Abstract
Background
Heat stress is one of the most crucial environmental factors, which reduces crop yield worldwide. In plants, the MYB family is one of the largest families of transcription factors (TFs). Although some wheat stress-related MYB TFs have been characterized, their involvement in response to high-temperature stress has not been properly studied.
Results
Six novel heat-induced MYB genes were identified by comparison with previously established de novo transcriptome sequencing data obtained from wheat plants subjected to heat treatment; genomic and complete coding sequences of these genes were isolated. All six TaMYBs were localized in the nucleus of wheat protoplasts. Transactivation assays in yeast revealed that all six proteins acted as transcriptional activators, and the activation domains were attributed to the C-termini of the six wheat MYB proteins. Phylogenetic analysis of the six TaMYBs and R2R3-MYBs from Arabidopsis revealed that all six proteins were in clades that contained stress-related MYB TFs. The expression profiles of TaMYB genes were different in wheat tissues and in response to various abiotic stresses and exogenous abscisic acid treatment. In transgenic Arabidopsis plants carrying TaMYB80 driven by the CaMV 35S promoter, tolerance to heat and drought stresses increased, which could be attributed to the increased levels of cellular abscisic acid.
Conclusions
We identified six heat-induced MYB genes in wheat. We performed comprehensive analyses of the cloned MYB genes and their gene products, including gene structures, subcellular localization, transcriptional activation, phylogenetic relationships, and expression patterns in different wheat tissues and under various abiotic stresses. In particular, we showed that TaMYB80 conferred heat and drought tolerance in transgenic Arabidopsis. These results contribute to our understanding of the functions of heat-induced MYB genes and provide the basis for selecting the best candidates for in-depth functional studies of heat-responsive MYB genes in wheat.
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1158-4) contains supplementary material, which is available to authorized users.
Keywords: Abiotic stress, Heat stress, MYB, Transgenic Arabidopsis, Wheat
Background
Heat stress is one of the most severe environmental factors restricting crop distribution and production [1, 2]. To overcome heat stress, plants adopt various strategies involving the perception of ambient temperature and the regulation of signaling networks [3, 4]. Gene regulation at the transcriptional level is one of the primary control points in biological processes in which transcription factors (TFs) play a key role by activating and/or repressing target genes. In vascular plant genomes, approximately 7% of the coding capacity is attributes to TFs [5], which implies complex gene regulation at the transcriptional level. Previous studies have revealed that several TF families, such as MYB, HSF, DREB, NAC, and bZIP, play important roles in response to stresses in plants [6–9]. For example, many of these TF genes were significantly induced by heat treatment in rice and wheat, as revealed by transcriptome analyses [10, 11].
MYB TFs are found in all eukaryotes. Based on the number of repeats of the MYB domain, the MYB TF family is divided into four major classes: 1R–MYB or MYB-related proteins (usually with a single repeat), R2R3-MYB (two repeats), 3R–MYB (three repeats), and 4R–MYB (four repeats). In plants, the MYB family is one of the largest families of TFs. So far, 204 MYB TFs have been identified in Arabidopsis, whereas there are 218 and 244 MYBs in rice (Oryza sativa) [12] and soybean (Glycine max) [13], respectively. Accumulated evidence has indicated that MYBs mediate stress-signaling pathways. AtMYB41, AtMYB74, AtMYB102, and AtMYB108 play critical roles in drought and salt tolerance [14]. For example, AtMYB41 transcripts are induced by drought and high salinity in Arabidopsis; and overexpression of AtMYB41 in transgenic Arabidopsis increases plant tolerance to drought and high salinity and the expression of several stress-related genes [15, 16]. Another type of MYB TF, AtMYB15, participates in the ABA signaling pathway by targeting ABA biosynthesis (ABA1, ABA2), signaling (ABI3), and responsive genes (AtADH1, RD22, RD29B, AtEM6) [17]. In rice, the expression of OsMYB2 is induced by cold, drought, and salt stresses. Analyses of transgenic rice plants overexpressing or repressing OsMYB2 showed that OsMYB2 increases tolerance to low temperature, salt, and drought treatments [18]. The rice OsMYB48–1 gene plays a positive role in stress tolerance, and overexpression of OsMYB48–1 provides salt and drought tolerance for transgenic rice plants [19]. GbMYB5 was isolated from cotton, and the role of GbMYB5 in response to drought stress has been recently characterized in cotton by VIGS and in tobacco by overexpression [20]. Silencing of GbMYB5 in cotton decreased proline content and antioxidant enzyme activities and compromised the tolerance of cotton plantlets to drought stress, whereas overexpression of GbMYB5 in tobacco increased tolerance to drought stress by decreasing water loss and elevating proline content and antioxidant enzyme activities. In wheat, several MYB genes involved in the response to multiple abiotic stresses have been identified, and transgenic Arabidopsis plants overexpressing TaMYB2A, TaMYB19, TaMYB30-B, or TaMYB33 show increased tolerance to multiple abiotic stresses compared with wild-type (WT) plants [21–24].
In comparison to the rapid progress made in studies on the role of MYBs in response to drought and high salinity stress, the functions of MYBs in response to heat stress are not as well known. So far, only AtMYB68, LeAN2, and OsMYB55 have been proposed to play a role in heat tolerance. In Arabidopsis, vegetative growth of Atmyb68 mutants was significantly inhibited under high temperature compared with WT plants [25]. The overexpression of LeAN2 in tomato caused anthocyanin accumulation and conferred increased tolerance to heat stress by maintaining a low level of reactive oxygen species and high non-enzymatic antioxidant activity [26]. In rice, overexpression analysis demonstrated that OsMYB55 improved high temperature tolerance in transgenic rice plants by increasing expression of the downstream genes OsGS1;2, GAT1, and GAD3, which are involved in amino acid metabolism [27]. Recently, the constitutive expression of rice OsMYB55 in maize induced several stress-related genes and resulted in improved plant growth and performance under high temperature and drought conditions [28]. Before the present study, wheat MYB genes involved in heat tolerance had not been identified.
Bread wheat (Triticum aestivum L.) is one of the most widely cultivated and important food and feed crops globally. High temperature affects the growth and productivity of wheat and reduces yields worldwide. To investigate the role of wheat MYB genes in plant tolerance to heat, we identified six heat-responsive MYB genes based on our previously published transcriptome data [10]. Further, we demonstrated that all six MYB TFs were transcriptional activators and we localized the positions of their activation domains (ADs) and we determined their subcellular locations. The expression levels of all six TaMYBs in different wheat tissues and under different stresses were analyzed by reverse transcription quantitative real-time PCR (RT-qPCR), and we also showed that the tolerance of transgenic Arabidopsis to heat and drought increased when TaMYB80 was constitutively overexpressed. This study provides a useful reference for the selection of heat-responsive MYB genes in wheat for further functional analyses of these genes and their products.
