Abstract
The principles governing evolution of tumors exposed to targeted therapy are poorly understood. Here we modeled the selection and propagation of BRAF amplification (BRAFamp) in patient-derived tumor xenografts (PDX) treated with a direct ERK inhibitor, alone or in combination with other pathway inhibitors. Single cell sequencing and multiplex-fluorescence in situ hybridization mapped the emergence of extra-chromosomal amplification in parallel evolutionary tracts, arising in the same tumor shortly after treatment. The evolutionary selection of BRAFamp is determined by the fitness threshold, the barrier subclonal populations need to overcome to regain fitness in the presence of therapy. This differed for ERK signaling inhibitors, suggesting that sequential monotherapy is ineffective and selects for a progressively higher BRAF copy number. Concurrent targeting of RAF, MEK and ERK, however, imposes a sufficiently high fitness threshold to prevent the propagation of subclones with high-level amplification. Administered on an intermittent schedule, this treatment inhibited tumor growth in 11/11-lung cancer and melanoma PDX without apparent toxicity in mice. Thus, gene amplification can be acquired and expanded through parallel evolution, enabling tumors to adapt while maintaining their intratumoral heterogeneity. Treatments that impose the highest fitness threshold will likely prevent the evolution of resistance-causing alterations and merit testing in patients.
BRAF mutations are found in approximately 7% of cancer patients, particularly those with melanoma, colorectal, thyroid or lung cancer1,2. The most frequent of these mutations, BRAFV600E, drives tumor growth by hyperactivating the extracellular signal regulated kinase (ERK) signaling pathway. Inhibition of RAF, alone or together with its downstream kinase MEK, is effective in slowing the progression of BRAFV600-mutant melanomas and lung cancers3-8. However, as tumors adapt to therapy, almost all patients succumb to the disease. Several mechanisms of resistance to these drugs have been reported, including NRAS mutations, BRAFV600E splice variants, BRAF amplification and MEK mutations9-13. Whether these are truly acquired or if they are selected during therapy remains under investigation. Durable suppression of ERK signaling is required for maximal antitumor effect and resistance to these drugs is often associated with reactivated ERK14,15. With this in mind, direct ERK inhibitors are entering clinical testing in order to improve the outcomes of such patients.
It is commonly viewed that the high mutational rate of cancer leads to diversification of the population, where one clone ultimately gains an advantageous mutation and is able to sweep or take over the tumor mass16-18. As selective pressures change, this process is repeated, enabling tumors to adapt to their environment. Single cell DNA sequencing is an emerging new technique that enables the identification of genomic alterations at the single cell level19-21, with the potential to yield a better resolution of the tumor's clonal architecture as compared to conventional bulk sequencing. Here we generated patient-derived xenograft (PDX) models and utilized single cell DNA sequencing to provide insight into the evolution of resistance during treatment with a direct ERK inhibitor (ERKi) and to identify therapeutic modalities that prevent this process.
Results
Effect of direct ERK inhibitor treatment in lung cancer and melanoma PDX models
PDX models were derived from patients with BRAFV600E-mutant lung cancer or melanoma. Lung cancer patients were previously treated with chemotherapy, as this is a standard management for stage IV disease. Melanoma patients were chemotherapy naïve, since this treatment is not effective for this disease and thus not utilized in the first-line treatment setting. The models were established from six patients who had just progressed on RAF or MEK inhibitor treatment, and from two patients who were treatment naïve (Table I and Supplementary Fig. 1a). For those patients who were previously on targeted therapy, the models were established from biopsy specimens or pleural effusions obtained at the time that the patient was found to have progressive disease. As noted above, ERK inhibitors are entering clinical testing in an effort to improve outcomes of patients who progressed on RAF inhibitor (RAFi) or MEK inhibitor (MEKi) therapy. In light of this, we tested the effect of an ATP-competitive inhibitor (SCH984), which inhibits the kinase activity of ERK and prevents its phosphorylation by MEK22,23. SCH984 inhibited growth in 3/6 PDX models tested (Fig. 1a), where the duration of response lasted several weeks. The tumors that grew on ERKi treatment had diminished sensitivity to this drug in subsequent passages (Supplementary Fig. 1b). Thus, ERKi-monotherapy in BRAFV600E-mutant cancer is limited by the emergence of resistance or de-novo insensitivity.
Table I.
Characteristics of lung cancer and melanoma patient-derived xenograft models.
| Pt | PDX | Age/Sex | Cancer | Stage | Site | BRAF | Chemotherapy | Targeted therapy |
|---|---|---|---|---|---|---|---|---|
| 1 | 1D | 65/M | LUAD | IV | Pericardium | V600E | Cis/Pem | RAFi |
| 1E | “ | LUAD | IV | Effusion | V600E | Cis/Pem | RAFi | |
| 2 | 7 | 57/M | LUAD | IV | RML | V600E, K601Δ | Pem/Bev | RAFi |
| 3 | 15 | 62/M | LUAD | IV | Effusion | WT | na | na |
| 4 | 17 | 73/F | LUAD | IIIA | RLL | WT | Cabo/Pem | na |
| 5 | 21 | 64/M | LUAD | IV | LN | V600E | Pem/Bev | RAFi |
| 6 | 23 | 42/F | Melanoma | IIIC | LN | V600E | na | na |
| 7 | 24 | 67/M | Melanoma | IV | Mesentery | V600E, V600M | na | RAFi |
| 8 | 25 | 42/F | Melanoma | IV | Spleen | V600E | na | MEKi |
| 9 | 27 | 66/F | Melanoma | IV | SubQ | V600E | na | RAFi |
| 10 | 28 | 39/F | Melanoma | IV | LN | V600E | na | na |
Abbreviations: Pt, patient; LUAD, lung adenocarcinoma; Cis, cisplatin; Carbo, carboplatin; Pem, pemetrexed; Bev, bevacizumab; na, not applicable.
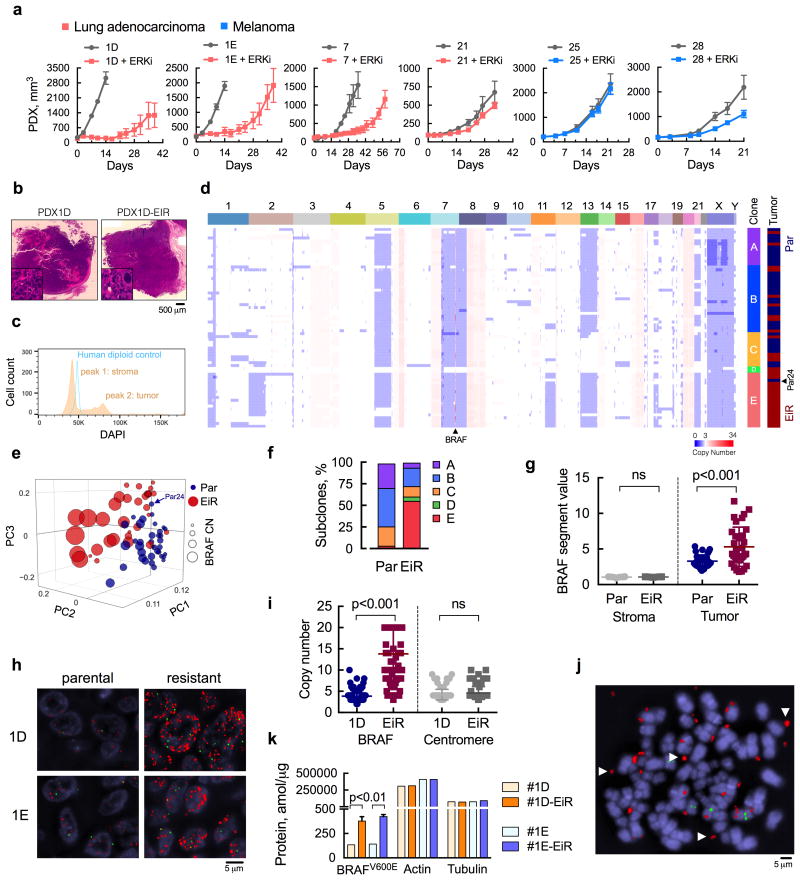
Figure 1. ERK inhibitor-resistant populations with extrachromosomal BRAF amplification.
