Abstract
Lipopolysaccharide (LPS), a glycolipid found in the outer membrane of Gram-negative bacteria, is a potent elicitor of innate immune responses in mammals. A typical LPS molecule is composed of three different structural domains: a polysaccharide called the O-antigen, a core oligosaccharide, and Lipid A. Lipid A is the amphipathic glycolipid moiety of LPS. It stimulates the immune system by tightly binding to Toll-like receptor 4. More recently, Lipid A has also been shown to activate intracellular caspase-4 and -5. An impressive diversity is observed in Lipid A structures from different Gram-negative bacteria, and it is well established that subtle changes in chemical structure can result in dramatically different immune activities. For example, Lipid A from Escherichia coli is highly toxic to humans, whereas a biosynthetic precursor called Lipid IVA blocks this toxic activity, and monophosphoryl Lipid A from Salmonella minnesota is a vaccine adjuvant. Thus, an understanding of structure-activity relationships in this glycolipid family could be used to design useful immunomodulatory agents. Here we review the biosynthesis, modification, and structure-activity relationships of Lipid A.
Lipopolysaccharide (LPS) is a complex glycolipid found in the outer membrane of Gram-negative bacteria. It acts as a barrier to entry of foreign molecules into the bacterium, and plays a role in maintaining the integrity of cell membrane. Lipid A, the conserved, lipid component of LPS, anchors LPS to the outer membrane. Canonical Lipid A, produced in E. coli, is a β-(1′,6)-linked disaccharide of glucosamine that is hexaacylated and bis-phosphorylated (Compound 2, Figure 4). The low pKa,1 value of its phosphate groups results in the compound having at least two negative charges at neutral pH, and contribute towards a net negative surface charge[1]. As a pathogen associated molecular pattern (PAMP), Lipid A induces a range of human innate immune responses upon binding to its receptors, namely Toll-like receptor (TLR4), caspase-4, and caspase-5. These responses result in the recruitment of immune cells and fluids to the site of infection to eliminate the foreign pathogen. In some situations, Lipid A can trigger systemic inflammation that cause tissue damage and occasionally death. To evade this immune response, some Gram-negative bacteria, such as Yersinia Pestis, have diversified their Lipid A structures to result in attenuated inflammation. In recent years, the structural diversity of Lipid A has been harnessed to develop a vaccine adjuvant (MPL®) that safely enhances a beneficial adaptive immune response against the co-inoculated antigen[2]. This review discusses the biosynthetic diversity of Lipid A, and the sensing of Lipid A analogs by components of the host defense systems. The consolidated information provides a landscape of our current understanding of Lipid A immune response and insights into the potential therapeutic use of this class of molecules.
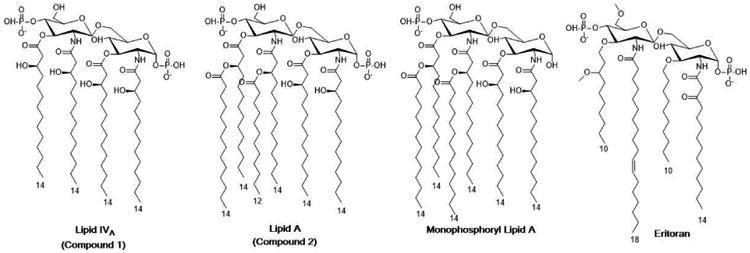
Figure 4. Structures of Lipid IVa, Lipid A, Monophosphoryl Lipid A, and Eritoran.
Biosynthesis of Lipid A in E. coli[3,*4]
In Escherichia coli, the biosynthesis of Lipid A is catalyzed by nine enzymes (LpxA, LpxC, LpxD, LpxH, LpxB, LpxK, KdtA, LpxL, LpxM), and requires three distinct protein-bound acyl donor substrates along with UDP-N-acetylglucosamine (UDP-GlcNAc), ATP, and CMP-3-deoxy-D-manno-octulosonic acid (CMP-Kdo). The natural acyl donor substrates are fatty acyl chains attached to the acyl carrier protein (acyl-ACPs), intermediates in fatty acid biosynthesis. Hence, the Lipid A biosynthetic pathway is considered to operate downstream of fatty acid biosynthesis (Figure 1). To facilitate the trafficking of LPS to the outer membrane, Lipid A biosynthetic enzymes (LpxA through LpxM) are either expressed in the cytoplasm or anchored to the inner membrane. After core oligosaccharide-Lipid A biosynthesis, the compound is enzymatically flipped and oriented towards the periplasm for the addition of O-antigenic polysaccharide, and it is subsequently transported to the outer membrane (reviewed in [3]).
Figure 1. Biosynthesis of Lipid A in E. coli.
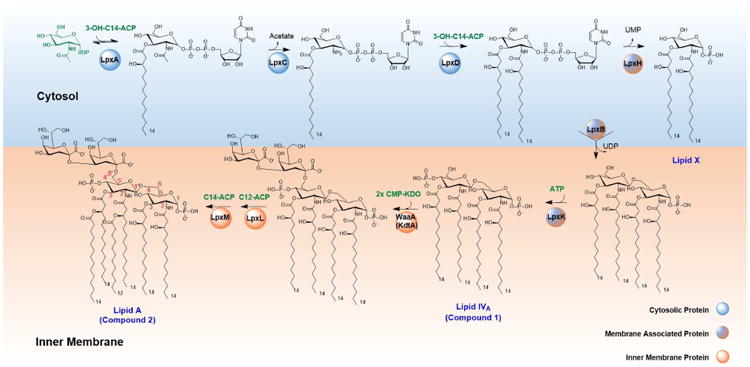
(Adapted from [4]) The enzymes LpxA, LpxC, and LpxD are located in the cytoplasm, whereas LpxH, LpxB, LpxK, WaaA, LpxL and LpxM are anchored to the inner membrane. Substrates for the biosynthetic are highlighted in green: acyl carrier protein (ACP)-bound acyl donors, UDP-N-acetylglucosamine (UDP-GlcNAc), ATP, and CMP-3-deoxy-D-manno-octulosonic acid (CMP-Kdo). The product of the pathway is further modified into variable lipopolysaccharide structures via further glycosylation of the Kdo sugars (not explicitly shown).
To initiate the biosynthetic pathway, cytosolic proteins LpxA, LpxC and LpxD catalyze a series of reactions converting UDP-GlcNAc into UDP-2,3-diacyl-glucosamine. First, LpxA transfers the acyl chain from (R)-β-hydroxymyristoyl-ACP to the 3-OH group of UDP-GlcNAc. This acyl transfer reaction is thermodynamically unfavorable. To drive the reaction forward, LpxC, a Zn2+-dependent metalloenzyme, hydrolyzes the 2-acetamido functionality, and thus catalyzes the first committed step in the pathway. LpxD then installs another (R)-β-hydroxymyristoyl moiety onto the 2-amino group, yielding UDP-2,3-diacyl-glucosamine.
