Abstract
With the availability of genome sequences for hundreds of microbial genomes, it has become possible to address several questions from a comparative perspective to understand the structure and function of regulatory systems, at least in model organisms. Recent studies have focused on topological properties and the evolution of regulatory networks and their components. Our understanding of natural networks is paving the way to embedding synthetic regulatory systems into organisms, allowing us to expand the natural diversity of living systems to an extent we had never before anticipated.
Keywords: Gene regulatory networks, Transcription, Protein-DNA interactions, Prokaryotes, Evolution
1. Introduction
One of the greatest challenges in the post-genomic era is to elucidate the complete set of gene expression programs in an organism for all possible stimuli to which it can respond. Although the number of completely sequenced genomes is mounting rapidly, our knowledge of transcription regulation is limited to a few model organisms. Organisms devote a considerable fraction of their DNA to encoding cis-regulatory elements, and a significant fraction of protein coding genes encode transcription factors (TFs), both of which play an important role in controlling and coordinating gene expression at the level of transcription. Unraveling the principles and organization of transcriptional programs is essential for understanding cellular responses to environmental perturbations and the molecular bases of many diseases caused by microbes.
It is now an accepted notion that transcriptional regulation can be visualized as a network consisting of TFs and their target genes (TGs) [34; 60; 64]. However, at a less abstract level, transcription involves a number of cis-regulatory elements like promoters, TF binding sites and transcription terminators, and trans-acting elements like TFs and sigma factors. The interplay between the cis and trans elements provides a plethora of transcriptional programs which ultimately control the state of every gene in the cell tailored for different conditions (see Fig. 1 for a genomic view of the gene regulatory network). Although it is now becoming increasingly evident that post-transcriptional control by small non-coding RNAs plays an important role in both prokaryotic and eukaryotic organisms, here we review recent advances in the computational analysis of transcriptional regulation in microbial organisms. We first discuss progress made at the level of trans and cis element identification and finally integrate our recent understanding of microbial transcriptional regulation from a network perspective.
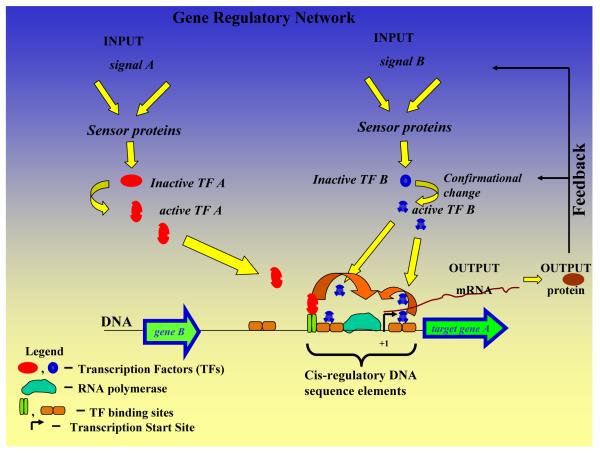
Fig. 1.
Genomic view of a gene regulatory network. Transcriptional regulation from a genomic perspective can be viewed as a complex interplay between cis-regulatory elements on the DNA and trans-acting factors like TFs and sigma factors. Depending on the set of input signals (extracellular or intracellular in nature), upon a cascade of events, transcription factors positively or negatively control transcription of the target gene. Often, transcription factors work in a combinatorial fashion to produce the final output. Protein products formed after transcription and translation of the gene regions are responsible for various cellular functions and ultimately feedback transcription factors at several levels to control their own transcription.
2. Evolution of trans-acting elements
At a trans-acting level, a number of protein families like sigma factors and TFs play important roles in controlling the regulation of gene expression. These elements contribute significantly to rewiring the network of transcriptional interactions depending on the environmental conditions and stimuli an organism is faced with. Although our understanding of the number and repertoire of the trans-acting elements has greatly increased in recent years, our knowledge of the functional roles played by these proteins is far from complete.