Methods
Plant material and stress treatments
Heat and drought tolerant wheat cv. TAM107, which was released by Texas A&M University in 1984, was used for this study. For stress treatment experiments, seeds were sterilized in a solution of 1% NaClO, and plants were grown hydroponically in a light chamber with 16/8-h photoperiod (photosynthetic photon flux density or PPFD of 100 μmol m−2 s−1) at 22 °C/18 °C (day/night)and 70% relative humidity. Seven-day-old wheat seedlings were treated by submerging the roots in 1/2 Hoagland’s solution with 200 μM ABA, 200 mM NaCl, or 20% PEG 6000 (polyethylene glycol 6000). After treatment, plants were grown under constant (24 h per day) illumination at 22 °C and 70% relative humidity. For heat stress, the seedlings were transferred to another growth chamber at a constant temperature of 40 °C and with the same settings of light and humidity as the previous chamber. For each treatment, five seedlings were separately sampled at 0, 0.5, 1, 2, 4, 6, 12, and 24 h after treatment. For analyses of tissue-specific expression patterns of TaMYBs, roots, stems, flag leaves, 10–20 mm developing spikes, and spikes at flowering were collected from wheat plants grown under the same field conditions. Root samples were quickly rinsed with running tap water to remove soil, then rinsed with deionized water and dried with absorbent paper. All collected samples were quickly placed into liquid nitrogen and stored at −80 °C for RNA extraction.
Extraction of genomic DNA and total RNA and cDNA synthesis
For cloning the genomic sequences of TaMYBs, genomic DNA was isolated from TAM107 young leaves using the CTAB method. To obtain the coding sequences of TaMYBs and to assess the expression patterns of cloned TaMYBs, total RNA was isolated from different samples using Trizol reagent according to the manufacturer’s protocol (TIANGEN, China) and treated with RNase-free DNase I (TaKaRa, Japan) to remove genomic DNA contamination. Two micrograms of total RNA were reverse transcribed using M-MLV reverse transcriptase (TaKaRa, Japan) following the manufacturer’s instructions.
Gene cloning and sequence analysis
Candidate MYB gene sequences were selected from the transcriptome data of the wheat cultivar TAM107 generated in our laboratory [10]. According to the obtained sequences, specific primers for the six TaMYBs were designed using DNAMAN software (www.lynnon.com). DNA and cDNA were used as templates to amplify the DNA sequences and cDNA sequences of wheat MYBs, respectively. PCR was performed using high-fidelity PrimeSTAR Polymerase (TaKaRa, Japan) under the following conditions: 98 °C for 3 min; 34 cycles of 98 °C for 15 s, 58 °C for 30 s, and 72 °C for 90 s; followed by an extension step of 72 °C for 10 min. The PCR amplification products were ligated into the pEASY-blunt Cloning Vector (TransGen, China), and the resulting ligation mixtures were transformed into E. coli Trans1-T1 chemically competent cells (TransGen, China). The positive clones verified by PCR were sequenced.
Protein domains of TaMYBs were predicted using the SMART tool (http://smart.embl-heidelberg.de/), and their gene structures were obtained using the GSDS tool (http://gsds.cbi.pku.edu.cn/). Theoretical molecular weight and isoelectronic point (pI) were calculated using the ProtParam tool (http://web.expasy.org/protparam/). For phylogenetic analysis, 126 Arabidopsis R2R3-MYB protein amino acid sequences were downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov/). The MYB protein amino acid sequences were aligned with the Clustal X version 2.0 program [29], and the phylogenetic tree was constructed using MEGA6 software with the neighbor-joining (NJ) method [30].
Gene expression analysis
TaMYB80 gene expression in transgenic Arabidopsis was analyzed by semi-quantitative reverse transcript (RT)-PCR. Actin2 was used as the internal control. Tissue-specific gene expression patterns of TaMYBs and gene expression patterns of TaMYBs under various stress treatments were determined by RT-qPCR analysis. The RT-qPCR was conducted using an SYBR® Green reaction kit (TaKaRa, Japan) and a Bio-Rad CFX96 real-time system (http://www.bio-rad.com/) with gene-specific primers. The relative expression was calculated using the 2-ΔΔCT method [31]. The wheat β-actin gene was used for standardization of target gene expression. The conditions for semi-quantitative RT-PCR and RT-qPCR have been described previously [32]. All experiments were performed with three biological replicates and three technical replicates. The primers are listed in Additional file 1: Table S1.
Subcellular localization analysis
The coding regions of the six TaMYBs without the stop codon were amplified using gene-specific primers and fused to the N-terminal of the green fluorescent protein (GFP) gene sequence under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. The subcellular localization of each TaMYB was assessed with transient expression after PEG-mediated transfection in wheat mesophyll protoplasts [33, 34] and was monitored 18 h after PEG-calcium transfection using a confocal microscope (FV1000, Olympus).
Transcriptional activation assays in yeast
The transactivation experiment was conducted according to the manual of Yeast Protocols Handbook (Clontech). The full-length and truncated coding sequences of TaMYBs obtained by PCR amplification were fused in-frame to the pGBKT7 vector (Invitrogen), which contained the coding sequence of the GAL4 DNA-binding domain (BD) and Trp reporter gene. Eco RI restriction sites were introduced into the forward and the reverse gene-specific primers (Additional file 1: Table S1). The constructs, positive control pGBKT7–53 and negative control pGBKT7 were respectively transformed into the Saccharomyces cerevisiae AH109 strain (harboring the HIS3 reporter gene) following the manufacturer’s recommended procedures (Clontech). After incubation at 28 °C for two days, positive clones were identified by PCR and plated onto synthetic defined (SD)/−Trp and SD/−Trp/−His media. The AH109 strain could not grow on the SD medium lacking Trp unless a functional TRP1 gene was introduced, and the strain could not grow on the SD/−His medium without activation of a HIS3 gene. Therefore, the growth of yeast cells on SD/−Trp medium showed that the pGBKT7 constructs were successfully transformed into yeast, and the growth of yeast on SD/−Trp/−His medium demonstrated the ability of the MYB proteins to activate the transcription of a reporter gene.
Arabidopsis transformation and stress treatment
The coding sequence of TaMYB80 was amplified from a plasmid by PCR using gene-specific primers containing attB sites. The PCR product was cloned downstream of the CaMV 35S promoter in the plant expression vector pB2GW7 using the Gateway cloning method. The construct was confirmed by sequencing. Arabidopsis (Col-0) transformation by the recombinant plasmid was performed via Agrobacterium tumefaciens-mediated floral-dip method [35]. Five-day-old transgenic seedlings were screened by spraying 0.1% (v/v) Basta, and the putative Basta-resistant plants were confirmed by RT-PCR. Homozygous T3 seeds of transgenic lines were used for phenotypic analyses.
For the germination assay under heat stress treatment, seeds of WT and transgenic lines were sterilized and subjected to 50 °C heat stress treatment for 1 h by submersion of the corresponding microtubes with seeds into a temperature-controlled water bath and then germinated on 1% agar plates. These plates were incubated at ambient temperature (22 °C) for 7 d before photographs were taken and the percentage of germinated seeds was calculated. Seeds were considered germinated when radicles had completely emerged from the seed coat. At least 50 seeds were assessed for each of five independent replicates. For the survival assay, 5-day-old seedlings were heat-treated at 45 °C for 1 h before being returned to 22 °C to grow for another two days after which photographs were taken and the survival rates were calculated. More than 40 plants of each line were used for analysis. For drought sensitivity assays, surface-sterilized WT and transgenic seeds were planted on a Murashige and Skoog medium and placed in a light chamber (22 °C, 16 h photoperiod). Seven-day-old seedlings were transferred to pots with mixed soil (rich soil:vermiculite = 2:1, v/v). Each pot contained four seedlings, which were grown under the same conditions for 21 days with sufficient watering before the application of drought. After two weeks of drought stress treatment by withholding water, the plants were re-watered and allowed to recover from the drought stress conditions.
