≤ k, where the ℓ0 “norm” ||β||0 counts the number of nonzero entries of the parameter vector β. The integer k serves as a tuning constant analogous to λ in LASSO and MCP regression. IHT can be viewed as a version of projected gradient descent tailored to sparse regression. Since these algorithms rely solely on gradients, they avoid computing and inverting large Hessian matrices and hence scale well to large datasets.
Like matching pursuit algorithms, IHT is fundamentally a greedy selection procedure. Distinguishing which greedy algorithm demonstrates superior performance is no simple task. Typical performance metrics include computational speed, signal recovery behavior, and convergence guarantees in noisy environments. Although from a theoretical point of view, no greedy algorithm is uniformly superior to the others, IHT demonstrates the best balance of these three criteria among greedy algorithms (Blanchard, Cartis, Tanner, & Thompson, 2011; Blanchard & Tanner, 2015). Implementation details such as memory management, hardware platform, and choice of computing environment can complicate this picture. In light of established results with greedy algorithms, we believe that a careful implementation of IHT can provide sparse approximation performance that is competitive or superior to current penalized regression procedures.
Our implementation of IHT addresses some of the specific concerns of GWAS. First, it accommodates genotypes presented in compressed PLINK format. Second, our version of IHT allows the user to choose the sparsity level k of a model. In contrast, LASSO and MCP penalized regression must choose model size indirectly by adjusting the tuning constant λ to match a given k. Third, our version of IHT is implemented in the package IHT.jl in the free Julia programming language. Julia works on a variety of hardware platforms, encourages prudent control of memory, exploits all available CPU cores, and interfaces with massively parallel graphics processing unit (GPU) devices. Finally, IHT performs more precise model selection than either LASSO or MCP penalized regression. We use “precise” in the information theoretic sense: given the sets of markers selected by each of the three algorithm, the set that IHT selects contains a higher proportion of causal markers than those of LASSO and MCP. While this does not mean that IHT consistently recovers all causal markers, the markers it does recover are more credible than the markers that LASSO and MCP recover. All of these advantages can be realized on a modern desktop computer. Although our current IHT implementation is limited to ordinary linear least squares, the literature suggests that logistic regression is within reach (Bahmani et al., 2013; Yuan et al., 2014).
It worth stressing that our focus is on parameter estimation and model selection. Historically IHT lacked a coherent inference framework for constructing valid post-selection confidence intervals and P-values. A recent paper (F. Yang, Barber, Jain, & Lafferty, 2016) tries to fill this gap for group IHT; its applicability to this work is tenuous. Post-selection inference theory for the LASSO (Lockhart, Taylor, Tibshirani, & Tibshirani, 2014; Taylor & Tibshirani, 2015; Lee, Sun, Sun, & Taylor, 2016) is implemented in the R package selectiveInference. Because this package lacks PLINK file support and parallel processing capabilities, its scalability to GWAS is problematic.
Before moving onto the rest of the paper, let us sketch its main contents. Section 2 describes penalized regression and greedy algorithms, including IHT. We dwell in particular on the tactics necessary for parallelization of IHT. Section 3 records our numerical experiments. The performance of IHT, LASSO, and MCP regression algorithms is evaluated by several metrics: computation time, precision, recall, prediction error, and heritability. The sparsity level k for a given dataset is chosen by cross-validation on both real and simulated genetic data. Our discussion in Section 4 summarizes results, limitations, and precautions.
2 Methods
2.1 Penalized regression
Consider a statistical design matrix X ∈ Rn×p, a noisy n-dimensional response vector y, and a sparse parameter vector β of regression coefficients. When y represents a continuous phenotype, then the residual sum of squares loss
| (1) |
is appropriate for a linear model y = Xβ*+ε with a Gaussian error vector ε with independent components. The goal of penalized regression is to recover the true vector β* of regression coefficients.
LASSO penalized regression imposes the convex ℓ1 penalty . In most applications, the intercept contribution |β1| is omitted from the penalty. Various approaches exist to minimize the objective f(β) + λ||β||1, including least angle regression (LARS) (Efron, Hastie, Johnstone, & Tibshirani, 2004), cyclic coordinate descent (Friedman, Hastie, Höfling, & Tibshirani, 2007; Wu, Chen, Hastie, Sobel, & Lange, 2009; Wu & Lange, 2008), and the fast iterative shrinkage and thresholding algorithm (FISTA) (Beck & Teboulle, 2009). The ℓ1 norm penalty induces both sparsity and shrinkage. Shrinkage per se is not an issue because selected parameters can be re-estimated omitting the non-selected parameters and the penalty. However, the severe shrinkage induced by the LASSO inflates false positive rates. In effect, spurious predictors enter the model to absorb the unexplained variance left by the shrinkage imposed on the true predictors.
The MCP penalty takes the form with
| (2) |
for positive tuning constants λ and γ. The penalty (2) attenuates penalization for large parameter values. Indeed, beyond βi = γλ, MCP does not subject βi to further shrinkage. Relaxing penalization of large entries of β ameliorates LASSO’s shrinkage. If one majorizes the MCP function q(βi) by a scaled absolute value function, then cyclic coordinate descent parameter updates resemble the corresponding LASSO updates (Jiang & Huang, 2011).
2.2 Greedy pursuit algorithms
The ℓ1 penalty is the smallest convex relaxation of the ℓ0 penalty. As mentioned earlier, one can obtain sparsity without shrinkage by directly minimizing f(β) subject to ||β||0 ≤ k. This subset selection problem is known to be NP-hard (Golub, Klema, & Stewart, 1976; Natarajan, 1995). Greedy pursuit algorithms (MP, OMP, CoSaMP, SP, HTP, IHT) can at best approximate the solution of the subset selection problem. Here we sketch the main idea of each approach and describe some subtle differences between them.
MP and OMP, which build β stagewise, are easy to describe. At stage k with reduced predictor matrix Xk and reduced parameter vector βk, OMP computes the least squares solution
| (3) |
to the normal equations, where denotes the pseudoinverse of Xk. Note that βk can be computed algebraically when k is small or iteratively when k is large. The next predictor to add is determined by the largest entry in magnitude of the gradient
| (4) |
The main difference between MP and OMP is that OMP re-estimates all currently selected regression coefficients once a new predictor is added. In contrast, MP fixes a regression coefficient once it is estimated. The more complex strategy of OMP gives it a slight edge in recovery performance at the cost of additional computation.
