Abstract
A(H3N2) influenza viruses have circulated in humans since 1968, and antigenic drift of the hemagglutinin (HA) protein continues to be a driving force that allows the virus to escape the human immune response. Since the major antigenic sites of the HA overlap into the receptor binding site (RBS) of the molecule, the virus constantly struggles to effectively adapt to host immune responses, without compromising its functionality. Here, we have structurally assessed the evolution of the A(H3N2) virus HA RBS, using an established recombinant expression system. Glycan binding specificities of nineteen A(H3N2) influenza virus HAs, each a component of the seasonal influenza vaccine between 1968 and 2012, were analyzed. Results suggest that while its receptor-binding site has evolved from one that can bind a broad range of human receptor analogs to one with a more restricted binding profile for longer glycans, the virus continues to circulate and transmit efficiently among humans.
Keywords: Influenza virus, hemagglutinin, H3N2, receptor specificity, glycan microarray
Introduction
Influenza A viruses are classified into subtypes according to the serological reactivity of their surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (World Health Organization, 1980). Currently, eighteen HA (H1–H18) and eleven NA (N1–N11) variants have been identified in aquatic birds and bats (Fouchier et al., 2005; Tong et al., 2012; Tong et al., 2013), and, of the 198 theoretical HA/NA combinations, only three influenza A subtypes in the last 100 years have successfully adapted to the human population to cause four pandemics, H1N1 in 1918 and 2009, H2N2 in 1957, and H3N2 in 1968 (Bean et al., 1992; Garten et al., 2009; Reid and Taubenberger, 2003; Scholtissek et al., 1978). This limited number suggests a virus must undergo a number of changes in its genes in order to successfully adapt to humans (Herfst et al., 2012; Imai et al., 2012). Several factors contribute to virus adaptation to humans and one is the ability of the HA virus coat protein to bind to sialic acid containing receptors on the host cell (Imai and Kawaoka, 2012). It is the nature of the sialic acid linkage preference of a virus, which varies in different host species, that helps maintain the species barrier and contributes to efficient infection and transmission between humans (Matrosovich et al., 2004; Nicholls et al., 2007; Rogers et al., 1983; Shinya et al., 2006; van Riel et al., 2006, 2007; Varki, 2007; Varki and Varki, 2007).
In humans, influenza A viruses utilize glycoproteins with complex carbohydrates containing terminal α2-6-linked sialic acids on the cell surface of upper respiratory tract epithelial cells in order to attach and initiate an infection (Baum and Paulson, 1990; Couceiro et al., 1993). Conversely, most aquatic bird viruses bind to α2-3-linked sialic acid containing receptors in the avian intestinal tract. Previous studies identified a number of key receptor binding site (RBS) mutations, responsible for switching avian/human receptor specificity in H1, H2 and H3 subtypes. In H1 subtypes, a Glu190Asp and Gly225Asp double mutation renders the HA capable of binding human α2-6 receptors (Matrosovich et al., 2000; Stevens et al., 2006a). For H2 and H3, two different mutations, Gln226Leu and Gly228Ser correlate with a shift to human receptor specificity (Connor et al., 1994; Rogers et al., 1983). As yet, the nature of the switch from other avian pandemic potential subtypes, such as H5 (Herfst et al., 2012; Imai et al., 2012), H7 (Gao et al., 2013), and H9 (Wan et al., 2008) necessary to cross the species barrier into the human population is unknown, although the recent H7N9 outbreak in China suggests this virus may prefer the H2/H3 route (Gao et al., 2013).
As seen with H1N1, H2N2 and H3N2 influenza A viruses, once established in the human population, the virus coat proteins hemagglutinin (HA) and to a lesser extent neuraminidase (NA) undergo continual change to evade the human immune system. This antigenic drift, as highlighted by the H3N2 influenza A virus (Pereira and Schild, 1971), can continue for many decades, whereby the human immune system targets specific antigenic sites (Daniels et al., 1983; Popova et al., 2012; Stray and Pittman, 2012; Webster and Laver, 1980; Wiley et al., 1981) in and around the virus RBS. The virus attempts to evade these responses by introducing escape mutations that successfully reduce host immune recognition, without compromising other functions, such as receptor binding. Due to the close proximity of the RBS and antigenic sites, the interplay between antigenic drift and potential changes in receptor binding is an important factor which has been studied by a number of groups for both H1N1 (Aytay and Schulze, 1991; Das et al., 2011; Hensley et al., 2009) and H3N2 viruses (Daniels et al., 1987; Lin et al., 2012; Underwood et al., 1987). Here, we have assessed structural changes in the HA, including changes in the receptor binding preferences of a number of H3 recombinant HA (recHA) proteins. Results suggest that its RBS has evolved from one that can bind a broad range of human receptor analogs to one with a more restricted binding to longer glycans alone.
Results
Sequence and structural evolution of the H3 HA ectodomain
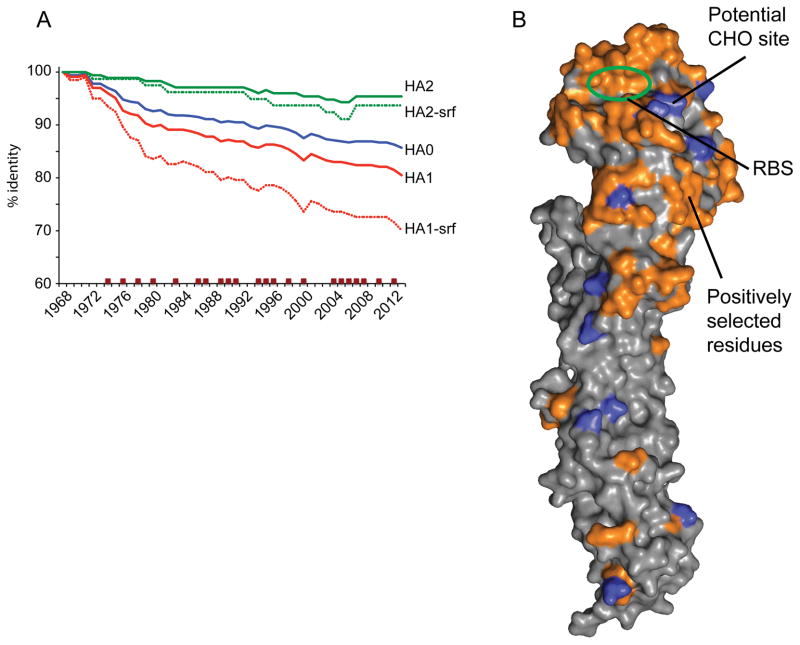
Comparison of the 2013 HA0 ectodomain consensus sequence with one from 1968 reveals that it still shares 87% identity to the 1968 virus HA (Fig 1A). With the HA2 component of the HA being 95% identical, the majority of the differences are therefore in the HA1 globular head region (81% identity). Structurally, the majority of these HA1 differences also belong to surface residues of the HA1 domain with only 70% identity (Fig 1A). Similarly, examination of residues in the A(H3N2) virus HA that have been subject to positive selection since 1968 also shows that the majority involved residues on the surface of the globular head of the HA (Fig 1A) (Bush et al., 1999a; Bush et al., 1999b; Smith et al., 2004; Tusche et al., 2012; Wiley and Skehel, 1987), with many surrounding the receptor binding site (RBS) (Fig 1B).
Fig 1.
A) To analyze HA evolution with time the HA sequences for A(H3N2) viruses were grouped according to year and consensus sequences were generated. Datasets corresponding to the HA0, HA1, HA2, as well as only surface residues (HA1-srf and HA2-srf) were also generated for comparison. Data was plotted as percentage change in identity to the 1968 consensus. Years where the H3 component of the Northern hemisphere vaccine was changed are marked with brown squares. B) Surface representation of the Victoria11 HA monomer highlighting surface residues (orange) that have undergone substitution and that were maintained in subsequent years. All structural figures were generated with MacPyMol (Delano, 2002).
To assess the structural consequences of evolutionary changes among these A(H3N2) virus HAs, we crystallized the full length recombinant HA (recHA) ectodomain proteins from HK68, PtChalmers73 and Victoria11, as well as the HA1 domains of Perth09 and Christchurch11. Upon removal of the foldon trimerization domain, all full-length recHAs purified and crystallized as trimers while the HA1 domain proteins were monomeric. Diffraction datasets were collected from native protein crystals of each HA, as described in the methods and Table 2. All crystal structures were determined by the molecular replacement method, using the previously published HK68 HA (PDB:2HMG) as a search model (Weis et al., 1990). Amino acid residues in each structure are numbered consecutively, according to the ectodomain fragments of the mature HA1 and HA2 subunits, respectively.
Table 2.
