Abstract
Aim:
This study aimed to examine pharmacogenomic test results and patient perspectives at an academic cardiovascular medicine clinic.
Patients & methods:
Test results for three common cardiovascular drug–gene tests (warfarin-CYP2C9-VKORC1, clopidogrel-CYP2C19 and simvastatin-SLCO1B1) of 208 patients in the Ohio State University-Coriell Personalized Medicine Collaborative were examined to determine the incidence of potentially actionable test results. A post-hoc, anonymous, patient survey was also conducted.
Results:
Potentially actionable test results for at least one of the three drug–gene tests were determined in 170 (82%) patients. Survey responses (n = 134) suggested that patients generally considered their test results to be important (median of 7.5 on a 10-point scale of importance) and were interested (median of 7.3 on a 10-point scale of interest) in a Clinical Pharmacogenomic Service.
Conclusion:
Attitudes toward pharmacogenomic testing were generally favorable, and potentially actionable test results were not uncommon in this cardiovascular medicine cohort.
Keywords: : actionable test results, cardiovascular drugs, clopidogrel, patient perspectives, personalized medicine, pharmacogenetics, pharmacogenomics, precision medicine, simvastatin, warfarin
Pharmacogenomic (PGx) testing, a cornerstone of personalized medicine [1], can inform drug- and dose-selection strategies to improve drug efficacy and decrease risk of adverse drug effects. The incorporation of PGx into routine clinical care, however, has been limited by several factors including the complexities of interpreting genotype data [2], lack of PGx education and training for prescribers [3], and limited evidence demonstrating improved clinical outcomes [4]. Difficulties related to delays in acquiring test results and limited financial reimbursement from health insurance companies have limited clinical implementation as well [2,5]. Studies examining attitudes toward PGx testing have focused generally on physicians and other health care providers (e.g., nurses, pharmacists and genetic counselors) [2,3,5–13]. Patient perspectives on PGx testing have also been reported [4,10,13–18]. Although attitudes have generally been favorable, concerns persist regarding privacy, financial reimbursement and proper use of test results. Assessment and consideration of the attitudes of patients and providers will remain critical as advancements continue in PGx and personalized medicine. This Short Communication adds to the findings of a previously published randomized controlled trial, the Ohio State University-Coriell Personalized Medicine Collaborative (OSU-CPMC) [19]. For this current analysis, the individual and collective incidences of potentially actionable results for three common cardiovascular drug–gene tests (warfarin-CYP2C9-VKORC1, clopidogrel-CYP2C19 and simvastatin-SLCO1B1) were determined, and the aggregate results from a post-hoc, anonymous, patient survey were evaluated.
Methods
Patients & participation
Two hundred and eight patients diagnosed with hypertension and/or congestive heart failure were enrolled in the OSU-CPMC. Oversight was provided by the Institutional Review Boards (IRBs) at both OSU and the Coriell Institute for Medical Research. Details regarding the design, recruitment, enrollment and implementation of the OSU-CPMC have been reported [19–22]. Briefly, consented participants provided medical, family, lifestyle and medication histories via a series of online surveys and provided saliva samples for genotyping. Genetic risk factors, family history and nongenetic risk influences (e.g., BMI) were utilized to generate personalized risk assessments for seven PGx tests (Supplementary Table 1). Participants were provided their results and were randomized to and received in-person genomic counseling from licensed board-certified genetic counselors in a hospital-based setting at the OSU academic medical center. Personalized reports were provided to participants (via mailed paper copies and via the online OSU-CPMC secure web portal) and to their health care team (via OSU's EPIC© electronic medical record system).