One potentially detrimental feature of OMP is that indices added to the active set remain there. CoSaMP and SP extend OMP by adding a debiasing step, thus permitting predictors to enter and exit during the model building process. At stage k debiasing is accomplished by taking the estimate βk derived from equation (3), computing its gradient ∇f(βk), and identifying the k largest components in magnitude of ∇f(βk). The identified components are then appended to the nonzero components of βk, and all 2k components are refit. The largest k components of β2k in magnitude then populate the revised k-sparse approximation βk. Once debiasing is complete, the sparsity level k is increased to k + 1, and the process is repeated.
IHT and HTP approximate the solution to the subset selection problem by taking the projected gradient steps
| (5) |
where μ denotes the step size of the algorithm, and PSk (β) denotes the projection of β onto the sparsity set Sk where at most k components of a vector are nonzero. For sufficiently small μ, the projected gradient update (5) is guaranteed to reduce the loss, but it forfeits stronger convergence properties because Sk is nonconvex. Projection is achieved by setting all but the k largest components of β in magnitude equal to 0. HTP projects by solving the normal equations in the form (3) on the active set supp(βk).
The pure gradient nature of IHT explains its the speed and scalability advantages over other greedy algorithms as k grows. Although the method of conjugate gradients can quickly compute the solution vector (3) when k is large and X is sparse, typical GWAS datasets involve dense predictor matrices X. Direct solution of the normal equations then has computational complexity 𝒪(k3). In contrast, the projection PSk in IHT succumbs to fast sorting algorithms with computational complexity of just 𝒪(k log p). For small k, IHT is not intrinsically faster than other greedy pursuit algorithms, but the performance gap increases quickly as k grows. This advantage is particularly relevant in heritability estimation since many complex traits depend on hundreds or thousands of SNPs with small individual effect.
2.3 Convergence of IHT
Convergence guarantees for IHT revolve around three criteria. Let β* denote the parameter vector under the true model, and let βk be the current estimate of β*. Convergence guarantees consider any or all of the following quantities: (a) ||f(βk) − f(β*) ||2, (b) ||βk − β* ||2, and (c) || supp(βk) − supp(β*)||1, where supp(β) denotes a 0/1 vector conveying the support of β. Convergence criteria (a) and (b) are better understood than criterion (c), so we first focus on (a) and (b).
The original convergence guarantees for IHT (Blumensath & Davies, 2008, 2010) relied on the restricted isometry property (RIP) (Candés, Romberg, & Tao, 2006) and the mutual coherence property (Donoho & Huo, 2001; Tropp, 2006) to show that criteria (a) and (b) converge to 0. RIP and mutual coherence together require that the normalized version of X approximate an orthonormal matrix whose columns are uncorrelated. However, RIP offers pessimistic worst-case bounds. Recent research has derived realistic guarantees of stable convergence and model recovery for HTP and by extension to to IHT. A combination of restricted strong convexity (RSC) (Dobson & Barnett, 2008; Loh & Wainwright, 2015) and restricted strong smoothness (RSS) (Agarwal, Negahban, & Wainwright, 2012; Jain, Tewari, & Kar, 2014) places local bounds on the curvature of the loss function. If f(β) satisfies RSC and RSS, then HTP converges to a k-sparse minimizer β provided the extreme eigenvalues of the Hessian matrix ∇2f(β♯) are bounded for any k-sparse approximation β ♯ near β. The adaptation of RSC and RSS to IHT was made by Bahmani, Raj, and Boufounos (Bahmani et al., 2013). They invoke the stable restricted Hessian (SRH) and the stable restricted linearization (SRL) conditions to bound the curvature of f(β) over a restricted subset of the domain. A key difference is that SRH and SRL relax RSC and RSS. Indeed, the former pair of conditions entail only local constraints, while the latter pair entail global constraints.
The case of criterion (c), which ensures the stability of the support, is more complicated. One can easily concoct a scenario in which ||β* ||0 = ||βk||0 and ||βk − β*||2 can be made arbitrarily small, but supp(β*) ≠ supp(βk). Recent research (Yuan, Li, & Zhang, 2016) directly addresses criterion (c) by requiring that the smallest nonzero entry βmin of β* exceed ||∇f(β*) ||∞. While a notable achievement, this result is of mainly academic interest since β* is rarely known in advance. Taken together, the results for the three convergence criteria suggest that IHT convergence is reasonably reliable for GWAS. In this context, we expect that IHT will recover the true model provided that the SNP markers are not in strong linkage disequilibrium and the magnitudes of the true regression coefficients are not too small.
2.3.1 IHT step sizes
Computing a reasonable step size μ is important for ensuring stable descent in projected gradient schemes. For the case of least squares regression, our implementation of IHT uses the “normalized” update of Blumensath and Davies (Blumensath & Davies, 2010). At each iteration m, this amounts to employing the step size
Convergence is guaranteed provided that μm < ωm, where
| (6) |
for some constant 0 < c ≪ 1. One can interpret ω as the normed ratio of the difference between successive iterates versus the difference between successive estimated responses.
2.3.2 Bandwidth optimization of IHT
Analysis of large GWAS datasets requires intelligent handling of memory and read/write operations. Our software reads datasets in PLINK binary format. The PLINK compression protocol stores each genotype in two bits of a 64-bit float, thus achieving 32× compression. Although PLINK compression facilitates storage and transport of data, it complicates linear algebra operations. On small datasets, we store the design matrix X in floating point. On large datasets, we store both a compressed X and a compressed transpose XT. The transpose XT is used to compute the gradient (4), while X is used to compute the predicted response Xβ. The counterintuitive tactic of storing both X and XT roughly doubles the memory required to store genotypes. However, it facilitates accessing all data in column-major and unit stride order, thereby ensuring that all linear algebra operations maintain full memory caches.
Good statistical practice dictates standardizing all predictors; otherwise, parameters are penalized non-uniformly. Standardizing nongenetic covariates is trivial. However, one cannot store standardized genotypes in PLINK binary format. The remedy is to precompute and cache vectors u and v containing the mean and inverse standard deviation, respectively, of each of the p SNPs. The product Xstβ invoking the standardized predictor matrix Xst can be recovered via the formula
where 1 is an n-vector of ones and diag(v) is a diagonal matrix with v on the main diagonal. Thus, there is no need to explicitly store Xst.
On-the-fly standardization is a costly operation and must be employed judiciously. For example, to calculate Xβ we exploit the structural sparsity of β by decompressing and standardizing just the submatrix Xk of X corresponding to the k nonzero values in β. We then use Xk for parameter updates until we observe a change in the support of β. Unfortunately, calculation of the gradient ∇f(β) offers no such optimization because it requires a fully decompressed matrix XT. Since we cannot store all n × p standardized genotypes in floating point format, the best that we can achieve is standardization on the fly every time we update the gradient.