Data collection and refinement statistics for the A(H3N2) HA crystal structures.
| Hong Kong/1/1968 (HK68) |
Port Chalmers/1/1973 (PtChalmers73) |
Perth/16/2009 (Perth09) HA1 domain |
Christchurch/28/ 2011 (Chrishchurch11) |
Victoria/361/2011 (Victoria11) | |||
|---|---|---|---|---|---|---|---|
| Apo | 3′SLN | 6′SLN | |||||
| Experimental | |||||||
| Protein Conc. (mg/ml) | 14 | 13 | 15 | 19 | 18 | ||
| Crystallization conditions | 0.1M Tris-HCl, pH7.9, 25% PEG 1000 | 0.1M HEPES, pH7.5, 25% PEG 550 MME | 0.1M Magnesium Chloride, 0.1M Tris-HCl, pH8.5, 25% PEG 3350 | 0.2M Calcium acetate, 0.1M MES, pH6, 20% PEG 8000 | 0.1M CAPS:NaOH, pH10.5, 30% (v/v) PEG 400 | ||
| Cryoprotectant | None | None | 20% glycerol | 20% Ethylene glycerol | None | ||
| Data collection | |||||||
| Beamline collected | APS, BM | SSRL, 9-2 | APS, 22-ID | APS, 22-ID | APS, 22-ID | ||
| Space group | I213 | I213 | P121 | P1211 | R32 | R32 | R32 |
| Cell dimensions (Å) | 155.51 Å, 155.51 Å, 155.51 Å | 153.13 Å, 153.13 Å, 153.13 Å | 42.47 Å, 50.57 Å, 182.26 Å | 48.30 Å, 214.90 Å, 101.81 Å | 100.91 Å, 100.91 Å, 383.36 Å | 101.14 Å, 101.14 Å, 384.93 Å | 100.73 Å, 100.73 Å, 382.61 Å |
| Cell angle (°) | 90°, 90°, 90° | 90°, 90°, 90° | 90°, 90.1°, 90° | 90°, 103.6°, 90° | 90°, 90°, 120° | 90°, 90°, 120° | 90°, 90°, 120° |
| Resolution (Å) | 50-2.35 (2.43-2.35)a | 50-2.1 (2.16-2.1) | 50-1.9 (1.97-1.9) | 50-2.5 (2.59-2.5) | 50-2.1 (2.16-2.1) | 50-2.2 (2.26-2.2) | 50-2.2 (2.26-2.2) |
| Rsym (%) | 7.3 (34.0) | 7.5 (86.8) | 8.4 (34.5) | 8.7 (62.3) | 7.5 (37.0) | 7.8 (31.3) | 7.1 (35.5) |
| I/sigma | 40.0 (7.1) | 27.3 (2.16) | 12.8/3.6 | 28.1 (2.3) | 36.8 (5.2) | 37.1 (4.5) | 43.5 (5.0) |
| Completeness (%) | 98.1 (99.3) | 99.5 (98.0) | 98.2 (100) | 98.1 (95.4) | 98.5 (95.1) | 99.4 (96.7) | 99.6 (96.6) |
| Redundancy | 10.8 (9.8) | 4.9 (4.5) | 3.0 (3.0) | 3.4 (3.0) | 4.8 (4.6) | 5.1 (4.1) | 6.9 (5.5) |
| Refinement | |||||||
| Resolution (Å) | 30.5-2.35 (2.45-2.35) | 30-2.10 (2.16-2.10) | 44.2-1.9 (1.93-1.90) | 43-2.5 (2.53-2.5) | 34.2-2.10 (2.15-2.10) | 42.7-2.20 (2.25-2.20) | 35.0-2.2 (2.26-2.20) |
| No. reflections (total) | 25624 | 34697 | 56707 | 67875 | 43789 | 38827 | 38431 |
| No. reflections (test) | 1307 | 1743 | 3023 | 3431 | 2218 | 1954 | 1922 |
| Rwork/ Rfree | 19.3/23.6 | 18.2/23.1 | 22.1/19.6 | 21.8/25.1 | 19.0/22.7 | 19.4/22/.6 | 19.0/22.3 |
| No. of atoms | 4484 | 4384 | 4954 | 8827 | 4280 | 4195 | 4187 |
| r.m.s.d.- bond length (Å) | 0.005 | 0.007 | 0.002 | 0.009 | 0.005 | 0.004 | 0.006 |
| r.m.s.d.- bond angle (°) | 0.923 | 1.041 | 1.168 | 1.264 | 0.931 | 0.838 | 1.021 |
| MolProbityb scores | |||||||
| Favored (%) | 95.7 | 94.5 | 97.8 | 95.2 | 96.3 | 95.3 | 95.1 |
| Outliers (%) | 0.2 | 0.6 | 0 | 0.2 | 0 | 0.4 | 0.6 |
| PDB Code | 4WE4 | 4WE5 | 4WE6 | 4WE7 | 4WE8 | 4WE9 | 4WEA |
Numbers in parentheses refer to the highest resolution shell.
Reference (Davis et al., 2007).
The full length PtChalmers73 H3 HA exhibited a high degree of amino acid sequence identity to HK68 (~97% across the entire HA), while Victoria11 was more distant (86%) (Table 3). Despite these sequence differences, the overall structures of PtChalmers73 and Victoria11 HAs are similar to that of HK68. Each HA is a trimer of identical monomeric subunits, each composed of a virus membrane-proximal α-helix-rich stem and a membrane-distal globular head. Comparison of the HA1/HA2 monomer of each structure with that of the HK68 virus HA as well as to two previously published HAs from A/Norway/807/2004 (Norway04) and A/Malaysia/33386/2005 (Malaysia05) revealed structurally similar molecules despite their 43 year separation (Lin et al., 2012). All Cα atoms in the HA1/HA2 monomers of PtChalmers73, Norway04, Malaysia05 and Victoria11 superimposed onto HK68 to give root mean square deviations (RMSD) of only 0.39 Å, 0.76 Å, 0.73 Å and 0.68 Å, respectively (Table 4). Similarly when only the HA1 domains are compared to HK68, including the HA1 only models for Perth09 and Christchurch11, RMSD values range between 0.27 – 0.8 Å (Table 5), indicating a high degree of structural homology in the configuration of these HAs, despite 43 years between the earliest and most recent HA.
Table 3.
Sequence identity (%) of recHAs used in structural analyses. Only the sequence of the HA fragments used in the expression constructs for the full-length ectodomain (FL) or HA1 domain (HA1) were used in the calculations.
| HK68 | PtChalmers73 | Perth09 | Christchurch11 | Victoria11 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FL | HA1 | FL | HA1 | FL | HA1 | FL | HA1 | FL | HA1 | |
| HK68 | 100 | 100 | 97 | 96 | 87 | 79 | 86 | 78 | 86 | 78 |
| PtChalmers73 | 97 | 96 | 100 | 100 | 88 | 81 | 88 | 80 | 87 | 80 |
| Perth09 | 87 | 79 | 88 | 81 | 100 | 100 | 98 | 96 | 98 | 97 |
| Christchurch11 | 86 | 78 | 98 | 80 | 98 | 96 | 100 | 100 | 98 | 96 |
| Victoria11 | 86 | 78 | 87 | 80 | 98 | 97 | 98 | 96 | 100 | 100 |
Table 4.
Comparison of RMSD (Å) for HA monomers. For analyses, the Cα atoms of residues, HA1: 8-319, HA2: 21-165 for HK68 were superimposed by sequence and structural alignment onto the equivalent regions of the other HAs.
| PtChalmers73 | Norway04 | Malaysia05 | Victoria11 | |
|---|---|---|---|---|
| HK68 | 0.39 | 0.76 | 0.73 | 0.68 |
| PtChalmers73 | 0.78 | 0.76 | 0.74 | |
| Norway04 | 0.13 | 0.52 | ||
| Malaysia05 | 0.45 |
Table 5.
Comparison of RMSD (Å) for HA1 domains. For analyses, the Cα atoms of residues, HA1: 8-319 for HK68 were superimposed by sequence and structural alignment onto the equivalent regions of the other HAs.