PGx testing & surveys
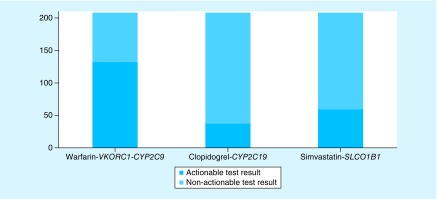
PGx testing, utilizing the Affymetrix Genome-Wide Human SNP 6.0 and DMET Plus genotyping arrays, was performed at a Clinical Laboratory Improvement Amendments (CLIA)-approved laboratory at the Coriell Institute for Medical Research (NJ, USA). The three cardiovascular drug–gene tests (warfarin-CYP2C9-VKORC1, clopidogrel-CYP2C19 and simvastatin-SLCO1B1) were selected for this analysis because these drugs are widely prescribed and because official guidance exists to readily determine whether test results were potentially actionable. Official guidance from the US FDA and/or the Clinical Pharmacogenetics Implementation Consortium (CPIC) was used to evaluate the drug–gene(s) test results, and a test result was determined to be potentially actionable if consideration for a dose adjustment or alternative treatment was recommended in the FDA-approved prescribing information. The ‘dosing recommendations with consideration of genotype’ table for warfarin provides dosing recommendations based on VKORC1 and CYP2C9 status [23]. For this analysis, warfarin-CYP2C9-VKORC1 drug–gene test results were considered potentially actionable when VKORC1/CYP2C9 genotype indicated a nonstandard (5 mg/day) starting dose of warfarin [23]. The lowest and highest ranges of daily warfarin dose recommended in the VKORC1-CYP2C9 table are 0.5–2 mg and 5–7 mg, respectively [23]. Official guidance for interpreting clopidogrel-CYP2C19 drug-genotype tests is provided in the FDA-approved drug label and more detailed information is provided in CPIC's table, ‘antiplatelet therapy recommendations based on CYP2C19 status when considering clopidogrel for acute coronary syndromes patients undergoing percutaneous intervention’ [24]. For this analysis, clopidogrel-CYP2C19 drug-genotype test results were considered potentially actionable when CYP2C19 genotype indicated either poor metabolizer (PM) or intermediate metabolizer (IM) phenotype status [24]. Prescribers are recommended to consider an alternate platelet P2Y12 inhibitor (e.g., prasugrel, ticagrelor) for those patents with genotype-defined IM or PM phenotype status [24]. Official guidance for interpreting simvastatin-SLCO1B1 drug-genotype tests is provided in CPIC's table, ‘recommended dosing of simvastatin based on SLCO1B1 phenotype’ [25]. For this analysis, simvastatin-SLCO1B1 test results were considered potentially actionable when the patient's SLCO1B1 genetic status was consistent with intermediate or high myopathy risk categories [25]. Per CPIC recommendations, prescribers should prescribe a lower dose of simvastatin or consider an alternative statin (e.g., pravastatin or rosuvastatin) in those patients with genotype-determined intermediate or high myopathy risk [25]. Individual (Figure 1) and summative counts of potentially actionable genetic test results for the three drug–gene(s) tests were determined.
Figure 1. . Incidence of actionable and nonactionable genetic test results.
The incidence of test results for three common cardiovascular drug–gene tests (warfarin-VKORC1-CYP2C9, clopidogrel-CYP2C19 and simvastatin-SLCO1B1) were examined in the Ohio State University-Coriell Personalized Medicine Collaborative (OSU-CPMC) chronic disease cohort (n = 208).
Patient perspectives toward PGx testing were queried with a six-question, anonymous, post-hoc survey (Supplementary Figure 1) that was mailed (via regular US postal service) to OSU-CPMC participants (n = 208) in January 2016. Developed by the authors to gain insight on patient attitudes toward PGx testing in this cohort, the post-hoc survey (not formally validated) was approved by the OSU Biomedical Sciences IRB. Responses to survey questions were examined, median and quartile values were determined for ordinal data (Questions 1–4), mean and standard deviation were determined for continuous data (Question 5), and count and percentage were determined for categorical data (Question 6).