Determination of the water loss rate in Arabidopsis
To measure water loss, rosette leaves from three-week-old transgenic and WT plants were detached and weighed immediately. The samples were placed on a laboratory bench (humidity 40–50%, 22–24 °C) and weighed at designated time points. The water loss rate was calculated based on the initial fresh weight of samples. Five plants of each transgenic or WT line were used for this measurement, and all tests were repeated three times.
Quantification of ABA levels in Arabidopsis
The ABA content was measured in rosette leaves of three-week-old transgenic and WT plants, with three biological replicates. For each replicate, at least 200 mg of Arabidopsis leaves were homogenized in liquid nitrogen, and 50 mg of the homogenized fresh weight of Arabidopsis leaves were used to measure ABA. ABA extraction and determination were performed as previously described [36].
Statistical analyses
Differences between each transgenic line and WT were analyzed using Student’s t-test in Excel (Microsoft Office 2016). Asterisks indicate significant differences (*P < 0.05, **P < 0.01).
Results
Cloning and sequence analysis of six heat-responsive MYB genes in wheat
We recently performed high-throughput transcriptome sequencing of wheat (TAM107) seedlings grown under normal conditions and subjected to heat stress for 6 h [10]. By scanning the transcriptome data, we identified multiple MYB TF genes up-regulated (fold change >2) by heat stress. Among these genes, we selected six novel heat-responsive MYB genes and isolated their genomic and coding sequences by PCR from wheat cultivar TAM107. According to Bi et al. [37], wheat MYB genes are named after TaMYB78; therefore, these six wheat genes were named TaMYB79–84. Details of the six cloned wheat MYB genes, including name, accession number, chromosomal location, molecular weight, and isoelectric point (pI), are summarized in Table 1. The domain structure analyses revealed that all six MYB sequences were in the R2R3-MYB subfamily, and each contained two adjacent, highly conserved SANT DNA-binding domains, a characteristic domain of MYB TFs and localized in the N-terminal portion of the proteins (Fig. 1a). The gene structure of the six wheat MYB genes was analyzed by pairwise comparison of their full-length cDNA and genomic DNA sequences. TaMYB79, TaMYB80, TaMYB81, TaMYB83, and TaMYB84 genes contained two introns and three exons, and TaMYB82 contained only one intron and two exons (Fig. 1b).
Table 1.
Cloned six wheat MYB genes
| Cloned genes | Accession number | Coding sequence | Chromosomal | Molecular | Isoelectric |
|---|---|---|---|---|---|
| length (bp) | location | weight | point (pI) | ||
| TaMYB79 | KY475607 | 837 | 5AS | 31.7 | 5.47 |
| TaMYB80 | KY475608 | 1050 | 2AS | 38.6 | 5.98 |
| TaMYB81 | KY475609 | 834 | 2AL | 30.6 | 6.16 |
| TaMYB82 | KY475610 | 873 | 4BS | 32.3 | 5.46 |
| TaMYB83 | KY475611 | 795 | 7BS | 29.1 | 5.98 |
| TaMYB84 | KY475612 | 849 | 4AL | 31 | 7.04 |
The chromosomal location of cloned genes was obtained by blasting against the International Wheat Genome Sequencing Consortium (IWGSC) database. The molecular weight and isoelectronic point (pI) were calculated using the ProtParam tool (http://web.expasy.org/protparam/)
Fig. 1.
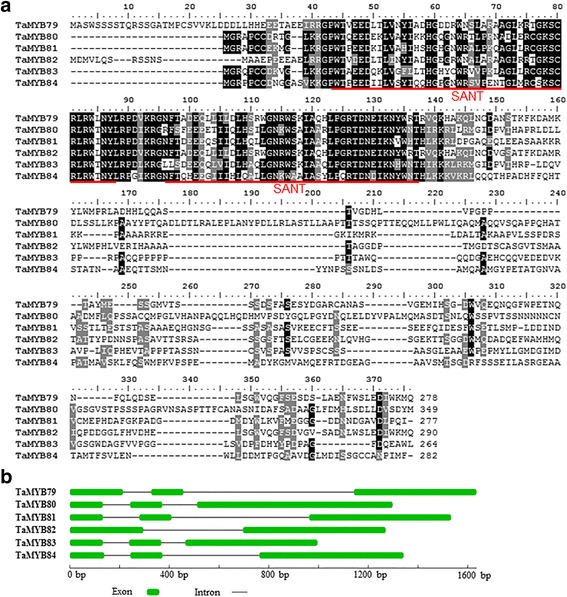
The protein sequence alignment and exon-intron structure of six wheat MYB genes responsive to heat. a The protein sequence alignment of wheat MYB proteins. Each of the six TaMYB proteins contained two SANT domains (underlined in red). b The gene structure of heat-responsive TaMYBs. Green boxes and black lines represent exons and introns, respectively. The scale bars on the line below indicate gene sizes
Nuclear localization of the six TaMYB proteins
To provide evidence of the role of all six selected TaMYB proteins in transcriptional regulation, we determined their subcellular localizations. The full-length sequence of each TaMYB was fused to the green fluorescent protein (GFP) gene sequence in the pJIT163-GFP vector. Each of the resulting constructs, designated TaMYB79-GFP, TaMYB80-GFP, TaMYB81-GFP, TaMYB82-GFP, TaMYB83-GFP, and TaMYB84-GFP, was transiently expressed in wheat protoplasts. As shown in Fig. 2, the control GFP, product of the construct 35S::GFP, was uniformly distributed throughout the mesophyll cell protoplast, whereas TaMYB fusion proteins were primarily localized in the nucleus.
Fig. 2.
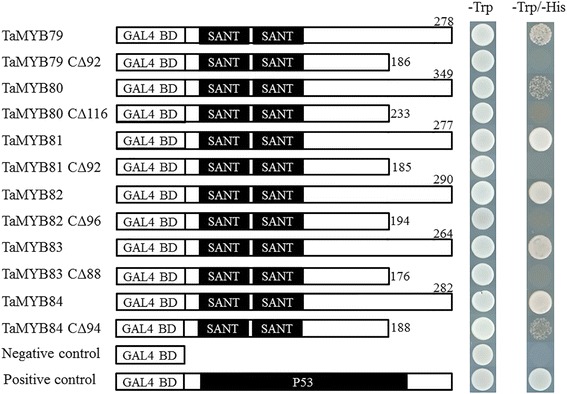
Subcellular localization of TaMYB proteins. The 35S::TaMYB79-GFP, 35S::TaMYB80-GFP, 35S::TaMYB81-GFP, 35S::TaMYB82-GFP, 35S::TaMYB83-GFP and 35S::TaMYB84-GFP constructs and the 35S::GFP control vector were transiently expressed in wheat protoplasts. Results were visualized under confocal microscopy 16 h after transformation. Bars = 20 μm
The C-terminal region of each TaMYB has transcriptional activation activity
To test whether TaMYBs had transcriptional activation activity, full-length coding sequences of TaMYBs were fused in-frame to the GAL4 DNA-binding domain of the pGBKT7 vector, and the fusion constructs were transformed into the yeast strain AH109. As shown in Fig. 3, the yeast transformants containing each of the pGBKT7-TaMYB plasmids or the positive control plasmid grew on an SD/−Trp-His plate, whereas yeasts harboring the pGBKT7-unloaded plasmid (negative control) did not grow.