| PtChalmers73 | Norway04 | Malaysia05 | Perth09 | Christchurch11 | Victoria11 | |
|---|---|---|---|---|---|---|
| HK68 | 0.27 | 0.66 | 0.65 | 0.8 | 0.7 | 0.58 |
| PtChalmers73 | 0.69 | 0.68 | 0.78 | 0.71 | 0.61 | |
| Norway04 | 1.03 | 0.56 | 0.38 | |||
| Malaysia05 | 1.01 | 0.55 | 0.37 | |||
| Perth09 | 0.81 | 0.61 | ||||
| Christchurch11 | 0.64 |
HA glycosylation
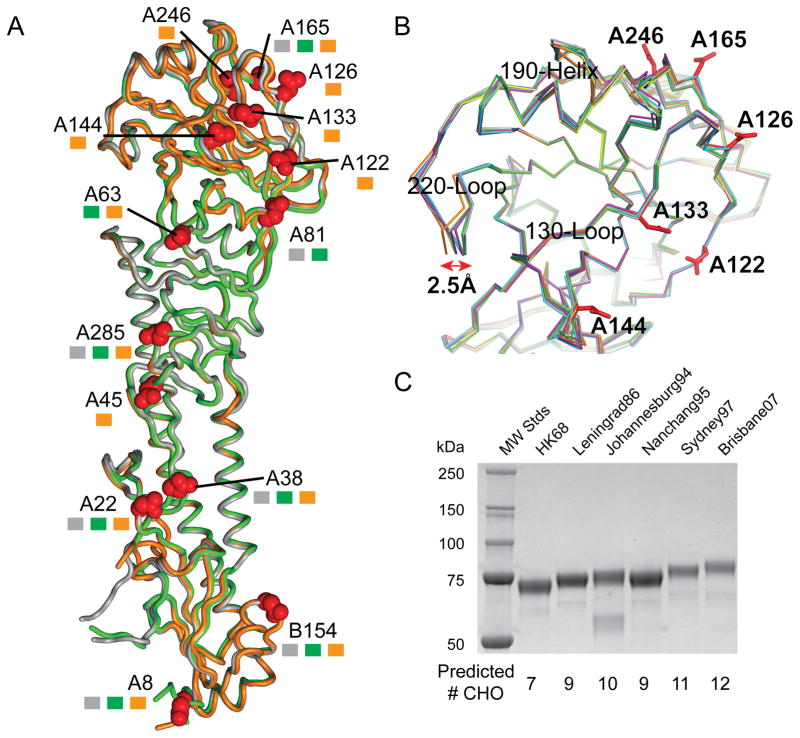
Since the A(H3N2) virus first adapted to humans in 1968, the virus HA has evolved from a protein with 7 potential glycosylation sites to one with 12–13 potential sites (Fig 2A), six of which (Asn122, Asn126, Asn133, Asn144, Asn165 and Asn246) are within or near to both the RBS and known antigenic sites (Fig 2B). All HAs were expressed in an insect expression system and thus should possess N-carbohydrates at these sites. However, occupied glycosylation sites are often difficult to resolve structurally due to carbohydrate flexibility in the crystals. In the models reported here, N-carbohydrates were clearly visible within the electron density at six of the seven potential sites in HK68 (HA1: Asn22, Asn38, Asn81, Asn165, Asn285 and HA2: Asn154; mature H3 HA numbering), at five of eight potential sites within PtChalmers73 (HA1: Asn38, Asn63, Asn81, Asn165 and Asn285) and only eight of potential thirteen glycosylation sites in Victoria11 (HA1: Asn22, Asn38, Asn63, Asn133, Asn165, Asn246, Asn285 and HA2: Asn154). SDS-PAGE also suggests occupancy of additional potential sites during the virus’ ~46 year circulation in humans (Fig 2C).
Fig 2.
A) Structural overlap of A(H3N2) HA monomers. HA monomers, HK68 (Grey), PtChalmers73 (Green) and Victoria11 (orange) are shown as tubes and the locations of glycosylation sites are highlighted with Asn side chain as red spheres. These sites if present on specific HAs are marked with squares colored according to the same HA coloring. B) RBS overlap of HK68 (Grey), PtChalmers73 (Green), Perth09 (magenta), Christchurch11 (cyan) and Victoria11 (orange). Previously published H3 HAs, Norway04 (blue) and Malaysia05 (yellow) (Lin et al., 2012) were also used in the alignment. C) SDS-PAGE of various recombinant H3 HA proteins indicates an increase in MW as the predicted number of glycosylation sites increase with time.
Evolution of the receptor binding site (RBS)
The RBS at the membrane distal end of each HA monomer (Fig 2A), comprises three main structural elements: a 190-helix (residues 183, 185–196), a 220-loop (residues 219–228), and a 130-loop (residues 131–138, 140) (Fig 2B). Additional residues (98, 144–146 and 153–159) flank these structural elements and contribute to the overall architecture of the RBS (Fig 3). While HA1 domains from recent virus HAs are 81% identical to the 1968 viruses (and 70% for just the surface residues), the RBS components are more divergent, at only 52% identity for the three structural elements combined (Fig 4A). An overlap of the RBS main chains of HK68, PtChalmers73, Norway04, Malaysia05, Perth09, Christchurch11 and Victoria11 highlights just how similar the recent RBSs are to the earlier HAs. While the only significant main chain difference is a 2.5 Å movement of the 220 loop of recent HAs away from the RBS (Fig 2B), side chain differences through sequence variation and/or antigenic drift (Smith et al., 2004), have affected both the shape as well as the overall charge of the RBS through the last 4 decades (Movie supplemental file and Fig 4B).
Fig 3.
Consensus sequence alignment of the A(H3N2) virus RBS by year. RBS residues that overlap onto recognized antigenic sites (AS) (Daniels et al., 1983; Popova et al., 2012; Stray and Pittman, 2012; Wiley et al., 1981) are colored red for AS-A, yellow for AS-B, and blue for AS-D.
Fig 4.
A) To assess HA RBS evolution with time, the HA sequences for A(H3N2) viruses were grouped according to year and consensus sequences of the RBS region of the protein were generated. Data was plotted as percentage change in identity to the 1968 consensus. Datasets were also divided into separate components of the RBS, corresponding to the 130/140-loop, the 190-helix and the 220-loop. Years where the H3 component of the Northern hemisphere vaccine was changed are marked with brown squares and a plot of the predicted number of glycosylation sites is also shown for comparison (black dotted line). B) Surface representation of the consensus RBS in 1968 compared to select examples from years (1979, 1992, 1997 and 2010) when the A(H3N2) component of the seasonal influenza vaccine was updated by the WHO. Each RBS model is colored by charge. A movie highlighting change by year can be found in the supplemental online material. C) Conserved residues in and around the H3 RBS. Throughout 46 years of circulation within the human population, sixteen residues in and around the RBS have remained conserved throughout and are represented as green sticks.
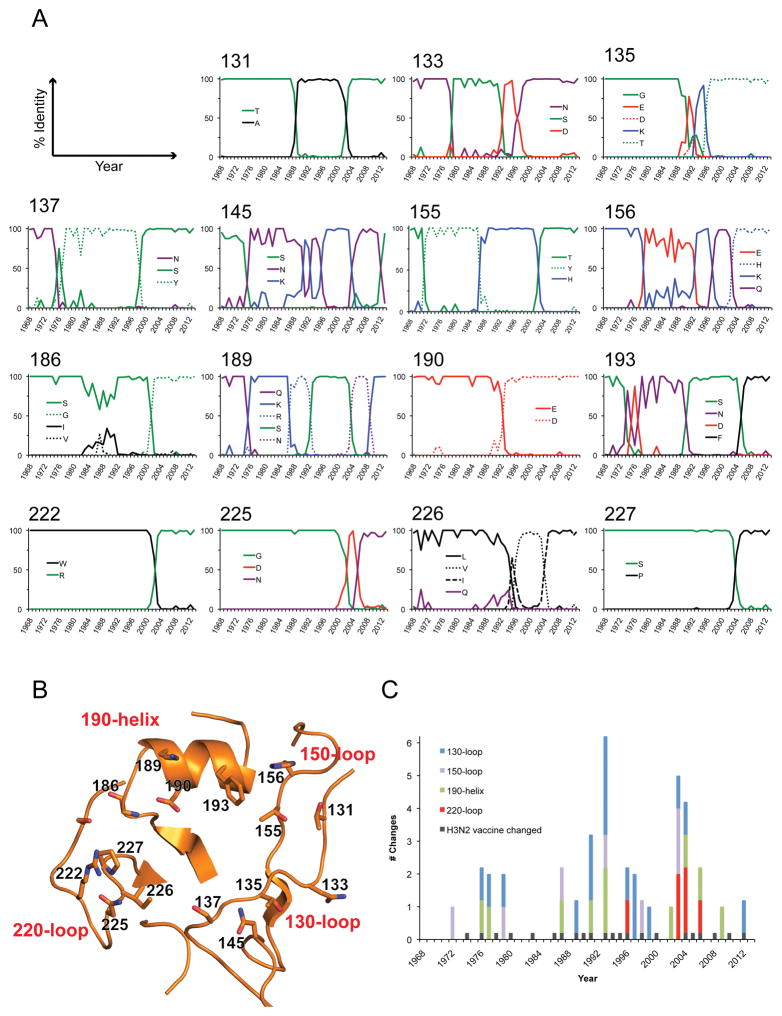
Of the sixteen residues in and around the RBS that have remained strictly conserved, 11 reside within, or point toward the sialic acid binding pocket (Fig 4C). Of these, Tyr98, Ser136, Trp153, His183, Leu194, Tyr195 are critical for the proper functioning of the RBS. Any mutations introduced at these sites either abolish or significantly reduce receptor binding (Skehel and Wiley, 2000). Fifteen variable residues in or near the RBS were selected and frequency diagrams were generated to assess variability with time at each position (Fig 5A). Results highlight that while all fifteen positions reside on structures that form the RBS (Fig 5B), they also belong to one of three identified antigenic sites: A, B or D (Daniels et al., 1983; Popova et al., 2012; Stray and Pittman, 2012; Wiley et al., 1981). From the data, residues in the 130-loop, 150-loop and 190-helix have been under selective pressure throughout the last 46 years and residues in these regions have changed frequently (Fig 5A and 5C). For the 220-loop however, changes occurred only during the 4 years; 1997, 2003, 2005 and 2006. Interestingly, of the 18 years where changes were incorporated, only ten of those years coincided with a decision by the WHO to change the H3N2 component of the influenza vaccine for the Northern hemisphere (Fig 5C). To assess whether changes at these variable sites has affected host receptor binding and specificity, nineteen human H3 recombinant HAs (Table 1) spanning the period from 1968 to 2011 were cloned, expressed and each was subjected to glycan microarray analysis. Results are presented in Fig 6 and summarized in Table 6 with full glycan information in the supplemental Excel file.
