Abstract
Using a family of cationic gold nanoparticles (NPs) with similar size and charge, we demonstrate that proper surface engineering can control the nature and identity of protein corona in physiological serum conditions. The protein coronas were highly dependent on the hydrophobicity and arrangement of chemical motifs on NP surface. The NPs were uptaken in macrophages in a corona-dependent manner, predominantly through recognition of specific complement proteins in the NP corona. Taken together, this study shows that surface functionality can be used to tune the protein corona formed on NP surface, dictating the interaction of NPs with macrophages.
Keywords: Gold nanoparticles, protein corona, macrophages, complement system, surface functionality
Proper control of nanoparticle (NP) surface functionality can dictate desired interactions of NPs with cells, maximizing their therapeutic efficacies. For example, cationic NPs are shown to interact with proteins,1 DNA,2 and siRNA3 in a charge complementary fashion and effectively deliver them both in vitro4 and in vivo.5 Cationic gold NPs, in particular, provide a suitable platform for targeted delivery due to their non-toxic core and functional versatility.6,7 The straightforward synthesis coupled with a tunable surface of gold NPs that can be engineered with a variety of chemical functional groups can further modulate their uptake efficiency.8 For instance, modification of gold NPs surface with lysine rendered tunable intracellular DNA delivery by simply changing the surface charge density of lysine residue, providing ~28 times higher transfection efficiency compared to polylysine analogue.9
It is well-recognized that after administration of NPs in blood, plasma/serum proteins adsorb on NP surface and form a protein layer, namely “protein corona”.10–16 There are several factors that can define the composition of protein corona. The surface chemistry of NPs largely dictates the thickness, decoration, and the identity of the protein corona, in addition to the physicochemical characteristics of the protein themselves together with incubation parameters (e.g., temperature).17,18 The protein corona gives NPs a new biological identity in contrast to their initial synthetic identity that determines NPs—cell interactions. Among various types of the cells, the interaction of NPs with macrophages is of significant interest because of macrophages’ crucial role in NP clearance from blood stream. For instance, we have demonstrated that blood half-life of cationic gold NPs is significantly lower compared to neutral and negative counterpart and accumulated primarily in the organs related to mononuclear phagocytic systems (MPS) including liver and spleen.19 This is mainly occurred due to the non-specific absorbance of certain plasma/serum proteins (opsonins) on cationic gold NP surface20,21 that helps MPS cells, i.e. macrophages, recognize and subsequent uptake of the NPs.22 Such a rapid clearance from the blood stream (after systemic administration via MPS) is the major hurdle for effective in vivo targeting applications of cationic gold NPs.23,24 In order to enhance the blood residency time of cationic gold NPs, it is of crucial importance to understand the effect of engineered surface functionality on protein corona composition and its recognition by macrophages.25, 26 However, interplay between surface structure and corona composition is still poorly understood, mainly because of the complexity of protein corona composition/structure.
Herein, we investigated the interplay of protein adsorption and macrophage recognition of surface engineered cationic gold NPs. We find that NP surface functionality dictates the protein corona formation in serum under physiological conditions. The uptake of NPs by macrophages was directly related to the amounts of key complement proteins adsorption on NP surface, suggesting a clear and crucial role of blood complement system in recognizing NP surface and subsequent phagocytosis. Importantly, NPs with similar hydrophobicity, but constitutional isomeric head-group, demonstrated altered corona identity and macrophage uptake, signifying the critical role of chemical design in nanomedicine.
RESULTS AND DISCUSSION
We synthesized a family of gold NPs of the same core size (~2 nm core) differing in their monolayer functionality (Figure 1) to probe the role of NP surface chemistry on protein corona formation and macrophage uptake.
Figure 1.
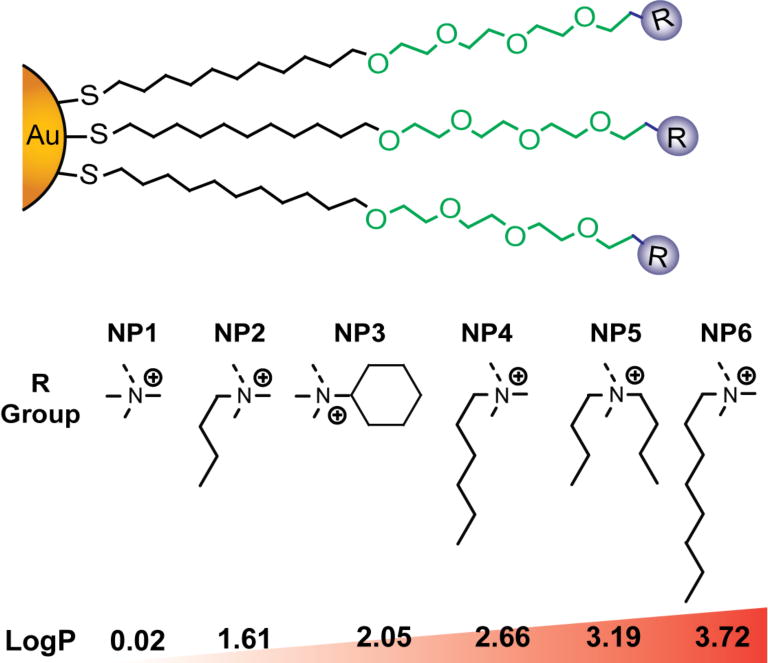
The gold NPs used in the present study with increasing hydrophobicity. Log P denotes relative hydrophobicity of the functional head-groups.