Fig. 3.
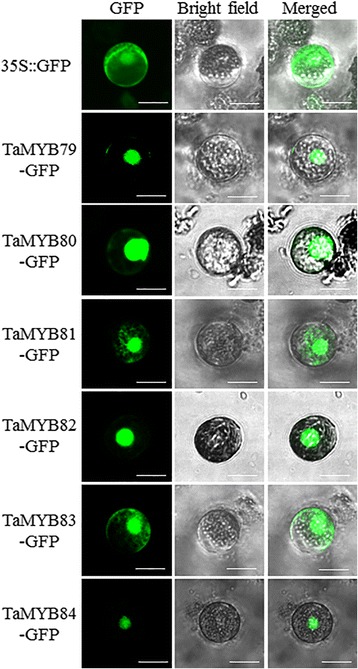
Transcriptional activity assay of the full-length and truncated heat-responsive TaMYB proteins in yeast. The length of each TaMYB protein and respective protein truncation are indicated. Fusion proteins of the GAL4 DNA-binding domain and full-length TaMYBs or truncated TaMYBs with deletion of X C-terminal amino acid residues (CΔX) were expressed in yeast strain AH109. The empty pGBKT7 vector and the pGBKT7-P53 construct were used as negative and positive controls, respectively. Equal amounts of liquid cultures of the transformed yeast cells were dropped onto SD plates containing no tryptophan (Trp) and onto SD plates without Trp and histidine (His)
According to a previous report, the C-terminal region of an MYB TF is required for transcriptional activation activity [38]. To identify the C-terminal region of the investigated TaMYBs responsible for transcriptional activation, CΔX (without the C-terminal X amino acids) regions of TaMYBs were fused to the GAL4 BD (Fig. 3). Yeasts containing CΔX regions of TaMYBs did not grow on an SD/−Trp-His plate, with the exception of TaMYB84, which led to weak growth of yeast.
Phylogenetic analysis of the six wheat MYBs
To further predict the functions of the six R2R3-MYB TFs, the evolutionary relationships of 132 members, including 126 Arabidopsis R2R3-MYB protein sequences and the 6 products of TaMYB genes cloned in this study, were inferred using the neighbor-joining method in MEGA6 (Fig. 4). The TaMYB TFs were clustered with different clades (indicated by red in Fig. 4), representing mostly Arabidopsis proteins with previously determined roles in responses to abiotic stress [39]. The G7 subgroup that contained TaMYB79, TaMYB82 and Arabidopsis proteins AtMYB2, AtMYB24, AtMYB78, AtMYB108, and AtMYB112 is involved in the regulation of anther development and responses to environmental signals [40, 41]. TaMYB80 was grouped into the G15 subgroup with AtMYB41, AtMYB74, and AtMYB102, which is involved in ABA-mediated responses to abiotic stress [14, 16]. TaMYB81 and TaMYB84 were in the G17 and G14 subgroups, respectively, with members that participate in abiotic stress responses in Arabidopsis [17, 42].
Fig. 4.
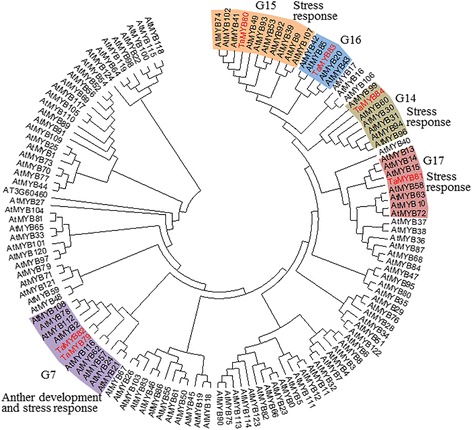
Phylogenetic relationships of the six wheat MYBs and Arabidopsis R2R3-MYB proteins. The complete amino acid sequences of TaMYB79–84 and 126 Arabidopsis R2R3-MYB proteins were aligned by ClustalW. The phylogenetic tree was constructed using MEGA6.0 with the neighbor-joining method. The bootstrap value was 1000 replicates. Each group containing a wheat MYB or wheat MYBs is indicated by a specific color. Wheat MYBs are highlighted in red
Expression of the six TaMYBs in different wheat tissues
Tissue-specific gene expression is generally associated with specific biological functions. Therefore, we characterized the expression patterns of the six TaMYBs in different tissues, including roots, stems, leaves, developing spikes, and spikes at flowering, using RT-qPCR. The six TaMYBs differed greatly in their expression profiles (Fig. 5). TaMYB79 showed a high level of expression in spikes at flowering, but very low levels of expression in other tissues (Fig. 5a). The highest levels of TaMYB80 and TaMYB81 expression were in the roots (Fig. 5b, c). TaMYB82 transcripts were abundant in roots and spikes at flowering, but expression levels of this gene in stems, leaves, and developing spikes were much lower (Fig. 5d). For TaMYB83, the highest level of transcripts was in developing spikes, with a moderate level in leaves (Fig. 5e), whereas TaMYB84 had the highest expression level in leaves, with low transcript levels in other tissues (Fig. 5f).
Fig. 5.
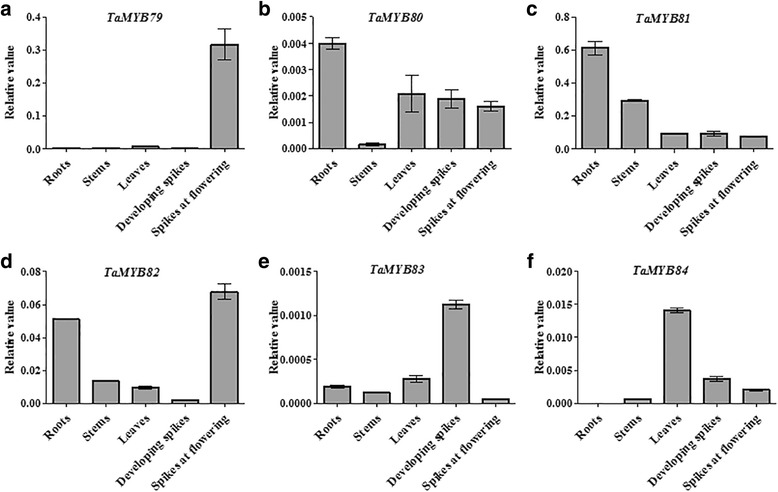
Tissue-specific expression patterns of the TaMYB79 (a), TaMYB80 (b), TaMYB81 (c), TaMYB82 (d), TaMYB83 (e), and TaMYB84 (f). Total RNA was isolated from roots, stems, leaves, developing spikes, and spikes at flowering. The wheat β-actin gene was used as the internal reference. Bars show standard deviations of three replicates
Expression of the six TaMYBs under different abiotic stresses and exogenous ABA treatment
The responses of TaMYBs to heat, drought, high salinity and exogenous ABA treatments were analyzed by RT-qPCR. As shown in Fig. 6, the expression of the six TaMYBs was dramatically up-regulated by heat stress (40 °C). The mRNA of four genes, TaMYB79, TaMYB80, TaMYB81, and TaMYB82, rapidly accumulated after 1 h of heat treatment. The expression of all TaMYBs, except for TaMYB81, was obviously up-regulated in wheat seedlings during the PEG treatment mimicking drought. Under the high salt treatment (200 mM), the transcripts of the TaMYBs were clearly enhanced after 4 h of treatment, with the exception of TaMYB84. In response to exogenous ABA, the expression of TaMYB79, TaMYB80, TaMYB82, and TaMYB83 was significantly up-regulated, whereas TaMYB84 showed weak up-regulation and the number of TaMYB81 transcripts initially decreased and then slightly increased. Overall, each TaMYB responded differently to the various treatments.