Fig. 5.
A) Frequency diagrams of variable RBS residues. For each RBS position a frequency diagram was generated to show the frequency of amino acid changes at the site from 1968 to 2013. Lines are colored according to the chemical properties of each residue: polar (Gly, Ser, Thr, Tyr, Cys) colored green, neutral (Gln, Asn) colored purple, basic (Lys, Arg, His) colored blue, acidic (Asp, Glu) colored red, and hydrophobic (Ala, Val, Leu, Ile, Pro, Trp, Phe, Met) colored black. Only residues that occurred at a frequency >15% in a year are plotted on the graphs. B) The positions for the 15 residues analyzed in Fig 4A are highlighted as sticks on the Vict11 RBS. C) The number of residue changes on the 190-helix (green), and 130- (blue), 150- (violet) and 220-loops (red) were totaled for each year and plotted on a stacked column graph. The years when the H3N2 component of the vaccine were changed are marked in black.
Table 1.
Recombinant A(H3N2) HA proteins used in this study
| Strain | GISAID accession Number | Propagation info in database | Abbreviation |
|---|---|---|---|
| A/Hong Kong/1/1968 | EPI240947 | ----* | HK68 |
| A/Port Chalmers/1/1973 | EPI365202 | Egg | PtChalmers73 |
| A/Victoria/3/1975 | EPI131278 | ---- | Victoria75 |
| A/Bangkok/1/1979 | EPI377537 | Egg | Bangkok79 |
| A/Leningrad/360/1986 | EPI385984 | Egg | Leningrad86 |
| A/Shanghai/11/1987 | EPI390002 | Egg | Shanghai87 |
| A/Beijing/32/1992 | EPI365898 | Cell | Beijing92 |
| A/Johannesburg/33/1994 | EPI390018 | Egg | Johannesburg94 |
| Nanchang/933/1995 | EPI362794 | Cell | Nanchang95 |
| A/Sydney/5/1997 | EPI362863 | Cell | Sydney97 |
| A/Panama/2007/1999 | EPI105036 | ----* | Panama99 |
| Wyoming/03/2003 | EPI152388 | Cell | Wyoming03 |
| A/California/7/2004 | EPI367105 | Cell | California04 |
| A/Hiroshima/52/2005 | EPI126805 | Cell | Hiroshima05 |
| A/Brisbane/10/2007 | EPI165489 | Cell | Brisbane07 |
| A/Perth/16/2009 | EPI182941 | Cell | Perth09 |
| A/Christchurch/28/2011 | EPI346305 | Cell | Christchurch11 |
| A/Victoria/361/2011 | EPI349103 | Cell | Victoria11 |
| A/Texas/50/2012 | EPI398417 | Cell | Texas12 |
No information availabl
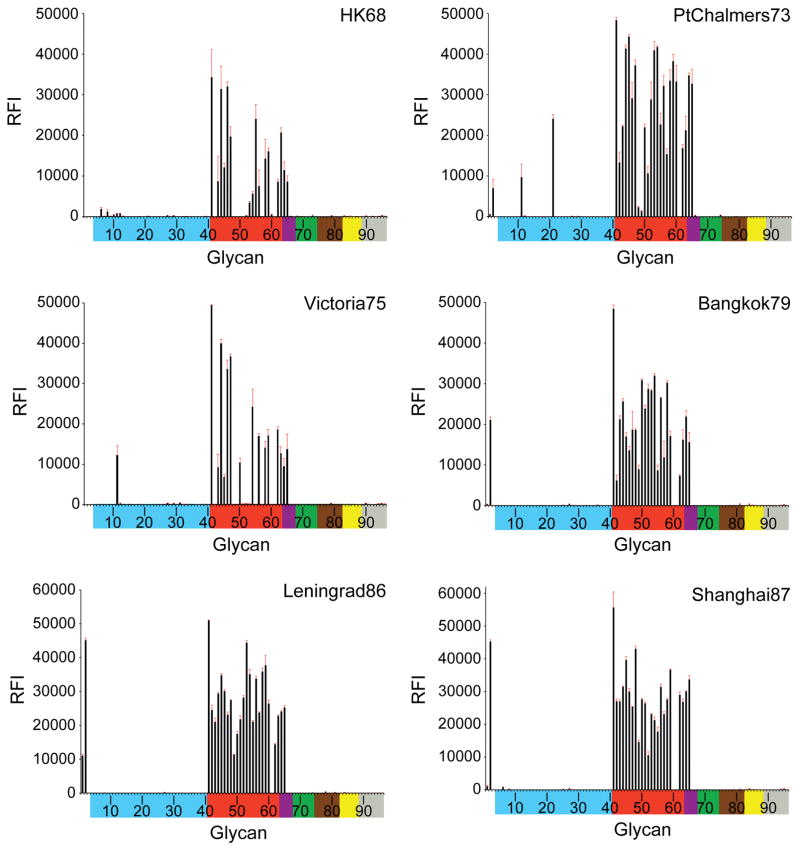
Fig. 6.
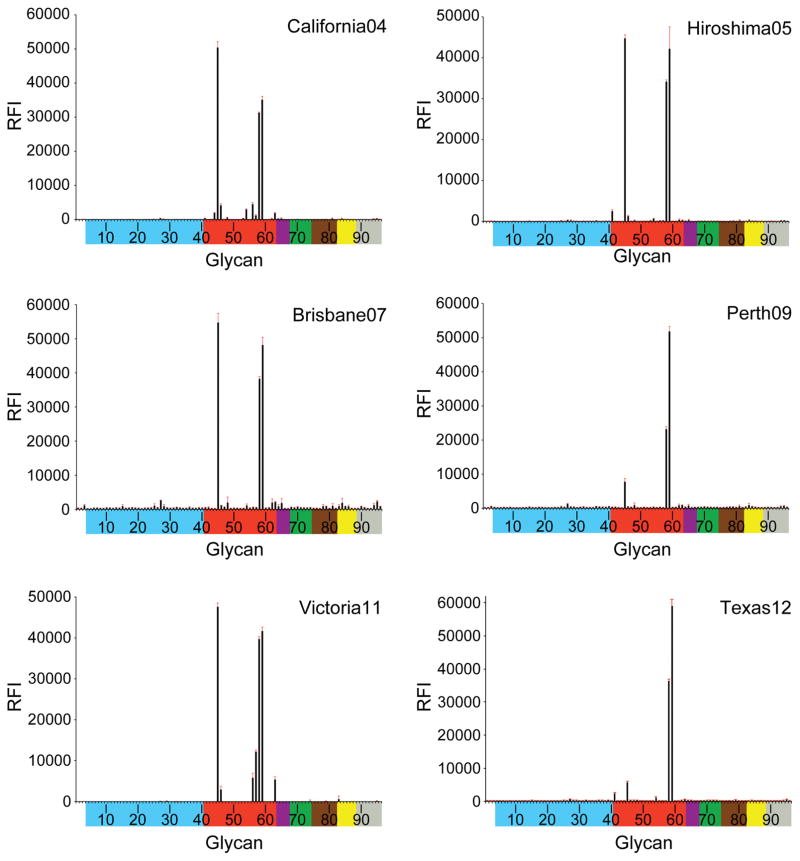
Receptor binding specificity of A(H3N2) influenza virus recHAs. Glycan microarray analysis of A(H3N2) influenza virus recHAs, as listed in Table 1. Different types of sialoglycans on the array (x-axis) are grouped according to SA linkage: α2-3 SA (blue), α2-6 SA (red), α2-6/α2-3 mixed SA (purple),N-glycolyl SA (green), α2-8 SA (brown), β2-6 and 9-O-acetyl SA (yellow), and asialo glycans (gray). The vertical bars denote relative fluorescence intensity (RFI) and error bars in each panel reflect the standard error in the signal for six independent replicates on the array. The structures of each of the numbered glycans are found in the supplemental Excel file. Specific glycan structures that were used in biosensor assays are represented on the array as glycan 22 (α2-3) and glycan 56 (α2-6).
Table 6.