3. Sigma factors
Sigma factors are a class of proteins forming essential dissociable subunits of prokaryotic RNA polymerase. The association of a sigma factor with core RNA polymerase provides the basis for transcriptional initiation and is an important step in the process of transcription. Sigma factors provide promoter recognition specificity to the polymerase and contribute to DNA strand separation, after which they dissociate from the RNA polymerase core enzyme following transcription initiation. The substitution of one sigma factor for another can redirect the polymerase to a different set of genes which would otherwise be transcriptionally silent, thereby determining the transcriptional response of a group of genes.
The number of sigma factors encoded in bacterial genomes is highly variable. Although the number of sigma factors generally increases with genome size, environmental bacteria and microorganisms that have developed differentiation programs like sporulation tend to have a higher number of sigma factors than most obligate pathogens. It is possible that the number of sigma factor genes is correlated with the diversity of lifestyles encountered by a bacterium. For instance, among the different mycobacterial species, Mycobacterium leprae has the lowest number of sigma factors and this seems to correlate with the fact that this organism has adapted to being an obligate pathogen, unlike other organisms of this phyla [13; 58].
Sigma factors can be classified into two structurally unrelated and phylogenetically distinct families: the σ70 and σ54 families [23]. While σ54 family members are relatively rare, σ70 family members are found in all bacterial genomes. σ70 factors typically consist of up to four conserved regions and are further classified into four different groups on the basis of their structure and physiological roles [23]. Structurally, σ70 family factors have four major regions, with the highest levels of conservation in regions 2 and 4. Subregions within region 2 are known to be involved in promoter melting (region 2.3) and -10 sequence recognition (region 2.4), while the well conserved subregion 4.2 of region 4 is involved in -35 recognition [54]. Within the σ70 family of sigma factors is a large, phylogenetically distinct subfamily called the extracytoplasmic function (ECF) factor, which typically contains only regions 2 and 4 of the σ70 family. These sigma factors are responsible for regulating a wide range of functions, all involved in sensing and reacting to conditions in the membrane, periplasm or extracellular environment [26]. Many bacteria contain multiple ECF factors and they generally outnumber all other types of sigma factors combined. ECF factors are often co-transcribed with one or more negative regulators [26].
Although no sequence conservation exists between σ54 and σ70 family members, both types bind to core RNA polymerase. However, the holoenzyme formed with the σ54 class has different properties from those of the σ70 holoenzyme. For instance, all σ54 species require a separate activator protein along with the core RNA polymerase to form an open promoter complex, and promoter structures recognized by σ54-RNAP differ from those recognized by σ70-RNAP. σ54 promoters generally are highly conserved, short sequences that are located at positions -24 and -12 upstream of the transcription start site, whereas σ70 promoter sites are typically located at -35 and -10 upstream [9].
4. DNA binding TFs as regulators of transcriptional control
Regulation of gene expression in an organism is predominantly controlled by DNA binding TFs. They form one of the largest protein groups in most genomes. TFs are proteins which are needed to activate or repress the transcription of a gene or operon. Most TFs form dimers and bind to the cis-regulatory elements on the DNA to control transcription initiation in bacterial genomes [8]. The fraction of TFs in bacterial genomes typically scales as the square of the total gene number of a genome [2; 65], with the maximum number of TFs observed in Streptomyces coelicolor among the publicly available completely sequenced genomes [7].
TFs can be classified as activators, repressors or dual regulators depending on their mode of action on a particular promoter [8; 55]. An activator stimulates the expression of its target gene by acting on a promoter to stimulate RNA polymerase. Activation is known to typically occur by binding of TFs upstream of the transcription start site and often upstream of the -35 promoter element [8; 40; 52]. For negative control of transcription, TFs act as repressors by binding to DNA to prevent RNA polymerase from initiating transcription. Repression normally occurs when TFs bind downstream of the transcription start site, causing DNA looping, or in between the -35 and -10 elements of the promoter, thereby blocking RNA polymerase by steric hindrance [8; 40]. Computational analyses suggest that repressors are dominant in both Escherichia coli and Bacillus subtilis, and are more likely to co-evolve with their target genes in closely related genomes [27; 52; 55].