All gold NPs were synthesized from the same batch of pentanethiol capped gold NPs27 followed by a place exchange reaction as described previously.28 A key feature of this NP design is the tetra(ethylene glycol) moiety in the NP monolayer that improves solubility and help expose the terminal functional groups on the surface of the NP, allowing us to probe the direct effect of surface chemistry on NP protein corona formation.29 In addition, the similar hydrodynamic radii and zeta potential of these NPs in buffer and cell culture media (Table 1) provide a suitable platform to directly probe the monolayer structure-property relationship (SAR) on corona formation and macrophage uptake. We further chose surface hydrophobicity as a representative variable due to its importance in a variety of critical delivery-related parameters including cellular uptake,30 hemolysis,31 biodistribution,32 and immune response33. Most importantly, the atomic level control of the design allowed us to fabricate NP bearing constitutional isomeric headgroups (e.g., NP5 and NP6) to probe their effect on the protein binding and macrophage uptake.
Table 1.
Nanoparticle physiochemical properties.
| Nanoparticle | Hydrodynamic size (nm) in 5mM PB pH 7.4a | Hydrodynamic size (nm) in cell culture mediaa | Zeta Potential (mV) in 5mM PB pH 7.4a | Calculated Log P of the ligand headgroup |
|---|---|---|---|---|
| NP1 | 10.9 ± 3.1 | 11.4 ± 3.3 | 15.2 ± 6.9 | 0.02 |
| NP2 | 12.8 ± 2.9 | 10.1 ± 3.0 | 17.8 ± 7.9 | 1.61 |
| NP3 | 9.9 ± 2.8 | 13.0 ± 2.7 | 21.7 ± 7.6 | 2.05 |
| NP4 | 10.4 ± 2.8 | 10.0 ± 3.0 | 19.7 ± 4.8 | 2.66 |
| NP5 | 11.0 ± 3.0 | 10.7 ± 2.4 | 24.9 ± 9.9 | 3.19 |
| NP6 | 9.9 ± 2.9 | 11.8 ± 3.8 | 18.2 ± 8.6 | 3.72 |
Uncertainty represents the standard deviation of three parallel measurements
Previously, we have demonstrated that cationic NPs can effectively bind to the anionic proteins at physiological pH.34–36 However, this ‘one binding site’ model cannot address the complex nature of NP-protein interaction in serum that contains thousands of proteins of wide variety of physicochemical properties. To this end, we have studied the NP-protein interaction in 10% and 50% serum, mimicking in vitro and in vivo conditions. Previous reports have shown that NP-bound serum proteins have two different layer, ‘soft’ corona (reversibly bound proteins) and ‘hard’ corona (irreversibly bound proteins).37–39 Hard corona proteins tend to stick with NP surface for longer time and can essentially dictate the NP biodistribution and macrophage uptake.40 We have studied the identity of ‘hard’ corona proteins using shotgun proteomics41,42 (liquid chromatography tandem mass spectrometry (LC-MS/MS)).43 Although other studies identified numerous proteins in the hard corona composition,44 we have successfully identified and semi-quantitatively analyzed corona composition of surface engineered NPs, comprising nearly 100 different types of proteins. The full list of NP-surface bound and control serum proteins identified including their molecular weight and relative abundance can be found in Supplementary Document S1 and S2. First, although there were no distinct protein-size dependent binding profile in NP1-NP6 (Figure 2a), we observed an enrichment of proteins with lower molecular weight compared to control serum (Supporting Information, Supplementary Figure S1) both in 10% and 50% serum conditions. Second, proteins bearing a net negative charge (pI<7) at pH 7.4 were significantly enriched the corona composition for NP1-NP6 irrespective of their relative abundance in serum, demonstrating that the binding of NP1-NP6 is predominantly electrostatic (Figure 2b). Significantly, the binding profile of proteins was greatly altered in different serum concentrations; proteins with pI>8 were enriched in 50% serum compared to 10% serum, depicting the dynamic nature of the corona evolution in higher serum concentration. Third, the hard corona protein found on the NP surface does not correspond to their abundance in serum, signifying a high degree of selectivity in NP-serum interaction. For example, serum albumin is the most abundant protein in serum (comprises ~60–70% of serum proteins) but it was not the highest abundant protein detected in the corona for any NPs. Instead, corona was significantly enriched with proteins that have very low abundance or below detection limit, e.g. complement component 3, but have major role in opsonization (Supplementary Document S1 and S2).45, 46
Figure 2.