Comparison of the detailed sialoside receptor specificity of H3 influenza virus recHAs.
| Glycans | Graph a | 1968 | 1973 | 1975 | 1979 | 1986 | 1987 | 1992 | 1994 | 1995 | 1997 | 1999 | 2003 | 2004 | 2005 | 2007 | 2009 | 2011 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α2–3 | ||||||||||||||||||
| Sulfated b | 4–8 | . | . | . | . | . | . | . | . | +++ | +++ | . | . | . | . | . | . | . |
| Branched di-sialyl | 9–12 | . | + | + | . | . | . | +++ | . | . | . | + | . | . | . | . | . | . |
| Linear | 13–26 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Fucosylated | 27–34 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Internal | 35–40 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| α2–6 | ||||||||||||||||||
| Sulfated | 41 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | . | +++ | +++ | +++ | . | . | . | . | . | . |
| Branched mono-sialyl | 42 | . | ++ | . | + | +++ | +++ | . | . | . | + | . | . | . | . | . | . | . |
| Branched di-sialyl | 43–47 | ++ | +++ | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | ++ | ++ | + | + | + | + |
| Linear (length) c | ||||||||||||||||||
| 2 | 48–49 | . | . | . | ++ | +++ | +++ | . | . | . | . | . | . | . | . | . | . | . |
| 3 | 52–54 | + | +++ | . | +++ | +++ | +++ | + | . | ++ | ++ | +++ | . | . | . | . | . | . |
| 4 | 55 | +++ | +++ | . | + | +++ | ++ | ++ | . | ++ | . | . | . | . | . | . | . | . |
| 5 | 56 | + | +++ | ++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | + | . | . | . | + |
| 6 | 57 | . | ++ | . | ++ | +++ | +++ | + | + | + | +++ | +++ | +++ | . | . | . | . | ++ |
| 7 | 58–59 | ++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Internal | 60–61 | . | + | . | . | + | . | + | . | . | . | . | . | . | . | . | . | . |
| Other | ||||||||||||||||||
| Sialic acid | 1–3 | . | + | . | +++ | +++ | +++ | + | . | . | . | . | . | . | . | . | . | . |
| Mixed α2-3/α2-6 | 64–67 | + | +++ | + | +++ | +++ | +++ | +++ | . | + | ++ | +++ | . | . | . | . | . | . |
| non-α2-3 or α2-6 | 68–88 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| asialo | 89–96 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
. No binding
Members of each group are identified according to the graph number used in the microarray data in Fig 5 and correspond to numbers in the complete glycan list (supplemental Excel file).
Binding of samples to glycan subclasses are qualitatively estimated based on relative strength of the signal for the data shown in Fig 5: strong >10,000 (+++), medium >5000 (++), weak >1250 (+), no binding <1249 (.).
Length refers to the number of monosaccharides in the glycan chain, including SA.
Glycan binding to recHAs
From 1968, only very weak binding to avian α2-3 sialosides was observed, highlighting the efficient adaptation of the H3 RBS binding to human receptors. Only sporadic binding signals for α2-3 sialosides in early recHAs, PtChalmers73 (glycan #11, 21) and Victoria 75 (#11), were apparent. Beijing92 gave strong binding signals to α2-3 branched di-sialosides (#9–12). While no avian receptor binding was observed in Johannesburg94, all three recHAs from the late 1990s: Nanchang95, Sydney97 and Panama99 bound to a subset of sulfated α2-3 glycans (#6–8), albeit with diminishing signal strength (Nanchang95 > Sydney97 ≫ Panama99). No binding to avian glycans was detected by glycan array to any recHA from the year 2000 onwards.
From 1968 until 1999, the majority of the recHAs analyzed bound well to sulfated (#41), branched di-sialosyl (#43–47) and linear (#50–59) α2-6 glycans represented on the microarrays. Indeed, even PtChalmers73 bound well to α2-6 glycans on the microarrays, which was surprising given the previously reported poor binding of viruses from this era (1972–74) (Gulati et al., 2013). Results from the all six post-2000 recHAs revealed a greatly reduced binding profile. Wyoming03 bound to only two branched di-sialoside glycans (#45, 46) and five long linear α2-6 glycans (#56–59, 63). The later recHAs, California04, Hiroshima05, Brisbane07, Perth09, Victoria11, were further restricted and strong binding signals were only observed for the longest branched (#45) and linear α2-6 glycans (#58 and 59) on the array.
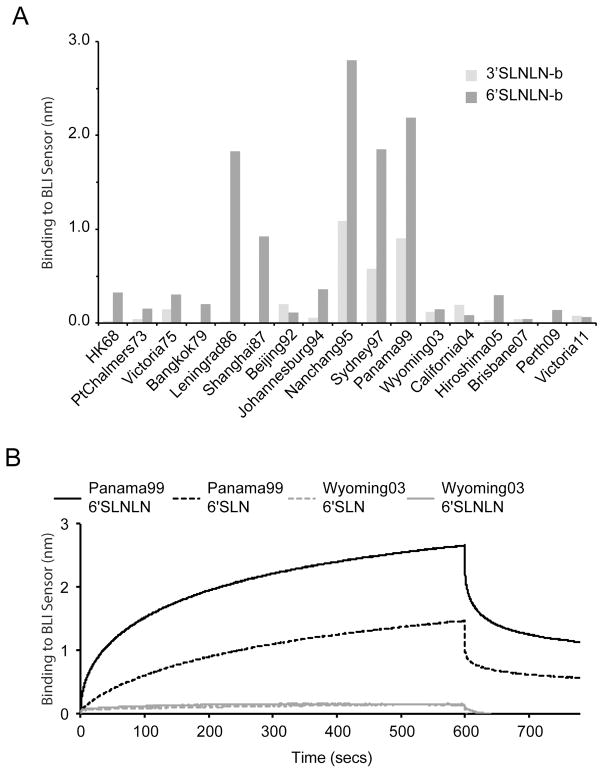
Glycan binding to recombinant HA protein was further analyzed by BLI using an Octet Red system (ForteBio Inc). This label free technology was used to measure recombinant HA binding to biotinylated glycans (3′SLNLN-b and 6′SLNLN-b) preloaded onto streptavidin-coated biosensors. Results for recHA proteins highlight the weak interactions of these glycans with many of the proteins analyzed (Fig 7A). While the 6′SLNLN-b analog bound preferentially to most of the proteins assayed, weaker binding to the 3′SLNLN-b was also observed for the majority of the recHAs from the 1990s. Strongest binding signals to 6′SLNLN-b were observed for Leningrad86, Shanghai87, Nanchang95, Sydney97 and Panama99. However, while 6′SLNLN binding was strong for Wyoming03 recHA, by glycan microarray analysis (#56), it bound poorly, if at all, to the 6′SLNLN-b glycan (Fig 7A and 7B).
Fig. 7.
A) Detection of recHA binding to biotinylated glycans by Bio-Layer Interferometry. RecHA binding to the biosensor tip containing either 3′SLNLN-b (light gray bars) or 6′SLNLN-b glycans (dark gray bars) was assessed. The level of recHA binding to the biosensor tip after 300 seconds, was measured as a wavelength shift (measured in nanometers) and was plotted for each protein. B) The full binding profile analysis of Panama99 and Wyoming03 binding to both single LacNAc (3′SLN-b, 6′SLN-b) and Di-LacNAc (3′SLNLN-b, 6′SLNLN-b) sialosides.
To probe the possible reason(s) for the loss of glycan binding between Panama99 and Wyoming03, RBS sequence differences for Wyoming03 HA were introduced onto the Panama99 HA framework and the resulting proteins were analyzed for glycan binding to microarrays (Fig 8). To simplify the number of targets, substitutions were combined according to the structural loops that comprise the HA RBS (i.e. the 130-loop, 150-loop, 190-helix and the 220-loop). While substitutions on the 130-loop (Thr128Ala and Ala131Thr) did not appear to affect binding to human receptor analogs, when compared to Panama99, an increase was observed in binding signals for the avian α2-3 bi-antennary glycans (#11, 12) on the array (Fig 8). Interestingly, combinations of substitutions in the 150-loop (His155Thr and Gln156His), 190-helix (Ser186Gly and Ile194Leu) and 220-loop (Ser220Arg, Trp222Arg and Gly225Asp) all resulted in a preference for α2-6 glycan binding similar to that of Wyoming03, with a loss of binding to the shorter linear sialosides (#52–54) on the array (Fig 8). This suggests multiple substitutions may be responsible for the observed enriched binding to the longer glycans represented on the array.
Fig. 8.
Effect of introducing Wyoming03 RBS residue differences onto the Panama99 recHA on glycan binding. RBS residue differences between Wyoming03 and Panama99 were grouped according to the RBS structural element they resided on and introduced collectively onto the Panama99 framework. The resulting Panama99 mutants were expressed and analyzed by glycan microarray. Results are presented as for Fig 5 and each graph is color coded according to the mutations introduced, as shown in the central image of an A(H3) HA RBS. Wild-type Panama99 and Wyoming03 controls were run and analyzed in parallel.
Attempts were also made to structurally analyze the glycan interactions of a recent 2011 HA, Victoria11. Diffraction data were obtained for Victoria11 HA alone (apo) as well in human (6′SLN) and avian (3′SLN) receptor analog complexes, and relevant crystallographic statistics are presented in Table 2. While the avian analog, 3′SLN, was not visualized at all in the density maps, the human analog, 6′SLN, was also very poorly visualized (data not shown), which is in agreement to recent structural data from 2005 HA (Lin et al., 2012).