DNA binding regions of prokaryotic TFs can be assigned to a number of families based on sequence and structural homologies [39; 55]. TF families classified based on structural domains are three-fold--the helix-turn-helix, the winged helix and the beta ribbon [30]--with the most abundant among TFs being the classical helix-turn-helix domain [2]. It has been proposed that about 75% of the TFs in E. coli are formed as a result of duplication and that TFs evolve faster than their respective TGs across genomes [36; 41].
Global transcription regulators have been defined as those TFs that have the ability to: regulate large number of genes belonging to diverse functional classes, control a complex regulatory cascade by both directly and indirectly effect expression of various cellular pathways and act on target promoters that use different sigma factors [46]. Based on this, seven global regulators in E. coli have been proposed, which control more than 50% of the genes in the entire transcriptional regulatory network. More recent studies used connectivity (see below) of the TF as a simplified measure to assess the global nature of a TF [36; 41].
5. Evolution of cis-regulatory elements
In recent years, due to accumulation of genome sequences of multiple strains of a single organism and those of phylogenetically close species, it has become possible to address a number of questions related to conservation of regulatory elements. The availability of genome sequences not only provides us with evolutionary insights into conservation of cis-regulatory elements like promoter regions, TF binding sites and terminator signals across organisms, but also enable us to predict them using a variety of comparative genome analysis techniques.
6. Promoter regions
Transcription initiation in bacteria requires that RNA polymerase (RNAP) recognize and bind specific DNA sequences upstream of transcription units called promoters. The recognition of promoter sequences by RNAP occurs when it associates with a small protein, known as sigma (σ) factor. The primary or housekeeping sigma factor in E. coli is encoded by the rpoD gene and is known as σ70 [16]. A bacterial promoter is defined as the segment of DNA that enables a gene or set of genes to be transcribed and is located immediately proximal (6–8 bp) to the transcription start site. Although there are several other condition-specific sigma factors besides the housekeeping ones, the most frequently studied, with extensive experimentally characterized information, remains σ70. In fact, E. coli has 6 other sigma factors which are encoded by genes rpoN (σ54), rpoS (σ38), rpoH (σ32), rpoF (σ28), rpoE (σ24) and fecI (σ19). The canonical model of the σ70 DNA promoter is characterized by two hexamers centered around positions -35 and -10 from the transcription start site and separated by 15–21 bp, with consensus sequences TTGACA and TATAAT, respectively. Although there is a direct relationship between promoter strength and similarity to the consensus sequence, a typical E. coli σ70 promoter sequence contains two mismatches within both the -35 and -10 hexa nucleotide elements [49]. In fact, variations of over three deviations from the consensus have been reported in σ70-dependent promoters from various studies. These variations can generate considerable differences in promoter efficiency during the transcription initiation reaction. All these factors make identification of functional promoter sequences a notoriously difficult task even in well studied model systems like E. coli, irrespective of the approach adopted [19; 28; 32; 66]. In fact, Huerta and Collado-Vides [28] show that several functional promoters are significantly different from the consensus and often occur in regulatory regions as dense overlapping signals. Further studies established that promoter densities are indeed different in the coding and regulatory regions of most bacterial genomes [28; 29; 31], following a regional rule which can distinguish organization of different adjacent gene pairs in bacterial genomes [50]. In contrast, certain genomes with significant size reduction were found not to show this tendency, which was attributed to a process of genome degradation resulting from the decreased efficiency of purifying selection in highly structured small populations [29]. Interestingly several of these genomes which deviate from this tendency were found to be intracellular parasites which exhibit severe reduction not only in their genome sizes, but also a disproportionate reduction in the number of TFs. These observations also suggest that the differential distribution of promoter-like signals between regulatory and non-regulatory regions detected in large bacterial genomes might confer a fitness advantage to these organisms in their natural habitats.