2.3.3 Parallelization of IHT
Our implementation of IHT for PLINK files relies on two parallel computing schemes. The first makes heavy use of multicore computing with shared memory arrays to distribute computations over all cores in a CPU. For example, suppose that we wish to compute in parallel the column means of X stored in a shared memory array. The mean of each column is independent of the others, so the computations distribute naturally across multiple cores. If a CPU contains four available cores, then we enlist four cores for our computations, one master and three workers. Each worker can see the entirety of X but only works on a subset of its columns. The workers compute the column means for the three chunks of X in parallel. Column-wise operations, vector arithmetic, and matrix-vector operations fit within this paradigm.
The two most expensive operations are the matrix-vector multiplications Xβ and XT (y − Xβ). We previously discussed intelligent computation of Xβ via Xkβk. Dense multithreaded linear algebra libraries such as BLAS facilitate efficient computation of Xkβk. Consequently, we obtain Xβ in 𝒪(nk) total operations. In contrast, the gradient criterion (4) requires a completely dense matrix-vector multiplication with a run-time complexity of 𝒪(np). We could lighten the computational burden by cluster computing, but communication between the different nodes then takes excessive time.
A reasonable alternative for acceleration is to calculate the gradient on a GPU running the OpenCL kernel code. An optimal GPU implementation must minimize memory transactions between the device GPU and the host CPU. Our solution is to push the compressed PLINK matrix X and its column means and precisions onto the device at the start of the algorithm. We also cache device buffers for the residuals and the gradient. Whenever we calculate the gradient, we compute the n residuals on the host and then push the residuals onto the device. At this stage, the device executes two kernels. The first kernel initializes many workgroups of threads and distributes a block of XT (y − Xβ) to each workgroup. Each thread handles the decompression, standardization, and computation of the inner product of one column of X with the residuals. The second kernel reduces across all thread blocks and returns the p-dimensional gradient. Finally, the host pulls the p-dimensional gradient from the device. Thus, after the initialization of the data, our GPU implementation only requires the host and device to exchange p + n floating point numbers per iteration.
2.4 Model selection
Given a regularization path computed by IHT, the obvious way to choose the best model along the path is to resort to simple q-fold cross-validation with mean squared error (MSE) as selection criterion. For a path of user-supplied model sizes k1, k2, . . . , kr, our implementation of IHT fits the entire path on the q−1 training partitions. We then view the qth partition as a testing set and compute its mean squared error (MSE). Finally, we determine the model size k with minimum MSE and refit the data subject to ||β||0 ≤ k.
3 Results
We tested IHT against LASSO and MCP on data from the Northern Finland Birth Cohort 1966 (NFBC1966) (Sabatti et al., 2009). These data contain several biometric phenotypes for 5402 patients genotyped at 370,404 SNPs. We imputed the missing genotypes in X with Mendel (Ayers & Lange, 2008) and performed quality control with PLINK 1.9 beta (Chang et al., 2015). Our numerical experiments include both simulated and measured phenotypes. For our simulated phenotype, we benchmarked the model recovery and predictive performance of our software against glmnet v2.0.5 and ncvreg v3.6.0 (Breheny & Huang, 2011; Friedman et al., 2010) in R v3.2.4. The statistical analysis summarized in Sections 3.2 and 3.3 includes as nongenetic covariates the SexOCPG factor, which we calculated per Sabatti et al., and the first two principal components of X, which we calculated with PLINK 1.9. All numerical experiments were run on a single compute node equipped with four 6-core 2.67Ghz Intel Xeon CPUs and two NVIDIA Tesla C2050 GPUs, each with 6Gb of memory. To simulate performance on a workstation, the experiment only used one GPU and one CPU. The computing environment consisted of 64-bit Julia v0.5.0 with the corresponding OpenBLAS library and LLVM v3.3 compiler.
3.1 Simulation
The goal of our first numerical experiment was to demonstrate the superior model selection performance of IHT versus LASSO and MCP. Here we used only the matrix Xchr1 of 23,695 SNPs from chromosome 1 of the NFBC1966 dataset. This matrix is sufficiently small to render PLINK compression and GPU acceleration unnecessary. Xchr1 uses the 5289 cases with observed BMI. Note that this number is larger than what we will use in Sections 3.2 and 3.3; no exclusion criteria were applied here since the phenotype was simulated. We standardized observed genotype dosages and then set unobserved dosages to 0. We simulated βtrue for true model sizes ktrue ∈ {100, 200, 300} and effect sizes independently drawn from the normal distribution N(0, 0.01). The simulated phenotypes were then formed by taking y = Xchr1βtrue +ε, where each component εi ~ N(0, 0.01). We will refer to this simulation scenario as having a signal-to-noise ratio (SNR) of 100%. To test more noisy scenarios, we simulated βtrue under three additional SNRs of 50%, 10%, and 5%. These successively lower SNRs were obtained by drawing each causal βj from N(0, 0.01/s), where s = 2, 10, 20, respectively. We approximated the (narrow-sense) heritability h2 for each combination of y and βtrue by taking the ratio of the sample variance of the predicted vector Xchr1βtrue to the sample variance of the response vector y. To assess predictive performance, we separated 289 individuals as a testing set and used the remaining 5000 individuals for 5-fold cross-validation. We generated 10 different models for each ktrue. For each replicate, we ran regularization paths of 100 model sizes k0, k0 +2, . . . , ktrue, ktrue +2, . . . , k0 +200 straddling ktrue and chose the model with minimum MSE.
The cross-validation choice of model size is straightforward under IHT. For cross-validation with LASSO and MCP, we used the cross-validation and response prediction routines in glmnet and ncvreg. To ensure roughly comparable lengths of regularization paths and therefore commensurate compute times, we capped the maximum permissible degrees of freedom at ktrue + 100 for both LASSO and MCP regression routines. The case of MCP regression is peculiar since ncvreg does not cross-validate the γ parameter. We modified the approach of (Breheny & Huang, 2011) to obtain a suitable γ for each model. Their protocol entails cross-validating λ once with the default γ = 3 and checking if the optimal λ, which we call λbest, exceeds the minimum lambda λmin guaranteeing a convex objective. Whenever λbest ≤ λmin, we incremented γ by 1 and cross-validated λ again. We repeated this process until λbest > λmin. The larger final γ then became the default for the next simulation, thereby amortizing the selection of a proper value of γ across all 10 simulations for a given ktrue. This procedure for selecting γ ensured model selection stability while simultaneously avoiding expensive cross-validation over a full grid of γ and λ values. The reported compute times for MCP reflect this procedure, though we never needed to increment γ in our simulations.