Discussion
Between 1976 and 2007, it was reported that for the U.S., the average mortality rates for those seasons during which influenza A(H3N2) was a prominent strain, which included four post-2000 flu seasons (2001–02, 2003–04, 2004–05 and 2005–06), were almost 3 times higher than for those seasons where other seasonal influenza viruses were dominant (Thompson et al., 2010). In order to maintain circulation within the human population, A(H3N2) viruses have continually evolved, by a process called antigenic drift, to escape from neutralizing antibodies produced in response to infection and mass vaccination. During the past 46 years there have been 158 amino acid changes identified at 60 surface residues in the HA1 domain, of which 121 of these changes, or 77%, are clustered within the five antigenic sites (A–E) previously described for A(H3N2) viruses (Daniels et al., 1983; Popova et al., 2012; Stray and Pittman, 2012; Webster and Laver, 1980; Wiley et al., 1981). Interestingly, three of these antigenic sites (A, B and D) also overlap significantly (48%, 79% and 36% of the antigenic site, respectively) with the HA RBS. Thus, any antigenic changes in these regions might also affect receptor binding. Indeed, while results show that the overall virus HA structure from 2011 is essentially the same as that from 1968, the residues comprising the RBS and antigenic sites have changed, affecting both the surface shape and charge (Movie supplemental file and Fig 4B).
With respect to influenza research and surveillance, ongoing changes within the RBS have been shown to affect established virus assays over time. For example, until the mid-1990s, human A(H3N2) and A(H1N1) influenza viruses agglutinated both human and chicken red blood cells (RBCs), after which both subtypes lost the ability to agglutinate chicken red blood cells (Grassauer et al., 1998; Medeiros et al., 2001; Nobusawa et al., 2000). Residue 226 was highlighted as a modulator of this effect, in that a progressive loss of HA binding to chicken erythrocytes was observed as viruses changed from Gln226 to Leu to Ile to Val (Medeiros et al., 2001). Even the residues identified for human adaptation of H1 subtypes, Glu190Asp and Gly225Asp (Glaser et al., 2005; Matrosovich et al., 2000; Nobusawa et al., 2000; Stevens et al., 2006a), were also shown to affect receptor binding to A(H3N2) viruses. A limited study using viruses with different receptor specificities was shown to correlate with RBC agglutination from different species (Ito et al., 1997), and a Glu190Asp substitution, which emerged around 1993 (Fig 3), was identified as a possible reason for this observed change (Nobusawa et al., 2000). This can be explained structurally since the longer Glu190 side chain (with an additional methylene group) can extend into the RBS and interact directly with the 9-hydroxyl group of a bound sialic acid. In contrast, Asp190 is too short to extend into the pocket and thus weakens the HA/receptor interaction. From 2000 onwards, researchers reported that isolated viruses grew poorly in both egg and cell culture systems used to propagate viruses due to the A(H3N2) virus having reduced affinity for both human and avian receptors (Lu et al., 2006; Nobusawa et al., 2000; Oh et al., 2008; Stevens et al., 2010). While a number of residue substitutions in the RBS (i.e. 131, 145, 155, 156, 159, 186, 189, 192, 193, 222, 225, 226 and 227) occurred during this time that could explain these observations, an Asp225Asn substitution that emerged during 2004–2005 was shown to affect virus binding in experiments with α2-6-sialyllactosamine (Lin et al., 2012).
In this study, we have assessed changes in receptor binding by glycan microarray analysis using recombinant HAs. Nineteen HAs selected from WHO vaccine strains and spanning the period from 1968 to 2012 were cloned, expressed and their glycan preferences were analyzed by glycan microarray (Blixt et al., 2004; Stevens et al., 2006a). Although HA sequence selections were made to minimize the effect of laboratory propagated strains, all of the older pre-1990 HA sequences were only available for viruses propagated from eggs (Table 1). Only one of the five recHA sequences used from the 90s, Johannesburg94, was listed in the database as egg propagated and it did possess an obvious egg adaptation marker (ie. Ser219Tyr) in its sequence (Fig S2). However, in 1994, 80% of the 124 HA sequences deposited in the GISAID database contained a Phe or Tyr (egg adaptation) at position 219, highlighting the difficulty of selecting suitable vaccine strain sequences for these types of studies.
Glycan microarrays can assess the binding of recombinantly produced HAs to multiple well-defined glycans simultaneously, and previous microarray binding analyses with influenza virions (both live and β-propiolactone inactivated) demonstrated a good correlation with receptor specificity, when compared to other methods of analysis (Blixt et al., 2004; Childs et al., 2009; Rogers and D’Souza, 1989; Rogers et al., 1985; Stevens et al., 2008; Stevens et al., 2006a; Stevens et al., 2006b; Stevens et al., 2006c). Indeed, unlike the virus studies performed previously, where no one glycan bound to every virus tested (Gulati et al., 2013), results here highlighted three long chain glycans (#45, 58 and 59) that bound to all 17 recHAs (Table 6). Results revealed that recHAs from the 1970s and 1980s bound to the majority of α2-6 glycans represented on the arrays (Fig 6). RecHAs selected from the 1990s however, not only revealed reduced binding to short linear α2-6 glycans (#48–51) on the array, but they also bound to a limited set of avian α2-3 glycans (#6–12). The reason for the binding of recHAs from the 1990’s to avian receptor analogs is unclear. While care was taken to reduce the selection of sequences of HA from egg adapted strains, passage history of these older strains cannot be confirmed with absolute confidence. RecHAs from 2000 onwards bound to an even more restricted set of glycans, comprising only the longest branched and linear glycans (#45, 58 and 59) represented on the array, which is in agreement with previous observations with H3N2 viruses (Gulati et al., 2013; Kumari et al., 2007; Walther et al., 2013).
One possible reason for a preference for binding to longer glycans is the accumulation of potential glycosylation sites on the HA1 chain of the viral HA. Since 1968, the number of potential glycosylation sites on the A(H3N2) HA head has increased from 3 to 8, while the HA2 domain has remained relatively constant with just four (Fig 2). Positioning of glycosylation sites can have a dramatic effect on the surface of the virus HA. For example, it can impact protein folding (Hebert et al., 1997; Land and Braakman, 2001), conformation (Meunier et al., 1999), and also affect distant parts of a protein through masking or conformational changes. Although accumulating carbohydrates has been suggested to effect immune evasion (Kobayashi and Suzuki, 2012; Seidel et al., 1991; Skehel et al., 1984; van Gils et al., 2011; Vigerust et al., 2007; Wang et al., 2010; Wanzeck et al., 2011), reducing the number of carbohydrate sites could also affect antibody accessibility through tighter packing of antigenic epitopes on the virus surface (Ye et al., 2000). Altering patterns of HA glycosylation has been shown to influence receptor binding in a number of subtypes including A(H3N2) viruses (Abe et al., 2004; Kaverin et al., 2002; Matrosovich et al., 1999; Owen et al., 2007; Stevens et al., 2008; Yang et al., 2013; Yang et al., 2012). Although this study cannot assess specific effects of changes to glycosylation sites on HA receptor specificity, results presented here suggest that there were no significant differences in binding signals between: Bangkok79 and Leningrad86 (gain of glycosylation site at 246), Nanchang95 and Sydney97 (gain of glycosylation sites: 122 and 133), Sydney97 and Panama99 (gain of glycosylation site at 144), and between California04, Hiroshima05 and Brisbane07 (loss and gain of glycosylation site at 165). While a more restricted binding profile was observed between Panama99 and Wyoming03, coinciding with a loss of glycosylation site at 126, the binding profile of California04 was not restored to that of Panama99, despite regaining the same glycosylation site.
Compared to Panama99, Wyoming03 possesses 9 substitutions around the RBS region of the molecule that may affect receptor binding (Fig 8). While previous research highlighted position 225 as a key determinant of decreased receptor binding in viruses circulating since 2005, our mutagenesis studies with Panama99 suggest more than one sequence change may be responsible for the loss of short chain glycan binding observed between Panama99 and Wyoming03 (Fig 8). Adding these Wyoming03 substitutions onto the 150-loop, 190-helix and 220-loop of the Panama99 framework all resulted in a loss of binding to the short chain glycans, suggesting that all three RBS structural elements can affect receptor binding.
Previous studies with viruses and purified HAs have shown clear differences in avidity between different A(H3N2) HAs to different glycan species (Asaoka et al., 2006; Lin et al., 2012; Nycholat et al., 2012; Oh et al., 2008). Our data complement these studies and highlight the complex nature of influenza virus host interactions. However, while HA receptor binding in these assays mostly correlate with one another, there are differences in results depending on the format of the HA under study (i.e. egg-propagated virus, cell-propagated or recombinant protein) or how they are analyzed (i.e. cell-based, microarray or biosensor assays). When considered together, one consistent feature through all these studies appears to be the maintenance of binding to the longer poly-N-acetyllactosamine sialosides. In line with previous research using viruses (Gulati et al., 2013), our recHA data presented here also reveals a loss of binding to shorter sialosides while the longer ones are maintained. While Lin et al. (Lin et al., 2012) have already highlighted a loss of avidity in glycan binding to 2004 and 2005 viruses, our data suggests that this loss in avidity had already started between 1999–2003 and 150-, 220-loops as well as the 190-helix of the RBS can effect this change to different degrees. The significance of these results remains unknown since the exact glycan profile on the respiratory tract epithelial cell used by influenza viruses to enter the cell and initiate an infection is still not known. Interestingly a new glycan array, containing endogenous natural glycans from the lungs of pigs, was recently developed to characterize influenza virus receptor interactions (Byrd-Leotis et al., 2014). The ‘shotgun glycomics’ approach used to isolate and characterize these natural swing glycans could be used in the future to develop specific human arrays containing a more natural repertoire of glycans that are present on the host respiratory tract.