The UDP-2,3-diacyl-glucosamine product of LpxA-C-D is transformed by three membrane associated enzymes, LpxH, LpxB and LpxK, into tetraacylated Lipid IVA (Compound 1, Figure 4). LpxH hydrolyzes UDP-2,3-diacylglucosamine to yield 2,3-diacylglucosamine 1-phosphate. LpxB catalyzes formation of a β-1′,6-glycosidic bond to generate the disaccharide backbone. In the final step leading up to Lipid IVA, LpxK phosphorylates the 4′-position of this disaccharide.
Prior to late-stage acylation, E. coli requires the addition of two 2-keto-3-deoxyoctonate (Kdo) sugars to Lipid IVA. A bifunctional enzyme KdtA catalyzes two successive glycosyl transfer reactions to form Kdo2-Lipid IVA. This tetraacyl tetrasaccharide is further modified by late acyl transferases LpxL and LpxM through the addition of lauroyl and myristoyl secondary acyl chains, respectively, to yield hexaacylated Lipid A.
Biosynthetic Diversity of Lipid A[3,*4]
Gram-negative bacteria have evolved an immense capacity for diversifying Lipid A structure (reviewed in [4]). The modification system typically involves enzymes within the biosynthetic pathway and/or downstream tailoring enzymes that recognize and modify Lipid A.
Within the Lipid A biosynthetic pathway, acyl transferase orthologs have variable substrate specificities, and thus contribute towards alterations in product structure (Table 1). These enzymes recognize fatty acyl chains of different lengths and oxidation patterns. For example, E. coli LpxA has 100-1000 times higher specificity for (R)-β-hydroxymyristoyl-ACP (C14:0) than C12 or C10 acyl-ACP substrates. While E. coli acyl transferases are chain length specific, orthologs from other Gram-negative bacteria are more tolerant, yielding a heterogeneous Lipid A composition of the outer membrane. These enzymes also show diversity with respect to the identity of their acyl acceptors. For example, E.coli LpxA transfers acyl chain to UDP-GlcNAc while Leptospira interrogans LpxA specifically modifies UDP 2-acetamido-3-amino-2,3-dideoxy-α-D-glucopyranose (UDP-GlcNAc3N) [5,6].
Table 1.
Differences in substrate specificity of LpxA, LpxD, LpxL and LpxM contribute to species specific acyl chain and disaccharide backbone variety in Lipid A variants.
| Protein Name | Organism | Related Protein (% identity) | Acyl Donor | Acyl Acceptor | Key Reference |
|---|---|---|---|---|---|
| LpxA | E. coli | Reference | (R)-β-hydroxymyristoyl-ACP | 3-OH group of UDP-GlcNAc | [55] |
| LpxA | C. trachomatis | 37.4% E. coli LpxA | Myristoyl-ACP | 3-OH group of UDP-GlcNAc | [54] |
| LpxA | P. aeruginosa | 53.4% E. coli LpxA | (R)-β-hydroxydecanoyl-ACP | 3-OH group of UDP-GlcNAc | [56] |
| LpxA | N. meningitidis | 47.3% E.coli LpxA | (R)-β-hydroxylauroyl-ACP | 3-OH group of UDP-GlcNAc | [57] |
| LpxA | H. pylori | 38.1% E. coli LpxA | (R)-β-hydroxypalmitoyl-ACP | 3-OH group of UDP-GlcNAc | [58] |
| LpxA | L. interrogans | 41.3% E.coli LpxA | (R)-β-hydroxylauroyl-ACP | 3-NH2 group of UDP-GlcNAc3N | [59] |
| LpxD | E. coli | Reference | (R)-β-hydroxymyristoyl-ACP | 2-NH2 group of UDP-3-O-(R-3-hydroxymyristoyl)-GlcN | [60] |
| LpxD | P.aeruginosa | 48.7% E. coli LpxD | (R)-β-hydroxydodecanoyl-ACP | 2-NH2 group of UDP-3-O-(R-3-hydroxydecanoyl)-GlcN | [61] |
| LpxL | E. coli | Reference | Lauroyl-ACP | β -OH group on 2′ acyl chain of Kdo2-LipidIVA | [62] |
| LpxM | E. coli | Reference | Myristoyl-ACP | β -OH group on 3′ position of Kdo2-(lauroyl) Lipid IVA | [63] |
| LpxP | E. coli | 54.1% E.coli LpxL | Palmitoleoyl-ACP | β -OH group on 2′ acyl chain of Kdo2-LipidIVA | [64] |
| LpxXL | R. leguminosarum | 20.4% E. coli LpxL, 23.5% E. coli LpxM | 27-hydroxyoctacosanoyl-ACPXL | Kdo2-LipidIVA | [65] |
| LpxL1 | B. pertussis | 23.9% E. coli LpxL, 20.6% E. coli LpxM | Not Available [2-hydroxylauroyl-ACP2] | Not Available [Observed as a secondary acyl chain at the 2-position of Lipid A] | [66] |
The abbreviations used are: UDP-GlcNAc, Uridine diphosphate N-acetylglucosamine; ACP, Acyl carrier protein; UDP-GlcNAc3N, Uridine diphosphate 2-acetamido-3-amino-2,3-dideoxy-α-D-glucopyranose; GlcN, Glucosamine; UDP, Uridine diphosphate; Kdo, 3-deoxy-D-manno-octulosonic acid
Inferred based on observed Lipid A structure
Beyond the enzymes in Lipid A biosynthesis, a variety of tailoring enzymes can further modify the Lipid A structure (Figure 2). Phosphatases LpxE and LpxF, anchored to the inner membrane, hydrolyze 1- and 4′- phosphate group in the periplasm, respectively. Group transferases such as LpxT and EptA use periplasmically accessible donors, such as undecaprenyl pyrophosphate and phosphatidyl ethanolamine, to further decorate the two phosphates on the disaccharide backbone. Similarly, group transferase ArnT transfers cationic sugars from undecaprenyl phosphate-L-Ara4N (4-amino-4-deoxy-alpha-L-arabinopyranosyl phosphate) to Lipid A. PagP and PagL, located in the outer membrane, catalyze acyl group transfer from phospholipids and deacylation, respectively. Some Gram-negative bacteria express multiple tailoring enzymes that work sequentially to bring yet more variety to Lipid A structures. For example, Capnocytophaga canimorsus and Helicobacter pylori have LpxE and EptA homologs that process Lipid A sequentially: first LpxE hydrolyzes the 1-phosphate, and then EptA transfers phosphoethanolamine directly onto the disaccharide backbone [7,8].
Figure 2. Modification of Lipid A by tailoring enzymes.