7. TF binding sites
TFs recognize the TGs, whose transcription they control, due to the presence of the binding sites in the promoter regions. Typically a TF, upon binding to the promoter regions of their target genes or transcription units, can control the expression of the genes positively or negatively. While repressor sites which can inhibit the transcription of genes are known to occur downstream of the transcription start site, activators generally attach to DNA upstream of the start site [22; 40; 52]. In E. coli, there is an enrichment for factors which act as transcriptional repressors and hence the majority of genes in the transcriptional network are negatively regulated [39; 55].
Two general computational approaches have emerged for inferring TF binding sites in promoter regions: i) analysis of co-regulated sets of genes; and ii) phylogenetic footprinting of the upstream regions of orthologous genes in closely related genomes, under the notion that selective pressure would sustain regulatory elements over the background non-coding DNA among organisms at short evolutionary distances [48; 56]. Both methods aim to identify statistically significant patterns which are conserved in the background of the remaining aligned intergenic regions. Since the identification of a putative set of co-regulated genes from genome sequences alone is not straightforward, the majority of computational approaches for inferring regulatory motifs use upstream regions of orthologous genes from phylogenetically close organisms as the seed set of sequences. However, Wang and Stormo used the conserved regions of orthologous sequences in multiple sequence alignments, and then compared profiles of nonorthologous sequences (genes with in a given organism) to generate sets of co-regulated genes [67], while others used co-expressed genes as a seed set to improve motif detection by incorporating phylogenetic conservation [51]. Another recent approach took into account the phylogenetic relationship between the species, in order to distinguish conservation due to the occurrence of functional sites from spurious conservation, which is due to evolutionary proximity, and they developed a Gibbs sampling algorithm for motif prediction from phylogenetic conservation [61].
Once regulatory elements are identified, they can either be compared with already known binding profiles for TFs, or subjected to experimental analyses to prove that they are functional and/or to determine binding factors. Some recent approaches also adopted the use of binding profiles obtained first using cross-species data and then generating genome-specific models through recursive training to attain higher specificity for identifying binding sites [20], or exploited the fact that most transcription factors bind to DNA as spaced dimers [35; 53], while others used the idea that TFs often bind cooperatively to their targets; hence, statistically overrepresented motif co-occurrence patterns can help identify novel TF-TG associations [10]. Another work attempted to integrate a variety of properties like proximity of TFs to their TGs, similarity in the binding properties of TFs which belong to the same family and phylogenetic correlation to develop a system for inferring regulatory interactions on a genomic scale [62].
8. Transcriptional termination
Transcription termination typically involves the release of the mRNA transcript and RNA polymerase from the template strand at the end of transcription. Proper termination is essential for bacteria, as the regions between transcription units are generally rather small. In bacterial genomes, termination generally occurs either spontaneously, which is termed intrinsic or Rho-independent termination, or involves the use of a set of trans-acting factors in conjunction with the cis-acting elements, referred to as Rho-dependent termination. Although Rho-dependent termination has been the focus of several experimental studies, our knowledge of defined rules which can be used to identify Rho-dependent termination signals from genome sequences is rather limited, believed to be due to the complex interplay of several auxiliary elements which occur at a specific Rho site depending on the local context of the terminator [12]. The only conserved element common to Rho-dependent terminators appears to be richness in cytosine residues. On the other hand, Rho-independent termination usually occurs due to the presence of a hairpin loop structure followed by a stretch of thymine residues. It is accepted that most Rho-independent terminators can be identified from sequences due to their dyad symmetry and poly-T tail [14; 18].
About half of the transcription terminators which are experimentally characterized in E. coli are Rho-dependent [59]. However, unlike the case in E. coli, the Rho protein is dispensable in B. subtilis, suggesting a limited role for Rho-dependent termination in the latter. In fact, recent work demonstrated that more than 90% of the termination signals in B. subtilis and other Firmicutes are Rho-independent in nature [15]. Another group developed an efficient algorithm for rapid determination of Rho-independent terminators and demonstrated that outside the Firmicutes division, Rho-independent termination is also found to be dominant in the Neisseria, Vibrio and Pasteurellaceae genera [33].