(a) Patient-derived xenograft (PDX) models from patients with BRAFV600E-mutant lung cancer or melanoma were treated with ERK inhibitor (ERKi) SCH984 over time (n = 5 mice, mean ± s.e.m). (b) H&E stained sections of the PDX models before and after ERKi treatment. (c) Single nuclei extracted from PDX1D tumors were analyzed by FACS to determine the distribution of cells according to their DNA content. A human diploid cell line was used as a control. (d) Copy number (CN) profiles of 69 single cells derived from parental (Par) and ERK inhibitor-resistant (EiR) PDX-1D tumors. (e), Projection of single cells into the top three principal components. (f) Subclonal distribution of parental and resistant tumors. (g) Segment values spanning the BRAF locus in tumor and stromal cells. For stromal cells, sequenced reads were mapped to the mouse genome (see Supplementary Fig. 1e). (h) Representative images of fluorescence in situ hybridization (FISH) analysis with probes spanning BRAF or chr7 centromere in red or green, respectively (a representative of five different fields is shown). (i) Probes were quantified by manual counting (n = 100 cells, all data are shown). (j) Representative image of extra-chromosomal localization of the BRAF gene (arrows) in an 1D-EiR cell undergoing metaphase. (k) The expression of BRAFV600E protein in matched PDX1D and 1E tumor sets was determined by mass spectrometry (n = 3, mean ± s.e.m). Actin and tubulin were used as controls.
Single cell copy number profiles in a parental and ERK inhibitor resistant PDX pair
Understanding the parameters that control the emergence of ERK inhibitor-resistance (EiR) might enable the identification of more effective therapies. To this end, we performed bulk and single-cell sequencing in a parental and EiR tumor pair derived from PDX1D (Fig. 1b and Supplementary Fig. 1c). This model was established from a patient who progressed on the RAFi dabrafenib and retained insensitivity to RAFi monotherapy in athymic mice (see below). Somatic variant analysis of these tumors revealed a close similarity to the patient material from which they were derived (Supplementary Fig. 1d). Mutant allele frequencies were not significantly affected by ERKi treatment and we subsequently focused on copy number (CN) alterations as a potential driver of resistance. For single cell sequencing, genomic DNA from flow-sorted single nuclei was amplified by whole-genome amplification and subjected to sparse massively parallel sequencing, as described19,21,24. Compared to a human diploid control, PDX nuclei distributed in near-diploid and polyploid populations, corresponding to mouse stromal cells and human tumor cells, respectively (Fig. 1c and Supplementary Fig. 1e).
The CN profiles of the human tumors were complex (Fig. 1d and Supplementary Fig. 1f, g), with almost all sequenced cells displaying chromosomal gains in 6p, 7p, 8q, 16q and 20, and losses in 1p, 7q and 8p, some of which are known to recur in lung adenocarcinoma genomes25. Heterogeneous alterations were identified on chromosomes 1, 2p, 3q, 11q, 13, 17q, 21 and X. This genetic diversity enabled the discrimination of parental from resistant cells in principal component analysis (Fig. 1e) and the inference of distinct subpopulations (A-E) through hierarchical clustering (Fig. 1d) and t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis (Supplementary Fig. 1h). While both parental and resistant tumors had a high Shannon diversity index (Supplementary Fig. 1i), their subclonal distribution differed due to the selective pressure of therapy (Fig. 1f). Parental cells were predominantly found in subpopulations A, B and C; intermixed with a few cells derived from the resistant tumor (Fig. 1d and f). In contrast, the majority of resistant cells clustered in subpopulation E, alongside a single parental cell (Par24), which is likely an earlier precursor of this dominant resistant clone.
BRAF amplification is selected and expanded through parallel evolution
While searching for CN alterations associated with resistance, we found that parental and resistant cells had progressively higher BRAF-segment counts compared to stromal cells (Fig. 1g). While nearly 50% of resistant cells had values greater than 6, no parental cells surpassed this threshold. Fluorescence in situ hybridization (FISH) confirmed a high-level BRAFamp in PDX1D- and PDX1E-EiR tumors, as well as the presence of cells with extra BRAF copies in the parental models (Fig. 1h). PDX1E was established from a separate site of progressive disease in patient 1 (Table 1). This model was also insensitive to RAFi (see below) and its ERKi-resistant derivative was established independently of PDX1D-EiR. In EiR cells the increase in BRAF CN was greater than the increase in centromere copies (Fig. 1i), and the BRAF gene was dispersed in extra-chromosomal regions (Fig. 1j). As expected, this amplification led to increased BRAFV600E protein expression (Fig. 1k) in resistant models. BRAF amplification was also identified in PDX25, a melanoma PDX model with de-novo ERKi insensitivity (Fig. 1a and Supplementary Table I).
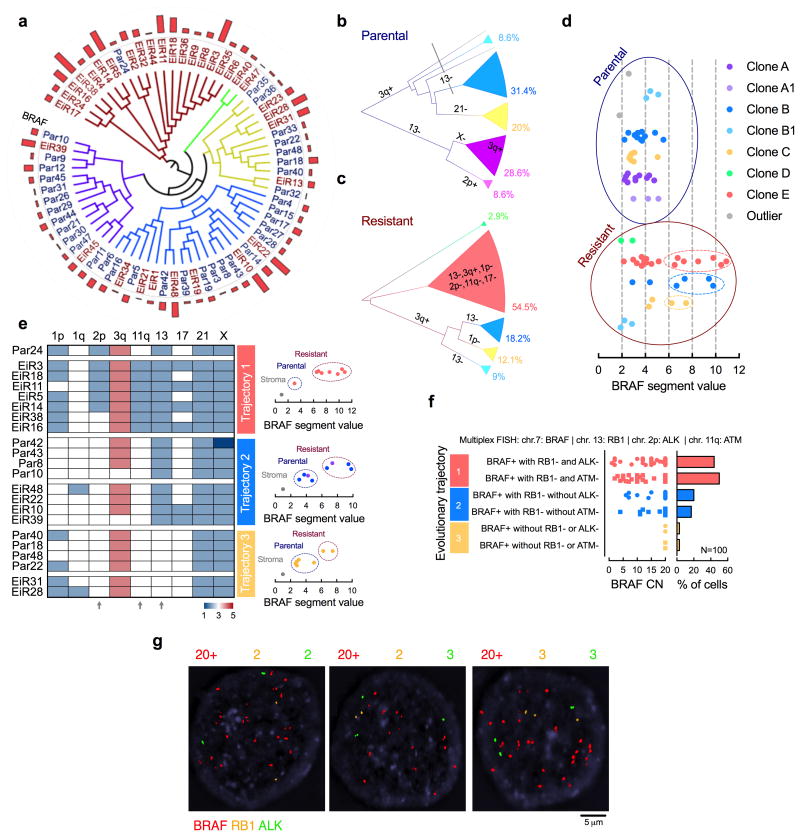
The clonal architecture in Fig. 1f suggests the emergence of a selective sweep by subclone E after ERKi-treatment. We were surprised, however, to find that resistant cells harboring high-level BRAFamp were also present in other subclones (Fig. 2a-d). A closer evaluation of chromosomes with heterogeneous CN profiles revealed three trajectories leading to high-level BRAFamp, defined by losses in chr.2p, chr.11q or chr.13 (Fig. 2e). Multiplex FISH with probes targeting genes on these chromosomal regions (Fig. 2f, g and Supplementary Fig. 2a) confirmed the presence of three distinct BRAFamp species in PDX1D-EiR: (i) BRAFamp with RB1 (chr.13) and ALK (chr.2p) loss, or, with RB1 and ATM (chr.11) loss; (ii) BRAFamp with RB1 loss only; or (iii) BRAFamp without these alterations. As shown in Fig. 2e, losses in 2p and 11q occur together and probing for ALK or ATM are orthogonal approaches to identify the various subclones. The same species were observed in the independently derived resistant tumor PDX1E-EiR (Fig. 1a and Supplementary Fig. 1c, d and 2b). Thus, under the selective pressure of ERKi treatment, BRAFamp is selected and propagated through parallel evolutionary trajectories that maintain intratumoral genetic heterogeneity.