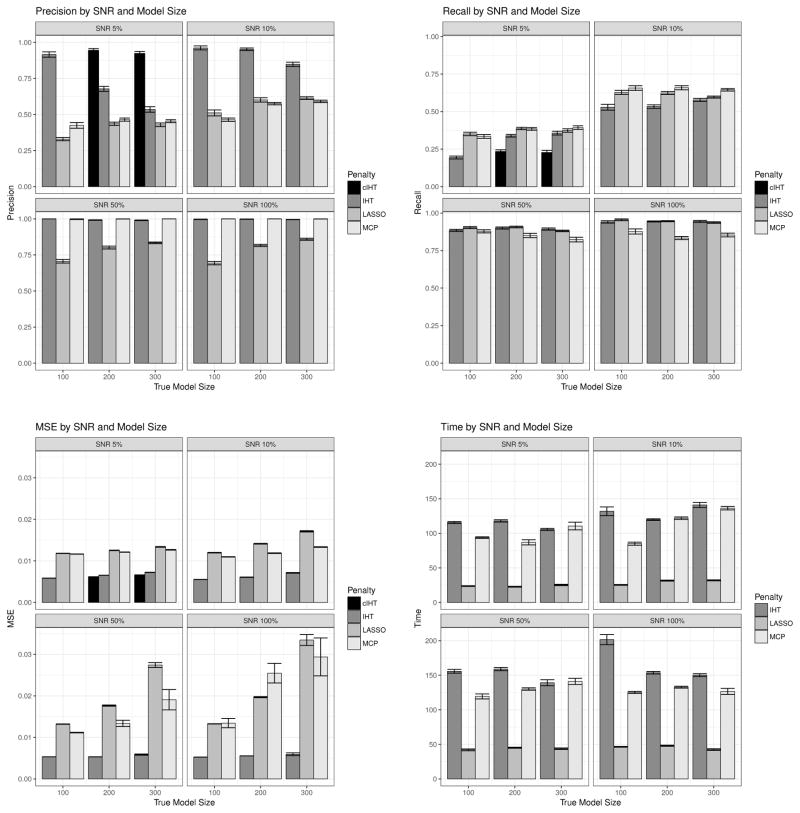
Figure 1 shows simulation results for precision, recall, prediction error (MSE), and compute time for each SNR and each model size ktrue. Here we compute precision as the ratio krecovered/k of recovered causal markers krecovered to the number of recovered markers k. Recall is computed as the ratio krecovered/ktrue of recovered causal markers to the true number ktrue of causal markers. We see that IHT consistently maintains good precision and superior prediction error (MSE), even with decreasing SNR. At high SNR, its precision is better than that of LASSO, while its recall is better than MCP. As SNR decreases, IHT cedes its advantage in recall. Despite these benefits, IHT pays only a modest price in computational speed versus LASSO and MCP. For example, IHT is only 4–6 times slower than LASSO, and it is often competitive with MCP in speed. We note that glmnet often exited before testing all 100 values of λ on its regularization path, so the timing values do not constitute truly rigorous performance comparisons.
Figure 1.
Careful readers may observe that the precision of IHT suddenly declines for k = 200 and k = 300 for SNR 5%. In these scenarios, IHT returns the minimum model size of the regularization path; for k = 200, the minimum is 100, while for k = 300 the minimum is 200. We suspected that this behavior was an artifact of our simulation scenario. Indeed, by testing every model k = 1, 2, . . . , ktrue +100, for these scenarios, we found that IHT estimated models sizes smaller than our simulation setup originally allowed. These results appear in Figure 1 for SNR 5% and k = 200, 300 as “cIHT” for “corrected IHT”. We discarded the timing results in this case since they are not comparable to our previous results. The corrected IHT results show IHT regaining its edge in precision over the other algorithms.
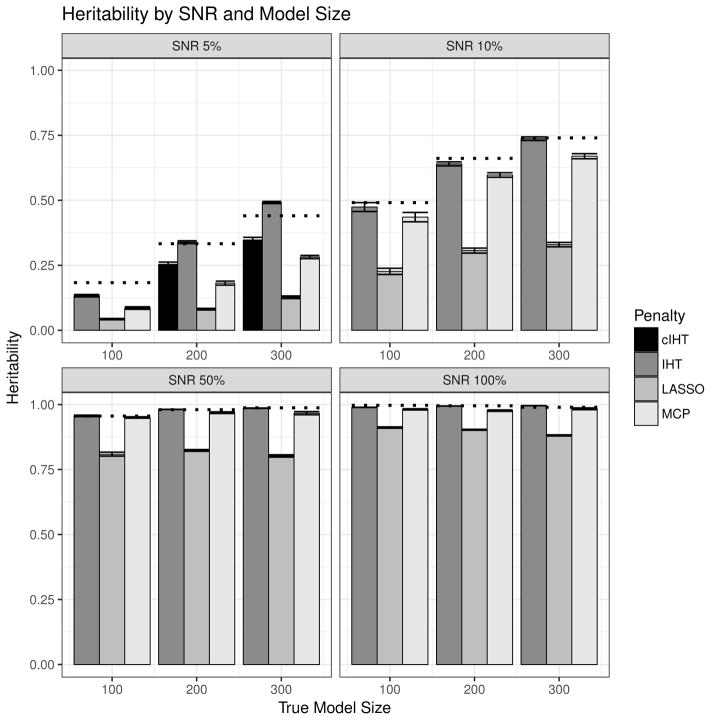
As noted previously, the heritability of the true model is given by . For each model β* selected by IHT, LASSO, or MCP, we obtain the estimated heritability similarly by substituting β* for βtrue. Figure 2 shows the heritability estimates from our simulation. In each case, we average the estimated heritability over all 10 simulation runs for each algorithm, model size, and SNR. A dotted line demarcates for each scenario, also averaged over all 10 simulation runs. We can see that as SNR decreases, IHT consistently estimates heritability better than either LASSO or MCP. For SNR 5% and k = 200, 300, the IHT estimates of h2 exceed and are obviously unrealistic. Over-estimation stems from our failure to allow for sufficiently small model sizes. Correcting this mistake eliminates the anomalies in estimated h2 and shows that IHT still estimates h2 better than the two competing algorithms in noisy scenarios.
Figure 2.