9. The link between cis- and trans-acting elements and the notion of transcriptional networks
An important notion that is emerging in post-genomic biology is that cellular components can be visualized as a network of interactions between different molecules like proteins, DNA and metabolites [5]. This has led to the application of network theory to biological problems, particularly in understanding the regulation of gene expression [60; 64]. In transcriptional networks typically trans-acting elements like TFs and sigma factors form one set of nodes and their target genes, of which they control the activity, form the other set of nodes. The links between them which have directionality from the trans-acting elements to their target genes, controlled by their cis-regulatory elements, form a complex and directional network of interactions (see Fig. 2 for a network view of transcriptional regulation).
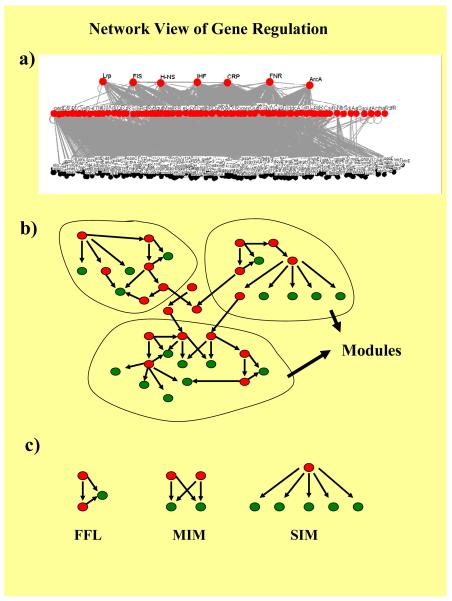
Fig. 2.
Network view of transcription regulation. Transcriptional regulatory interactions at a genomic level can be visualized as a network between TFs (shown in red) and target genes (shown in green). a) The transcriptional regulatory network is a multi-layer hierarchical modular structure without feedback regulation at the transcription level [38; 68], with the global regulators at the top of this layout and local TFs at the bottom, regulating a few genes. b) Modules are interconnected clusters which divide the network of transcriptional interactions into subnetworks. Modules have been identified using a variety of approaches [17; 38; 57] and have been found to be semi-independent in nature. Modules are in turn formed by one or more different types of network motifs. c) Motifs are patterns of interconnections which are overrepresented in transcriptional networks. Known transcriptional regulatory networks were found to have feed forward loops (FFLs), multiple input modules (MIMs) and SIMs, with each kind of motif playing a different role [1].
10. Network structure
One of the most important and obvious pieces of information that can be obtained is the distribution of connectivity, i.e how many connections a node has and how many nodes have a particular number of connections. In the case of transcriptional networks, these parameters actually have two sides, as incoming and outgoing connections must be considered separately. The incoming connectivity is the number of transcription factors regulating a target gene, which gives a sense of the combinatorial effect of gene regulation. The fraction of target genes with a given incoming connectivity was observed to follow an exponential distribution in both E. coli and Saccharomyces cerevisiae [24; 64]. The exponential behavior indicates that most target genes are regulated by a similar number of factors and apparently reflects limits in the size of multiprotein complexes that can be bound near the promoter, as well as by the amount of DNA sequences in upstream regions of genes. On the other hand, outgoing connectivity, which is the number of target genes regulated by each transcription factor, was found to be distributed according to a power law, contrary to incoming connectivity distribution. This is indicative of a hub-containing network structure, in which a select set of transcription factors participate in the regulation of a disproportionately large number of target genes.