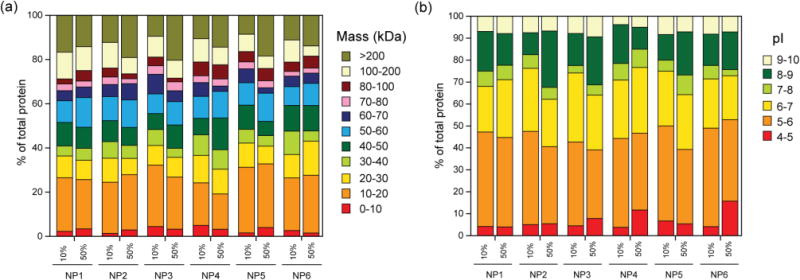
Classification of surface bound proteins for NP1-NP6 according to their (a) molecular weight and (b) calculated isoelectric point (pI). Corona was enriched with proteins across all molecular weights for both 10% and 50% serum. In contrast, proteins with pI<7 were adsorbed predominantly to NP1-NP6 at pH 7.4 for both 10% and 50% serum, demonstrating that binding of NP1-NP6 with serum proteins is principally electrostatic in nature.
To understand the role of specific chemical group on corona formation and subsequent effect on NP fate in blood, we have further employed bioanalytical tools47 to classify the identified proteins according to their function in blood namely complement activation, immune response, coagulation, acute phase response, and lipid metabolism (Figure 3). In our analysis, immunoglobulins and apolipoproteins were major protein types (~60% of total protein) that were found in corona composition, regardless of NP surface composition, when incubated in 10% (Figure 3) and 50% serum (Figure 4). Notably, the number of identified proteins in 10% serum was higher than that of in 50% serum. This is presumably happened due to the fact that in diluted serum samples the interaction between NP and serum proteins is predominantly reversible in nature while at high serum concentrations the abundant serum proteins can mask NP surfaces, thereby decreasing the access of low abundant proteins to NP surfaces. Apolipoproteins are involved in a variety of cardiovascular and neurodegenerative diseases;48 thus, their binding to the surface of NPs can affect the NP biodistribution.49 Notably, the amount of apolipoproteins was decreased with increasing NP surface hydrophobicity, confirming the key role of NP hydrophobicity in apolipoprotein formation in 10% serum. However, at higher amount of proteins (50% serum), we did not observe a significant correlation between the amounts of apolipoproteins and NP hydrophobicity, demonstrating the critical role of protein amount in variation of protein-NP interactions (Figure 4b).
Figure 3.
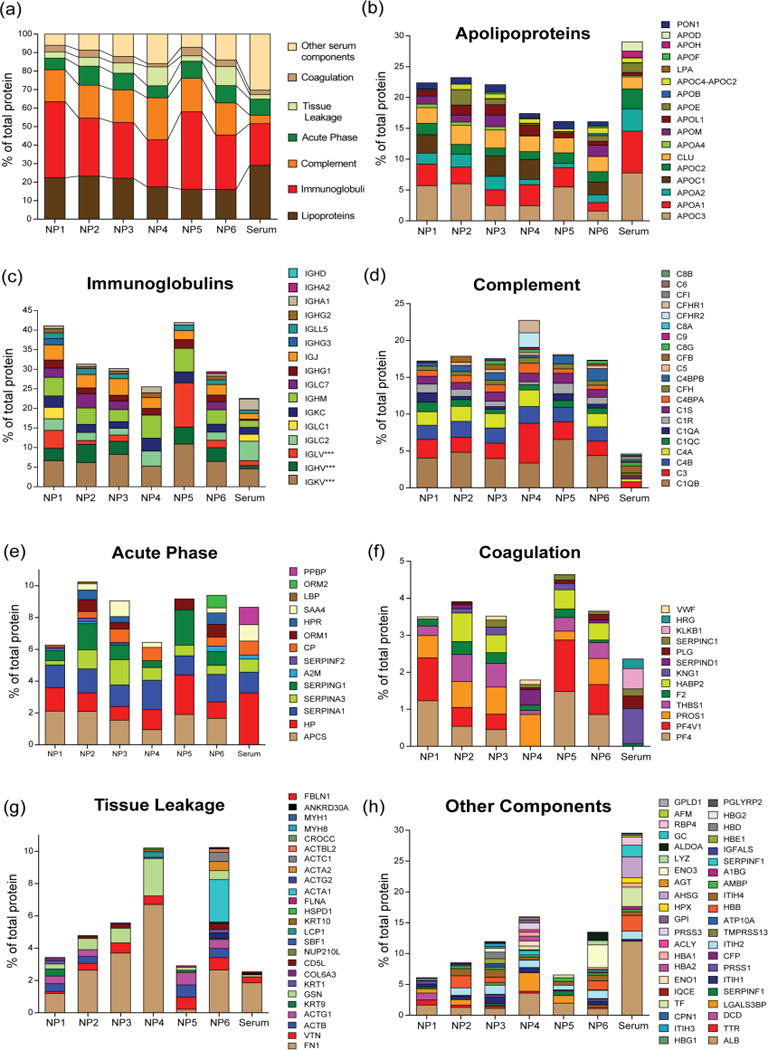
Classification of identified corona on NP1-NP6 according to their physiological functions in 10% serum.