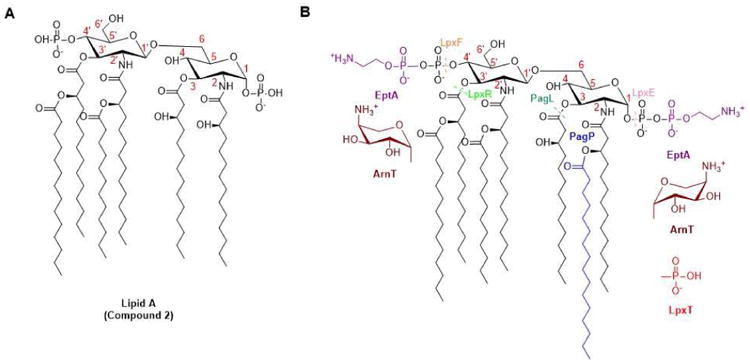
(Adapted from [4]) (A) The canonical structure of Lipid A from E. coli is shown absent the two Kdo sugars, which are not essential for the immunological activities of Lipid A. (B) Modification(s) introduced by each representative tailoring enzyme is indicated in a different color, while components of E. coli Lipid A remain in black. Tailoring enzymes such as ArnT, EptA, LpxT, and PagP catalyze group transfer reactions, while enzymes such as PagL, LpxE, LpxF, and LpxR promote hydrolysis (dashed line across scissile bond). LpxE and/or LpxF homologs are observed in Francisella tularensis[67], Helicobacter pylori [68], Porphyromonas gingivalis [69], Capnocytophaga canimorsus [7], and Bacteroides thetaiotaomicron [70]. LpxR homologs are found in Salmonella typhimurium [71], Yersinia enterocolitica [72], and Helicobacter pylori [73]. PagL is found in Salmonella typhimurium [74] and Pseudomonas aeruginosa [75]. LpxT is found in Escherichia coli [76]. EptA in Escherichia coli [77] and Campylobacter jejuni [78] has broad specificity for both 1 and 4′ positions, while P. aeruginosa EptA only modifies the 4′-phosphate group [79]. PagP in Pseudomonas aeruginosa transfers a palmitoyl chain to the 3-OH of the 3′- acyl residue [80], whereas its homologs in Escherichia coli [81] and Salmonella enterica recognize the 3-OH of the 2-position acyl chain [82,83], and Bordetella parapertussis PagP can modify either hydroxyl group [84]. ArnT in Salmonella typhimurium can decorate the 1- and 4′-phosphates of Lipid A with 4-amino-4-deoxy-L-arabinose (L-Ara4N) [85,86], whereas ArnT in Francisella tularensis adds a galactosamine unit onto the 1-phosphate [87] and ArnT in Bordetella pertussis can add a glucosamine unit on either phosphate groups [88].
In an effort to further diversify the Lipid A scaffold, several studies have combinatorially modified this biosynthetic pathway in different host bacteria. For example, Escherichia coli [9] and Neisseria meningitides[10] have been tested with constructs expressing one to three of the above-mentioned modifying enzymes. Interestingly, most strains expressing one modifying enzyme displayed a single dominant Lipid A species in their outer membranes, whereas strains expressing two or more modifying enzymes displayed a heterogeneous mixture of glycolipids. This observation highlights the substrate specificity of modifying enzymes.
Extracellular sensing of Lipid A by Toll-Like Receptors (TLRs)
Innate immune receptors such as TLRs recognize conserved pathogen-associated molecular patterns (PAMPs) and trigger signaling cascades in response to these PAMPs, often leading to the release of pro-inflammatory cytokines. As a major component of the outer membrane of Gram-negative bacteria, LPS is recognized by Toll-like receptor 4 (TLR4) as a PAMP. A landmark series of experiments by Beutler and coworkers first revealed that a strain of mice harboring a missense mutation in the TLR4 gene was resistant to otherwise fatal doses of LPS [11]. Subsequent studies reported that LPS recognition involves cooperation of multiple accessory proteins, including the lipopolysaccharide binding protein (LBP), CD14, and the TLR4-MD2 complex [12-14].
Due to the amphiphilic nature of LPS, it forms aggregates in blood. LBP separates a LPS monomer from its aggregated form and transfers the monomer to CD14, which in turn delivers the glycolipid molecule to the TLR4-MD2 complex [15-18]. The Lipid A portion of LPS binds to the extracellular domain of TLR4-MD2 and recruits another TLR4-MD2 complex to form α2β2 homodimer (Figure 3) [1]. Although TLR4-MD2 complex is sufficient for signaling by Lipid A, it has been discovered that CD14 is essential for more elaborate Lipid A variants, such as “smooth LPS” in which Lipid A is conjugated to the O-antigen of E. coli [19].
Figure 3. Non-canonical NLRP3 inflammasome activation.
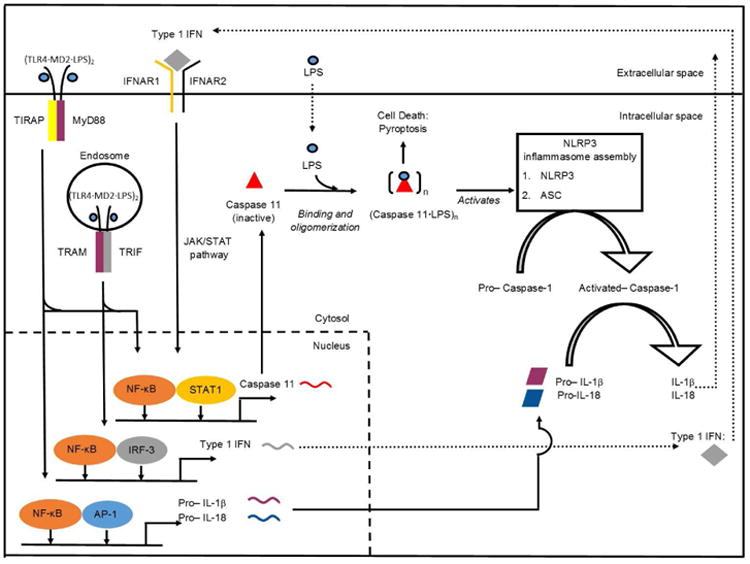
Extracellular LPS is recognized by TLR4/MD2 complex, and triggers MyD88- and TRIF-dependent signaling cascades. MyD88- and TRIF- signaling contribute to the transcriptional upregulation of pro-inflammatory cytokines (purple and blue rhombi) and Type I IFN (grey diamond), respectively. Secreted type I IFN binds the IFNAR1/IFNAR2 receptor and leads to the activation of JAK/STAT pathway. Both type I IFN signaling through JAK/STAT pathway, and LPS signaling through TRIF and MyD88 contribute to the expression of pro-caspase 11 (red triangle). Pro-caspase-11 directly binds cytosolic LPS through its CARD domain. Upon LPS binding, pro-caspase-11 oligomerizes to trigger its activation through proximity induced proteolytic cleavage. The activated caspase 11 results in cell pyroptosis and also NLRP3 inflammasome activation to trigger IL-1β and IL-18 release. Abbreviations: TLR, toll-like receptors; MD, myeloid differentiation; LPS, lipopolysaccharide; MyD88, adaptor molecules myeloid differentiation primary response 88; TIRAP, toll-interleukin 1 receptor domain containing adaptor protein;TRIF, toll/ IL-1 receptor homology (TIR)-domain-containing adapter-inducing interferon-β; NF-κB, nuclear factor- κB; TRAM, TRIF- related adaptor molecule; IRF, interferon regulatory factor; IFN, interferon; IL, interleukin; IFNAR, interferon-α/β receptor; STAT, signal transducer and activator of transcription; AP, activator protein; NLRP3, nucleotide-binding oligomerization domain, leucine rich repeat and pyrin domain-contain protein 3; ASC, apotosis-associated speck-like protein containing CARD; JAK/STAT, janus kinase/signal transducers and activators of transcription.