At a local level, in transcriptional networks, certain subnetworks appear more often than expected by chance and have been referred to as motifs, analogous to sequence motifs which occur repeatedly in sequences. Motifs were originally described in an E. coli transcriptional regulatory network, but were subsequently found in yeast and other organisms [1; 60]. Three network motifs were found to predominantly occur in most transcriptional networks: 1) a feed-forward loop (FFL), in which a transcription factor regulates expression of another transcription factor which, in turn, regulates a gene that is also regulated by the first transcription factor; 2) a single-input module (SIM), in which a single transcription factor regulates several genes, usually also referred to as a simple regulon [25]; 3) dense overlapping regulons (DORs) in which several TFs regulate overlapping sets of genes; these groups are also called a complex regulon. FFL appears to be the most abundant motif among the best studied transcriptional networks. FFLs have been further classified into eight motif subtypes (see Fig. 3) and two of them, namely coherent type-1 and incoherent type-1 FFL, appear to be much more predominant than others [1; 43]. The former was shown to act as a sign-sensitive delay element and a persistence detector, while the latter was demonstrated to function as a pulse generator and response accelerator [44; 45]. Although motifs form overrepresented subgraphs in the entire network of transcriptional regulation, they do not appear independently but rather integrate to form superstructures or modules that carry out a common biological function by sharing some of their edges [17; 57].
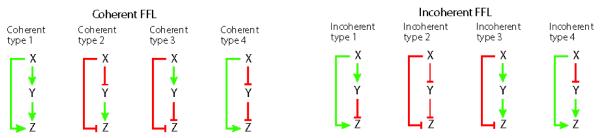
Fig. 3.
Different subtypes of feed forward loop motifs. Directed arrow represents positive regulation and is shown in green, while negative transcriptional control is shown in red.
11. Network conservation
With the availability of documented information on transcriptional regulation, based on experimental evidence in model organisms for a significant fraction of the TFs, it has now become possible to address questions on the evolution of the structure and components of regulatory systems across bacterial genomes [42; 59]. From the perspective of the evolution of transcriptional networks in a given organism, it was proposed that duplication of TFs and their TGs could have given rise to a significant proportion of the currently known regulatory networks in both E. coli and S. cerevisiae [63]. However, from a cross-genome perspective, it was found that TFs are poorly conserved across genomes in comparison to their TGs, and hence are likely to evolve faster than their TGs [36; 41]. It was also found that global regulators are not more conserved than general TFs, suggesting a possible scenario for rapid evolution of gene regulatory mechanisms across bacteria. The latter group also showed that regulatory interactions within a network motif do not show any preference for evolving together and that organisms with a similar lifestyle are likely to preserve equivalent regulatory interactions and network motifs.
12. Network dynamics
Despite several studies which focus on regulatory networks at a static level, it should be noted that the regulatory network of an organism is highly dynamic and different sections of the network could be active under different conditions [21]. In fact, it has been shown in yeast, by integrating expression and regulatory interaction data, that the regulatory subnetworks for different conditions vary significantly [37]. In particular, it was demonstrated that in multistage processes like the cell cycle or sporulation, there are extensive variations in the regulatory networks. In an attempt to extend this work to bacterial systems, another study systematically identified topological units called origons under the notion that different subnetworks from the completely known transcriptional regulatory network could be active under different experimental conditions depending on the environmental signals sensed by the sensor TFs [4]. In another recent work using a static regulatory network of E. coli, the authors classified the complete set of TFs in the currently known regulatory network as those sensing endogenous or exogenous signals. Curiously enough, global regulators often correspond to those sensing internal signals, and TFs sensing internal signals were found to direct the activity of the regulatory network in E. coli [47].
13. Conclusions and perspectives
Although our understanding of the design principles of complex transcriptional regulatory networks is far from complete, we are beginning to design biological circuits and predict their behavior. Progress in sequencing technologies, high throughput experimental techniques like chromatin immunoprecipitation, and advances such as noise filters [6] and oscillators that can combine repressor functionality with that of a two-component system [3], together with improvements in measuring cellular quantities at high resolution [69], should not only enable us to design synthetic circuits for maneuvering bacterial systems [11], but also enable us to address several fundamentally unanswered questions in the years to come.