Figure 4.
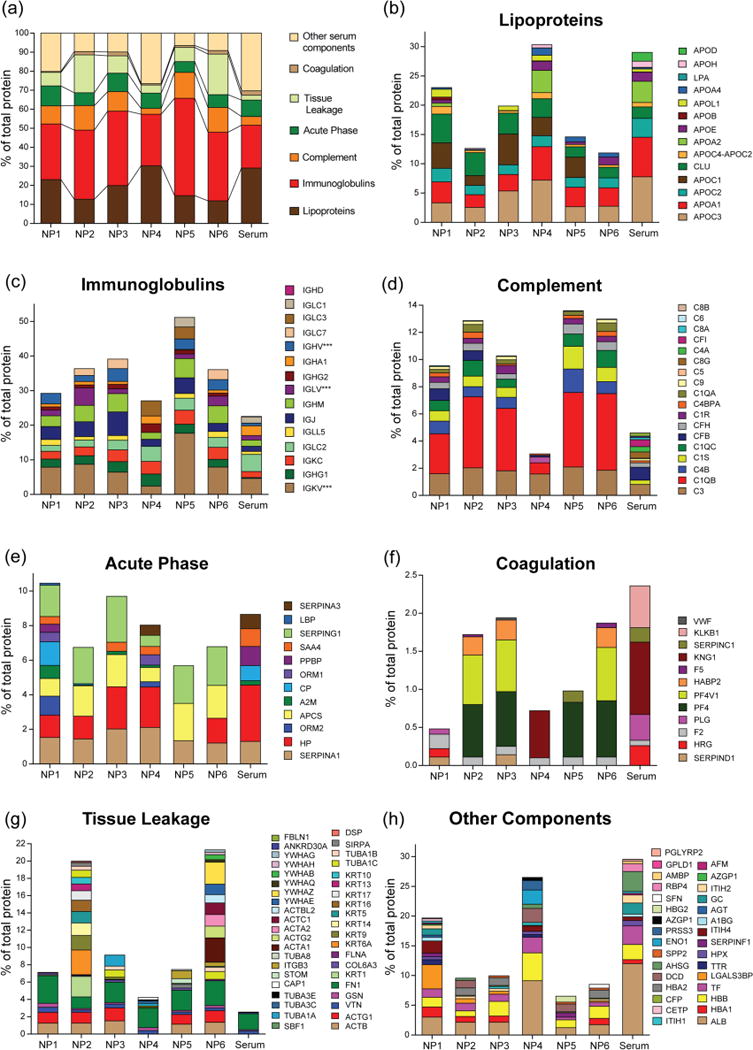
Classification of identified corona according to their physiological functions in 50% serum.
As such, all NPs strongly interacted with immunoglobins that are highly abundant in blood, play critical role in immune reaction, and promote phagocytosis.50 In 10% serum with increasing hydrophobicity, the amount of immunoglobulins were decreased on NP surface (Figure 3c), however, in 50% serum the opposite trend was observed (Figure 4c), mirroring significant change in behavior of NP-protein interaction at different protein concentrations.51 However, a 3–4 fold enrichment of complement proteins were observed for all NPs in both 10% and 50% serum compared to control (Figure 3d and 4d), indicating complement system plays a crucial role in interacting with NP surfaces and might be responsible of clearing these 2 nm cationic gold NPs from the systemic circulation via reticuloendothelial system. Importantly, NP surface design also played a vital role in determining the interaction with particular complement proteins. For example, NP3 demonstrated a 2.5 fold increase in complement component 3 (C3) on its surface compared to control serum sample, however, NP4 that bears a similar number of carbon atoms showed ~7 times enrichment of C3 in its corona in 10% serum (Supplementary Document S1), signifying that a small change in chemical group arrangement can greatly alter the corona composition on NP surface.