Once activated by Lipid A or LPS, the TLR4-MD2 dimer recruits adapter proteins for functional signaling (Figure 3). Among all TLRs, TLR4 uniquely signals through two distinct pathways, the MyD88-dependent and TRIF-dependent pathways in a spatial and sequential order. The early-stage response, within the first 6 hours, occurs mainly on the cell surface. Activated TLR4-MD2 complex engages MyD88 and TIRAP (TIR-domain-containing adaptor protein); it activates transcription factors including nuclear factor-κB (NF-κB), and activator protein-1 (AP-1) and leads to rapid production of potent inflammatory cytokines such as interleukins -6 and -1β [20-23]. The delayed response involves endocytosis of TLR4-MD2 complex and recruitment of TRIF (TIR-domain-containing adaptor protein inducing IFN-β) and TRAM (TRIF-related adaptor molecule). Interferon regulatory factor 3 (IRF3) is then activated, leading to the production of interferon (IFN)-β [21]. As a type I interferon, IFN-β leads to maturation of antigen presenting cells; thus it provides a critical link between the innate and the adaptive immune responses [24-27].
Although the TLR4-MD2 complex can bind to a broad range of Lipid A variants, its downstream signaling activity is qualitatively and quantitatively influenced by the precise structure of the glycolipid ligand. The differences can be measured at both transcriptional levels as well as the subsequent cytokine profiles. Thus, different glycolipids have alternative cytokine fingerprints characteristic of MyD88-dependent and TRIF-dependent signaling cascades.
The number and length of acyl chains have strong effects, as do the identity and substituent pattern of the disaccharide scaffold. To begin with, tetra-, penta-, and hexa-acylated E. coli Lipid A differ in the number of acyl chains, and this results in different signaling activities. The canonical E. coli (4+2) hexa-acylated Lipid A, comprising of two acyl chains on one GlcN and four on the other, is a potent agonist that activates both the MyD88-dependent and TRIF-dependent signaling cascades [28]. In contrast, penta-acylated Lipid A (3+2) and tetra-acylated Lipid IVA are weaker agonists and antagonists, respectively, compared to the hexa-acylated version (Table 2a) [9,29]. Second, the acyl chain length (Table 2c) can also modulate the severity of TLR4 mediated immune responses [30]. Systematic evaluation of the effect of chain length of secondary fatty acyl substituents (4 to 14 carbons) revealed that a threshold chain length of 10 carbon atoms was required to induce cytokine production by human monocytes, and that Lipid A derivatives with 10 carbon chains have the highest cytokine induction and greatest pyrogenicity [30,31]. Third, phosphorylation state of the 1 and 4′-position on the disaccharide backbone also has an effect on TLR4 activation (Table 2b). For instance, recombinant E. coli's LPS containing penta-acylated, 1-phosphorylated Lipid A is more TLR4 stimulatory than LPS containing penta-acylated, 4′-phosphorylated Lipid A [32]. In addition, the 1-dephosphorylated lipid A congeners from Salmonella minnesota RE595, a mixture of tetra, penta and hexa-acylated species, promote T-cell response and can be safely used as an adjuvant [33]. Finally, disaccharide backbone variations, usually entailing incorporation of different sugar molecules, impact TLR4 activity. Although most Lipid A species contain a di-glucosamine backbone, Campylobacter jejuni and L. interrogans Lipid A possess a 2,3-diamino-2,3-dideoxy-D-glucopyranose backbone The presence of the two additional amide linkages compromises TLR-4 agonism, as demonstrated by a decreased level of tumor necrosis factor (TNF-α) induction in THP-1 cells, a human leukemia monocytic cell line [34]. Therefore, these one-factor-at-a-time changes to Lipid A show that the number and distribution of both acyl chains and substituent groups have a profound effect on TLR4 stimulating activity.
Table 2.
Structure-activity relationships of Lipid A analogs binding to TLR4.
| 2 a) TLR4 activity as a function of number and distribution of acyl chains | ||||||
|---|---|---|---|---|---|---|
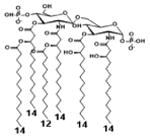
|
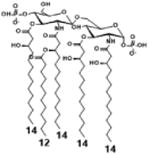
|
|
|
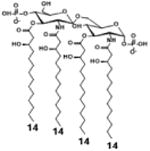
|
||
| Description | Lipid A [Benchmark] Compound 2 | Penta-acylated Lipid A | Penta-acylated Lipid A | Tetra-acylated Lipid A | Lipid IVA Compound 1 | |
| NF-κB | Activation | +++++ | ND | +++++ | ++++ | ND |
| MyD88 | TNF-α | +++++ | ND | ++ | ++ | N/A |
| IL-1β | +++++ | + | +++ | ++ | N/A | |
| IL-8 | +++++ | ++ | +++++ | +++++ | N/A | |
| TRIF | G-SCF | +++++ | ND | ++ | + | N/A |
| RANTES | +++++ | + | ++ | ++ | N/A | |
| MCP-1 | +++++ | + | +++ | +++ | N/A | |
| Conditions | Lipid A analog cone. | 100ng/mL | 100ng/mL | 100ng/mL | 100ng/mL | 100ng/mL |
| Cell type | THP-1 | THP-1 | THP-1 | THP-1 | THP-1 | |
| Reference | [9] | [9] | [9] | [9] | [29] | |
| 2 b) TLR4 activity as a function of number and distribution of phosphate substituents | ||||
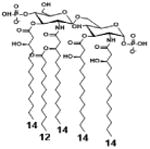
|
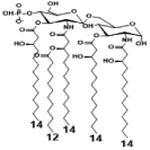
|
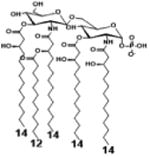
|
||
| Description | Bis-phosphorylated Lipid A [Benchmark] | 4′- phosphorylated Lipid A | 1- phosphorylated Lipid A | |
| NF-κB | Activation | ++ | ++ | +++ |
| Conditions | Lipid A analog cone. | 100ng/mL | 100ng/mL | 100ng/mL |
| Cell type | HEK293 hTLR4/hMD-2 | HEK293 hTLR4/hMD-2 | HEK293 hTLR4/hMD-2 | |
| Reference | [32] | [32] | [32] | |
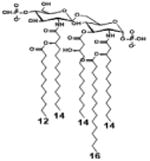
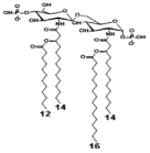
| 2 c) TLR4 activity as a function of secondary acyl chain length | ||||||