Figure 2. BRAFamp emerges in parallel evolutionary tracts.
(a) The clonal relationship of single cells, as inferred by Manhattan-Ward clustering of integer CN. The bar graph shows BRAF CN. (b, c) Single cells derived from parental (b) or resistant (c) tumors were subjected to hierarchical clustering and subclonal analysis independent of each other. Phylogenetic inference was established using a heuristic maximal parsimony approach. Subclones A and B were subdivided on the basis of additional CN alterations and their inferred phylogeny. (d) Single cells with high-level BRAFamp were found in three distinct resistant clones. (e) The CN state of select chromosomal regions with heterogeneous profiles. Note the emergence of BRAFamp cells in three distinct evolutionary trajectories depending on co-occurring losses in chr2p, 11q and/or 13 (arrows). (f, g) Multiplex FISH with probes targeting the indicated chromosomal regions in PDX1D-EiR. Manual quantification (f) and representative images (g) are shown.
BRAF amplification provides a selective growth advantage in the presence of ERK inhibitor treatment
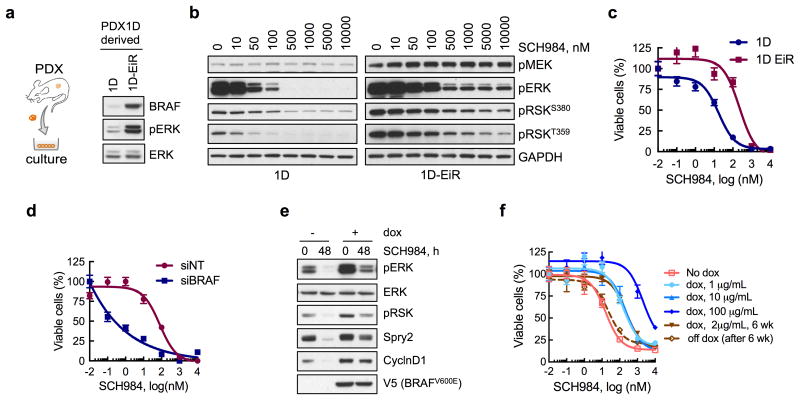
To determine if BRAFamp is sufficient to confer a fitness advantage during ERKi-treatment, we established cell lines from PDX1D and PDX1D-EiR (Fig. 3a). The inhibitor suppressed signaling and proliferation less potently in 1D-EiR than in 1D cells, as evidenced by the residual phosphorylation of ERK and its substrate RSK, as well as a right-shift in proliferation dose-response curves (Fig. 3b, c). The effect of another ERK inhibitor, Vx11e, was attenuated in a similar manner (Supplementary Fig. 3a, b). siRNAs targeting BRAF in 1D-EiR cells enabled a more potent inhibition of signaling (Supplementary Fig. 3c) and proliferation (Fig. 3d) by the drug. Furthermore, inducing the expression of BRAFV600E in melanoma cells (A375) engineered to express BRAFV600E under a doxycycline (dox)-inducible promoter26 diminished the inhibition of pERK and pRSK immediately after ERKi treatment or after longer treatment intervals (Supplementary Fig. 3d-g). The expression of two ERK-dependent signaling markers27, CyclinD1 and Spry2, was restored to near baseline levels after 48h of ERKi treatment in dox-induced cells (Fig. 3e). Due to these direct and adaptive changes, increased BRAFV600E expression attenuated the anti-proliferative effect of ERKi-treatment in a dose-dependent and reversible manner (Fig. 3f). Inducing the expression of BRAFV600E conferred a growth advantage only during treatment with the ERKi (Supplemental Fig. 3h), and when PDX1D-EiR tumors were grown in the absence of such treatment, there was a decrease in BRAF copy number (Supplemental Fig. 3i).
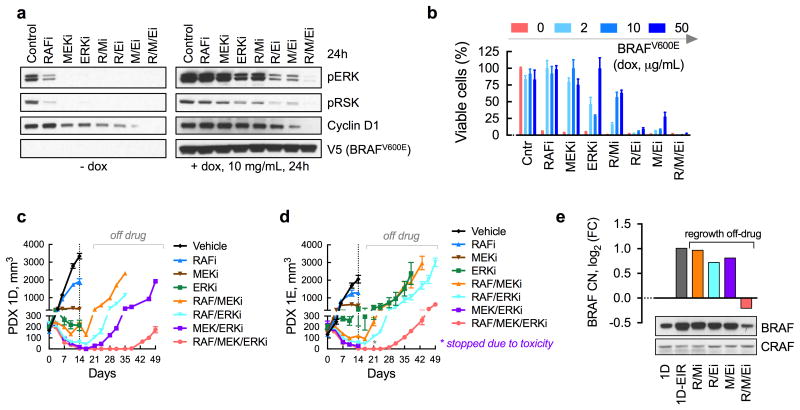
Figure 3. BRAFamp is sufficient to confer a selective advantage in the presence of ERKi treatment.
(a) Immunoblot analysis of cell lines derived from parental (1D) or ERK inhibitor-resistant (1D-EiR) PDX. (b) Immunoblot analysis of signaling intermediates in 1D and 1D-EiR cells treated for 1h with SCH984. (c) Cell viability at 72h after treatment. (d) Cell viability of 1D-EiR cells transfected with BRAF specific or control siRNAs followed by drug treatment as in c. (e & f) A375 cells, engineered to express BRAFV600E under a doxycycline (dox)-induced promoter, were treated as shown (dox, 2μg/mL; SCH984, 500 nM) to determine the effect on signaling by immunoblotting (e) or viability (f). Withdrawal of dox after a 6-week stimulation restored sensitivity to the ERKi. A representative of at least two independent experiments is shown for the immunoblots in this figure. In viability experiments, n = 3, mean ± s.e.m.