These results show that while the exact mechanism for loss of virus binding avidity to shorter glycans with time may not be attributed to any one specific residue change in the RBS, it is clear that RBS changes that have occurred significantly overlap onto the recognized antigenic sites on the A(H3N2) virus. Thus, antigenic drift will continue to have a major impact on receptor interactions. Under pressure for the virus to evade the host immune system without compromising its fitness, even after 46 years, the virus continues to be successful, maintaining its efficiency in infection and transmission within the human population.
Materials and Methods
Recombinant HA cloning and expression
cDNA encoding for residues 1 to 329 of the HA1 chain and 1–174 of the HA2 chain of the mature ectodomain from each HA listed in Table 1 was cloned into the baculovirus transfer vector, pAcGP67-B (BD Biosciences), in-frame with an N-terminal baculovirus GP67 signal peptide and a C-terminal thrombin cleavage site, a T4 fibritin sequence for generating functional trimers (Frank et al., 2001), and a His-Tag to aid purification (Stevens et al., 2004). For HA1 domain only constructs, residues 41 to 313 of the mature HA ectodomain from A/Perth/16/2009 (Perth09) and A/Christchurch/28/2011 (Christchurch11) were used. Wyoming03 mutations were introduced onto the Panama99 framework by mutagenesis of the wild-type Panama99 construct, using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Transfection and virus amplification were carried out according to the baculovirus expression system manual (BD Biosciences, San Jose, CA).
Protein expression and purification
Secreted soluble recombinant HA (recHA) protein was recovered from the cell culture supernatant by metal affinity chromatography and gel filtration chromatography and subjected to either thrombin digestion as described previously (Yang et al., 2012) or trypsin digestion (1:1000 w/w ratio of trypsin:protein). For Victoria11 HA trimers a further treatment with EndoH was required to obtain material suitable for crystallization (Lin et al., 2012). All HAs were buffer-exchanged into 10 mM Tris-HCl, 50 mM NaCl, pH 8.0 and concentrated to 13–19 mg/ml for crystallization trials.
Crystallization, ligand soaking and data collection
Initial nanoscale crystallization trials were set up using a TopazTM Free Interface Diffusion (FID) Crystallizer system (Fluidigm Corporation, San Francisco, CA). Conditions in which crystals were observed were optimized at 20 °C using a modified method for microbatch under oil (Chayen, 2007). For receptor analog complexes, crystals were soaked for 3 hours in the crystallization buffer containing 10 mM 3′-Sialyl-N-acetyllactosamine (3′SLN) or 6′-Sialyl-N-acetyllactosamine (6′SLN) (V-labs Inc., Covington, LA). All crystals were flash-cooled at 100 K, and datasets were collected and processed with the DENZO-SACLEPACK suite (Otwinowski and Minor, 1997). More specific information for each HA is included in Table 2.
Structure determination and refinement
All H3 HA structures were determined by molecular replacement with Phaser (McCoy et al., 2007) using the HA structure from A/Aichi/2/1968, PDB: 2HMG (Weis et al., 1990). Models were then “mutated” to their correct sequences, rebuilt by Coot (Emsley et al., 2010), and refined with Phenix (Adams et al., 2010) and REFMAC using TLS refinement (Winn et al., 2001). The final models were assessed using MolProbity (Davis et al., 2007). Statistics on data processing and refinement are presented in Table 2.
Glycan Binding Analyses
Glycan microarray printing and recombinant HA analyses have been described previously (Blixt et al., 2004; Stevens et al., 2008; Stevens et al., 2006a; Stevens et al., 2006c) (see Supplemental Excel file of glycans used in these studies). For biosensor studies, biotinylated glycans: Neu5Ac(α2-3)Gal(β1-4)GlcNAcβ-Biotin (3′SLN-b), Neu5Ac(α2-3)Gal(β1-4)GlcNAc(β1-3)Gal(β1-4)GlcNAcβ-Biotin (3′SLNLN-b), Neu5Ac(α2-6)Gal(β1-4)GlcNAcβ-Biotin (6′SLN-b) and Neu5Ac(α2-6)Gal(β1-4)GlcNAc(β1-3)Gal(β1-4)GlcNAcβ-Biotin (6′SLNLN-b), obtained from the Consortium for Functional Glycomics (www.functionalglycomics.org) through the resource request program, were coupled to streptavidin coated biosensors (Fortebio Inc.). Recombinant HA was diluted to 4.42 μM trimer in kinetics buffer (PBS containing 0.02% Tween 20, 0.005% sodium azide and 100 μg/ml bovine serum albumin). Bio-Layer Interferometry (BLI) analyzed binding on an Octet Red instrument (Fortebio Inc.) according to the manufacturer’s instructions and data were analyzed using the system software.
PDB accession codes
The atomic coordinates and structure factors of each HA model described herein are available from the RCSB PDB database (www.pdb.org) under the accession codes listed in Table 2.
Supplementary Material
Acknowledgments
This work was funded by the Centers for Disease Control and Prevention. The authors would like to thank the WHO Global Influenza Surveillance and Response System (GISRS), and all submitting laboratories that have deposited their H3N2 HA sequences to the GISAID database. The authors would also like to thank the staff of both SER-CAT sector-22, as well as SSRL BL9-2, for their help for data collection. The U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No: DE-AC02-06CH11357, support use of the Advanced Photon Source at Argonne National Laboratory. Use of the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory is operated for the U.S. Department of Energy Office of Science by Stanford University and is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The Consortium for Functional Glycomics (CFG) funded by National Institute of General Medical Sciences Grant GM62116, produced Glycan microarrays under contract, for the Centers for Disease Control and Prevention. We also thank the CFG for supplying the biotinylated glycans used here, through their resource request program. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
References
- Abe Y, Takashita E, Sugawara K, Matsuzaki Y, Muraki Y, Hongo S. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol. 2004;78:9605–9611. doi: 10.1128/JVI.78.18.9605-9611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka N, Tanaka Y, Sakai T, Fujii Y, Ohuchi R, Ohuchi M. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes and infection / Institut Pasteur. 2006;8:511–519. doi: 10.1016/j.micinf.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Aytay S, Schulze IT. Single amino acid substitutions in the hemagglutinin can alter the host range and receptor binding properties of H1 strains of influenza A virus. J Virol. 1991;65:3022–3028. doi: 10.1128/jvi.65.6.3022-3028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum LG, Paulson JC. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- Bean WJ, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, Webster RG. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J Virol. 1992;66:1129–1138. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999a;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- Bush RM, Fitch WM, Bender CA, Cox NJ. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol Biol Evol. 1999b;16:1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, Steinhauer DA, Cummings RD. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci U S A. 2014;111:E2241–E2250. doi: 10.1073/pnas.1323162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayen NE. Optimization techniques for automation and high throughput. Methods Mol Biol. 2007;363:175–190. doi: 10.1007/978-1-59745-209-0_9. [DOI] [PubMed] [Google Scholar]
- Childs RA, Palma AS, Wharton S, Matrosovich T, Liu Y, Chai W, Campanero-Rhodes MA, Zhang Y, Eickmann M, Kiso M, Hay A, Matrosovich M, Feizi T. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- Daniels PS, Jeffries S, Yates P, Schild GC, Rogers GN, Paulson JC, Wharton SA, Douglas AR, Skehel JJ, Wiley DC. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 1987;6:1459–1465. doi: 10.1002/j.1460-2075.1987.tb02387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RS, Douglas AR, Skehel JJ, Wiley DC. Analyses of the antigenicity of influenza haemagglutinin at the pH optimum for virus-mediated membrane fusion. J Gen Virol. 1983;64(Pt 8):1657–1662. doi: 10.1099/0022-1317-64-8-1657. [DOI] [PubMed] [Google Scholar]
- Das SR, Hensley SE, David A, Schmidt L, Gibbs JS, Puigbo P, Ince WL, Bennink JR, Yewdell JW. Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc Natl Acad Sci USA. 2011;108:E1417–E1422. doi: 10.1073/pnas.1108754108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL. The Pymol Molecular Graphics System. 2002 http://www.pymol.org.