Although acute phase response proteins present in high amount in serum, no significant enrichment was observed in both 10% and 50% serum (Figures 3e and 4e). However, a similar and/or decreased affinity of coagulation protein was observed in case of coagulation proteins when incubated with different concentrations of serum (Figures 3f and 4f). The “tissue leakage” proteins, that are very low abundance in serum but are involved in many diseases,52 were also significantly enriched on NP surface. Significantly, the long carbon tail of NP6 amassed more “tissue leakage” proteins compared to NP5 with branched end group (Figures 3g and 4g), again mirroring the fact that branched/cyclic structural design on NP instead of linear chemical functionality can potentially decrease the protein corona formation on NP surface, in 10% serum. Finally, the other major components of serum including albumin displayed a lower affinity for these NPs (Figures 3h and 4h).
In our analysis, complement factors were shown to be significantly enhanced on NP surface and are expected to play a major role in NP phagocytosis. To understand the role of NP surface functionality on the phagocytosis, we performed cellular uptake of NP1-NP6 upon incubation with RAW 264.7 (murine macrophage) in 10% and 50% human serum (Figure 5). In general, hydrophobic NPs demonstrated lower uptake compared to hydrophilic NPs in both serum concentrations whereas control experiment in serum-free media demonstrated high non-specific uptake for all NPs (Supplementary Figure S2), a feature that has been observed in prior studies.30. Significantly, NP5 and NP6 bearing branched and linear alkyl tail with same carbon atoms showed similar uptake pattern in serum free media (Supplementary Figure S2) but showed significantly different uptake in 10% serum, further attesting the fact that both type and arrangement of NP surface functionality53 can dictate specific protein adsorption leading to differential macrophage recognition. However, the uptake of these two particles was similar in 50% serum, presumably due to the fact that higher serum concentration can mask the minute changes in NP chemical functionality and alter its behavior for macrophage recognition. It is also important to note that all NPs were non-toxic at the concentration (50 nM) tested for this experiment (Supporting Information, Supplementary Figure S3).
Figure 5.
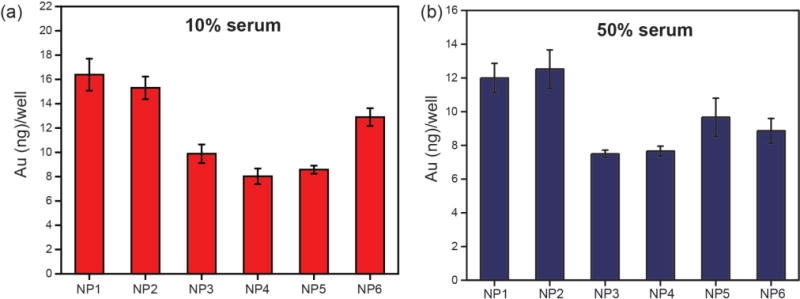
Uptake of NP1-NP6 (50 nM) in RAW cells after 3 h in (a) 10% and (b) 50% serum. Error bars represent standard deviation.
We further investigated the correlation between the uptake pattern and corona formation of NP1-NP6 to determine the type of proteins that in general contributed to macrophage recognition. Figure 6 shows the Pearson correlation coefficients (r) of proteins found in corona with uptake values in 50% serum. In general, proteins from complement, lipoprotein, and coagulation categories showed high positive correlation with the macrophage uptake, confirming that these three protein category has significant influence on macrophage uptake. Significantly, the presence of complement proteins in the corona has favored the uptake of NPs in macrophages while a reverse behavior has been observed in case of Immunoglobulins. This analysis clearly demonstrates that although immunoglobulins were found in high amount on NP1-NP6 surfaces in 50% serum, there is a significant competition/reorganization of low abundant protein moieties (e.g. complement factors, coagulation) on NP surfaces that eventually led to macrophage recognition. Analysis in 10% serum yielded similar negative correlation with NP uptake and immunoglobulins, however, a positive correlation was observed with a number of lipoproteins, demonstrating protein concentration plays a critical role in corona dependent macrophage uptake (see Supporting Information, Supplementary Figure S4).
Figure 6.