3.2 Analysis of compressed data with IHT
Our next numerical experiment highlights the sacrifice in computational speed that IHT incurs with compressed genotypes. The genotype matrixXchr1 is now limited to patients with both BMI and weight directly observed, a condition imposed by (Sabatti et al., 2009). The response vector y is the log body mass index (BMI) from NFBC1966. As mentioned previously, we included the SexOCPG factor and the first two principal components as nongenetic covariates. We then ran three different schemes on a single compute node. The first used the floating point version of Xchr1. We did not explicitly enable any multicore calculations. For the second run, we used a compressed copy of Xchr1 with multicore options enabled, but we disabled the GPU acceleration. The third run used the compressedXchr1 data with both multicore and GPU acceleration. We ran each algorithm over a regularization path of model sizes k = 5, 10, 15, . . . , 100 and averaged compute times over 10 runs. For all uncompressed arrays, we used double precision arithmetic.
Table I shows the compute time statistics. The floating point variant is clearly the fastest, requiring about 8 seconds to compute all models. The analysis using PLINK compression with a multicore CPU suffers a 17× slowdown, clearly demonstrating the deleterious effects of repeated decompression and on-the-fly standardization. Enabling GPU acceleration leads to an 8× slowdown and fails to entirely bridge the performance gap imposed by compressed data. In defense of GPU computing, it is helpful to make a few remarks. First, the computational burden of analyzing just a single chromosome does not fully exploit GPU capabilities; hence, Table I does not demonstrate the full potential of GPU acceleration. Second, the value of GPU acceleration becomes more evident in cross-validation: a 5-fold cross-validation on one machine requires either five hexcore CPUs or one hexcore CPU and one GPU. The latter configuration lies within modern workstation capabilities. Third, our insistence on the use of double precision arithmetic dims the luster of GPU acceleration. Indeed, in our experience using compressed arrays and a GPU with single precision arithmetic is only 1.7× slower than the corresponding floating point compute times. Furthermore, while we limit our computations to one CPU with six physical cores, including additional physical cores improves compute times even for compressed data without a GPU.
Table I.
Computational times in seconds on NFBC1966 chromosome 1 data.
| Data type | Mean Time | Standard Deviation |
|---|---|---|
| Uncompressed Data | 8.27 | 0.056 |
| Compressed Data, no GPU | 141.28 | 0.998 |
| Compressed Data with CPU + GPU | 65.48 | 0.040 |
3.3 Application of IHT to lipid phenotypes
For our final numerical experiment, we embarked on a genome-wide search for associations based on the full n × p NFBC1966 genotype matrix X. In addition to BMI, this analysis considered three additional phenotypes from (Sabatti et al., 2009): HDL cholesterol (HDL), LDL cholesterol (LDL), and triglycerides (TG). HDL, LDL, and TG all rely on SexOCPG and the first two principal components as nongenetic covariates. Quality control on SNPs included filters for minor allele frequencies below 0.01 and Hardy-Weinberg P-values below 10−5. Subjects with missing traits were excluded from analysis. We applied further exclusion criteria per Sabatti et al. (Sabatti et al., 2009); for analysis with BMI, we excluded subjects without direct weight measurements, and for analysis of TG, HDL, and BMI, we excluded subjects with nonfasting blood samples and subjects on diabetes medication. These filters yield different values of n and p for each trait. Table II records problem dimensions and trait transforms. To select the best model size, we performed 5-fold cross-validation over a path of sparsity levels k = 1, 2, . . . , 50. Refitting the best model size yielded effect sizes. Table II records the compute times and best model sizes, while Table III shows the SNPs recovered by IHT.
Table II.
Dimensions of data used for each phenotype in GWAS experiment. Here n is the number of cases, p is the number of predictors (genetic + covariates), and kbest is the best cross-validated model size. Note that kbest includes nongenetic covariates.
| Phenotype | n | p | Transform | kbest | Compute Time (Hours) |
|---|---|---|---|---|---|
| BMI | 5122 | 333,656 | log | 2 | 1.12 |
| HDL | 4729 | 333,774 | none | 9 | 1.28 |
| LDL | 4715 | 333,771 | none | 6 | 1.32 |
| TG | 4728 | 333,769 | log | 10 | 1.49 |
Table III.
Computational results from the GWAS experiment. Here β is the calculated effect size. Known associations include the relevant citation.
| Phenotype | Chromosome | SNP | Position | β | Status |
|---|---|---|---|---|---|
| BMI | 6 | rs6917603 | 30125050 | −0.01995 | Unreported |
|
| |||||
| HDL | 6 | rs6917603 | 30125050 | 0.10100 | Reported |
| 6 | rs9261256 | 30129920 | −0.06252 | Nearby | |
| 11 | rs7120118 | 47242866 | −0.03351 | Reported | |
| 15 | rs1532085 | 56470658 | −0.04963 | Reported | |
| 16 | rs3764261 | 55550825 | −0.02808 | Reported | |
| 16 | rs7499892 | 55564091 | 0.02625 | Reported | |
|
| |||||
| LDL | 1 | rs646776 | 109620053 | 0.09211 | Reported |
| 2 | rs693 | 21085700 | −0.08544 | Reported | |
| 6 | rs6917603 | 30125050 | −0.07536 | Reported | |
|
| |||||
| TG | 2 | rs676210 | 21085029 | 0.03633 | Nearby |
| 2 | rs1260326 | 27584444 | −0.04088 | Reported | |
| 6 | rs7743187 | 25136642 | 0.03450 | Unreported | |
| 6 | rs6917603 | 30125050 | −0.08215 | Unreported | |
| 7 | rs2286276 | 72625290 | 0.01858 | Reported | |
| 7 | rs11974409 | 72627326 | 0.01759 | Nearby | |
| 8 | rs10096633 | 19875201 | 0.03781 | Reported | |
| 13 | rs3010965 | 60937883 | 0.02828 | Unreported | |
| 19 | rs2304130 | 19650528 | 0.03039 | Reported | |
One can immediately see that IHT does not collapse causative SNPs in strong linkage disequilibrium. IHT finds the adjacent pair of SNPs rs6917603 and rs9261256 for HDL. For TG, rs11974409 is one SNP separated from rs2286276, while SNP rs676210 is one SNP separated from rs673548. Note that rs673548 is not in Table II since IHT does not flag rs673548. However, its association with TG in NFBC1966 is known. (Sabatti et al., 2009) Common sense suggests treating each associated pair of SNPs as a single predictor.
Our analysis not only replicates several associations from the literature but also finds new ones as well. For example, Sabatti et al. found associations between TG and the SNPs rs1260326 and rs10096633, while rs2286276 was identified elsewhere. The SNPs rs676210, rs7743187, rs6917603, and rs3010965 are new associations with TG. We find that SNP rs6917603 is associated with all four traits; the BMI connection was missed by Sabatti et al..