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Gou Y, Lustig A, Bächinger HP, Engel J. Stabilization of short collagen-like triple helices by protein engineering. Journal of molecular biology. 2001;308:1081–1089. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human Infection with a Novel Avian-Origin Influenza A (H7N9) Virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassauer A, Egorov AY, Ferko B, Romanova I, Katinger H, Muster T. A host restriction-based selection system for influenza haemagglutinin transfectant viruses. The Journal of general virology. 1998;79(Pt 6):1405–1409. doi: 10.1099/0022-1317-79-6-1405. [DOI] [PubMed] [Google Scholar]
- Gulati S, Smith DF, Cummings RD, Couch RB, Griesemer SB, St George K, Webster RG, Air GM. Human H3N2 Influenza Viruses Isolated from 1968 To 2012 Show Varying Preference for Receptor Substructures with No Apparent Consequences for Disease or Spread. PLoS One. 2013;8:e66325. doi: 10.1371/journal.pone.0066325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, Viswanathan K, Raman R, Sasisekharan R, Bennink JR, Yewdell JW. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2:160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology. 1997;227:493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- Kaverin NV, Rudneva IA, Ilyushina NA, Varich NL, Lipatov AS, Smirnov YA, Govorkova EA, Gitelman AK, Lvov DK, Webster RG. Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of escape mutants. J Gen Virol. 2002;83:2497–2505. doi: 10.1099/0022-1317-83-10-2497. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Suzuki Y. Evidence for N-Glycan Shielding of Antigenic Sites during Evolution of Human Influenza A Virus Hemagglutinin. J Virol. 2012;86:3446–3451. doi: 10.1128/JVI.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J. 2007;4:42. doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land A, Braakman I. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie. 2001;83:783–790. doi: 10.1016/s0300-9084(01)01314-1. [DOI] [PubMed] [Google Scholar]
- Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ, Gamblin SJ, Hay AJ, Daniels RS, McCauley JW. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Zhou H, Chan W, Kemble G, Jin H. Single amino acid substitutions in the hemagglutinin of influenza A/Singapore/21/04 (H3N2) increase virus growth in embryonated chicken eggs. Vaccine. 2006;24:6691–6693. doi: 10.1016/j.vaccine.2006.05.062. [DOI] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros R, Escriou N, Naffakh N, Manuguerra JC, van der Werf S. Hemagglutinin residues of recent human A(H3N2) viruses that affect agglutination of chicken erythrocytes. International Congress Series. 2001;1219:369–374. doi: 10.1006/viro.2001.1121. [DOI] [PubMed] [Google Scholar]
- Meunier JC, Fournillier A, Choukhi A, Cahour A, Cocquerel L, Dubuisson J, Wychowski C. Analysis of the glycosylation sites of hepatitis C virus (HCV) glycoprotein E1 and the influence of E1 glycans on the formation of the HCV glycoprotein complex. The Journal of general virology. 1999;80(Pt 4):887–896. doi: 10.1099/0022-1317-80-4-887. [DOI] [PubMed] [Google Scholar]
- Nicholls JM, Chan MC, Chan WY, Wong HK, Cheung CY, Kwong DL, Wong MP, Chui WH, Poon LL, Tsao SW, Guan Y, Peiris JS. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med. 2007;13:147–149. doi: 10.1038/nm1529. [DOI] [PubMed] [Google Scholar]
- Nobusawa E, Ishihara H, Morishita T, Sato K, Nakajima K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 2000;278:587–596. doi: 10.1006/viro.2000.0679. [DOI] [PubMed] [Google Scholar]
- Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk W, Wilson IA, Paulson JC. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angewandte Chemie. 2012;51:4860–4863. doi: 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DY, Barr IG, Mosse JA, Laurie KL. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol. 2008;46:2189–2194. doi: 10.1128/JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Scalepack Manual. 1997. [Google Scholar]
- Owen RE, Yamada E, Thompson CI, Phillipson LJ, Thompson C, Taylor E, Zambon M, Osborn HM, Barclay WS, Borrow P. Alterations in receptor binding properties of recent human influenza H3N2 viruses are associated with reduced natural killer cell lysis of infected cells. J Virol. 2007;81:11170–11178. doi: 10.1128/JVI.01217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MS, Schild GC. An antigenic variant of the Hong Kong-68 influenza A 2 virus. J Hyg (Lond) 1971;69:99–103. doi: 10.1017/s0022172400021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova L, Smith K, West AH, Wilson PC, James JA, Thompson LF, Air GM. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS ONE. 2012;7:e41895. doi: 10.1371/journal.pone.0041895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Taubenberger JK. The origin of the 1918 pandemic influenza virus: a continuing enigma. J Gen Virol. 2003;84:2285–2292. doi: 10.1099/vir.0.19302-0. [DOI] [PubMed] [Google Scholar]
- Rogers GN, D’Souza BL. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Rogers GN, Daniels RS, Skehel JJ, Wiley DC, Wang XF, Higa HH, Paulson JC. Host-mediated selection of influenza virus receptor variants. Sialic acid-α2,6Gal-specific clones of A/duck/Ukraine/1/63 revert to sialic acid-α2,3Gal-specific wild type in ovo. J Biol Chem. 1985;260:7362–7367. [PubMed] [Google Scholar]
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza hemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- Seidel W, Kunkel F, Geisler B, Garten W, Herrmann B, Dohner L, Klenk HD. Intraepidemic variants of influenza virus H3 hemagglutinin differing in the number of carbohydrate side chains. Archives of virology. 1991;120:289–296. doi: 10.1007/BF01310484. [DOI] [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- Skehel JJ, Stevens DJ, Daniels RS, Douglas AR, Knossow M, Wilson IA, Wiley DC. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci USA. 1984;81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Ann Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Chen LM, Donis RO, Paulson JC, Wilson IA. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. Journal of molecular biology. 2008;381:1382–1394. doi: 10.1016/j.jmb.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. Journal of molecular biology. 2006a;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Microbiol. 2006b;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006c;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Stevens J, Chen LM, Carney PJ, Garten R, Foust A, Le J, Pokorny BA, Manojkumar R, Silverman J, Devis R, Rhea K, Xu X, Bucher DJ, Paulson J, Cox NJ, Klimov A, Donis RO. Receptor Specificity of Influenza A H3N2 Viruses Isolated in Mammalian Cells and Embryonated Chicken Eggs. J Virol. 2010 doi: 10.1128/JVI.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Stray SJ, Pittman LB. Subtype- and antigenic site-specific differences in biophysical influences on evolution of influenza virus hemagglutinin. Virol J. 2012;9:91. doi: 10.1186/1743-422X-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MG, Shay DK, Zhou H, Bridges CB, Cheng PY, Burns E, Bresee JS, Cox NJ. Estimates of deaths associated with seasonal influenza - United States, 1976–2007. MMWR Morb Mortal Wkly Rep. 2010;59:1057–1062. [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New World Bats Harbor Diverse Influenza A Viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusche C, Steinbruck L, McHardy AC. Detecting patches of protein sites of influenza A viruses under positive selection. Mol Biol Evol. 2012;29:2063–2071. doi: 10.1093/molbev/mss095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood PA, Skehel JJ, Wiley DC. Receptor-binding characteristics of monoclonal antibody-selected antigenic variants of influenza virus. J Virol. 1987;61:206–208. doi: 10.1128/jvi.61.1.206-208.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gils MJ, Bunnik EM, Boeser-Nunnink BD, Burger JA, Terlouw-Klein M, Verwer N, Schuitemaker H. Longer V1V2 region with increased number of potential N-linked glycosylation sites in the HIV-1 envelope glycoprotein protects against HIV-specific neutralizing antibodies. J Virol. 2011;85:6986–6995. doi: 10.1128/JVI.00268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigerust DJ, Ulett KB, Boyd KL, Madsen J, Hawgood S, McCullers JA. N-linked glycosylation attenuates H3N2 influenza viruses. J Virol. 2007;81:8593–8600. doi: 10.1128/JVI.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T, Karamanska R, Chan RW, Chan MC, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, Haslam SM, Nicholls JM. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013;9:e1003223. doi: 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS ONE. 2008;3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu B, Zhou H, Suguitan AL, Jr, Cheng X, Subbarao K, Kemble G, Jin H. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol. 2010;84:6570–6577. doi: 10.1128/JVI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzeck K, Boyd KL, McCullers JA. Glycan shielding of the influenza virus hemagglutinin contributes to immunopathology in mice. American journal of respiratory and critical care medicine. 2011;183:767–773. doi: 10.1164/rccm.201007-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Laver WG. Determination of the number of nonoverlapping antigenic areas on Hong Kong (H3N2) influenza virus hemagglutinin with monoclonal antibodies and the selection of variants with potential epidemiological significance. Virology. 1980;104:139–148. doi: 10.1016/0042-6822(80)90372-4. [DOI] [PubMed] [Google Scholar]
- Weis WI, Brunger AT, Skehel JJ, Wiley DC. Refinement of the influenza virus hemagglutinin by simulated annealing. Journal of molecular biology. 1990;212:737–761. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]
- Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- World Health Organization. A revision of the system of nomenclature for influenza viruses: a WHO memorandum. Bull, WHO. 1980;58:585–591. [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol. 2013;22:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney PJ, Donis RO, Stevens J. Structure and Receptor Complexes of the Hemagglutinin from a Highly Pathogenic H7N7 Influenza Virus. J Virol. 2012;86:8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Si ZH, Moore JP, Sodroski J. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J Virol. 2000;74:11955–11962. doi: 10.1128/jvi.74.24.11955-11962.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.