Fig. 6.
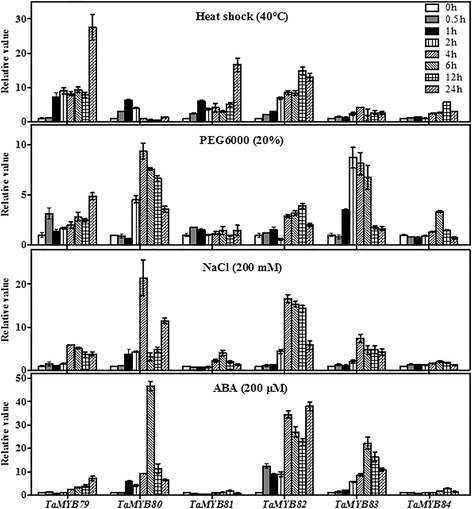
RT-qPCR assessment of the expression of six wheat MYBs under different stress conditions. Total RNAs were extracted from 7-day-old wheat seedlings subjected to either heat shock at 40 °C, 20% PEG 6000, 200 mM NaCl or 200 μM ABA treatment for the indicated time periods. The expression levels of genes at 0 h (not treated) were set to “1”. The wheat β-actin gene was used as the internal reference. Bars represent standard deviations of three biological replicates. X-axes show the time courses of abiotic stress treatments for each gene. Y-axes indicate the scales of the relative expression levels
TaMYB80 confers heat and drought tolerance in transgenic Arabidopsis
As described above, the TaMYB80 gene was strongly induced by multiple abiotic stresses. To confirm the functions of TaMYB80 in the abiotic stress response, we generated transgenic Arabidopsis plants overexpressing TaMYB80 driven by the CaMV 35S promoter (Fig. 7a). Three T3 homozygous transgenic lines (L1, L2, and L3) containing a single insertion of the TaMYB80 were selected for phenotypic analyses. The transcript levels of TaMYB80 expression in these lines were determined by RT-PCR (Fig. 7b).
Fig. 7.
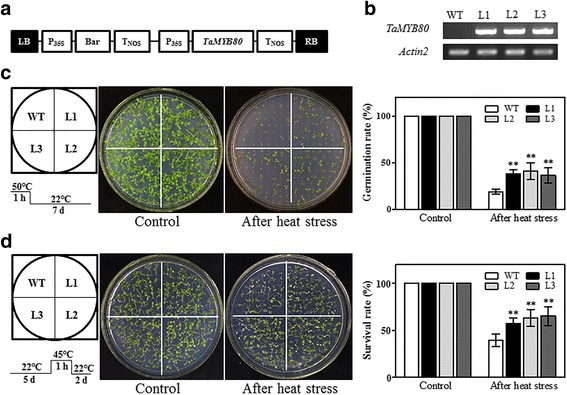
Overexpression of TaMYB80 confers heat tolerance to transgenic Arabidopsis plants. a Schematic diagram of the TaMYB80 overexpression construct driven by the 35S promoter. b Analysis of TaMYB80 transcript levels in WT Arabidopsis and transgenic lines (L1, L2, and L3). Total RNA was isolated from 1-week-old plants. Actin2 was used as the control gene. c Germination rates (%) of TaMYB80-overexpression plants after treatment with heat stress. Seeds of WT and transgenic lines were sterilized and subjected to 50 °C treatment for 1 h and then germinated on 1% agar plates. The plates were placed at ambient conditions (22 °C) for 7 d before photographs were taken. At least 50 seeds were assessed per replicate for five independent replicates. d Survival rates (%) of WT and transgenic seedlings after treatment with heat stress. Five-day-old seedlings were heat-treated at 45 °C for 1 h before returning to 22 °C to grow for two days, and then photographs were taken and the survival rates were calculated. More than 40 plants of each line were used. Bars represent standard deviations
The germination rate was not significantly different between the WT and the transgenic lines under normal temperature (22 °C) conditions. However, all three transgenic lines exhibited significantly higher germination percentages than the WT plants when seeds were germinated after 1 h of exposure at 50 °C on 1% agar plates (Fig. 7c). For the survival assay under heat stress, 5-day-old seedlings of WT and transgenic lines were heat-treated at 45 °C for 1 h followed by 2 days of recovery under normal conditions (22 °C). The survival rates of all transgenic lines (57%, 63% and 65%) were significantly higher than that of the WT plants (40%; Fig. 7d).
To validate that the TaMYB80 gene was also involved in drought stress tolerance, 14-day-old WT and transgenic plants were grown in soil and subjected to a water-withholding treatment for 14 days, followed by re-watering for 3 days. Under normal growth conditions (22 °C), the transgenic lines showed no obvious difference in performance compared with the WT plants (Fig. 8a). However, after withholding water for 14 days, the WT leaves were severely dehydrated, whereas the transgenic plants showed much less wilting (Fig. 8a). After the 3-day recovery, only 30% of the WT plants survived, whereas more than 70% of the transgenic plants survived in lines L1, L2, and L3 (Fig. 8b). Additionally, under drought stress, the transgenic plants showed lower rates of water loss than the WT plants (Fig. 8c).
Fig. 8.
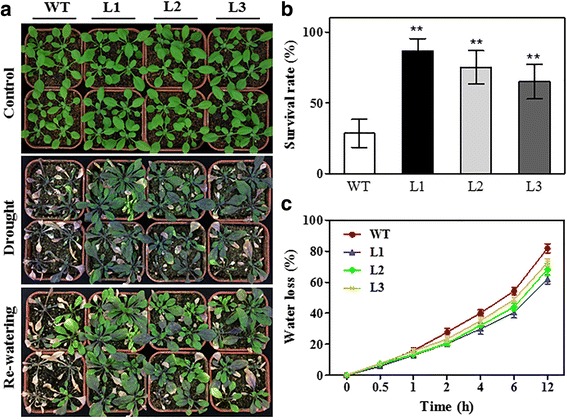
Phenotypic differences between transgenic lines overexpressing TaMYB80 and WT plants under drought treatment. a Photographs showing plants before and after dehydration stress. Four-week-old seedlings of WT and three transgenic lines were subjected to drought stress treatment by withholding irrigation for 14 days, followed by resumed watering and plant growth for another two days. Three independent experiments were conducted. b Survival rates of WT and three transgenic lines after drought treatment. Bars represent standard deviations (n = 60) of three biological replicates. c Water loss from detached leaves of 4-week-old plants measured at room temperature. Error bars represent the standard deviations of three replicates with 5 seedlings per replicate
Overexpression of TaMYB80 increases cellular ABA content and expression of ABA-dependent stress-related genes
As shown above, overexpression of the TaMYB80 gene in Arabidopsis increased drought tolerance and reduced water loss. ABA plays a role in decreasing water loss via the induction of stomatal closure and some other mechanisms [43, 44]. Therefore, we hypothesized that the transgenic plants might have elevated amounts of endogenous ABA in the absence of stress. To test this hypothesis, we measured the content of endogenous ABA using a UPLC–MS/MS system. The results of three independent experiments showed that levels of ABA in transgenic plants (L1, L2, L3) were increased compared with that of WT plants (Fig. 9a). These data clearly demonstrate that the overexpression of the TaMYB80 gene increases cellular ABA levels.