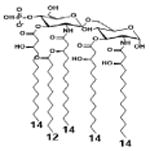
|
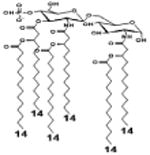
|
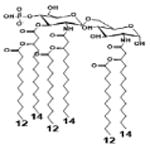
|
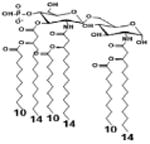
|
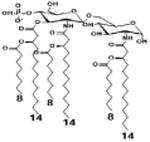
|
||
| Description | Secondary acyl chain length | 4′- phosphorylated Lipid A [Benchmark] | 14-14-14 | 12-12-12 | 10-10-10 | 8-8-8 |
| MyD88 | TNF-α | + | + | +++ | ++ | ND |
| IL-1β | + | ++ | ++ | +++++ | ND | |
| Conditions | Lipid A analog conc. | 10μg/mL | 10μg/mL | 10μg/mL | 10μg/mL | 10μg/mL |
| Cell type | Human whole blood | Human whole blood | Human whole blood | Human whole blood | Human whole blood | |
| Reference | [30] | [30] | [30] | [30] | [30] | |
| 2 d) TLR4 activity as a function of combined structural changes | ||||||
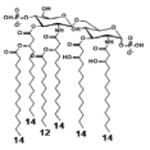
|
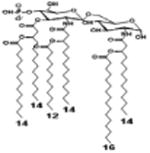
|
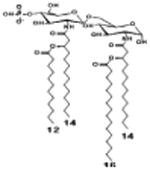
|
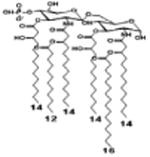
|
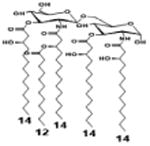
|
||
| Description | Lipid A [Benchmark] Compound 2 | Hexa-acylated, monophosphorylated Lipid A | Tetra-acylated, monophosphorylated Lipid A | Hexa-acylated, monophosphorylated Lipid A | Penta-acylated Lipid A without phosphates | |
| Modification enzymes involved | Benchmark | PagP, PagL, LpxE | LpxE, PagL, LpxR | LpxM, LpxE, PagP | LpxM, LpxE, LpxF | |
| VF-κB | Activation | +++++ | +++++ | +++++ | ++++ | ++ |
| MyD88 | TNF-α | +++++ | ++ | + | + | ND |
| IL-1β | +++++ | +++ | + | + | ND | |
| IL-9 | +++++ | +++++ | ++++ | +++++ | +++ | |
| TRIF | G-SCF | +++++ | ++ | + | ND | ND |
| RANtes | +++++ | ++ | + | + | ND | |
| MCP-1 | +++++ | ++ | + | ++ | + | |
| Conditions | Lipid A analog conc, | 100ng/ml | l00ng/mL | 100ng/mL | 100ng/mL | 100ng/mL |
| Cell type | THP-1 | THP-1 | THP-1 | THP-1 | THP-1 | |
| Reference | [9] | [9] | [9] | [9] | [9] | |
‘+’ symbols correspond to the signaling output intensity. All activity intensities are normalized with respect to the benchmark (Column 1, Tables (2a, 2b, 2c, 2d)). The greater the number of ‘+’ symbols, the greater the signaling intensity.
Signaling intensity of ‘ND’ corresponds to baseline/ negligible activity output
N/A: not available
Several analogs of Lipid A have been investigated for use in clinical settings (Figure 4). Eritoran, an analog of Lipid IVA, has undergone extensive evaluation in clinical trials for sepsis [35]. Monophosphoryl Lipid A from Salmonella minnesota R595 (MPL®)is gaining widespread use as an adjuvant in modern vaccines, including the hepatitis B virus (HBV) vaccine Fendrix and the human papillomavirus (HPV) vaccine Cervarix [36]. Recently, a structural similar MPLA analog G100 is in several clinical trials for cancer immuno-oncology therapies [37]. The X-ray crystal structures of the TLR4-MD2 complex bound to rough LPS [1] and Eritoran [38], and MD-2 bound to Lipid IVA [39] have been solved, and provide some insight into the mechanistic logic of the structure-function relationships (reviewed in [40]).
Intracellular sensing of Lipid A by Caspases
For a long time, the TLR pathway remained the only known mechanism by which higher eukaryotes recognized and responded to the Lipid A family of glycolipids. Unexpectedly however, caspase-11-deficient mice showed resistance to LPS-induced septic shock [41]. It took another two decades before two independent groups made the discovery that intracellular LPS (as well as Lipid A) activated caspase-11 in a TLR4-independent manner, thereby adding a caspase to a growing list of high-affinity receptors of this remarkable family of bacterial glycolipids [42,43]. This novel caspase-mediated inflammatory response to LPS was coined as the “non-canonical inflammasome”[43] (Figure 3), contrasting it to the canonical TLR-mediated pathways.
Both canonical and non-canonical inflammasome signaling exhibit auto-amplificatory behavior. TLR4 priming triggers NF-κB activity, thereby increasing cellular expression of both inflammasome components [44–47]. Subsequent ligand binding triggers proteolytic activation of the caspase, leading to pyroptosis as well as proteolytic maturation and release of IL-1β and IL-18 [48]. Importantly, other TLRs such as TLR2 [43] and TLR3 [42] can also prime the non-canonical inflammasome. Moreover, CD14, MD-1, and MD-2 are dispensable for non-canonical inflammasome activity [43]. Recently, it has been demonstrated that LPS directly binds to mouse caspase-11 and its human counterparts, caspase-4/5, thereby triggering activation in an oligomeric state [49,50]. The binding site is the caspase recruitment domain (CARD), and the Kd of glycolipid binding is exceptionally low [49].
Notably, all of the above studies used non-physiological methods for intracellular delivery of Lipid A, such as transfection, electroporation or cholera toxin subunit B mediated transport (which requires the core oligosaccharide of LPS). As such, the available structure-activity data for non-canonical inflammasome activity must be treated with caution. Nonetheless, it is clear that the Lipid A portion of LPS is a potent ligand of CARD and is sufficient for caspase activation [42,49]. Under-acylated Lipid A species, such as E. coli Lipid IVA, tretra-aclylated F. novicida Lipid A and penta-acylated Lipid A from R. sphaeroides' LPS (LPS-RS), failed to activate the non-canonical inflammasome pathway [42,43,49]. Subsequent experiments investigating mechanistic logic revealed that Lipid IVA and LPS-RS could bind caspase 4/11 but fail to induce their oligomerization and activation [49]. Further analysis of both the canonical and non-canonical inflammasome pathways rests critically on the availability of a systematic series of Lipid A analogs.