Results
As shown in Figure 1, 132 (63%), 37(18%) and 59 (28%) participants had potentially actionable test results for warfarin-CYP2C9-VKORC1, clopidogrel-CYP2C19 and simvastatin-SLCO1B1 drug–gene(s) tests, respectively. Summative analysis determined that 170 (82%) participants had potentially actionable test results for at least one of the three drug–gene tests. Sixty-nine patients (33.2%) had two potentially actionable test results, and 17 patients (8.2%) had three potentially actionable test results.
One-hundred thirty-four (64%) patients of the OSU-CPMC cohort participated in the anonymous, post-hoc survey. Analysis of the survey responses from survey Questions 1–5 is presented in Table 1. Patient survey responses suggested participants generally considered their PGx test results important (median 7.5 on a 10-point scale of importance) and believed their physician had a good understanding of their PGx test results (median 7.1 on a 10-point scale of understanding). In addition, patient survey participants were interested (median 7.3 on a 10-point scale of interest) in the development of a Clinical PGx Service at the OSU academic medical center. The mean out-of-pocket amount that survey participants were willing to pay for utilizing a Clinical PGx Service was 56.30 USD (standard deviation of 81.30 USD). Responses to Question 6 (relative amount participants were willing to pay) indicated that most (76%) were willing to pay an amount equal to or greater than that paid for other specialty health care services.
Table 1. . Responses to post-hoc survey.
| Question | n | Mean | Standard deviation | 1st quartile | Median | 3rd quartile |
|---|---|---|---|---|---|---|
| 1 | 135 | – | – | 6.5 | 7.5 | 8.8 |
| 2 | 134 | – | – | 5.0 | 7.0 | 8.1 |
| 3 | 105 | – | – | 4.9 | 7.1 | 8.4 |
| 4 | 131 | – | – | 5.2 | 7.3 | 8.5 |
| 5 | 75 | 56.30 | 81.30 | – | – | – |
Questions #1–4 were scored using a 10-point visual analog scale. Question #5 was answered in US Dollars.
Discussion & future perspective
The frequencies of VKORC1, CYP2C9, CYP2C19 and SLCO1B1 polymorphisms observed in this analysis were consistent with those reported in other cohorts [25–28]. As polymorphisms in these genes do not affect disease risk (i.e., they affect only the drug metabolism or transport or pharmacologic target), neither enrichment nor deficit of the polymorphisms was expected in this patient cohort. It is important to recognize this was not an enriched cohort and the frequent occurrence of potentially actionable PGx test results suggests routine testing of these three cardiovascular drug–gene(s) tests may prove worthwhile in similar patient populations. Even though only about a third of this patient cohort was prescribed one or more of the target medications at baseline, diagnoses of hypertension and/or heart failure increase the likelihood that the remaining participants also may be prescribed one or more of the target medications in the future.
Although the post-hoc survey was not formally validated and anonymity did not allow for examining whether potential actionability of the test results influenced patient's attitudes toward PGx testing, responses to the survey suggest that patients generally held favorable views of PGx testing. This finding is consistent with previous reports that examined patient perspectives in the USA and Europe [13–17]. Interestingly, most respondents reported that they were willing to pay an out-of-pocket amount equal to or greater than the amount paid for other health care specialty services. The mean value patients indicated they were willing to pay 56.30 USD, but the randomness of the responses (large standard deviation, 81.30 USD) limits the interpretation of this finding. Future queries may yield more meaningful results if answer options containing appropriate cost ranges were provided (e.g., 0–25, 25–50, 50–75, 75–100 USD).
The adoption of routine PGx testing remains limited. Although multifactorial, some barriers (e.g., prescriber buy-in, complexities of interpreting PGx test results, limited familiarity and knowledge of PGx training among health care professionals), might be overcome, at least in part, by further inclusion of genetics, genomics and hands-on PGx training in medical schools and other medical education programs [29,30]. Integrated team approaches (e.g., genetic counselors and pharmacists to provide support to both the patient and the prescriber) to clinical implementation of PGx may help also to move clinical PGx applications forward [31]. Importantly, the findings from recent studies involving two institutions in the Clinical Sequencing Exploratory Research consortium and involving the several institutions of the Electronic Medical Records and Genomics network strongly suggest that improved access to genomic data in electronic medical records systems remains vital to supporting clinical implementation of PGx testing [32–34].