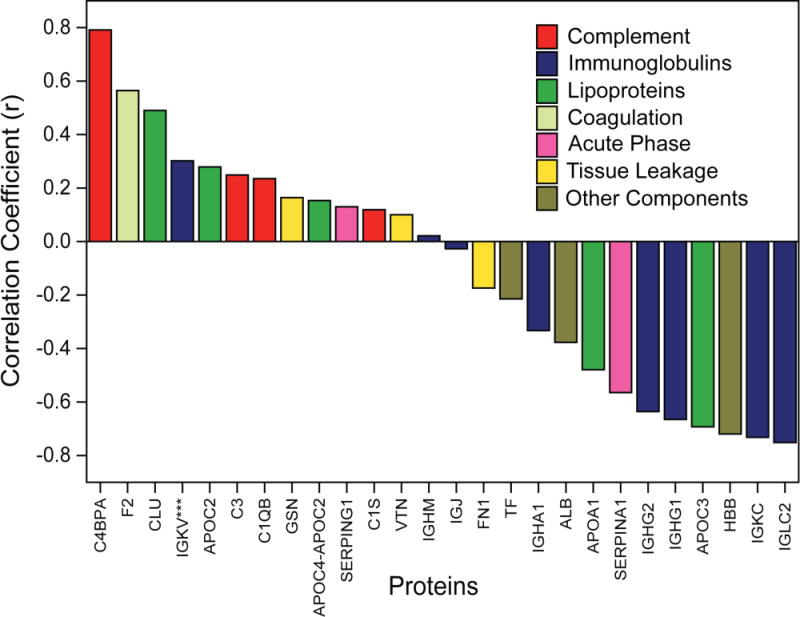
Correlation between the macrophage uptake and type of corona protein found in NP1-NP6 in 50% serum condition. The proteins have been selected form different categories that showed large positive or negative correlation.
C4BPA and IGLC2 showed highest positive and negative correlation coefficient (r) with macrophage uptake (Figure 6). C4BPA has the capability to bind to apoptotic/necrotic cells to identify them for immune system for cleaning up.54, 55 Interestingly, some of the bacterial (e.g., Streptococcus pyogenes) and fungal pathogens would be established as infection, through binding of C4BP proteins to their membrane.56–58 Therefore, one can expect that the association of C4BPA-based proteins in the composition of protein corona may cause higher uptake of NPs by macrophages. To understand the specific role of NP surface functionality that yielded this behavior, we further studied the uptake behavior of the NPs with respect to the amount of proteins that have the highest correlation with the NP uptake pattern (namely C4BPA and IGLC2). Figure 7 shows the correlation between macrophage uptake of NP1-NP6 and these corona proteins in 50% serum. The results show that hydrophillic NPs (NP1 & NP2) showed high C4BPA adsorption (Figure 7a; Pearson correlation coefficient of 0.80 with P-Value <0.1) and consequent high uptake while hydrophobic NPs showed moderate to low adsorption with lower macrophage uptake, demonstrating an effective way to reduce macrophage recognition by modulating NP surface hydrophobicity. On the other hand, higher adsorption of IGLC2 (Figure 7b; Pearson correlation coefficient of 0.82 with P-Value <0.05) on hydrophobic NP surface consequently decreased their uptake by macrophages, demonstrating simple engineering of NP surface can either attract or repel particular serum proteins that eventually lead to macrophage recognition. Taken together, the amount adsorbed and subsequent uptake were dictated by the type of NP surface used, providing an essential design parameter for regulating macrophage recognition via surface chemistry of NPs.
Figure 7.
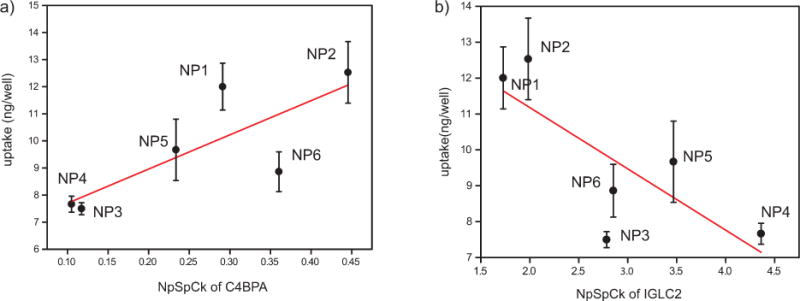
Correlation between macrophage uptake of NP1-NP6 and particular corona proteins in 50% serum a) C4BPA and b) IGLC2. Line is added to lead the eye.
CONCLUSION
In summary, we have demonstrated that NP surface functionality dictate the formation of protein corona in both in vitro and physiological serum concentrations. Significantly, this differential evolution of corona on NP surface was altered not only with NP surface hydrophobicity but also with the arrangement of organic end group on NP surface. For example, NPs featuring branched or cyclic end groups, in general, adsorbed less proteins compared to linear structures with similar hydrophobicity. This formation of protein corona further dictated the uptake in macrophages and was shown to be highly correlated to specific complement proteins in both 10% and 50% serum conditions. Moreover, the uptake was both positively or negatively correlated to specific type of proteins and dictated by type of NP surface functionality, demonstrating significant contribution stemming from NP surface to organize the corona proteins and subsequent macrophage recognition. This study provides critical guidelines for engineering the NP surface that can either avoid or harness key protein components from serum for different therapeutic applications and attests the importance of chemical design in controlling the in situ formation of NP protein corona.