Acknowledgements
We thank Arthur Wuster for critically reading and providing comments on a previous version of this manuscript. We also thank Agustino Martinez-Antonio for providing assistance in the generation of Fig. 2. This work was partially supported by NIH grant RO1 GM 071962.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- [2].Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- [3].Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- [4].Balazsi G, Barabasi AL, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc Natl Acad Sci U S A. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- [6].Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- [7].Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- [8].Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- [9].Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bulyk ML, McGuire AM, Masuda N, Church GM. A motif cooccurrence approach for genome-wide prediction of transcription-factor-binding sites in Escherichia coli. Genome Res. 2004;14:201–208. doi: 10.1101/gr.1448004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chin JW. Modular approaches to expanding the functions of living matter. Nat Chem Biol. 2006;2:304–311. doi: 10.1038/nchembio789. [DOI] [PubMed] [Google Scholar]
- [12].Ciampi MS. Rho-dependent terminators and transcription termination. Microbiology. 2006;152:2515–2528. doi: 10.1099/mic.0.28982-0. [DOI] [PubMed] [Google Scholar]
- [13].Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- [14].d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- [15].de Hoon MJ, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput Biol. 2005;1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].deHaseth PL, Nilsen TW. Molecular biology. When a part is as good as the whole. Science. 2004;303:1307–1308. doi: 10.1126/science.1095483. [DOI] [PubMed] [Google Scholar]
- [17].Dobrin R, Beg QK, Barabasi AL, Oltvai ZN. Aggregation of topological motifs in the Escherichia coli transcriptional regulatory network. BMC Bioinformatics. 2004;5:10. doi: 10.1186/1471-2105-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. Prediction of transcription terminators in bacterial genomes. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- [19].Eskin E, Keich U, Gelfand MS, Pevzner PA. Genome-wide analysis of bacterial promoter regions. Pac Symp Biocomput. 2003:29–40. [PubMed] [Google Scholar]
- [20].Espinosa V, Gonzalez AD, Vasconcelos AT, Huerta AM, Collado-Vides J. Comparative studies of transcriptional regulation mechanisms in a group of eight gamma-proteobacterial genomes. J Mol Biol. 2005;354:184–199. doi: 10.1016/j.jmb.2005.09.037. [DOI] [PubMed] [Google Scholar]
- [21].Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gralla JD, a.Collado.-Vides J. Organization and Function of Transcription Regulatory Elements. In: Neidhardt FC, Ingraham J, Lin ECC, Low KB, Magasanik B, Reznikoff W, Schaechter M, Umbarger HE, Riley M, editors. Cellular and Molecular Biology: Escherichia coli and Salmonella, C.I.R. American Society for Microbiology; Washington, D.C.: 1996. pp. 1232–1245. [Google Scholar]
- [23].Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- [24].Guelzim N, Bottani S, Bourgine P, Kepes F. Topological and causal structure of the yeast transcriptional regulatory network. Nat Genet. 2002;31:60–63. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- [25].Gutierrez-Rios RM, Rosenblueth DA, Loza JA, Huerta AM, Glasner JD, Blattner FR, Collado-Vides J. Regulatory network of Escherichia coli: consistency between literature knowledge and microarray profiles. Genome Res. 2003;13:2435–2443. doi: 10.1101/gr.1387003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- [27].Hershberg R, Margalit H. Co-evolution of transcription factors and their targets depends on mode of regulation. Genome Biol. 2006;7:R62. doi: 10.1186/gb-2006-7-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J Mol Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- [29].Huerta AM, Francino MP, Morett E, Collado-Vides J. Selection for unequal densities of sigma70 promoter-like signals in different regions of large bacterial genomes. PLoS Genet. 2006;2:e185. doi: 10.1371/journal.pgen.0020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr Opin Struct Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- [31].Janga SC, Lamboy WF, Huerta AM, Moreno-Hagelsieb G. The distinctive signatures of promoter regions and operon junctions across prokaryotes. Nucleic Acids Res. 2006;34:3980–3987. doi: 10.1093/nar/gkl563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kanhere A, Bansal M. A novel method for prokaryotic promoter prediction based on DNA stability. BMC Bioinformatics. 