Abstract
Hypoplastic Myelodysplastic Syndrome (h-MDS) comprises 15% of all MDS and has traditionally been difficult to distinguish from aplastic anemia (AA) by current testing. Accurate differentiation is important because treatment and prognosis differ. Since the publication of the 2008 World Health Organization classification of MDS, next-generation DNA sequencing has discovered novel mutations strongly associated with AA and MDS. Recent research supports the utility of identifying these mutations in the diagnosis and management of MDS; however, use of next-generation sequencing is not yet recommended in guidelines and the study is not routinely performed. We present a case where next-generation sequencing performed on a peripheral blood specimen aided the diagnosis and management of a 74-year-old man with h-MDS. This case adds to the growing body of evidence supporting the utility of next-generation DNA sequencing in the evaluation of MDS and h-MDS, particularly when diagnosis remains unclear after standard testing.
Keywords: Anemia, Aplastic/genetics
Introduction
Approximately 15% of all myelodysplastic syndromes (MDS) is the hypoplastic subtype (h-MDS). H-MDS is a clinically distinct entity from MDS because it is characterized by a hypocellular marrow rather than the hyperprolific/dysplastic hematopoietic cells found in MDS. The etiology of h-MDS is not well understood but it is related to autoimmune activity of increased oligoclonal T-cell expansion and suppression of hematopoietic precursors by cytotoxic T-cells.1 Aplastic anemia (AA) is a distinct clinical entity from h-MDS that can result in pancytopenia and severely hypocellular marrow. Morphologically h-MDS and AA are very difficult to differentiate by pathologists. There are criteria for diagnosing and classifying AA, and there are 2008 WHO guidelines for diagnosis and classification of MDS, however there are not clearly defined criteria to diagnose the h-MDS subtype. Although some criteria have been proposed to diagnose h-MDS, they have not been tested extensively.2 As a result, dependence on morphology alone is prone to interobserver variability and error. This is a problem because accurate differentiation is important as prognosis differ between h-MDS and AA. h-MDS can progress to AML more frequently than AA. In addition, the treatments of h-MDS and AA are different. While first line treatment with immunosuppressive therapy (IST) is similar for both diseases, second and third-line therapies are different and the supportive therapies are different. Hematopoietic growth factors such as GM-CSF and erythropoietin can benefit patients with MDS but will not work for patients with AA. In addition, there is evidence suggesting that response rates may be lower for IST's in h-MDS compared to AA.3
A large body of research the last decade has demonstrated cytogenetic abnormalities found in both AA and h-MDS. These cytogenetic abnormalities have been documented and incorporated into the IPSS-R for calculation of risk and survival for MDS, however new cytogenetic abnormalities, and molecular genetic abnormalities have been discovered since these criterions were established. Using targeted gene sequencing and single nucleotide polymorphism (SNP) multiple genes have been discovered that are related to MDS showing different prognostic information independent of the IPSS-R scoring system. The prognostic information of these gene mutations can range from benign to poor overall survival. For example, mutations in MDS driver genes (TP53, ASXL1, DNMT3A, EZH2 and RUNX1) were associated with lower overall survival.4 In addition, some mutations can help classify the subtype of MDS because they are associated with clinical phenotypes such as SF3B1 which is strongly associated with ringed sideroblasts.
Next-generation DNA Sequencing is a molecular genetic tool that uses probes to sequence specific targeted areas. This technology has previously discovered novel AA and MDS associated mutations, but their utility in the clinical setting remains unclear. We present a case where next-generation sequencing performed on a peripheral blood specimen aided the diagnosis and management of a patient with h-MDS.
Case
A 74-year-old man with three-year history of asymptomatic pancytopenia presented with fatigue, anorexia, and 10 lb. weight loss over 6-months. Vital signs and physical exam were unremarkable. CBC revealed progression of pancytopenia compared to previously stable three-year ranges. A new mild normocytic anemia with reticulocyte index indicative of inappropriate marrow response was noted. Blood smear showed rare teardrop cells and elliptocytes without dysplasia. Bone marrow evaluation revealed low normal cellularity at 25% with diminished myeloid maturation, erythroid hyperplasia, and decreased and dysmorphic megakaryocytes. These findings raised concern for aplastic anemia or hypoplastic MDS, but neither were definitively diagnosed. Chromosome analysis and MDS panel (FISH) were normal. Infectious, autoimmune, nutritional, metabolic and other malignant etiologies of pancytopenia were also excluded. Repeat bone marrow showed a progressive decline in cellularity (10%) compared to the study 6-months earlier with otherwise similar cellular characteristics. Repeat MDS panel was normal and additional testing for MDS/AML associated gene mutations was negative (table 1).
Table 1.