A fitness threshold model to explain the selection and propagation of BRAF amplification during treatment
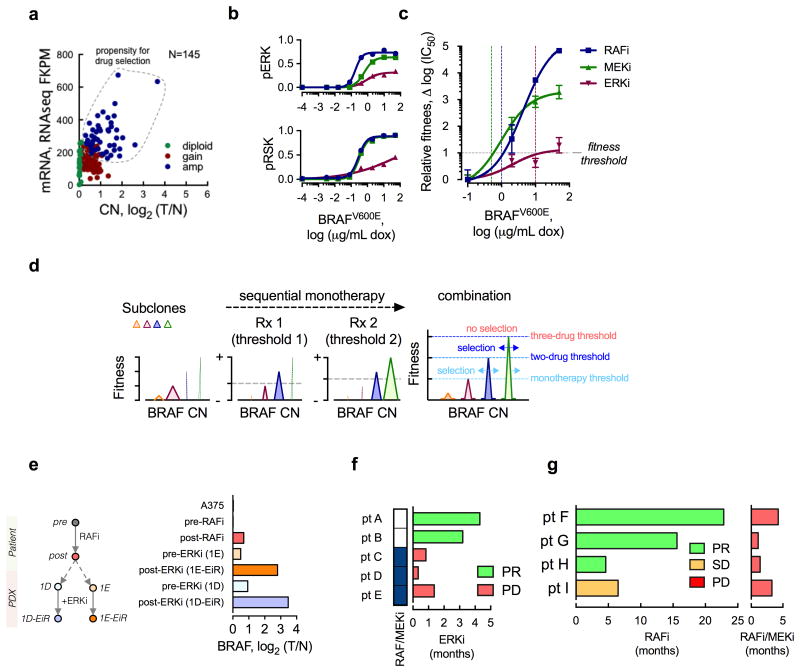
In addition to attenuating direct ERK inhibition, BRAF amplification is a frequent cause of resistance in melanoma patients treated with RAF and/or MEK inhibitors10-13. Interestingly, BRAF CN gains or amplifications were present in a significant proportion of melanoma patients even before exposure to targeted therapy (Fig. 4a). This suggests that selection of BRAFamp during therapy is a widespread phenomenon in these tumors. To define how this evolutionary selection is determined, we compared the fitness effect of BRAFamp in the presence of RAFi-, MEKi- or ERKi-treatment. BRAFV600E expression attenuated signaling inhibition (Fig. 4b and Supplementary Fig. 4a-c) and conferred a fitness advantage, i.e. continued proliferation, in the presence of each drug (Fig. 4c and Supplementary Fig. 4d). The level of BRAFV600E expression required to confer a similar fitness increment in the presence of RAFi- or MEKi-treatment, however, was lower compared to that of ERKi-treatment (Fig. 4c and Supplementary Fig. 4d). Thus, the magnitude of amplification required for continued pathway activity and proliferation in the presence of the drug, a cut-off that we refer to as fitness threshold, is drug-dependent, and higher levels of BRAFamp are needed to bypass the effect of direct ERKi.
Figure 4. Fitness threshold model.
(a) The BRAF mRNA expression as a function of CN in 145 untreated BRAFV600E-mutant melanomas. The data were obtained from TCGA. The dotted area represents tumors at risk for selective propagation of BRAFamp during drug treatment. (b, c) A375 cells were stimulated with increasing concentrations of dox (24h), followed by treatment with ERK signaling inhibitors (RAFi: vemurafenib, 1 μM; MEKi: trametinib, 25 nM; or ERKi: SCH984, 500 nM) for 1h (b) or 72h (c), to determine the effect on signaling (b, quantification of representative immunoblots of 2 independent experiments) or relative fitness (c, n = 3, mean ± s.e.m.). The effect on signaling was adjusted for the effect of dox alone. Relative fitness is the change in log(IC50) with increasing concentrations of dox. (d) A schematic representation of the fitness threshold model. Sequential monotherapy is predicted to impose a selective gradient for the propagation of high level BRAFamp. In contrast, combination therapy is predicted to maximally elevate the fitness threshold, thus suppressing the selection and propagation of BRAFamp subclones. (e) Genomic DNA extracted from the patient biopsies before and after RAFi treatment (pre, post) or their derivative PDX models, before and after exposure to the ERKi, were sequenced to determine the BRAF CN. Diploid A375 cells were used as a control. (f) The duration of ERKi treatment response in patients who were either targeted therapy-naïve (pt. A and B) or pretreated with a RAF/MEKi combination (pt. C, D, E). See also Supplementary Table II. (g) The duration of treatment response in lung cancer patients treated first with a RAFi (left) followed by the addition of a MEKi (right). By comparison, the RAF/MEKi combination therapy has nearly a 60% response rate in treatment-native lung cancer patients9. PR: partial response, SD: stable disease, PD: progressive disease.
Sequential therapy may select for progressive increases in BRAF copy number
The data suggest that sequential exposure to ERK signaling inhibitors is ineffective, serving as a selective gradient for the propagation of tumor subpopulations with a progressively higher BRAF CN (Fig. 4d). Indeed, bulk sequencing of patient biopsy specimens after treatment with a RAFi and matched PDX exposed to an ERKi revealed a progressive increase in BRAF CN (Fig. 4e). By comparison, a normal BRAF copy number was observed in the sample obtained prior to RAFi treatment but after exposure to chemotherapy. Thus, while exposure to chemotherapy may create a permissive environment, this alone appears to be insufficient for the expansion of BRAFamp subpopulations.
The data above suggest that sequential treatment with these drugs is suboptimal in achieving maximal or durable response in patients. To provide some evidence in support of this, we evaluated the responses of several patients who were treated sequentially with ERK signaling inhibitors. Three patients who were previously on a RAF/MEK combination therapy failed to respond to ERKi treatment, whereas two targeted therapy-naïve patients responded to this agent (Fig. 4f and Supplementary Table II). Addition of a MEKi after progression on RAFi therapy was also ineffective in four lung cancer patients (Fig. 4g and Supplementary Table II), a finding that is in agreement with previous reports in melanoma patients28. While more clinical work is needed to prospectively test and validate these observations, these data suggest that sequential therapy is not an optimal therapeutic approach.
Derivation of an intermittent combination therapy to suppress the expansion of BRAF amplified clones
While ineffective as monotherapy, ERK signaling inhibitors given in combination may raise the fitness threshold to prevent the expansion of heterogeneous BRAFamp subclones (Fig 4d). Combined RAFi, MEKi and ERKi treatment durably inhibited signaling and proliferation in A375 cells induced to express intermediate- or high-level BRAFV600E (Fig. 5a, b and Supplementary Fig. 5a). A durable target inhibition was also observed in 1D- and 1D-EiR cells (Supplementary Fig. 5b) and in other PDX models in vivo (Supplementary Fig. 5c). As expected, the three-drug combination produced the strongest antitumor effect against PDX1D and PDX1E, which was most apparent after drug-withdrawal (Fig. 5c, d). To determine if the treatment is sufficient to suppress the growth of BRAFamp subclones, we intentionally stopped treatment early and allowed the tumors to regrow in the absence of treatment. Tumors that regrew following discontinuation of two-drug regimens had elevated BRAF CN and protein expression, compared to untreated tumors (Fig. 5e). By comparison, those that regrew after discontinuation of the three-drug regimen had lower BRAF CN and protein expression. Thus, the three-drug regimen imposed the highest fitness threshold to prevent the propagation of BRAFamp-tumor subpopulations (Fig. 4d). By coincidence, the triple drug combination had a lower toxicity profile compared to the MEK/ERKi combination (Supplementary Fig. 5d, e). Addition of the RAFi diminished the inhibitory effect of the other drugs in normal tissue (Supplementary Fig. 5f), suggesting that the ability of the RAFi to paradoxically activate ERK in BRAFWT cells ameliorates the toxicity of therapy.
Figure 5. Identification of a treatment to suppress the evolution of BRAF amplified clones.
(a) Immunoblot analysis (n = 2 independent experiments) of dox-induced A375 cells treated as shown (doses as in Fig. 4b) for 24h to determine the effect on ERK signaling intermediates. (b) As in a, but cells were treated for 72h to determine the effect on viability (n = 3, mean ± s.e.m). (c, d) Mice bearing PDX1D (c) or PDX1E (d) were treated with dabrafenib (RAFi), trametinib and/or SCH984 daily for 14 days followed by discontinuation of treatment to determine the effect on tumor growth (n = 5 mice, mean ± s.e.m). A vemurafenib analogue (PLX4720), alone or in combination, had a similar effect to dabrafenib (see below). Mice treated with the MEK/ERKi combination experienced significant toxicity leading to discontinuation of the experiment in (d). (e) Tumors that regrew after discontinuation of drug treatment from (c) were analyzed to determine BRAF CN by sequencing and BRAF protein expression by immunoblot analysis.