Fig. 9.
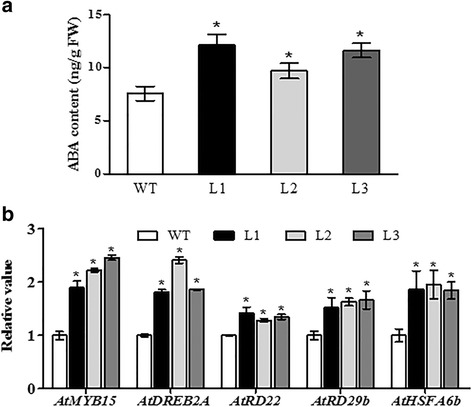
Endogenous ABA content and expression of stress-related genes of WT and transgenic plants under normal conditions. a ABA levels were determined in 21-day-old seedlings. FW, Fresh weight. b Expression of ABA-dependent stress-related genes in WT and TaMYB80 transgenic lines. The total RNA was extracted from 21-day-old seedlings. The Arabidopsis Actin2 gene was used as the internal control. The gene expression in WT was regarded as standard, and transgenic lines (L1, L2, and, L3) values were compared with it. Error bars represent the standard deviations of three biological replicates
To investigate the molecular basis of the stress tolerance provided by the TaMYB80 gene for transgenic Arabidopsis, we analyzed the expression of several stress-related genes in three independent transgenic lines (L1, L2, and L3) and WT plants via RT-qPCR. We found that the expression levels of several genes positively regulated by ABA, including AtMYB15, AtHSFA6b, AtDREB2A, AtRD22, and AtRD29b, were upregulated in the transgenic lines (Fig. 9b).
Discussion
High temperature negatively affects plant growth and crop productivity. Cloning and functional characterization of heat responsive genes in crops is the first step in providing valuable information for crop thermotolerance breeding. MYB transcription factors constitute one of the largest transcription factor families, and some members of the family play crucial roles in the regulation of stress responses, particularly in response to high temperatures [26, 28]. However, data on heat-responsive MYB genes in wheat are limited [10]. In this study, six putative heat-induced MYB genes were cloned from a heat and drought tolerant wheat cultivar TAM107. The putative functions of the six TaMYBs were investigated by phylogenetic analysis and expression profile analyses in different wheat tissues and under multiple stress treatments. We also showed that constitutive overexpression of TaMYB80 in transgenic Arabidopsis improved plant tolerance to high temperatures and drought.
Transactivation assays in yeast revealed that the tested TaMYBs all had transcriptional activation activity. In particular, TaMYB81, TaMYB82, TaMYB83, and TaMYB84 showed strong transcriptional activation, whereas weak transcriptional activation was observed for TaMYB79 and TaMYB80 (Fig. 3). Additionally, yeasts containing CΔX regions of individual TaMYBs (except TaMYB84) could not grow on an SD/−Trp-His plate, suggesting that the excised C-terminal region of each tested TaMYB (except TaMYB84) was essential for the transcriptional activation of a gene. In the case of truncated TaMYB84, clear but weak growth was observed (Fig. 3), and so a significant part of the AD was in the C-terminal region.
As previously reported, 126 Arabidopsis R2R3-MYB TFs are divided into 17 groups based on complete amino acid sequence, with the group often comprising TFs with similar functions [39]. Five wheat MYB proteins characterized in this study were clustered into four Arabidopsis groups that contained products of stress-responsive genes. The remaining TaMYB83 was clustered in group G16, which is not a stress response group but shares a close evolutionary relationship with AtMYB20, which is involved in stress response (Fig. 4). Previous studies have indicated that the overexpression of AtMYB20 in transgenic Arabidopsis plants increased salt tolerance [45], implying that TaMYB83 may also play a role in stress response of wheat. Overall, the results suggested that the six TaMYB proteins are involved in wheat responses to abiotic stresses.
Gene expression patterns are indications of gene functions [46]. In this study, tissue- and developmental stage-specific expression of TaMYB genes suggested diverse functions of their products. Preferred or specific expression of TaMYB79 in spikes at flowering, TaMYB81 in roots, TaMYB83 in developing spikes, and TaMYB84 in leaves indicated that these MYBs might be involved in the development of different organs and tissues. Likewise, expression analysis of the six TaMYBs in leaves of seedlings under different stresses showed that each of the tested TaMYBs differentially responded to 2–4 abiotic stresses and treatments, indicating the involvement of these MYB TFs in the physiological processes associated with multiple stress responses in wheat. According to previous studies, many MYB TFs respond to abiotic stresses via the ABA-mediated signaling pathway. For example, AtMYB2 regulates the ABA-dependent expression of salt- and dehydration-responsive genes, and its interaction with a calmodulin increases salt tolerance [47]. Another MYB TF, AtMYB96, mediates ABA signals via RD22 in plant resistance responses to water deficit by reducing stomatal openings [48]. By contrast, AtMYB20, a negative regulator of ABA signaling, increased plant salt tolerance by down-regulating the expression of PP2C genes [45]. Gene expression analysis showed that TaMYB79, TaMYB80, TaMYB82, and TaMYB83 were strongly induced under ABA treatment (Fig. 4), suggesting that each of these genes may be part of an ABA-dependent signaling transduction pathway in response to abiotic stress. Exogenous ABA levels did not affect the expression patterns of the other two heat-inducible TaMYB genes (TaMYB81 and TaMYB84).
Ectopic expression of wheat genes in the model plant Arabidopsis is a fast and effective approach to investigate gene functions [49]. In the present study, transgenic Arabidopsis plants overexpressing TaMYB80 were generated; these plants showed increased tolerance to heat and drought stresses and had higher ABA content (Figs. 7, 8 and 9a). As shown in Fig. 8, plant growth and survival increased significantly for the transgenic plants during drought treatment. All WT plants showed severe wilting, and some even died, whereas the rosette leaves of some of the transgenic plants remained green. Abogadallah et al. found that the overexpression of HARDY improved the growth of Trifolium alexandrinum under drought stress, which may have resulted from higher rates of photosynthesis [50]. Thus, the rates of photosynthesis in TaMYB80 transgenic plants could have been higher than those in WT plants, resulting in the increased level of drought tolerance. Further studies are required to test this hypothesis. ABA has an important role in the response of plants to water deficit by regulating stomatal closure and the expression of stress-related genes [51]. For example, in transgenic plants overexpressing AtLOS5, AtHSPR, or OsASR5 genes, stomatal apertures decrease via increased ABA content in response to drought stress [44, 52, 53]. Previously, ABA-dependent stomatal closure was thought to be disadvantageous for plant acclimation to heat stress because closure could prevent leaf cooling via transpiration [54]; however, increases in endogenous ABA level and ABA-dependent stomatal closure have been reported to participate in protection from heat damage [54–57]. For example, a recent study demonstrated that the RING finger ubiquitin E3 ligase OsHTAS positively regulated ABA biosynthesis and induced stomatal closure during heat stress, consequently increasing the heat tolerance of rice [57]. Additionally, temporal-spatial interaction between ABA and reactive oxygen species signals plays a key role in the regulation of systemic acquired acclimation of plants to heat stress [58]. Thus, we hypothesized that the increased tolerance of TaMYB80 overexpression plants to heat and drought stresses was most likely due to the increased level of endogenous ABA.