Summary of patient's studies to include CBC, Marrow exam, cytogenetic studies, molecular studies.
| REFERENCE | RESULTS | ASSESSMENT |
| Baseline Past 3-Yrs |
• WBC 2.8-4.0, Hgb 13.1-14.3, Plt 100-125 • ANC 1200-2500 |
Pancytopenia |
| Presentation | • CBC WBC 2.7, Hgb 12.4, Plt 82 • ANC 760 , RPI 1.23 • Blood smear - pancytopenia, no blasts or dysmorphic cells • Bone Marrow - 25% cellularity, low myeloid maturation, dysmorphic megakaryocytes. • Karyotype wnl, FISH for MDS wnl |
h-MDS > AA |
| 6-Month Follow-Up |
• WBC 1.9, Hgb 11.9, Plt 68 • ANC 470, RPI 1.22 • Blood smear - unchanged • Bone Marrow - 10% cellularity, low myeloid maturation, erythroid/megakaryocytic dysplasia. • Karyotype wnl, FISH for MDS/AML wnl • Flow cytometry for PNH - non-diagnostic |
AA > h-MDS |
| Next-Gen Sequencing |
• RUNX1 and SF3B1 mutations | h-MDS > AA |
At this point, an evolving AA was favored over h-MDS, but diagnostic criteria were not met for either condition. A peripheral blood specimen was submitted for next-generation DNA sequencing of 42 leukemia associated genes (Table 2). These genes were known to have associations with leukemia, prognostic information, and response to therapy. Pathologic mutations in the RUNX1 and SF3B1 genes were identified which strongly favored h-MDS over AA. Treatment with immunosuppressive therapy was then selected based on likelihood of response in h-MDS patients with HLA-DR15 subtype which the patient had.
Table 2.
List of 42 target genes sequenced by Next Generation Sequencing
| ABL1 | GATA2 | NPM1 |
| ASXL1 | HRAS | NRAS |
| BCOR | IDH1 | PAX5 |
| CBL | IDH2 | PTPN11 |
| CBLB | IKZF1 | RUNX1 |
| CEBPA | IL7R | SF3B1 |
| CREBBP | JAK1 | SRSF2 |
| CSF3R | JAK2 | STAT3 |
| DNMt3A | JAK3 | SUZ12 |
| ETV6 | KDM6A | TET2 |
| E2H2 | Kit | TP53 |
| FBXW7 | KRAS | U2AF1 |
| FLT3 | MPL | WT1 |
| GATA1 | NOTCH1 | ZRSR2 |
Discussion
Modern molecular genetics has dramatically advanced our understanding of MDS. Dozens of MDS-associated genes have been identified since the publication of the 2008 WHO guidelines. However, its use in the clinical setting has been unclear. Currently, diagnosis of MDS hinges on morphologic assessment, which relies on subjectively assessing for dysplasia and quantification of blast forms. This contributes to diagnostic uncertainty and error in morphologically similar processes that cannot be ruled out such as nutrition deficiencies, toxin exposures, other myeloid neoplasms, h-MDS and AA.5, 6 Recent studies identified at least one MDS-related mutation in 50% of suspected MDS cases that failed to meet morphologic criteria. When morphologic assessment cannot diagnose MDS then cytogenetic tests are utilized to help distinguish MDS from other processes that may mimic MDS, however it is well documented that there are cases of MDS without blasts or chromosomal abnormalities on karyotype as presented in our case. When morphologic assessment fails to differentiate a patient with h-MDS versus AA, then molecular genetic techniques should be considered to aid in the diagnosis.
Next-generation sequencing will undoubtedly become a part of routine MDS care and opens the door for possible targeted therapies and agents to avoid during therapy. It has already been demonstrated that MDS patients with TP53 mutations and del(5q) have poor responses to lenalidomide therapy.7 It was also found that patients with TP53 mutations have increased risk of mortality and relapse after Hematopoietic Stem Cell Transplantation.8 Our case is an example of next-generation sequencing as a powerful diagnostic, prognostic, and therapeutic planning tool in the management of a patient with h-MDS. Treatment with Immunosuppressive therapy and supportive therapy with hematopoietic growth factors were initiated based on this patient's MDS mutations.
Conclusion
The differentiation of AA and h-MDS using mainstream histopathologic evaluation and cytogenetic studies alone is difficult, prone to interobserver variability and error. Next generation gene sequencing for MDS/AML associated mutations may significantly improve diagnostic accuracy and should be considered when diagnosis remains unclear after standard evaluation.
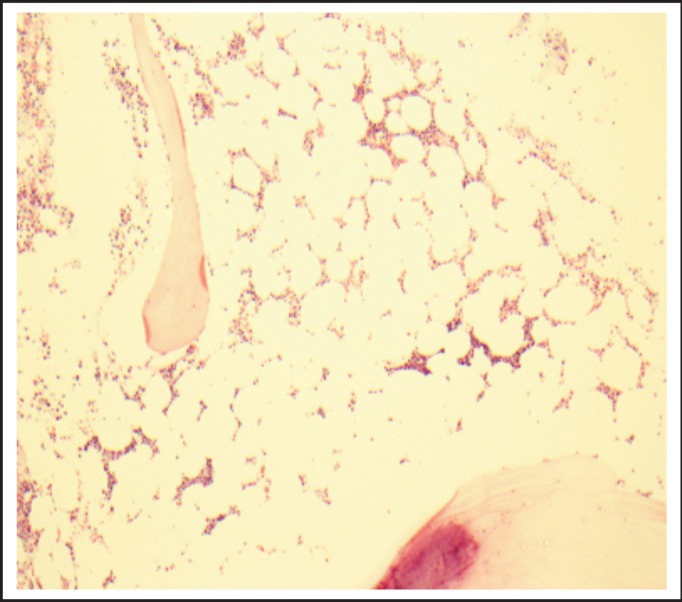
Figure 1.