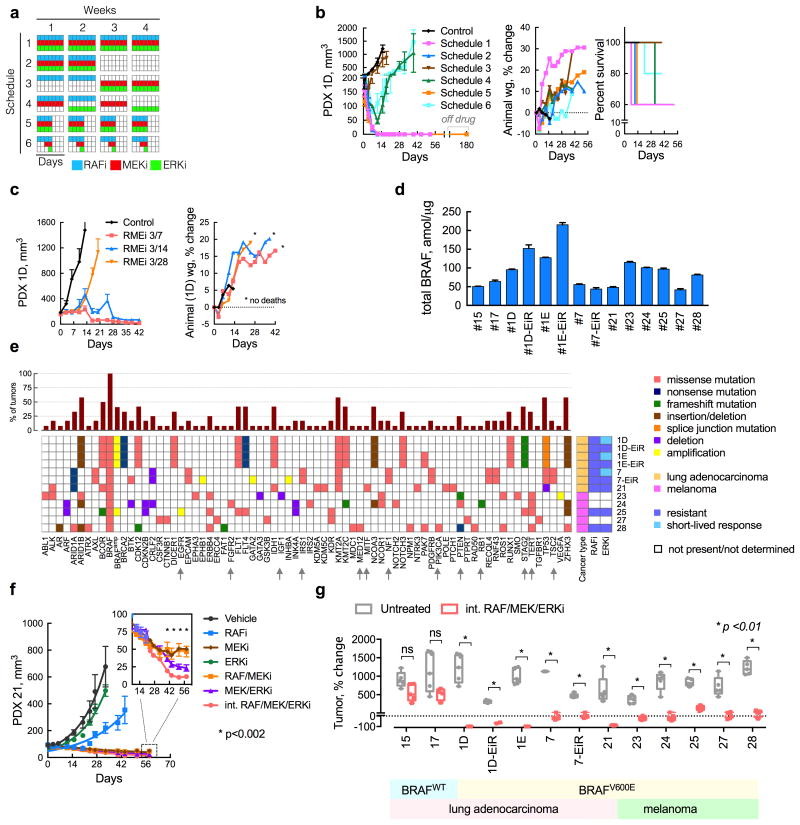
In order to further reduce the toxicity associated with maximal inhibition of ERK (Supplementary Fig. 5d, e), we evaluated the potential benefit of intermittent drug administration schemes (Fig. 6a). Several administration schedules were compared to the effect of continuous administration of the three drugs (Schedule 1). The three drugs are administered together for 2 weeks every month (Schedule 2) or for four days every week (Schedule 5). The inhibitors were also administered in an alternating fashion (Schedules 3 and 4) or dosed in a sequential intermittent fashion (Schedule 6). Concurrent administration of the three drugs for 2/4-weeks (Schedule 2) or 4/7-days (Schedule 5) had a similar antitumor effect as the continuous schedule (Fig. 6b). Regimens where the drugs were not given concurrently were less effective. To determine if treatment on Schedule 5 completely suppressed tumor growth, we discontinued treatment after six cycles and monitored tumor growth for 180 days. No re-growth was observed during this time. Schedule 5 was further optimized by increasing the off-drug interval to produce a regimen consisting of three-days-on treatment followed by four-days-off (3/7), which maximally inhibited tumor growth without measurable toxicity in mice (Fig. 6c and Supplementary Fig. 6a).
Figure 6. An intermittent combination treatment inhibits tumor growth in lung cancer and melanoma BRAFV600E PDX models.
(a, b) A schematic representation (a) of several three-drug combination treatment schedules and their effect on the growth of PDX1D tumors in athymic mice (b, n = 5 mice, mean ± s.e.m, RAFi: vemurafenib analogue PLX4270, MEKi: trametinib, ERKi: SCH984). Treatment related toxicity was determined by monitoring animal weight or mortality. Mice treated on Schedule 5 remained free of tumor for up to 180 days after drug discontinuation. (c) Additional optimization of the off-drug interval in order to minimize toxicity, while retaining maximal tumor growth inhibition. (d) The expression of total BRAF in the PDX models was determined using mass spectrometry (n = 3, mean ± s.e.m). (e) The profile of genetic alterations in the BRAFV600E PDX models utilized in this study. (f) Effect of the intermittent regimen in a model with de-novo insensitivity to ERKi treatment. (g) The effect of the intermittent three drug combination treatment (administered on a 3/7-day schedule) in lung and melanoma PDX models (n = 5 mice, for each untreated or treated arm, mean ± range; ns: p>0.05; primary data are shown in Supplementary Fig. 6).
Broader testing of the intermittent drug regimen
By imposing a high fitness threshold, the intermittent regimen may have a strong antitumor effect in a broader panel of PDX models. To this end, we tested 13 lung cancer and melanoma PDX models with varying levels of BRAF expression (Fig. 6d) and multiple concurrent genetic co-alterations (Fig. 6e). The intermittent treatment produced statistically significant tumor growth inhibition in 11/11 BRAFV600-mutant PDX models (Fig. 6f, g and Supplementary Fig. 6a-i). Specifically, of the 55 tumors tested, 42 (76%) regressed and 55 (100%) were inhibited during treatment. This approach inhibited growth in models with acquired (PDX1D-EiR and PDX7-EiR), or de-novo (PDX21, PDX25 and PDX28) resistance to ERKi-treatment (Fig. 6f and Supplementary Fig. 6b, g, i). It also inhibited tumors harboring various other alterations reported to confer resistance to RAFi or MEKi therapy29-34, including NF1, PTEN, IRS, EGFR and TSC2 (Fig. 6e, arrows). Finally, the treatment had minimal antitumor effects in BRAFWT PDX (Fig. 6g and Supplementary Fig. 6c, d) and did not produce toxicity in mice (Supplementary Fig. 6c-i). These data suggest that the parameters that control the evolution of BRAFamp also regulate the selection of other resistance-causing alterations.
Discussion
Parallel evolution has been described in hematologic malignancies35,36 and when comparing primary and metastatic lesions in solid tumors37. In our study, single cell DNA sequencing revealed that parallel evolutionary tracts enable the selection and propagation of distinct BRAF amplified subclones. These occur in the same tumor shortly after drug treatment, allowing the tumor to adapt while maintaining its intratumoral heterogeneity.
To explain the process driving the evolutionary selection of this alteration, we derived the fitness threshold model, where fitness threshold refers to the barrier subclonal populations need to overcome to regain fitness in the presence of drug treatment. Drugs targeting different nodes of the same pathway have distinct mechanisms of actions and, as a consequence, they exert a different evolutionary selective pressure. As such, the level of BRAFamp required to overcome the effect of the drug differs between RAF, MEK and ERK inhibitors, with the latter tolerating higher levels of BRAFamp. This is probably why the ERKi produced a short-lived response in PDX models harboring low level BRAFamp.
The fitness threshold model links the effect of the drug on its target with the evolutionary selection of resistance causing alterations and has two immediate implications for the treatment of cancer patients. The model predicts that sequential treatment is ineffective, a prediction that is supported by our findings that treatment with a RAFi followed by an ERKi led to a progressive increase in BRAF copy number and that patients who were pre-treated with ERK signaling inhibitors did not respond well to subsequent treatment with another inhibitor of the pathway. As noted above, a concentrated effort is required to prospectively evaluate these observations in the clinic, as they may reshape how patients are enrolled into clinical trials.
The model also predicts that at a sufficiently high fitness threshold, a broader range of BRAFamp subclones, including those with high-level amplification, are at a fitness disadvantage and prevented from propagation. One way to achieve this is with concurrent targeting of RAF, MEK and ERK kinases. It remains to be seen if newer ERK signaling inhibitors with distinct mechanisms of actions (particularly inhibitors that target RAF dimers, or phosphorylated ERK) are able to sufficiently raise the fitness threshold during monotherapy. We went a step further and identified an intermittent administration scheme that retains the negative effect of the three-drug combination on fitness, while minimizing its toxicity in preclinical models. The intermittent administration has the dual benefit of providing a recovery window, allowing for the drugs to be partially cleared, as well as removing the strong positive selective pressure applied on the tumor by therapy. The intermittent three-drug combination caused regressions (∼75%) and suppressed tumor growth (100%) in PDX models harboring diverse co-alterations alongside BRAFV600E. These findings are important not only because they serve as proof-of-principle that intermittent administration enables concurrent delivery of multiple targeted therapies, but also because they suggest that the fitness threshold model explains how other resistance-causing alterations are propagated during targeted therapy.