The association of SNP rs6917603 with BMI may be secondary to its association to lipid levels. Indeed, a multivariate analysis of the combined traits BMI, HDL, LDL, and TG reveals a much stronger association to rs6917603 than BMI alone does. In the former case P = 9.88 × 10−105 while in the latter case P = 1.19 × 10−15. The four traits exhibit fairly high correlations, with the correlation coefficient ρ ranging between 0.25 and 0.35. We conjecture that some of the observed pleiotropic effect of rs6917603 may be explained by these trait correlations. A more extensive analysis that incorporates nearby genetic markers in LD may clarify the association pattern displayed by these correlated traits (Fan et al., 2013, 2015; Wang et al., 2015). Note that the aforementioned P-values are meant to be compared to each other, not to previous GWAS associations. Our pleiotropic analysis of rs6917603 makes no assertion of a genome-wide significant association for any of the phenotypes since the P-values reported here are likely inflated by selection bias (Taylor & Tibshirani, 2015).
IHT flags an association between SNP rs2304130 and TG. This association was validated in a large meta-analysis of 3,540 cases and 15,657 controls performed after (Sabatti et al., 2009) was published. Finally, some of the effect sizes in Table III are difficult to interpret. For example, IHT estimates effects for rs10096633 (β = 0.03781) and rs1260326 (β = −0.04088) that are both smaller and in opposite sign to the estimates in (Sabatti et al., 2009).
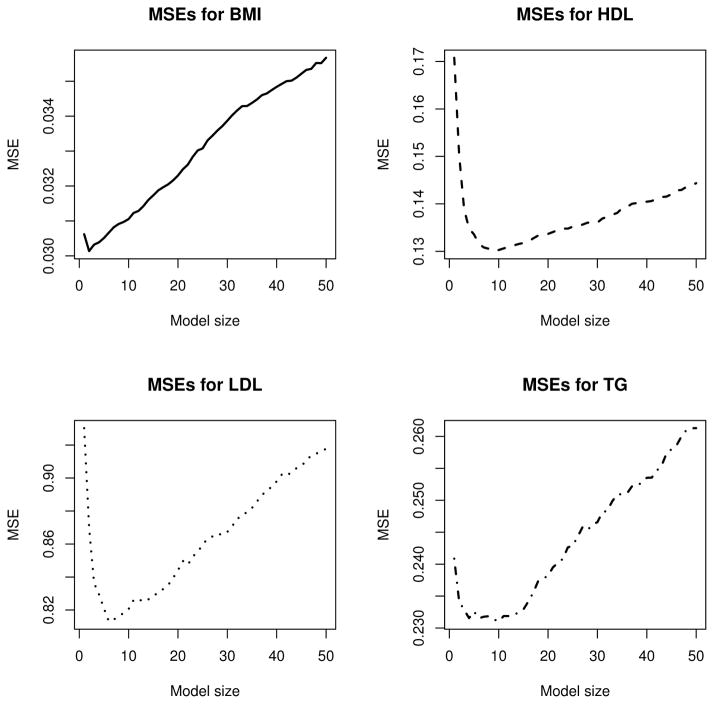
The potential new associations of TG with SNPs rs7743187 and rs3010965 are absent from the literature. Furthermore, our analysis misses borderline significant associations identified by Sabatti et al (Sabatti et al., 2009), such as rs2624265 for TG and rs9891572 for HDL; these SNP associations went unreplicated in later studies. In this regard it is worth emphasizing that the best model size kbest delivered by cross-validation is a guide rather than definitive truth. Figure 3 shows that the difference in MSE between kbest and adjacent model sizes can be quite small. Models of a few SNPs more or less than kbest predict trait values about as well. Thus, TG with ktrue = 10 has MSE = 0.2310, while TG with ktrue = 4has MSE = 0.2315. Refitting the TG phenotype with k = 4 yields only three significant SNPs: rs1260326, rs6917603, and rs10096633. The SNPs rs7743187 and rs3010965 are absent from this smaller model, so we should be cautious in declaring new associations. This example also highlights the value in computing many model sizes, which univariate regression schemes typically overlook.
Figure 3.
In light of the simulation results from Section 3.1, an obvious question is how our IHT results on real data compare to those from LASSO and MCP. To this end, we ran LASSO and MCP on each of the four phenotypes and the full genotype matrix. For LASSO, we used PLINK 1.9 (Chang et al., 2015) after tuning the regularization path using heritability estimates from GCTA (J. Yang et al., 2010; J. Yang, Lee, Goddard, & Visscher, 2011). For MCP, we used the GWAS module in Mendel (Lange et al., 2013; Wu et al., 2009; Wu & Lange, 2008; Zhou, Sehl, Sinsheimer, & Lange, 2010). Since Mendel requires the user to specify the desired model size k, we simply used the best cross-validated k from IHT for each phenotype. A rigorous comparison of model selection performance would use a predictive measure such as out-of-sample predictive accuracy. Unfortunately, neither PLINK or Mendel return MSEs, so we cannot compare predictive power directly. Instead, we compare to which extent the marker sets selected by IHT agree with corresponding sets selected by LASSO or MCP, similar in spirit to our previous validation of IHT results with the literature. Tables showing the markers recovered by LASSO and MCP appear as supplementary material. In general, LASSO returns a superset of the markers selected by IHT, as expected from our simulations. MCP and IHT usually but not always select similar markers. Given our limited ability to compare the three methods, the set of markers selected by IHT seems reasonable in light of results from LASSO and MCP, but we refrain from drawing conclusions about comparative predictive performance among the three methods.
Finally, we comment on compute times. IHT requires about 1.5 hours to cross-validate the best model size over 50 possible models using double precision arithmetic. Obviously, computing fewer models can decrease this compute time substantially. If the phenotype in question is scaled correctly, then analysis with IHT may be feasible with single precision arithmetic, which yields an additional speedup as suggested in Section 3.2. Analyses requiring better accuracy will benefit from the addition of double precision registers in newer GPU models. Thus, there is further room for speedups without sacrificing model selection performance.
4 Discussion
The mathematical literature on big data analysis is arcane to the point of being nearly inaccessible to geneticists. In addition to absorbing the obvious mathematical subtleties, readers must be wary of the hype that infects many papers. This manuscript represents our best effort to sort through the big data literature and identify advances most pertinent to the analysis of GWAS data. Once we decided that the iterative hard thresholding (IHT) algorithm had the greatest potential, we set about adapting it to the needs of geneticists and comparing it to existing methods. This paper is a synopsis of our experience in carrying out these tasks.