Importantly, the adoption of routine PGx testing has been hindered by substantial discord regarding whether implementation truly improves clinical outcomes. Considerable evidence exists to suggest potential improvements, but incontrovertible evidence (e.g., prospective randomized controlled trials, RTCs) of improvements in specific clinical outcomes is lacking for many potentially important drug(s)–gene(s) tests. For the three PGx tests considered in this article, the neutral perspectives of the FDA recommendations reflect the lack of consensus among and within expert, clinician, research and regulatory groups. Currently, the FDA-approved drug labels provide prescribing recommendations for consideration when PGx test results are known. Advances in clinical implementation of PGx testing are expected to continue, but the rate of advancement remains attenuated largely due to the paucity of RCT data supporting improved clinical outcomes and cost/benefit relationships.
Conclusion
In this analysis of 208 patients with hypertension and/or heart failure in the OSU-CPMC cohort, potentially actionable results among three drug–gene(s) tests (warfarin-CYP2C9-VKORC1, clopidogrel-CYP2C19 and simvastatin-SLCO1B1) were frequent. Specifically, 170 (82%) had a potentially actionable test result for at least one of the three, and 69 (33.2%) and 17 (8.2%) had two and three potentially actionable results, respectively. The majority of patients (64%) responded to a post-hoc survey, and responses suggested that patients’ attitudes toward PGx testing were generally favorable. Future studies regarding the perspectives of patients, prescribers and other health care workers and future studies examining clinical impact (e.g., incidence of actionable test results, effect on prescriber dose and medication selections, impact on specific clinical outcomes) are warranted.
Executive summary.
Potentially actionable results among three drug–gene(s) tests (warfarin-CYP2C9-VKORC1, clopidogrel-CYP2C19 and simvastatin-SLCO1B1) were frequent in this cohort of 208 patients with hypertension and/or heart failure.
Eighty-two percent of the patients had potentially actionable test results for at least one pharmacogenomic (PGx) test when considering the results of all three cardiovascular drug–gene(s) tests.
Most studies on attitudes toward PGx testing have focused on physicians and other health care providers (e.g., nurses, pharmacists and genetic counselors), and some have included also the perspectives of patients.
A post-hoc, anonymous patient survey (response rate of 64%) was conducted to gain insight on attitudes toward PGx testing in a cohort of patients with hypertension and/or heart failure.
Survey responses indicated that patients generally felt that PGx testing was important (median 7.5 on a 10-point scale of importance), had high confidence in their physician's understanding of the PGx test results (median 7.1 on a 10-point scale of understanding), and had a high level of interest in the development of a Clinical PGx Service (median 7.3 on a 10-point scale of interest).
Survey responses indicated that most (76%) were willing to spend an out-of-pocket amount equal to or greater than the amount paid for other specialty care services.
Future studies regarding the perspectives of health care providers and patients toward PGx testing are needed, especially as advancements continue in PGx and personalized medicine.
Prospective randomized controlled trials examining clinical impact (e.g., incidence of actionable test results, effect on prescriber dose and medication selections, impact on specific clinical outcomes) are warranted to better characterize the clinical utility of PGx testing.
Supplementary Material
Footnotes
Financial & competing interests disclosure
The Coriell Personalized Medicine Collaborative was funded by the William G. Rohrer Foundation, the RNR Foundation and a grant from the endowment of the Coriell Institute for Medical Research. This work was supported by the NIH (grant awards R21HG006575, K23GM100372, L32MD006365, L30HL110279, UL1TR001070) and the American Heart Association (grant award 14POST20100054). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the American Heart Association. This work was also supported in part by the Ohio State University Comprehensive Cancer Center. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Alessandrini M, Chaudhry M, Dodgen TM, Pepper MS. Pharmacogenomics and global precision medicine in the context of adverse drug reactions: top 10 opportunities and challenges for the next decade. OMICS. 2016;20(10):593–603. doi: 10.1089/omi.2016.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin. Genet. 2012;82(4):388–394. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A national survey of primary care physicians demonstrating that the majority anticipate pharmacogenetic testing would soon become a valuable tool to inform drug response, but only a minority felt comfortable ordering pharmacogenetic tests.