Further study revealed that increases in endogenous ABA levels modified the expression of several ABA-dependent stress-related genes, including AtMYB15, AtHSFA6b, AtDREB2A, AtRD22, and AtRD29b (Fig. 9b). AtMYB15 increases the expression of genes involved in ABA biosynthesis and when overexpressed in transgenic plants, improves salt and drought tolerance [17]. HSFA6b plays a primary role in an ABA-mediated signaling network related to heat stress responses [59], and DREB2A is a crucial regulatory element involved in drought stress response [60]. RD22 and RD29b encode low molecular hydrophilic proteins and have a cellular protection function [17]. In this study, the expression levels of these genes were significantly higher in the transgenic plants than in the WT plants. This result suggested that the overexpression of TaMYB80 enhanced the expression levels of ABA-dependent stress-related genes, thereby conferring heat and drought tolerance in Arabidopsis. Future studies on the possible roles of TaMYB80 in ABA biosynthesis and/or ABA signaling pathways would be of interest.
Conclusions
In this study, we identified and cloned six heat-induced MYB genes. All the genes were localized in the nucleus and were demonstrated to function as transcriptional activators. The ADs of all six MYB TFs were localized at the C-terminus of the molecules. Phylogenetic analysis assigned all six proteins to clades containing stress-related MYB TFs. Different expression profiles of MYB genes were revealed in various wheat tissues and under multiple stress treatments. We demonstrated that the overexpression of TaMYB80 in transgenic Arabidopsis increased plant tolerance to high temperatures and drought. Our study provides useful information for breeders and genetic engineers working to improve the heat and drought tolerance of wheat.
Acknowledgements
We thank Dr. Sergiy Lopato at the University of Adelaide for critical reading of this manuscript. We are grateful to the editor and reviewers for their valuable comments and constructive suggestions on the manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (31571747) and the National Transgenic Research Project (2016ZX08002–002). The funding bodies did not participate in study design, collection, analysis or interpretation of data, or writing the manuscript.
Availability of data and materials
The datasets supporting the results of this article are included within the article and its additional files. Sequence data used in this manuscript can be found in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under the following accession numbers: KY475607 (TaMYB79), KY475608 (TaMYB80), KY475609 (TaMYB81), KY475610 (TaMYB82), KY475611 (TaMYB83), KY475612 (TaMYB84). The phylogenetic tree in Fig. 4 and related sequences were deposited in TreeBASE (https://treebase.org/treebase-web/home.html) under the following URL: http://purl.org/phylo/treebase/phylows/study/TB2:S21690.
Abbreviations
- ABA
Abscisic acid
- AD
Activation domain
- BD
Binding domain
- CaMV
Cauliflower mosaic virus
- DREB
Dehydration-responsive element-binding protein
- GFP
Green fluorescent protein
- HSF
Heat shock factor
- PEG
Polyethylene-glycol
- PP2C
Protein phosphatase 2C
- RD
Responsive to dehydration
- RT-qPCR
Reverse transcription quantitative real-time polymerase chain reaction
- SD
Synthetic defined
- TFs
Transcription factors
- UPLC–MS/MS
Ultra-performance liquid chromatography tandem mass spectrometry
- WT
Wild-type
Additional file
Primer sequences used in this study (DOCX 24 kb)
Authors’ contributions
HP, MX and QS conceived the research and revised the manuscript. YZ and XT carried out most of the experiments and drafted the manuscript. FW and LZ grew the plant samples, collected all the phenotypic data, and contributed to revise the manuscript with constructive discussion. ZH, YY, and ZN helped design the research work, analyze the data, and revise the manuscript. HP finalized the manuscript. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1158-4) contains supplementary material, which is available to authorized users.
Contributor Information
Yue Zhao, Email: zwb1013@126.com.
Xuejun Tian, Email: tianxuejun1@163.com.
Fei Wang, Email: wangfeihh@163.com.
Liyuan Zhang, Email: zhangliyuan1226@163.com.
Mingming Xin, Email: mingmingxin@cau.edu.cn.
Zhaorong Hu, Email: hzr803@163.com.
Yingyin Yao, Email: yingyin@cau.edu.cn.
Zhongfu Ni, Email: wheat3392@cau.edu.cn.
Qixin Sun, Email: qxsun@cau.edu.cn.
Huiru Peng, Email: penghuiru@cau.edu.cn.
References
- 1.Lobell DB, Schlenker W, Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333(6042):616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- 2.Lesk C, Rowhani P, Ramankutty N. Influence of extreme weather disasters on global crop production. Nature. 2016;529(7584):84–87. doi: 10.1038/nature16467. [DOI] [PubMed] [Google Scholar]
- 3.Walbot V. How plants cope with temperature stress. BMC Biol. 2011;9(1):79. doi: 10.1186/1741-7007-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu A, Ding Y, Jiang Q, Zhu C. Molecular mechanisms of the plant heat stress response. Biochem Bioph Res Co. 2013;432(2):203–207. doi: 10.1016/j.bbrc.2013.01.104. [DOI] [PubMed] [Google Scholar]
- 5.Rushton PJ, Bokowiec MT, Laudeman TW, Brannock JF, Chen X, Timko MPTOBFAC. The database of tobacco transcription factors. BMC Bioinformatics. 2008;9:53. doi: 10.1186/1471-2105-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldoni E, Genga A, Cominelli E, Plant MYB. Transcription factors: their role in drought response mechanisms. Int J Mol Sci. 2015;16(7):15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25(12):1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 8.Shao H, Wang H, Tang XNAC. Transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci. 2015;6:902. doi: 10.3389/fpls.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee A, Roychoudhury A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma. 2017;254(1):3–16. doi: 10.1007/s00709-015-0920-4. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, et al. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.) BMC Plant Biol. 2015;15:152. doi: 10.1186/s12870-015-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Rerksiri W, Liu A, Zhou X, Xiong H, Xiang J, et al. Transcriptome profile reveals heat response mechanism at molecular and metabolic levels in rice flag leaf. Gene. 2013;530(2):185–192. doi: 10.1016/j.gene.2013.08.048. [DOI] [PubMed] [Google Scholar]
- 12.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 13.Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu R, Wang Y, Zheng H, Lu W, Wu C, Huang J, et al. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J Exp Bot. 2015;66(19):5997–6008. doi: 10.1093/jxb/erv312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 2008;53(1):53–64. doi: 10.1111/j.1365-313X.2007.03310.x. [DOI] [PubMed] [Google Scholar]
- 16.Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, et al. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 2009;149(4):1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding Z, Li S, An X, Liu X, Qin H, Wang D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics. 2009;36(1):17–29. doi: 10.1016/S1673-8527(09)60003-5. [DOI] [PubMed] [Google Scholar]
- 18.Yang A, Dai X, Zhang WHA. R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63(7):2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, et al. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS One. 2014;9(3):e92913. doi: 10.1371/journal.pone.0092913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T, Li W, Hu X, Guo J, Liu A, Zhang BA. Cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015;56(5):917–929. doi: 10.1093/pcp/pcv019. [DOI] [PubMed] [Google Scholar]