Conclusions
Gram-negative bacteria have developed an immense capacity to synthesize structurally diverse Lipid A analogs. These analogs differentially modulate human immune response through interactions with extracellular TLR and intracellular caspase pathways. While considerable effort has been directed to investigate their structure-activity relationships, many of these studies go back to the early days of discovery of these biological pathways, and have therefore not taken sufficient advantage of more recent insights into Lipid A chemistry and biosynthesis. Meanwhile, a number of recent studies involving mutant [51,52] or engineered [9,53,54] Gram-negative bacteria have vividly demonstrated the potential for discovery of novel immune-modulatory agents via engineered biosynthesis and semisynthesis. The stage is set for the development of more advanced chemical biology platforms that exploit the resurgence of interest into the biology of one of nature's oldest and most powerful natural products.
Highlights.
The glycolipid Lipid A is the conserved, amphipathic moiety of LPS
Lipid A binds to certain toll like receptors and caspases with high affinity
Bacteria alter their Lipid A structures to modulate immune activities
Non-toxic Lipid A congeners are effective immunomodulatory agents
Acknowledgments
Research on Lipid A glycolipids in the authors' laboratory has been supported by a grant from the NIH (U19 AI109662).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 2.MacLeod MKL, McKee AS, David A, Wang J, Mason R, Kappler JW, Marrack P. Vaccine adjuvants aluminum and monophosphoryl lipid A provide distinct signals to generate protective cytotoxic memory CD8 T cells. Proc Natl Acad Sci U S A. 2011;108:7914–7919. doi: 10.1073/pnas.1104588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 4*.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A Modification Systems in Gram-Negative Bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. This comprehensive review describes Lipid A modification systems in Gram-negative bacteria and their ability to confer selective advantages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweet CR, Preston A, Toland E, Ramirez SM, Cotter RJ, Maskell DJ, Raetz CRH. Relaxed Acyl Chain Specificity of Bordetella UDP-N-acetylglucosamine Acyltransferases. J Biol Chem. 2002;277:18281–18290. doi: 10.1074/jbc.M201057200. [DOI] [PubMed] [Google Scholar]
- 6.Bainbridge BW, Karimi-Naser L, Reife R, Blethen F, Ernst RK, Darveau RP. Acyl Chain Specificity of the Acyltransferases LpxA and LpxD and Substrate Availability Contribute to Lipid A Fatty Acid Heterogeneity in Porphyromonas gingivalis. J Bacteriol. 2008;190:4549–4558. doi: 10.1128/JB.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renzi F, Zähringer U, Chandler CE, Ernst RK, Cornelis GR, Ittig SJ. Modification of the 1-Phosphate Group during Biosynthesis of Capnocytophaga canimorsus Lipid A. Infect Immun. 2016;84:550–561. doi: 10.1128/IAI.01006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran AX, Karbarz MJ, Wang X, Raetz CRH, McGrath SC, Cotter RJ, Trent MS. Periplasmic Cleavage and Modification of the 1-Phosphate Group of Helicobacter pylori Lipid A. J Biol Chem. 2004;279:55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. Modulating the innate immune response by combinatorial engineering of endotoxin. Proc Natl Acad Sci U S A. 2013;110:1464–1469. doi: 10.1073/pnas.1218080110. Combinatorial synthesis of Lipid A analogs through heterologous expression of Lipid A modifying enzymes in E. coli. Altered Lipid A structures led to changes in their immune activities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zariri A, Pupo E, van Riet E, van Putten JPM, van der Ley P. Modulating endotoxin activity by combinatorial bioengineering of meningococcal lipopolysaccharide. Sci Rep. 2016;6:srep36575. doi: 10.1038/srep36575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 12.Miyake K. Roles for accessory molecules in microbial recognition by Toll-like receptors. J Endotoxin Res. 2006;12:195–204. doi: 10.1179/096805106X118807. [DOI] [PubMed] [Google Scholar]
- 13.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 14.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a Molecule that Confers Lipopolysaccharide Responsiveness on Toll-like Receptor 4. J Exp Med. 1999;189:1777. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 16.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a Molecule that Confers Lipopolysaccharide Responsiveness on Toll-like Receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essential role of MD-2 in LPS responsiveness and TLR4 distribution, (n.d.).
- 18.Schumann RR, Leong GW, Flaggs PW, Gray SD, Wright JC, Mathison PS, Tobias RJ. Ulevitch, Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 19.Huber M, Kalis C, Keck S, Jiang Z, Georgel P, Du X, Shamel S, Sovath S, Mudd B, Beutler C, Galanos MA. Freudenberg, R-form LPS, the master key to the activation ofTLR4/MD-2-positive cells. Eur J Immunol. 2006;36:701–711. doi: 10.1002/eji.200535593. [DOI] [PubMed] [Google Scholar]
- 20.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 23.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 24.O'Neill LAJ, Golenbock D, Bowie AG. The history of Toll-like receptors — redefining innate immunity. Nat Rev Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 25.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, Tough DF. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99:3263–3271. doi: 10.1182/blood.v99.9.3263. [DOI] [PubMed] [Google Scholar]
- 26.Kaisho T, Takeuchi O, Kawai T, Hoshino K, Akira S. Endotoxin-Induced Maturation of MyD88-Deficient Dendritic Cells. J Immunol. 2001;166:5688–5694. doi: 10.4049/jimmunol.166.9.5688. [DOI] [PubMed] [Google Scholar]
- 27.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 28.Bryant CE, Spring DR, Gangloff M, Gay NJ. The molecular basis of the host response to lipopolysaccharide. Nat Rev Microbiol. 2010;8:8–14. doi: 10.1038/nrmicro2266. [DOI] [PubMed] [Google Scholar]
- 29.Golenbock DT, Hampton RY, Qureshi N, Takayama K, Raetz CR. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J Biol Chem. 1991;266:19490–19498. [PubMed] [Google Scholar]
- 30.Johnson DA, Keegan DS, Sowell CG, Livesay MT, Johnson CL, Taubner LM, Harris A, Myers KR, Thompson JD, Gustafson GL, Rhodes MJ, Ulrich JT, Ward JR, Yorgensen YM, Cantrell JL, Brookshire VG. 3-O-Desacyl Monophosphoryl Lipid A Derivatives: Synthesis and Immunostimulant Activities. J Med Chem. 1999;42:4640–4649. doi: 10.1021/jm990222b. [DOI] [PubMed] [Google Scholar]
- 31.Johnson DA. Synthetic TLR4-active glycolipids as vaccine adjuvants and stand-alone immunotherapeutics. Curr Top Med Chem. 2008;8:64–79. doi: 10.2174/156802608783378882. [DOI] [PubMed] [Google Scholar]