Our experiments clearly demonstrate the utility of IHT in large-scale GWAS analysis. It exhibits better model selection than more popular and mature tools such as LASSO- and MCP-regression. Despite its nonconvex nature, IHT enjoys provable convergence guarantees under ideal regularity conditions. We prefer IHT to other greedy algorithms because it provides the best balance of computational speed, model recovery, and convergence behavior (Blanchard et al., 2011; Blanchard & Tanner, 2015). Our software directly and intelligently handles the PLINK compression protocol widely used to store GWAS genotypes. Finally, IHT can be accelerated by exploiting shared-memory and massively parallel processing hardware. As a rule of thumb, computation times with IHT scale as 𝒪(np) or somewhat worse if more predictors with small effect sizes come into play.
Lack of general support for PLINK binary genotype data, poor memory management, and primitive parallel capabilities limit the use of software such as glmnet and ncvreg in GWAS. Our IHT package IHT.jl enables analysis of models of any sparsity. In contrast, gpu-lasso is designed solely for very sparse models. Both IHT.jl and gpu-lasso cross-validate for the best model size over a range of possible models. IHT.jl has the edge in scalability and model selection. Despite these advantages, IHT is hardly a panacea for GWAS. Geneticists must still deal with perennial statistical issues such as correlated predictors, sufficient sample sizes, and population stratification. IHT can handle population stratification by inclusion of principal components as nongenetic covariates, but the onus falls upon the analyst to decide the appropriate number of PCs to use. Furthermore, while the estimation properties of greedy pursuit algorithms are well understood, the theory of inference with IHT is immature (F. Yang et al., 2016). More progress has been made in understanding postselection inference with LASSO penalties (Lockhart et al., 2014; Taylor & Tibshirani, 2015; Lee et al., 2016; Loftus & Taylor, 2015; Loftus, 2016). The rapid pace of research in selective inference makes us hopeful that inference with IHT will soon become routine.
Our analysis framework neglects the wealth of data available from whole genome sequencing. As sketched in Section 2.1, the mutual coherence condition for convergence of IHT discourages the use of IHT on strongly correlated predictors such as those that arise from analyzing genome sequences base-by-base. Users interested in rare variant analysis could feasibly put IHT to their advantage by lumping correlated rare SNPs into univariate predictors and performing model selection on these measures of genetic burden (Li & Leal, 2008).
As formulated here, the scope of application for IHT is limited to linear least squares regression. Researchers have begun to extend IHT to generalized linear models, particularly logistic regression (Bahmani et al., 2013; Yuan et al., 2014). We anticipate that IHT will eventually join LASSO and MCP in the standard toolbox for sparse linear regression and sparse generalized linear regression. In our opinion, gene mapping efforts clearly stand to benefit from application of the IHT algorithm.
Supplementary Material
Acknowledgments
Funding
This work was supported by grants from the National Human Genome Research Institute [HG006139] and the National Institute of General Medical Sciences [GM053275] to K.L. and fellowship support from the National Human Genome Research Institute [HG002536] and the National Science Foundation [DGE-0707424] to K.L.K.
The authors are grateful to Aditya Ramdas for his guidance on IHT and to Dennis Sun for discussions about general model selection. We benefited from discussions about IHT at the American Institute of Mathematics. The authors thank the two anonymous reviewers whose comments substantially improved the quality of the manuscript. Finally, we thank the Stanford University Statistics Department for hosting us as sabbatical guests during the 2014–2015 academic year.
Footnotes
Conflict of Interest Disclosure
The authors declare no competing interests.
Availability: Source code is freely available at https://github.com/klkeys/IHT.jl.
References
- Abraham G, Kowalczyk A, Zobel J, Inouye M. SparSNP: Fast and memory-efficient analysis of all SNPs for phenotype prediction. BMC Bioinformatics. 2012;13(1):88. doi: 10.1186/1471-2105-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A, Negahban S, Wainwright MJ. Fast global convergence rates of gradient methods for high-dimensional statistical recovery. The Annals of Statistics. 2012;40(5):2452–2482. [Google Scholar]
- Ayers K, Lange K. Penalized estimation of haplotype frequencies. Bioinformatics. 2008;24:1596–1602. doi: 10.1093/bioinformatics/btn236. [DOI] [PubMed] [Google Scholar]
- Bahmani S, Raj B, Boufounos PT. Greedy sparsity-constrained optimization. Journal of Machine Learning Research. 2013;14(3):807–841. [Google Scholar]
- Beck A, Teboulle M. A fast iterative shrinkage thresholding algorithm for linear inverse problems. SIAM Journal of Imaging Sciences. 2009;2(1):183–202. [Google Scholar]
- Blanchard JD, Cartis C, Tanner J, Thompson A. Phase transitions for greedy sparse approximation algorithms. Applied and Computational Harmonic Analysis. 2011;30(2):188–203. [Google Scholar]
- Blanchard JD, Tanner J. Performance comparisons of greedy algorithms in compressed sensing. Numerical Linear Algebra with Applications. 2015;22(2):254–282. [Google Scholar]
- Blumensath T. Accelerated iterative hard thresholding. Signal Processing. 2012;2(1):183–202. [Google Scholar]
- Blumensath T, Davies ME. Iterative hard thresholding for sparse approximation. Journal of Fourier Analysis and Applications. 2008;14:629–654. [Google Scholar]
- Blumensath T, Davies ME. Iterative hard thresholding for compressed sensing. Applications of Computational and Harmonic Analysis. 2009;27:265–274. [Google Scholar]
- Blumensath T, Davies ME. Normalized iterative hard thresholding: Guaranteed stability and performance. IEEE Journal of Selected Topics in Signal Processing. 2010;4(2):298–309. [Google Scholar]
- Breheny P, Huang J. Coordinate descent algorithms for nonconvex penalized regression, with applications to biological feature selection. Annals of Applied Statistics. 2011;5(1):232–253. doi: 10.1214/10-AOAS388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candés E, Romberg JK, Tao T. Stable signal recovery from incomplete and inaccurate measurements. Communications on Pure and Applied Mathematics. 2006;59(8):1207–1223. [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4(7) doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GK. A scalable and portable framework for massively parallel variable selection in genetic association studies. Bioinformatics. 2012;28:719–720. doi: 10.1093/bioinformatics/bts015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Donoho DL. Basis pursuit. Stanford, CA: Department of Statistics, Stanford University; 1994. (Tech. Rep.) [Google Scholar]