- 3.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin. Pharmacol. Ther. 2012;91(3):450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]; •• A survey of 10,303 physicians across the USA demonstrating that early and future adopters of pharmacogenetic testing were more likely to have received training in pharmacogenomics, but only 29% of physicians had pharmacogenetic education.
- 4.Shields AE, Najafzadeh M, Schachter AB. Bumps along the translational pathway: anticipating uptake of tailored smoking cessation treatment. Per. Med. 2013;10(8):813–825. doi: 10.2217/pme.13.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haga SB, LaPointe NM. The potential impact of pharmacogenetic testing on medication adherence. Pharmacogenomics J. 2013;13(6):481–483. doi: 10.1038/tpj.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoop JG, Lapid MI, Paulson RM, Roberts LW. Clinical and ethical considerations in pharmacogenetic testing: views of physicians in 3 “early adopting” departments of psychiatry. J. Clin. Psychiatry. 2010;71(6):745–753. doi: 10.4088/JCP.08m04695whi. [DOI] [PubMed] [Google Scholar]
- 7.Overby CL, Erwin AL, Abul-Husn NS, et al. Physician attitudes toward adopting genome-guided prescribing through clinical decision support. J. Pers. Med. 2014;4(1):35–49. doi: 10.3390/jpm4010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghavan S, Vassy JL. Do physicians think genomic medicine will be useful for patient care? Per. Med. 2014;11(4):424–433. doi: 10.2217/pme.14.25. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Review of the literature on physician attitudes regarding the usefulness and limitations of genomic testing.
- 9.Bernhardt BA, Zayac C, Gordon ES, Wawak L, Pyeritz RE, Gollust SE. Incorporating direct-to-consumer genomic information into patient care: attitudes and experiences of primary care physicians. Per. Med. 2012;9(7):683–692. doi: 10.2217/pme.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fargher EA, Eddy C, Newman W, et al. Patients’ and healthcare professionals’ views on pharmacogenetic testing and its future delivery in the NHS. Pharmacogenomics. 2007;8(11):1511–1519. doi: 10.2217/14622416.8.11.1511. [DOI] [PubMed] [Google Scholar]
- 11.Mountcastle-Shah E, Holtzman NA. Primary care physicians’ perceptions of barriers to genetic testing and their willingness to participate in research. Am. J. Med. Genet. 2000;94(5):409–416. doi: 10.1002/1096-8628(20001023)94:5<409::aid-ajmg13>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 12.Zachry WM, Armstrong EP. Health care professionals’ perceptions of the role of pharmacogenomic data. J. Manag. Care Pharm. 2002;8(4):278–284. doi: 10.18553/jmcp.2002.8.4.278. [DOI] [PubMed] [Google Scholar]
- 13.Mai Y, Koromila T, Sagia A, et al. A critical view of the general public's awareness and physicians’ opinion of the trends and potential pitfalls of genetic testing in Greece. Per. Med. 2011;8(5):551–561. doi: 10.2217/pme.11.48. [DOI] [PubMed] [Google Scholar]
- 14.Rogausch A, Prause D, Schallenberg A, Brockmoller J, Himmel W. Patients’ and physicians’ perspectives on pharmacogenetic testing. Pharmacogenomics. 2006;7(1):49–59. doi: 10.2217/14622416.7.1.49. [DOI] [PubMed] [Google Scholar]
- 15.Haga SB, Mills R, Moaddeb J, Allen Lapointe N, Cho A, Ginsburg GS. Patient experiences with pharmacogenetic testing in a primary care setting. Pharmacogenomics. 2016;17(15):1629–1636. doi: 10.2217/pgs-2016-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson ML, Hohmeier KC, Smith CT. Pharmacogenomics testing in a community pharmacy: patient perceptions and willingness-to-pay. Pharmacogenomics. 2017;18(3):227–233. doi: 10.2217/pgs-2016-0161. [DOI] [PubMed] [Google Scholar]