- 21.Qin Y, Wang M, Tian Y, He W, Han L, Xia G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol Biol Rep. 2012;39(6):7183–7192. doi: 10.1007/s11033-012-1550-y. [DOI] [PubMed] [Google Scholar]
- 22.Mao X, Jia D, Li A, Zhang H, Tian S, Zhang X, et al. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct Integr Genomics. 2011;11(3):445–465. doi: 10.1007/s10142-011-0218-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Liu G, Zhao G, Xia C, Jia J, Liu X, et al. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol. 2014;55(10):1802–1812. doi: 10.1093/pcp/pcu109. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Zhao G, Xia C, Jia J, Liu X, Kong XA. Wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J Exp Bot. 2012;63(16):5873–5885. doi: 10.1093/jxb/ers237. [DOI] [PubMed] [Google Scholar]
- 25.Feng C, Andreasson E, Maslak A, Mock HP, Mattsson O, Mundy J. Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 2004;167(5):1099–1107. doi: 10.1016/j.plantsci.2004.06.014. [DOI] [Google Scholar]
- 26.Meng X, Wang JR, Wang GD, Liang XQ, Li XD, Meng QW. An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J Plant Physiol. 2015;175:1–8. doi: 10.1016/j.jplph.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 27.El-Kereamy A, Bi YM, Ranathunge K, Beatty PH, Good AG, Rothstein SJ. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One. 2012;7(12):e52030. doi: 10.1371/journal.pone.0052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casaretto JA, El-Kereamy A, Zeng B, Stiegelmeyer SM, Chen X, Bi YM, et al. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics. 2016;17:312. doi: 10.1186/s12864-016-2659-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30(12):2725–2729. [DOI] [PMC free article] [PubMed]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Tian X, Li Y, Zhang L, Guan P, Kou X, et al. Molecular and functional characterization of wheat ARGOS genes influencing plant growth and stress tolerance. Front Plant Sci. 2017;8:170. doi: 10.3389/fpls.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 34.Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9(10):2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]
- 35.Bechtold N, Pelletier G. Planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82(10):259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Wang C, Liu X, Song J, Li H, Sui Z, et al. Up-regulating the abscisic acid inactivation gene ZmABA8ox1b contributes to seed germination heterosis by promoting cell expansion. J Exp Bot. 2016;67(9):2889–2900. doi: 10.1093/jxb/erw131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi H, Luang S, Li Y, Bazanova N, Morran S, Song Z, et al. Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J Exp Bot. 2016;67(18):5363–5380. doi: 10.1093/jxb/erw298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66(1):94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen N, Yang Q, Pan L, Chi X, Chen M, Hu D, et al. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.) Gene. 2014;533(1):332–345. doi: 10.1016/j.gene.2013.08.092. [DOI] [PubMed] [Google Scholar]
- 40.Yu L, Chen H, Guan Q, Ma X, Zheng X, Zou C, et al. AtMYB2 transcription factor can interact with the CMO promoter and regulate its downstream gene expression. Biotechnol Lett. 2012;34(9):1749–1755. doi: 10.1007/s10529-012-0961-0. [DOI] [PubMed] [Google Scholar]
- 41.Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 2015;169(3):1862–1880. doi: 10.1104/pp.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HG, Choi YR, Seo PJ. Increased STM expression is associated with drought tolerance in Arabidopsis. J Plant Physiol. 2016;201:79–84. doi: 10.1016/j.jplph.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Sirichandra C, Wasilewska A, Vlad F, Valon C, Leung J. The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J Exp Bot. 2009;60(5):1439–1463. doi: 10.1093/jxb/ern340. [DOI] [PubMed] [Google Scholar]
- 44.Yue Y, Zhang M, Zhang J, Tian X, Duan L, Li Z. Overexpression of the AtLOS5 gene increased abscisic acid level and drought tolerance in transgenic cotton. J Exp Bot. 2012;63(10):3741–3748. doi: 10.1093/jxb/ers069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, et al. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013;587(12):1773–1778. doi: 10.1016/j.febslet.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 46.Guo M, JP L, Zhai YF, Chai WG, Gong ZH, Genome-wide LMH. Analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.) BMC Plant Biol. 2015;15:151. doi: 10.1186/s12870-015-0512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15(1):63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo PJ, Xiang F, Qiao M, Park J, Lee YN, Kim S, et al. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151(1):275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koornneef M, Meinke D. The development of Arabidopsis as a model plant. Plant J. 2010;61(6):909–921. doi: 10.1111/j.1365-313X.2009.04086.x. [DOI] [PubMed] [Google Scholar]
- 50.Abogadallah GM, Nada RM, Malinowski R, Quick P. Overexpression of HARDY, an AP2/ERF gene from Arabidopsis, improves drought and salt tolerance by reducing transpiration and sodium uptake in transgenic Trifolium alexandrinum L. Planta. 2011;233(6):1265–1276. doi: 10.1007/s00425-011-1382-3. [DOI] [PubMed] [Google Scholar]
- 51.Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol. 2007;10(3):296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 52.Yang T, Zhang L, Hao H, Zhang P, Zhu H, Cheng W, Wang Y, et al. Nuclear-localized AtHSPR links abscisic acid-dependent salt tolerance and antioxidant defense in Arabidopsis. Plant J. 2015;84(6):1274–1294. doi: 10.1111/tpj.13080. [DOI] [PubMed] [Google Scholar]
- 53.Li J, Li Y, Yin Z, Jiang J, Zhang M, Guo X, et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol J. 2017;15(2):183–196. doi: 10.1111/pbi.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki N, Bassil E, Hamilton JS, Inupakutika MA, Zandalinas SI, Tripathy D, et al. ABA is required for plant acclimation to a combination of salt and heat stress. PLoS One. 2016;11(1):e147625. doi: 10.1371/journal.pone.0147625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larkindale J, Knight MR. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002;128(2):682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zandalinas SI, Balfagon D, Arbona V, Gomez-Cadenas A, Inupakutika MA, Mittler RABA. Is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J Exp Bot. 2016;67(18):5381–5390. doi: 10.1093/jxb/erw299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Zhang C, Wei C, Liu X, Wang M, Yu F, et al. The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol. 2016;170(1):429–443. doi: 10.1104/pp.15.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki N, Miller G, Salazar C, Mondal HA, Shulaev E, Cortes DF, et al. Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell. 2013;25(9):3553–3569. doi: 10.1105/tpc.113.114595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang YC, Niu CY, Yang CR, Jinn TL. The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016;172(2):1182–1199. doi: 10.1104/pp.16.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiang L, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10(8):1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the results of this article are included within the article and its additional files. Sequence data used in this manuscript can be found in the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) under the following accession numbers: KY475607 (TaMYB79), KY475608 (TaMYB80), KY475609 (TaMYB81), KY475610 (TaMYB82), KY475611 (TaMYB83), KY475612 (TaMYB84). The phylogenetic tree in Fig. 4 and related sequences were deposited in TreeBASE (https://treebase.org/treebase-web/home.html) under the following URL: http://purl.org/phylo/treebase/phylows/study/TB2:S21690.