Over the nearly five years that ERK inhibitors have been available for clinical testing alongside RAF and MEK inhibitors, no clinical trials have evaluated the effect of the three-drug combination in patients. The intermittent regimen identified in this study warrants clinical testing as it may halt the evolution of resistance and improve clinical outcomes in patients whose tumors harbor BRAFV600 mutations.
Online Methods
Cell culture and reagents
All cell lines used in this study were maintained in DMEM medium supplemented with 10% FBS, penicillin, streptomycin and L-glutamine. A375 cells were obtained from ATCC. A375 dox-inducible BRAFV600E cells were established by Zhan Yao and validated by the presence of fluorescence and/or BRAF expression upon dox treatment. The cell lines tested negative for mycoplasma. The inhibitors used in this study, including vemurafenib, dabrafenib, trametinib, SCH984 (similar to MK8353, Phase I) and VTx11e (similar to BDV523, Phase I) were obtained from Selleckem. The in vivo studies were carried out with the following inhibitors at their maximal tolerated dose in mice: PLX4720 (an analogue of vemurafenib that has been commonly used in preclinical in vivo studies), 50 mpk, dabrafenib, 30 mpk, trametinib, 3 mpk and SCH984, 35-75 mpk (this drug was not well tolerated in some mice strains thus requiring dose reduction to avoid toxicity).
Patient derived xenograft models
These were established as described38,39, in accordance with the Memorial Sloan Kettering Cancer Center Institutional Review Board. Informed consent was obtained in all cases. All animal studies were done in accordance with protocols approved by the MSKCC Institutional Animal Care and Use Committee (IACUC). Melanoma models were generated previously40. Patient derived tissue (biopsy or surgical resection) or pleural fluid was used to establish the lung cancer models in Table I. For biopsy or resection specimens, the tumor sample was minced under aseptic conditions, vigorously washed in 1× PBS, passed through a 60-μm filter, centrifuged, and then re-suspended in 500 μL of Matrigel (BD Biosciences) at 4°C. For pleural effusions, the fluid was centrifuged in order to isolate the cellular fraction and washed several times in cold PBS. Cells were then injected subcutaneously in the flanks of NSG mice and monitored for tumor growth. When the tumors reached 1 cm in diameter, the mouse was sacrificed and the tumor was divided into sections for snap freezing, formalin fixation or serial passage.
Establishment of cell lines from PDX tumors
PDX tumors were dissociated using a gentleMACS automated dissociator and human tumor dissociation kit (Miltenyi) as described39. Single-cell suspensions were filtered through a 70-μm mesh, washed twice with wash buffer (PBS, 2% FBS and 1 mM EDTA) and red blood cells were lysed with ACK buffer (Crystalgen Inc.). Approximately 1 × 106 viable cells were seeded in 10 ml of DMEM supplemented with 10% FBS, L-glutamine and antibiotics. Alternatively, tumors were manually dissected and minced to a near single cell suspension and cultured as above. DNA sequencing was used to confirm that the genotype of the cultured cells matched that of PDX tumors.
Tumor lysate extraction
Tumors were dissected with a clean scalpel on dry ice, snap frozen in liquid nitrogen, and grinded with a Micro Sample Pestle (Thomas Scientific). 300 μL of RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) containing protease and phosphatase inhibitors was added per 5 mg piece of tissue. The samples were then sonicated in the Bioruptor® sonication device (Diagenode) for 5 cycles of 30 sec ON/30 sec OFF at 4°C, and centrifuged for 10 min at 16,000g at 4°C to remove any remaining insoluble material.
Immunoblotting
Harvested cells were lysed in NP40 lysis buffer (50 mM Tris (pH 7.5), 1% NP40, 150 mM NaCl, 10% Glycerol, and 1mM EDTA) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific) for 10 min on ice. Lysates were centrifuged at 16,000g for 10 min, quantified using BCA assay (Thermo Fisher Scientific), resolved on 4-12% SDS–PAGE gels (Thermo Fisher Scientific), and transferred to nitrocellulose membranes (GE Healthcare). Blots were probed with primary antibodies overnight at 4°C and visualized using horseradish peroxidase (HRP)-conjugated secondary antibodies and ECL (Thermo Fisher Scientific). Antibodies detecting BRAF (sc-5284), CyclinD1 (sc-718), Spry2 (sc-18601) or GAPDH (sc-32233) were obtained from Santa Cruz Biotechnology. Those detecting pMEK (9121), pERK (9101), ERK (4696), pRSK T359 (8753) or pRSK S380 (12032) were obtained from Cell Signaling Technology. Antibody detecting V5 (R960-25) was obtained from Thermo Fisher Scientific. Immunoblots were quantified using Image J. Uncropped blots are shown in Supplementary Fig. 7.
Targeted exome sequencing
DNA derived from patients or PDX frozen tissue was subjected to targeted capture massively parallel sequencing using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) sequencing assay as previously described41. Briefly, this assay involves hybridization of barcoded libraries to custom oligonucleotides (Nimblegen SeqCap) designed to capture all protein-coding exons and select introns of 410 commonly implicated oncogenes, tumor suppressor genes, and members of pathways deemed actionable by targeted therapies. Barcoded sequence libraries were prepared using 100-250 ng genomic DNA (Kapa Biosystems) and combined into equimolar pools of 13-21 samples. The captured pools were subsequently sequenced on an Illumina HiSeq 2000 as paired-end 100-base pair reads, producing a median of 588-fold coverage per tumor. Sequence data were demultiplexed using CASAVA, and reads were aligned to the reference human genome (hg19) using BWA and post-processed using the Genome Analysis Toolkit (GATK) according to GATK best practices. MuTect and GATK were used to call single-nucleotide variants and small indels, respectively. Candidate mutations were manually reviewed using the Integrative Genomics Viewer (IGV) to eliminate likely false positive calls. Because matched normal DNA was not available, tumors were compared to a pool of 10 unmatched normal samples to eliminate common polymorphisms and systematic sequencing artifacts.
Single cell sequencing
Nuclei preparation from tumor samples and whole-genome amplification
Frozen tumor specimens were processed as previously described19,21. Briefly, tumors were minced using a single edge razor blade in 400 μl NST buffer (146 mM NaCl, 10 mM Tris base at pH 7.8, 1 mM CaCl2, 21 mM MgCl2, 0.05% BSA, 0.2% Nonidet P-40) supplemented with 4′6-diamidino-2-phenylindole (DAPI; 10 μg/mL), 0.1% DNase-free RNase A (Life Technologies) and incubated on wet ice for 1 h. Nuclei suspension were washed twice with NST-DAPI (800 μl wash, 7 min at 5000 rpm centrifugation), then filtered twice through a strainer mesh (35 μm) and collected into a 5 ml Polystyrene round-bottom tube. Samples were rested on wet ice for immediate sorting or frozen in dry ice for transportation or supplemented with 10% DMSO and placed in a freezing container at -80°C to obtain a ∼1°C/min cooling rate for nuclear integrity cryopreservation of nuclei overnight. Single nuclei were sorted by FACS using the BD Biosystems Aria II flow cytometer by gating cellular distributions with differences in their total genomic DNA content according to DAPI intensity. First, a small amount of prepared nuclei from each tumor sample was mixed with a diploid control sample (derived from a lymphoblastoid cell line of a healthy individual, 315A) to accurately determine the diploid peak position within the tumor and establish FACS collection gates. Before sorting single nuclei, a few thousand cells were sorted to determine the DNA content distributions for gating. Visual inspection of the nuclei using DAPI staining was performed to ensure the integrity of the nuclei sorted. Single nuclei were deposited into individual wells in the 96-well plate containing 9 μl of lysis solution in each well from the GenomePlex WGA4 kit (Sigma-Aldrich). Whole-genome amplification (WGA) was performed on single flow-sorted nuclei as described in the GenomePlex WGA4 kit protocol. WGAs were assessed on a 1.5% agarose gel to confirm amplification. The WGA products were then cleaned using QIAamp DNA Mini Kit (Qiagen) and eluted in 50 μl EB buffer.