- 32.Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect Immun. 2011;79:203–210. doi: 10.1128/IAI.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mata-Haro V, Cekic C, Martin M, Chilton PM, Casella CR, Mitchell TC. The Vaccine Adjuvant Monophosphoryl Lipid A as a TRIF-Biased Agonist of TLR4. Science. 2007;316:1628–1632. doi: 10.1126/science.1138963. doi:10.1126/science. 1138963. [DOI] [PubMed] [Google Scholar]
- 34.Stephenson HN, John CM, Naz N, Gundogdu O, Dorrell N, Wren BW, Jarvis GA, Bajaj-Elliott M. Campylobacter jejuni lipooligosaccharide sialylation, phosphorylation, and amide/ester linkage modifications fine-tune human Toll-like receptor 4 activation. J Biol Chem. 2013;288:19661–19672. doi: 10.1074/jbc.M113.468298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opal SM, Laterre PF, Francois B, LaRosa SP, Angus DC, Mira JP, Wittebole X, Dugernier T, Perrotin D, Tidswell M, Jauregui L, Krell K, Pachl J, Takahashi T, Peckelsen C, Cordasco E, Chang CS, Oeyen S, Aikawa N, Maruyama T, Schein R, Kalil AC, Nuffelen MV, Lynn M, Rossignol DP, Gogate J, Roberts MB, Wheeler JL, Vincent JL for the A.S. Group. Effect of Eritoran, an Antagonist of MD2-TLR4, on Mortality in Patients With Severe Sepsis: The ACCESS Randomized Trial. JAMA. 2013;309:1154–1162. doi: 10.1001/jama.2013.2194. [DOI] [PubMed] [Google Scholar]
- 36.Casella CR, Mitchell TC. Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell Mol Life Sci. 2008;65:3231. doi: 10.1007/s00018-008-8228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Temizoz B, Kuroda E, Ishii KJ. Vaccine adjuvants as potential cancer immunotherapeutics. Int Immunol. 2016;28:329–338. doi: 10.1093/intimm/dxw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal Structure of the TLR4-MD-2 Complex with Bound Endotoxin Antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Ohto U, Fukase K, Miyake K, Satow Y. Crystal Structures of Human MD-2 and Its Complex with Antiendotoxic Lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 40*.Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. 2013;45:e66. doi: 10.1038/emm.2013.97. This review summarizes the crystal structures of CD 14 and TLR4-MD-2 complex. It also offers structural insights into Lipid A triggered agonistic versus antagonistic behavior by comparing and contrasting different TLR4-MD2 complex structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Miura M, Jung Y, Zhu H, Li E, Yuan J. Murine Caspase-11, an ICE- Interacting Protease, Is Essential for the Activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/S0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 42**.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. This reference and the next demonstrate the existence of a TLR4 independent mechanism to detect cytoplasmic LPS. It goes on to shown that the activity of non-canonical inflammasome pathway depends on the precise Lipid A structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, Dixit VM. Noncanonical Inflammasome Activation by Intracellular LPS Independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. A contemporaneous and independent study to that in ref. 42. [DOI] [PubMed] [Google Scholar]
- 44.Guo H, Callaway JB, Ting JPY. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, Hornung V, Latz E. Cutting Edge: NF-κB Activating Pattern Recognition and Cytokine Receptors License NLRP3 Inflammasome Activation by Regulating NLRP3 Expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang HJ, Wang M, Wang L, Cheng BF, Lin XY, Feng ZW. NF-κB Regulates Caspase-4 Expression and Sensitizes Neuroblastoma Cells to Fas-Induced Apoptosis. PLOS ONE. 2015;10:e0117953. doi: 10.1371/journal.pone.0117953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schauvliege R, Vanrobaeys J, Schotte P, Beyaert R. Caspase-11 Gene Expression in Response to Lipopolysaccharide and Interferon-γ Requires Nuclear Factor-κB and Signal Transducer and Activator of Transcription (STAT) 1. J Biol Chem. 2002;277:41624–41630. doi: 10.1074/jbc.M207852200. [DOI] [PubMed] [Google Scholar]
- 48.Stowe I, Lee B, Kayagaki N. Caspase-11: arming the guards against bacterial infection. Immunol Rev. 2015;265:75–84. doi: 10.1111/imr.12292. [DOI] [PubMed] [Google Scholar]
- 49**.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. This investigation showed that intracellular caspases 4, 5 and 11 directly bind Lipid A. Structure-activity relationship (SAR) analysis of this novel Pattern Recognition Receptor (PRR) showed that Lipid IVA and LPS-RS fail to trigger the non-canonical inflammasome pathway. [DOI] [PubMed] [Google Scholar]
- 50.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingram BO, Masoudi A, Raetz CRH. Escherichia coli mutants that synthesize dephosphorylated lipid A molecules. Biochemistry (Mosc) 2010;49:8325–8337. doi: 10.1021/bi101253s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mamat U, Wilke K, Bramhill D, Schromm AB, Lindner B, Kohl TA, Corchero JL, Villaverde A, Schaffer L, Head SR, Souvignier C, Meredith TC, Woodard RW. Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb Cell Factories. 2015;14:57. doi: 10.1186/s12934-015-0241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawasaki K, Ernst RK, Miller SI. Purification and characterization of deacylated and/or palmitoylated lipid A species unique to Salmonella enterica serovar Typhimurium. J Endotoxin Res. 2005;11:57–61. doi: 10.1177/09680519050110010101. [DOI] [PubMed] [Google Scholar]
- 54.Sweet CR, Lin S, Cotter RJ, Raetz CR. A Chlamydia trachomatis UDP-N-acetylglucosamine acyltransferase selective for myristoyl-acyl carrier protein. Expression in Escherichia coli and formation of hybrid lipid A species. J Biol Chem. 2001;276:19565–19574. doi: 10.1074/jbc.M101868200. [DOI] [PubMed] [Google Scholar]
- 55.Anderson MS, Raetz CR. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J Biol Chem. 1987;262:5159–5169. [PubMed] [Google Scholar]
- 56.Dotson GD, Kaltashov IA, Cotter RJ, Raetz CRH. Expression Cloning of a Pseudomonas Gene Encoding a Hydroxydecanoyl-Acyl Carrier Protein-Dependent UDP-GlcNAc Acyltransferase. J Bacteriol. 1998;180:330–337. doi: 10.1128/jb.180.2.330-337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Odegaard TJ, Kaltashov IA, Cotter RJ, Steeghs L, van der Ley P, Khan S, Maskell DJ, Raetz CRH. Shortened Hydroxyacyl Chains on Lipid A of Escherichia coli Cells Expressing a Foreign UDP-N-Acetylglucosamine O-Acyltransferase. J Biol Chem. 1997;272:19688–19696. doi: 10.1074/jbc.272.32.19688. [DOI] [PubMed] [Google Scholar]