METHODS
Nanoparticle synthesis and characterization
Gold nanoparticles cores (d~2nm) stabilized with a monolayer of 1-pentanethiol were synthesized following the Brust-Schiffrin methodology.23 All the impurities were removed according to previous protocol.59 Murray place-exchange reaction24 was performed by dissolving the synthesized thiolated ligand60 in dry dichloromethane (DCM) with the pentanethiol-capped gold cores, stirring for 3 days at room temperature (5:1 w/w ratio of ligand/gold cores). DCM was then evaporated under reduced pressure and the black residue was dissolved in distilled water. Dialysis was performed for 5 days (membrane MWCO = 10,000) to remove excess ligand and salts remaining in the nanoparticle solution. After dialysis, the particles were lyophilized and dissolved in deionized MQ water. NP hydrodynamic radii and zeta potential were measured using Zetasizer instrument (Zetasizer Nano ZS, Malvern) according to the previous reports.31,33
Nanoparticle incubation in human serum and protein isolation
Human serum was collected/purchased from healthy voluntaries. Written informed consent was obtained from all participants. The methods were carried out in accordance with the approved guidelines. This was approved by the Medical Ethical Committee of the Academic Medical Center (W11_084/#11.17.864). 100 μL of AuNPs (1μM) were mixed with 400 μL of human serum (for 10%: 50 μL of human serum and 350 μL of PBS; for 50%: 250 μL of human serum and 150 μL of PBS) and incubated for 1h at 37 °C. The mixture were then centrifuged at 13 000g at 15 °C for 30 min. The supernatant was removed and the collected corona coated AuNPs was redispersed in 500 μL of cold (15 °C) phosphate buffered saline (PBS). The solution was centrifuged again (at the same condition) and the collected particles were redispersed in 500 μL of cold (15 °C) PBS. After another centrifugation process, the hard corona coated particles were introduced to LC MS/MS procedure.
Mass spectrometric detection of NP-bound proteins
The nano liquid chromatography (n-LC) combined with tandem mass spectrometry (nLC-MS/MS) was used to identify the NP-bound proteins (corona layer of NPs), calculate the total spectra count of all peptides matched to a specific protein, and finally normalized for molecular weight (Mw) of each protein to assess relative quantitative concentration of each protein present on the corona of NP. The equation used for the semi-quantitative assessment of each protein present on the corona is as follows which was described in previous studies:43
NpSpCk is the normalized percentage of the spectral count for protein k, SpC is the spectral count identified, and Mw is the molecular weight (kDa) of protein k. The SpC of each identified protein was normalized to the protein mass and expressed as the Semi-quantitative assessment of protein. This correction is based on the protein size and evaluates the relative contribution of each protein present in the corona of NPs.
Nanoparticle uptake in macrophages
RAW 264.7 cells (murine macrophage) were cultured at 37 °C under a humidified atmosphere of 5 % CO2. The cells were grown in RPMI media containing 10 % fetal bovine serum (FBS) and 1% antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). For the uptake experiment, 250,000 cells/well were plated in a 48-well plate prior to the experiment. After 24h, cells were washed one time with PBS followed by NP treatment (50 nM/well) in either 10% or 50% serum for 3 h. Following incubation, cells were washed three times with PBS, lysed and the intracellular gold amount was measured using inductively coupled plasma mass spectrometry (Elan 6100, Perkin-Elmer, Shelton, CT, USA). Each cell uptake experiment was done using at least five parallel replicates and each replicate was measured 5 times by ICP-MS. ICP-MS operating conditions are as below: rf power 1600 W; plasma Ar Flow rate, 15 ml/min, nebulizer Ar flow rate, 0.98 ml/min and dwell time, 45 ms.
Statistical analysis
All quantitative measurements were collected at least three parallel replicates in each group and data were expressed as mean ± standard deviation. Pearson Correlation Coefficient (PCC) were calculated to determine the relationship of uptake and corona protein of interest. The Pearson Correlation can be any value between −1 and 1 depending on the extent of collinearity. Negative values of r show a negative correlation between data sets.61 Correlation matrix was calculated using IBM SPSS Statistics software; (IBM Corp. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp.)
Supplementary Material
Acknowledgments
This work was supported by the grants from the NIH (GM GM077173) and the Center for Hierarchical Manufacturing (CMMI-1025020). M.M. would like to thank “Saramadan Elmi” federation for grant number: 11/66332.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: …
Notes
The authors declare no competing financial interest.