- Dai W, Milenkovic O. Subspace pursuit for compressive sensing signal reconstruction. IEEE Transactions on Information Theory. 2009;55(5):2230–2249. [Google Scholar]
- Dobson AJ, Barnett AG. An introduction to generalized linear models. Vol. 3. Chapman and Hall/CRC Press; 2008. [Google Scholar]
- Donoho DL, Huo X. Uncertainty principles and ideal atomic decomposition. IEEE Transactions on Information Theory. 2001;47(7):2845–2862. [Google Scholar]
- Donoho DL, Tsaig Y, Drori I, Starck JL. Sparse solution of underdetermined systems of linear equations by stagewise orthogonal matching pursuit. IEEE Transactions on Information Theory. 2012;58(2):1094–1121. [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. The Annals of Statistics. 2004;32(2):407–499. [Google Scholar]
- Fan R, Wang Y, Boehnke M, Chen W, Li Y, Ren H, … Xiong M. Gene level meta-analysis of quantitative traits by functional linear models. Genetics. 2015;200:1089–115. doi: 10.1534/genetics.115.178343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Wang Y, Mills JL, Wilson AF, Bailey-Wilson JE, Xiong M. Functional linear models for association analysis of quantitative traits. Genetic Epidemiology. 2013;37(7):726–742. doi: 10.1002/gepi.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucart S. Hard thresholding pursuit: An algorithm for compressive sensing. SIAM Journal on Numerical Analysis. 2011;49(6):2543–2563. [Google Scholar]
- Friedman J, Hastie T, Höfling H, Tibshirani R. Pathwise coordinate optimization. Annals of Applied Statistics. 2007;1(2):302–332. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- Golub G, Klema V, Stewart GW. Rank degeneracy and least squares problems. Stanford, CA: Stanford University Department of Computer Science; 1976. (Tech. Rep.) [Google Scholar]
- Hastie T, Friedman J, Tibshirani R. The Elements of Statistical Learning. Vol. 2. Springer; 2009. [Google Scholar]
- Jain P, Tewari A, Kar P. On iterative hard thresholding methods for high-dimensional Mestimation. Advances in Neural Information Processing Systems. 2014:685–693. [Google Scholar]
- Jiang D, Huang J. Majorization minimization by coordinate descent for concave penalized generalized linear models. Iowa City, IA: Department of Statistics and Actuarial Science, The University of Iowa; 2011. Oct, (Tech. Rep. No. 412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K. Numerical Analysis for Statisticians. Springer Science & Business Media; 2010. [Google Scholar]
- Lange K, Papp JC, Sinsheimer JS, Sobel EM. Next generation statistical genetics: Modeling, penalization, and optimization in high-dimensional data. Annual Review of Statistics and Its Application. 2014;1(1):279–300. doi: 10.1146/annurev-statistics-022513-115638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Papp JC, Sinsheimer JS, Sripracha R, Zhou H, Sobel EM. Mendel: The Swiss army knife of genetic analysis programs. Bioinformatics. 2013;29:1568–1570. doi: 10.1093/bioinformatics/btt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JD, Sun DL, Sun Y, Taylor JE. Exact post-selection inference, with application to the lasso. The Annals of Statistics. 2016;44(3):907–927. [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. The American Journal of Human Genetics. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart R, Taylor J, Tibshirani RJ, Tibshirani R. A significance test for the lasso. The Annals of Statistics. 2014 Apr;42(2):413–468. doi: 10.1214/13-AOS1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus JR. Selective inference after cross-validation. 2016 arXiv preprint arXiv:1511.08866. [Google Scholar]
- Loftus JR, Taylor JE. Selective inference in regression models with groups of variables. 2015 arXiv preprint arXiv:1511.01478. [Google Scholar]
- Loh PL, Wainwright MJ. Regularized M-estimators with nonconvexity: statistical and algorithmic theory for local optima. The Journal of Machine Learning Research. 2015;16(1):559–616. [Google Scholar]
- Mallat SG, Zhang Z. Matching pursuits with time-frequency dictionaries. IEEE Transactions on Signal Processing. 1993;41(12):3397–3415. [Google Scholar]
- Natarajan BK. Sparse approximate solutions to linear systems. SIAM Journal on Computing. 1995;24(2):227–234. [Google Scholar]
- Needell D, Tropp JA. CoSaMP: iterative signal recovery from incomplete and inaccurate samples. Applied and Computational Harmonic Analysis. 2009;26(3):301–321. [Google Scholar]
- Pati YC, Rezaiifar R, Krishnaprasad P. Orthogonal matching pursuit: Recursive function approximation with applications to wavelet decomposition. Proceedings of the 27th Asilomar Conference on Signals, Systems, and Computers; 1993. pp. 40–44. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, … Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, … Peltonen L. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nature Genetics. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Tibshirani RJ. Statistical learning and selective inference. Proceedings of the National Academy of Sciences. 2015;112(25):7629–7634. doi: 10.1073/pnas.1507583112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society, Series B. 1996;58(1):267–288. [Google Scholar]
- Tropp JA. Just relax: Convex programming methods for identifying sparse signals in noise. IEEE Transactions on Information Theory. 2006;52(3):1030–1051. [Google Scholar]
- Tropp JA, Gilbert AC. Signal recovery from random measurements via orthogonal matching pursuit. IEEE Transactions on Information Theory. 2007;53(12):4655–4666. [Google Scholar]
- Wang Y, Liu A, Mills JL, Boehnke M, Wilson AF, Bailey-Wilson JE, … Fan R. Pleiotropy analysis of quantitative traits at gene level by multivariate functional linear models. Genetic Epidemiology. 2015;39(4):259–275. doi: 10.1002/gepi.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25(6):714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Lange K. Coordinate descent algorithms for lasso penalized regression. Annals of Applied Statistics. 2008;2(1):224–244. doi: 10.1214/10-AOAS388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Barber RF, Jain P, Lafferty J. Selective inference for group-sparse linear models. Advances in Neural Information Processing Systems. 2016:2469–2477. [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, … Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88(1):76. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Li P, Zhang T. Gradient hard thresholding pursuit for sparsity-constrained optimization. Journal of Machine Learning Research. 2014:32. [Google Scholar]
- Yuan X, Li P, Zhang T. Exact recovery of hard thresholding pursuit. Advances in Neural Information Processing Systems. 2016:3558–3566. [Google Scholar]
- Zhang CH. Nearly unbiased variable selection under minimax concave penalty. Annals of Statistics. 2010;38(2):894–942. [Google Scholar]
- Zhou H, Sehl ME, Sinsheimer JS, Lange K. Association screening of common and rare genetic variants by penalized regression. Bioinformatics. 2010;26(19):2375. doi: 10.1093/bioinformatics/btq448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.