- 58.Stead CM, Beasley A, Cotter RJ, Trent MS. Deciphering the Unusual Acylation Pattern of Helicobacter pylori Lipid A. J Bacteriol. 2008;190:7012–7021. doi: 10.1128/JB.00667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sweet CR, Williams AH, Karbarz MJ, Werts C, Kalb SR, Cotter RJ, Raetz CRH. Enzymatic Synthesis of Lipid A Molecules with Four Amide-linked Acyl Chains. J Biol Chem. 2004;279:25411–25419. doi: 10.1074/jbc.M400597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartling CM, Raetz CRH. Crystal Structure and Acyl Chain Selectivity of Escherichia coli LpxD, the N-Acyltransferase of Lipid A Biosynthesis. Biochemistry (Mosc) 2009;48:8672–8683. doi: 10.1021/bi901025v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williamson JM, Anderson MS, Raetz CR. Acyl-acyl carrier protein specificity of UDP-GlcNAc acyltransferases from gram-negative bacteria: relationship to lipid A structure. J Bacteriol. 1991;173:3591–3596. doi: 10.1128/jb.173.11.3591-3596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clementz T, Bednarski JJ, Raetz CRH. Function of the htrB High Temperature Requirement Gene of Escherichia coli in the Acylation of Lipid A HtrB CATALYZED INCORPORATION OF LAURATE. J Biol Chem. 1996;271:12095–12102. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 63.Clementz T, Zhou Z, Raetz CRH. Function of the Escherichia coli msbB Gene, a Multicopy Suppressor of htrB Knockouts, in the Acylation of Lipid A ACYLATION BY MsbB FOLLOWS LAURATE INCORPORATION BY HtrB. J Biol Chem. 1997;272:10353–10360. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- 64.Carty SM, Sreekumar KR, Raetz CRH. Effect of Cold Shock on Lipid A Biosynthesis inEscherichia coli INDUCTION AT 12 °C OF AN ACYLTRANSFERASE SPECIFIC FOR PALMITOLEOYL-ACYL CARRIER PROTEIN. J Biol Chem. 1999;274:9677–9685. doi: 10.1074/jbc.274.14.9677. [DOI] [PubMed] [Google Scholar]
- 65.Basu SS, Karbarz MJ, Raetz CRH. Expression Cloning and Characterization of the C28 Acyltransferase of Lipid A Biosynthesis in Rhizobium leguminosarum. J Biol Chem. 2002;277:28959–28971. doi: 10.1074/jbc.M204525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geurtsen J, Angevaare E, Janssen M, Hamstra HJ, ten Hove J, de Haan A, Kuipers B, Tommassen J, van der Ley P. A Novel Secondary Acyl Chain in the Lipopolysaccharide of Bordetella pertussis Required for Efficient Infection of Human Macrophages. J Biol Chem. 2007;282:37875–37884. doi: 10.1074/jbc.M706391200. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CRH. MsbA Transporter-dependent Lipid A 1-Dephosphorylation on the Periplasmic Surface of the Inner Membrane TOPOGRAPHY OF FRANCISELLA NOVICIDA LpxE EXPRESSED IN ESCHERICHIA COLI. J Biol Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J Bacteriol. 2006;188:4531–4541. doi: 10.1128/JB.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol. 2009;11:1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CRH, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reinés M, Llobet E, Dahlström KM, Pérez-Gutiérrez C, Llompart CM, Torrecabota N, Salminen TA, Bengoechea JA. Deciphering the Acylation Pattern of Yersinia enterocolitica Lipid A. PLOS Pathog. 2012;8:e1002978. doi: 10.1371/journal.ppat.1002978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stead CM, Beasley A, Cotter RJ, Trent MS. Deciphering the Unusual Acylation Pattern of Helicobacter pylori Lipid A. J Bacteriol. 2008;190:7012–7021. doi: 10.1128/JB.00667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trent MS, Pabich W, Raetz CR, Miller SI. A PhoP/PhoQ-induced Lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem. 2001;276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 75.Geurtsen J, Steeghs L, ten Hove J, van der Ley P, Tommassen J. Dissemination of Lipid A Deacylases (PagL) among Gram-negative Bacteria IDENTIFICATION OF ACTIVE-SITE HISTIDINE AND SERINE RESIDUES. J Biol Chem. 2005;280:8248–8259. doi: 10.1074/jbc.M414235200. [DOI] [PubMed] [Google Scholar]
- 76.Touzé T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol. 2008;67:264–277. doi: 10.1111/j.1365-2958.2007.06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herrera CM, Hankins JV, Trent MS. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol. 2010;76:1444–1460. doi: 10.1111/j.1365-2958.2010.07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci U S A. 2010;107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nowicki EM, O'Brien JP, Brodbelt JS, Trent MS. Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol Microbiol. 2015;97:166–178. doi: 10.1111/mmi.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thaipisuttikul I, Hittle LE, Chandra R, Zangari D, Dixon CL, Garrett TA, Rasko DA, Dasgupta N, Moskowitz SM, Malmström L, Goodlett DR, Miller SI, Bishop RE, Ernst RK. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol Microbiol. 2014;91:158–174. doi: 10.1111/mmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brozek KA, Bulawa CE, Raetz CR. Biosynthesis of lipid A precursors in Escherichia coli. A membrane-bound enzyme that transfers a palmitoyl residue from a glycerophospholipid to lipid X. J Biol Chem. 1987;262:5170–5179. [PubMed] [Google Scholar]
- 82.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CRH. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/19.19.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janusch H, Brecker L, Lindner B, Alexander C, Gronow S, Heine H, Ulmer AJ, Rietschel ET, Zähringer U. Structural and biological characterization of highly purified hepta-acyl lipid A present in the lipopolysaccharide of the Salmonella enterica sv. Minnesota Re deep rough mutant strain R595. J Endotoxin Res. 2002;8:343–356. doi: 10.1179/096805102125000678. [DOI] [PubMed] [Google Scholar]
- 84.Hittle LE, Jones JW, Hajjar AM, Ernst RK, Preston A. Bordetella parapertussis PagP Mediates the Addition of Two Palmitates to the Lipopolysaccharide Lipid A. J Bacteriol. 2015;197:572–580. doi: 10.1128/JB.02236-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CRH. An Inner Membrane Enzyme in Salmonellaand Escherichia coli That Transfers 4-Amino-4-deoxy-l-arabinose to Lipid A INDUCTION IN POLYMYXIN-RESISTANT MUTANTS AND ROLE OF A NOVEL LIPID-LINKED DONOR. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 86.Trent MS, Ribeiro AA, Doerrler WT, Lin S, Cotter RJ, Raetz CR. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J Biol Chem. 2001;276:43132–43144. doi: 10.1074/jbc.M106962200. [DOI] [PubMed] [Google Scholar]
- 87.Wang X, Ribeiro AA, Guan Z, McGrath SC, Cotter RJ, Raetz CRH. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida, Biochemistry (Mosc) 2006;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marr N, Tirsoaga A, Blanot D, Fernandez R, Caroff M. Glucosamine Found as a Substituent of Both Phosphate Groups in Bordetella Lipid A Backbones: Role of a BvgAS-Activated ArnT Ortholog. J Bacteriol. 2008;190:4281–4290. doi: 10.1128/JB.01875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