2005;6:1. doi: 10.1186/1471-2105-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- [35].Li H, Rhodius V, Gross C, Siggia ED. Identification of the binding sites of regulatory proteins in bacterial genomes. Proc Natl Acad Sci U S A. 2002;99:11772–11777. doi: 10.1073/pnas.112341999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34:3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- [38].Ma HW, Buer J, Zeng AP. Hierarchical structure and modules in the Escherichia coli transcriptional regulatory network revealed by a new top-down approach. BMC Bioinformatics. 2004;5:199. doi: 10.1186/1471-2105-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Babu M. Madan, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Babu M. Madan , Teichmann SA. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 2003;19:75–79. doi: 10.1016/S0168-9525(02)00039-2. [DOI] [PubMed] [Google Scholar]
- [41].Babu M. Madan, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- [42].Makita Y, Nakao M, Ogasawara N, Nakai K. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 2004;32:D75–77. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- [45].Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- [46].Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- [47].Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J. Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol. 2006;14:22–27. doi: 10.1016/j.tim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- [48].McCue L, Thompson W, Carmack C, Ryan MP, Liu JS, Derbyshire V, Lawrence CE. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res. 2001;29:774–782. doi: 10.1093/nar/29.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mitchell JE, Zheng D, Busby SJ, Minchin SD. Identification and analysis of ‘extended -10’ promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mitchison G. The regional rule for bacterial base composition. Trends Genet. 2005;21:440–443. doi: 10.1016/j.tig.2005.06.002. [DOI] [PubMed] [Google Scholar]
- [51].Monsieurs P, Thijs G, Fadda AA, De Keersmaecker SC, Vanderleyden J, De Moor B, Marchal K. More robust detection of motifs in coexpressed genes by using phylogenetic information. BMC Bioinformatics. 2006;7:160. doi: 10.1186/1471-2105-7-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Moreno-Campuzano S, Janga SC, Perez-Rueda E. Identification and analysis of DNA binding transcription factors in Bacillus subtilis and other Firmicutes--a genomic approach. BMC Genomics. 2006;7:147. doi: 10.1186/1471-2164-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mwangi MM, Siggia ED. Genome wide identification of regulatory motifs in Bacillus subtilis. BMC Bioinformatics. 2003;4:18. doi: 10.1186/1471-2105-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Perez-Rueda E, Collado-Vides J. The repertoire of DNA binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rajewsky N, Socci ND, Zapotocky M, Siggia ED. The evolution of DNA regulatory regions for proteo-gamma bacteria by interspecies comparisons. Genome Res. 2002;12:298–308. doi: 10.1101/gr.207502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Resendis-Antonio O, Freyre-Gonzalez JA, Menchaca-Mendez R, Gutierrez-Rios RM, Martinez-Antonio A, Avila-Sanchez C, Collado-Vides J. Modular analysis of the transcriptional regulatory network of E. coli. Trends Genet. 2005;21:16–20. doi: 10.1016/j.tig.2004.11.010. [DOI] [PubMed] [Google Scholar]
- [58].Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol Rev. 2006;30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- [59].Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, Segura-Salazar J, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- [61].Siddharthan R, Siggia ED, van Nimwegen E. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput Biol. 2005;1:e67. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tan K, McCue LA, Stormo GD. Making connections between novel transcription factors and their DNA motifs. Genome Res. 2005;15:312–320. doi: 10.1101/gr.3069205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- [64].Thieffry D, Huerta AM, Perez-Rueda E, Collado-Vides J. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays. 1998;20:433–440. doi: 10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [65].van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- [66].Wang H, Benham CJ. Promoter prediction and annotation of microbial genomes based on DNA sequence and structural responses to superhelical stress. BMC Bioinformatics. 2006;7:248. doi: 10.1186/1471-2105-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wang T, Stormo GD. Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- [68].Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci U S A. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]