Library preparation and sequencing
800 ng of WGA products were diluted to 75 μL with EB buffer (Qiagen) and acoustically sonicated using the Covaris E210 focus acoustics system with a target base pair peak of 300 (i.e. Duty Cycle: 10%, Intensity: 4, Cycle per Bust: 200 and Time: 80 sec). The sonicated WGA was end repaired using NEBNext End Repair module following manufacturer's protocol (New England Biolabs). The end-repaired DNA was cleaned with QIAamp DNA Mini Kit (Qiagen), eluted in 42 μL EB buffer and subjected to dA-Tailing using the NEBNext dA-Tailing module following manufacturer's protocol (New England Biolabs). Then, the dA-tailed DNA was cleaned with QIAamp DNA Mini Kit (Qiagen), eluted in 35 μL EB buffer, and exactly 34 μL of eluate was combined with 10 μL of 2× Quick Ligation Reaction Buffer, 4 μL of 10 μM barcoded adapter and 2 μL of Quick T4 DNA Ligase (New England Biolabs), and incubated at 20°C for 15 min. The ligated product was then combined with 26.25 μL Agencourt AMPure XP magnetic beads (Beckman Coulter) (ligated product/magnetic bead ratio 0.35), thoroughly mixed and incubated at RT for 10 min. The magnetic beads-DNA complexes were washed twice with freshly prepared 80% ethanol, dried for 10 min at RT, eluted in 30 μL of EB buffer and quantified using a Qubit Fluorometer. The indexed/barcoded libraries were then pooled mixing equal amounts (∼20 ng each), quantitated and PCR-enriched in duplicate using NEBNext High-Fidelity 2× PCR Master Mix (New England Biolabs) containing up to 80 ng of pooled library and 2.5 μL of enrichment primers. The reactions were incubated for 30 sec at 98°C and then 5 cycles of 10 sec at 98°C, 30 sec at 60/65°C (depending on primer set) and 30 sec at 72°C, with a final 5 min incubation at 72°C to ensure polished ends. Individual replicates were then combined, cleaned using the QIAamp DNA Mini Kit (Qiagen) and eluted in 50 μL EB buffer. Enriched libraries were assessed on a Bioanalyzer instrument (Agilent Technologies), quantified and sequenced on a HiSeq4000 instrument using PE 2×150 bp (Illumina).
CN analysis of single cells
Multiplexed single-cell sequencing libraries were split according to their unique barcode identifiers specified by the first seven bases of the sequencing reads. Single-cell sequencing data were aligned to the human reference genome hg19 (or to the mouse genome mm10 in the case of stromal cells) using Bowtie42. Sequencing reads were sorted, followed by removal of PCR duplicates, and then indexed using SAMtools43. CN assessment of single cells was performed using the Ginkgo5 pipeline20 (http://qb.cshl.edu/ginkgo) using the following settings: variable bin size of 250 kb, bins based on simulations of 101 bp, and CBS segmentation. Bad bins and Y-chr pseudo-autosomal regions were masked and the clustering was done using Manhattan distance and Ward linkage algorithms on integer copy number values.
Subclonal diversity index
Clusters of genotypes were identified by hierarchical clustering, as noted above. The proportion of cells that belonged in each group (p) was used to calculate the Shannon diversity index with the formula: Dc = −Σi(pi × lnpi), embedded in the R package “vegan”, as described44. A Shannon index greater than 1 represents a high clonal diversity.
Dimensionality reduction
This was achieved by using eigenvector-based principal component analysis (PCA) on single cell copy numbers with the “xlstat” package. t-Distributed Stochastic Neighbor Embedding (t-SNE) was also used. For this, integer copy number states of single cells were assembled in a Manhattan distance matrix followed by analysis with the tSNE package in R (perplexity 4, iterations 1000, epoch 100). A third approach involved identifying chromosomal regions with heterogeneous copy number alterations in parental and resistant tumors, defined as those with an abundance of 20-80% (Supplementary Fig. 1f). These were then used to distinguish BRAFamp subclonal populations.
Fluorescence in situ hybridization
FISH analysis was performed on formalin fixed paraffin embedded (FFPE) sections or cell line suspension, as described45. Cell lines were harvested and fixed in methanol:acetic acid (3:1) as per standard procedures. Three separate probe-sets were designed to confirm copy number change detected by single cell sequencing: 2-color BRAF/Cen7 (Control) probe, 3-color BRAF/RB1/ALK probe and 3-color BRAF/RB1/ATM probe. The BAC or plasmid clones used in the probe-mix were as follows: BRAF (RP11-788O6, RP11-1065D4, and RP11-133N19; labeled with Red dUTP), Centromere 7 (p7t1; labeled with Green dUTP), RB1 (RP11-795F23 and RP11-305D15; labeled with Orange dUTP), ALK (RP11-701P18, RP11-644H8, and RP11-229K3; labeled with Green dUTP), ATM (RP11-56J3 and RP11-241D13; labeled with Green dUTP). Probe labeling, tissue processing, hybridization, post-hybridization washing, and fluorescence detection were performed according to standard laboratory procedures. Slides were scanned using a Zeiss Axioplan 2i epifluorescence microscope equipped with a megapixel CCD camera (CV-M4+CL, JAI) controlled by Isis 5.5.9 imaging software (MetaSystems Group Inc, Waltham, MA). Metafer and VSlide module within MetaSystems were used to generate the virtual image of H&E and DAPI-stained sections.
Each probe was hybridized on a separate slide or section. For the cell lines, the entire hybridized area was scanned through 63× or 100× objective, representative cells/regions imaged and a minimum of 50-200 discrete nuclei and 25 metaphases were scored. For paraffin tissue, the entire section was scanned under 63× or 100×objective, intratumoral heterogeneity was assessed, and representative regions imaged through the depth of the tissue (compressed/merged stack of 12 z-section images taken at 0.5 micron intervals). At least 10 images per representative region were captured and a minimum of 50-200 discrete nuclei scored for each distinct region or sample. Amplification was defined as Gene:Control ratio of ≥2.0, >10 copies of Gene (independent of control locus) or at least one small cluster of Gene (≥4 signals resulting from tandem repeat/duplication). In cells with high-level amplification, signals ≥20 cannot be accurately counted and therefore given a score of 20. Cells with 3∼5 and 6∼10 discrete copies of Gene/Control were considered to be polysomic and high-polysomic, respectively.
Mass spectrometry detection of BRAF protein expression in vivo
BRAF (Total or V600E) protein was quantitated by SRM-MS as previously described46. Briefly, tissue sections (10 μM) from FFPE blocks were placed onto DIRECTOR® microdissection slides followed by deparaffinization and hematoxylin staining. Tumor areas were marked by a board-certified pathologist and a 12 mm2 section containing nearly 50,000 malignant cells was microdissected and solubilized to tryptic peptides using Liquid Tissue® technology. The solution was subjected to SRM-MS analysis using stable isotope-labeled internal standard peptides for BRAFV600E and total BRAF quantitation. Actin and tubulin quantitation was monitored to verify sample quality and efficiency of microdissection. On-column injection resulted in 5 fmol of isotopically labeled internal standard peptides and 1 μg (∼4000 cells) of total tumor protein as measured by microBCA (Thermo Fisher Scientific). Instrumental analyses were performed on TSQ Quantiva triple quadrupole mass spectrometer (Thermo Fisher Scientific), as previously described47.