First Marrow Biopsy showing hypocellular bone marrow mostly composed of myeloid cells.
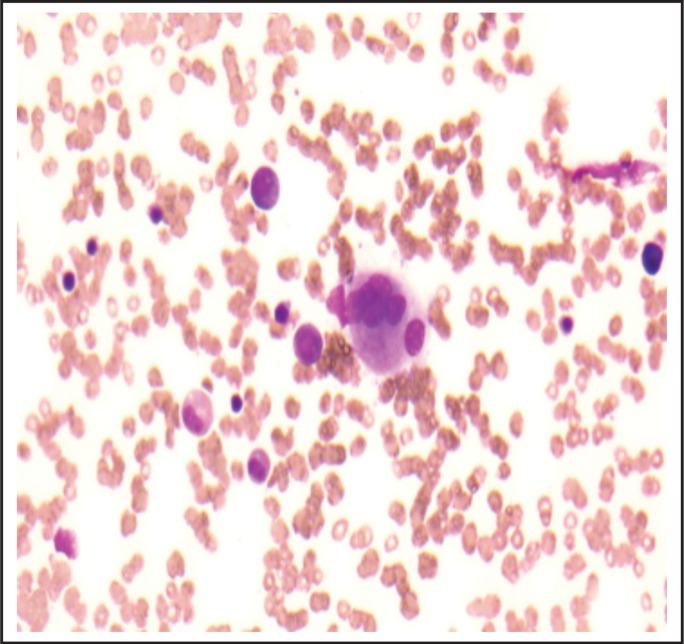
Figure 2.
High powered view of first marrow biopsy showing paucity of megakaryocytes and one multinucleated dysmorphic megakaryocyte slightly favoring MDS over AA.
Disclaimer
The views expressed in this abstract/manuscript are those of the authors and do not reflect the official policy or position of the Department of the Army, Department of Defense, or the US Government.
Conflict of Interest
All of the above authors have no financial disclosures or conflicts of interests related to this study.
References
- 1.Melenhorst JJ, Eniafe R, Follmann D, et al. Molecular and flow cytometric characterization of the CD4 and CD8 T-cell repertoire in patients with myelodysplastic syndrome. Br J Haematol. 2002;119:97–105. doi: 10.1046/j.1365-2141.2002.03802.x. [DOI] [PubMed] [Google Scholar]
- 2.Bennett JM, Orazi A. Diagnostic criteria to distinguish hypocellular acute myeloid leukemia from hypocellular myelodysplastic syndrome and aplastic anemia: recommendations for a standardized approach. Haematologica. 2009;94:264–268. doi: 10.3324/haematol.13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koh Y, Lee HR, Kim HK, Kim I, Park S, Park MH, Kim BK, Yoon SS, Lee DS. Hypoplastic myelodysplastic syndrome (h-MDS) is a distinctive clinical entity with poorer prognosis and frequent karyotypic and FISH abnormalities compared to aplastic anemia (AA) Leukemia Res. Oct;34(10):1344–1350. doi: 10.1016/j.leukres.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Bejar R, Stevenson KE, Caughey BA, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndrome. J Clin Oncol. 2012;30(27):3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mufti GJ, Bennett JM, Goasguen J, et al. International Working Group on Morphology of Myelodysplastic Syndrome. Diagnosis and classification of myelodysplastic syndrome: International Working Group on Morphology of myelodysplastic syndrome (IWGM-MDS) consensus proposals for the definition and enumeration of myeloblasts and ring sideroblasts. Haematologica. 2008 Nov;93(11):1712–1717. doi: 10.3324/haematol.13405. [DOI] [PubMed] [Google Scholar]
- 6.Steensma DP. Dysplasia has a differential diagnosis: distinguishing genuine myelodysplastic syndromes (MDS) from mimics, imitators, copycats and imposters. Curr Hematol Malig Rep. 2012;7(4):310–320. doi: 10.1007/s11899-012-0140-3. [DOI] [PubMed] [Google Scholar]
- 7.Jadersten M, Saft L, Smith A, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29(15):1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- 8.Luger SM, Ringden O, Zhang MJ, et al. Similar outcomes using myeloablative vs reduced-intensity allogeneic transplant preparative regimens for AML or MDS. Bone Marrow Transplant. 2012;26(3):381–389. doi: 10.1038/bmt.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]