- 17.Olson JE, Rohrer Vitek CR, Bell EJ, et al. Participant-perceived understanding and perspectives on pharmacogenomics: the Mayo Clinic RIGHT protocol (Right Drug, Right Dose, Right Time) Genet. Med. 2017;19(7):819–825. doi: 10.1038/gim.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bielinski SJ, St Sauver JL, Olson JE, et al. Are patients willing to incur out-of-pocket costs for pharmacogenomic testing? Pharmacogenomics. 2017;17(1):1–3. doi: 10.1038/tpj.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweet K, Sturm AC, Schmidlen T, et al. Outcomes of a randomized controlled trial of genomic counseling for patients receiving personalized and actionable complex disease reports. J. Genet. Couns. 2016 doi: 10.1007/s10897-017-0073-z. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller MA, Gordon ES, Stack CB, et al. Coriell personalized medicine collaborative: a prospective study of the utility of personalized medicine. Per. Med. 2010;7(3):301–317. doi: 10.2217/pme.10.13. [DOI] [PubMed] [Google Scholar]
- 21.Sturm AC, Sweet K, Manickam K. Implementation of a clinical research pharmacogenomics program at an academic medical center: role of the genetics healthcare professional. Pharmacogenomics. 2013;14(7):703–706. doi: 10.2217/pgs.13.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sweet K, Gordon ES, Sturm AC, et al. Design and implementation of a randomized controlled trial of genomic counseling for patients with chronic disease. J. Pers. Med. 2014;4(1):1–19. doi: 10.3390/jpm4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bristol-Myers Squibb. Coumadin (warfarin sodium) Prescribing Information. September 2016. packageinserts.bms.com/pi/pi_coumadin.pdf
- 24.Scott SA, Sangkuhl K, Stein CM, et al. The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for cytochrome P450–2C19 (CYP2C19) genotype and clopidogrel therapy: 2013 Update. Clin. Pharm. Ther. 2013;94(3):317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramsey LB, Johnson SG, Caudle KE, et al. The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin. Pharm. Ther. 2014;96(4):423–428. doi: 10.1038/clpt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1(6):e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 28.Jakobsson M, Scholz SW, Scheet P, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 29.Lee KC, Hudmon KS, Ma JD, Kuo GM. Evaluation of a shared pharmacogenomics curriculum for pharmacy students. Pharmacogenomics. 2015;16(4):315–322. doi: 10.2217/pgs.14.181. [DOI] [PubMed] [Google Scholar]
- 30.Luzum JA, Luzum MJ. Physicians’ attitudes toward pharmacogenetic testing before and after pharmacogenetic education. Per. Med. 2016;13(2):119–127. doi: 10.2217/pme.15.57. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Novel report of the positive impact of pharmacogenetic education for practicing physicians.
- 31.Crews KR, Cross SJ, McCormick JN, et al. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am. J. Health Syst. Pharm. 2011;68(2):143–150. doi: 10.2146/ajhp100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freimuth RR, Formea CM, Hoffman JM, et al. Implementing genomic clinical decision support for drug-based precision medicine. CPT Pharmacometrics Syst. Pharmacol. 2017;6(3):153–155. doi: 10.1002/psp4.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen LV, Overby CL, Connolly J, et al. Practical considerations for implementing genomic information resources. Experiences from eMERGE and CSER. Appl. Clin. Inform. 2016;7(3):870–882. doi: 10.4338/ACI-2016-04-RA-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin. Pharmacol. Ther. 2017;102(2):254–264. doi: 10.1002/cpt.707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.