Viability and clonogenic assays
For viability assays, 2 × 103 cells were seeded per well in 96 well plates and grown in the presence or absence of each inhibitor for 72 h. Viable cells were determined using the CellTiter-Glo® (Promega) assay as described previously48. For siRNA studies, cells were grown in the presence of siRNA for 72 h before drug treatment. siRNAs used: BRAF (Dhamarcon SMARTpool: ON-TARGETplus BRAF siRNA #L-003460-00) and non-targeting (Santa Cruz Control siRNA-A #sc-37007). For clonogenic assays, cells were seeded at a density of 2 to 3 × 103 cells per well in triplicate into six-well plates. They were cultured in the absence or presence of doxycycline and/or drug as indicated in complete media for 10 days, with media change every other day. The plates were fixed with cold methanol and stained with 0.05% crystal violet.
Animal studies
Nu/nu athymic or NSG mice were obtained from the Harlan Laboratories and maintained in compliance with IACUC guidelines. Animals implanted with xenografts were chosen for efficacy studies in an unbiased manner. In rare instances, animals were excluded if the subcutaneous tumors failed to engraft. Tumor bearing animals were treated in a random fashion with drug or the appropriate vehicle control. Subcutaneous xenografts and tumor measurements were performed as described15 in a non-blinded manner by a research technician not involved in the rest of the study. Treatment-related toxicity was determined by measuring animal weight and survival. Weight was reported either as absolute value or as % change relative to the weight of the animal prior to treatment. Animal survival was reported in Kaplan-Meier plots. In figures where the animal weight is reported alone, no animal mortality was observed during treatment. All animal studies were performed in compliance with institutional guidelines under an IACUC approved protocol (Memorial Sloan-Kettering Cancer Center No. 09-05-009).
Statistics and data analysis
Prism (GraphPad Software Inc.) was used for data analysis unless otherwise specified. The average tumor volume of each study arm was plotted over time. For 5 mice per cohort, the power to detect an odds parameter of 14.0 for each pairwise comparison, with two-sided α level of 0.05, was 80%. Doubling times were calculated by fitting tumor volumes into exponential growth curves and determining their rate constants in Prism. Negative doubling times indicate tumor regression compared to the pre-treatment size. Statistically significant differences in rate constants were determined by using the extra-sum-of-squares F test (with p<0.05) embedded in Prism. Unless otherwise stated, groups were compared using non-parametric tests. This included the comparisons of BRAF segment, copy number, and protein expression analysis.
Data availability
The sequencing data generated in this project are available as supplemental files.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (K08 CA191082-01A1 to P.L.), the Uniting Against Lung Cancer Foundation (P.L.), the Damon Runyon Clinical Investigator Award (P.L.), the Josie Robertson Investigator Program at MSKCC (P.L.), the Druckenmiller Center for Lung Cancer Center at MSKCC (P.L.) and the Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program (Y.X.). E.d.S and S.L are supported in part by the MSKCC Pilot Center for Precision Disease Modeling program (U54 OD020355). T.B. is supported by the William C. and Joyce C. O'Neil Charitable Trust, Memorial Sloan Kettering Single Cell Sequencing Initiative. J.N. is supported by the Knut & Alice Wallenberg Foundation. The authors also acknowledge the MSKCC Support Grant/Core Grant program (P30 CA008748). The authors thank David Solit, Nancy Bouvier and the Center for Molecular Oncology at MSKCC for assistance with next generation sequencing, and Zhan Yao (MSKCC) for providing A375 dox-inducible BRAFV600E cells. The authors are grateful to Charles Sawyers, Charles Rudin, John Poirier and Megan Mroczkowski for reviewing the manuscript.
Competing Financial Interests: NR is on the SAB and has received grant support from Chugai Pharmaceutical and is on the SAB of Astra-Zeneca, Beigene and Kura. S.S., T.H., K.S. and F.C are employees of NantOmics.
Footnotes
Author Contributions: Y.X. and P.L. were the principal writers of the manuscript. All authors reviewed the manuscript and contributed in writing. Y.X., L.M., A.V., M.S. T.T.M. and N.C. performed experiments. L.M., T.B. and J.S.R.-F. performed, analyzed and helped interpret single cell sequencing. M.F.B. analyzed bulk-sequencing data. G.J.R. and B.T.L. provided patient samples. J.N. and U.S. provided PDX models. E.d.S. performed animal experiments. K.S., F.C., T.H. and S.S. performed mass spectrometry experiments. KC performed FISH experiments and GN analyzed the results. N.R. and S.L. provided key scientific insight and reagents. P.L. conceived and supervised the study, designed/performed experiments and interpreted data.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Planchard D, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. The Lancet Oncology. 2016;17:642–650. doi: 10.1016/S1470-2045(16)00077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. The New England journal of medicine. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larkin J, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. The New England journal of medicine. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 7.Long GV, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. The New England journal of medicine. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 8.Planchard D, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. The Lancet Oncology. 2016;17:984–993. doi: 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das Thakur M, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer discovery. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nature medicine. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 14.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lito P, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipinski KA, et al. Cancer Evolution and the Limits of Predictability in Precision Cancer Medicine. Trends in cancer. 2016;2:49–63. doi: 10.1016/j.trecan.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nature reviews Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 19.Baslan T, et al. Genome-wide copy number analysis of single cells. Nature protocols. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garvin T, et al. Interactive analysis and assessment of single-cell copy-number variations. Nature methods. 2015;12:1058–1060. doi: 10.1038/nmeth.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer discovery. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 23.Wong DJ, et al. Antitumor activity of the ERK inhibitor SCH772984 [corrected] against BRAF mutant, NRAS mutant and wild-type melanoma. Molecular cancer. 2014;13:194. doi: 10.1186/1476-4598-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baslan T, et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome research. 2015;25:714–724. doi: 10.1101/gr.188060.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao Z, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer cell. 2015;28:370–383. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KB, et al. Phase II study of the MEK1/MEK2 inhibitor Trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:482–489. doi: 10.1200/JCO.2012.43.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 31.Shi H, et al. A novel AKT1 mutant amplifies an adaptive melanoma response to BRAF inhibition. Cancer discovery. 2014;4:69–79. doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 33.Maertens O, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer discovery. 2013;3:338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. doi:310.1038/nature09650. Epub 02010 Dec 09615. [DOI] [PubMed] [Google Scholar]
- 36.Melchor L, et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28:1705–1715. doi: 10.1038/leu.2014.13. doi:1710.1038/leu.2014.1713. Epub 2014 Jan 1713. [DOI] [PubMed] [Google Scholar]
- 37.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. doi:1110.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen A, et al. PKLR promotes colorectal cancer liver colonization through induction of glutathione synthesis. The Journal of clinical investigation. 2016;126:681–694. doi: 10.1172/JCI83587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poirier JT, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015;34:5869–5878. doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Einarsdottir BO, et al. Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Oncotarget. 2014;5:9609–9618. doi: 10.18632/oncotarget.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao R, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nature genetics. 2016;48:1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanjala J, et al. Identifying actionable targets through integrative analyses of GEM model and human prostate cancer genomic profiling. Molecular cancer therapeutics. 2015;14:278–288. doi: 10.1158/1535-7163.MCT-14-0542-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hembrough T, et al. Application of selected reaction monitoring for multiplex quantification of clinically validated biomarkers in formalin-fixed, paraffin-embedded tumor tissue. The Journal of molecular diagnostics : JMD. 2013;15:454–465. doi: 10.1016/j.jmoldx.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Catenacci DV, et al. Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue. PloS one. 2014;9:e100586. doi: 10.1371/journal.pone.0100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this project are available as supplemental files.