Author Contributions
K.S. and V.M.R. conceived the concept. K.S., S.T.K., V.M.R., and M.M. designed the experiments. D.F.M. synthesized and characterized the nanoparticles. M.R. and F.R. performed the mass spectrometric identification of protein corona formed on nanoparticles. K.S. and S.T.K. performed cellular uptake and viability studies. S.H. and R.D. performed ICP-MS analysis. K.S., M.Y., and R.M. analyzed the data. K.S. wrote the paper with input from M.M. and V.M.R.
References
- 1.Ghosh P, Yang X, Arvizo R, Zhu Z-J, Agasti SS, Mo Z, Rotello VM. Intracellular Delivery of a Membrane-Impermeable Enzyme in Active Form Using Functionalized Gold Nanoparticles. J Am Chem Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Gao S, Ye W-H, Yoon HS, Yang Y-Y. Co-Delivery of Drugs and DNA from Cationic Core-Shell Nanoparticles Self-Assembled from a Biodegradable Copolymer. Nat Mater. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG. Degradable Lipid Nanoparticles with Predictable In Vivo siRNA Delivery Activity. Nat Commun. 2014;5:4277–4287. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majzoub RN, Chan C-L, Ewert KK, Silva BFB, Liang KS, Jacovetty EL, Carragher B, Potter CS, Safinya CR. Uptake and Transfection Efficiency of PEGylated Cationic Liposome–DNA Complexes with and without RGD-Tagging. Biomaterials. 2014;35:4996–5005. doi: 10.1016/j.biomaterials.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-Viral Vectors for Gene-Based Therapy. Nat Rev Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 6.Saha K, Bajaj A, Duncan B, Rotello VM. Beauty is Skin Deep: A Surface Monolayer Perspective on Nanoparticle Interactions with Cells and Bio-Macromolecules. Small. 2011;7:1903–1918. doi: 10.1002/smll.201100478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boisselier E, Astruc D. Gold Nanoparticles in Nanomedicine: Preparations, Imaging, Diagnostics, Therapies and Toxicity. Chem Soc Rev. 2009;38:1759–1782. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- 8.Ding Y, Jiang Z, Saha K, Kim CS, Kim ST, Landis RF, Rotello VM. Gold Nanoparticles for Nucleic Acid Delivery. Mol Ther. 2014;22:1075–1083. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh PS, Kim C-K, Han G, Forbes NS, Rotello VM. Efficient Gene Delivery Vectors by Tuning the Surface Charge Density of Amino Acid-Functionalized Gold Nanoparticles. ACS Nano. 2008;2:2213–2218. doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein−Nanoparticle Interactions: Opportunities and Challenges. Chem Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 11.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DWH, Cohen Y, Emili A, Chan WCW. Protein Corona Fingerprinting Predicts the Cellular Interaction of Gold and Silver Nanoparticles. ACS Nano. 2014;8:2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 12.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 13.Kah JCY, Chen J, Zubieta A, Hamad-Schifferli K. Exploiting the Protein Corona around Gold Nanorods for Loading and Triggered Release. ACS Nano. 2012;6:6730–6740. doi: 10.1021/nn301389c. [DOI] [PubMed] [Google Scholar]
- 14.Cifuentes-Rius A, de Puig H, Kah JCY, Borros S, Hamad-Schifferli K. Optimizing the Properties of the Protein Corona Surrounding Nanoparticles for Tuning Payload Release. ACS Nano. 2013;7:10066–10074. doi: 10.1021/nn404166q. [DOI] [PubMed] [Google Scholar]
- 15.Kah JCY, Grabinski C, Untener E, Garrett C, Chen J, Zhu D, Hussain SM, Hamad-Schifferli K. Protein Coronas on Gold Nanorods Passivated with Amphiphilic Ligands Affect Cytotoxicity and Cellular Response to Penicillin/Streptomycin. ACS Nano. 2014;8:4608–4620. doi: 10.1021/nn5002886. [DOI] [PubMed] [Google Scholar]
- 16.Hamad-Schifferli K. Exploiting the Novel Properties of Protein Coronas: Emerging Applications in Nanomedicine. Nanomed. 2015;10:1663–1674. doi: 10.2217/nnm.15.6. [DOI] [PubMed] [Google Scholar]
- 17.Sakulkhu U, Mahmoudi M, Maurizi L, Salaklang J, Hofmann H. Protein Corona Composition of Superparamagnetic Iron Oxide Nanoparticles with Various Physico-Chemical Properties and Coatings. Sci Rep. 2014;4:5020–5029. doi: 10.1038/srep05020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoudi M, Abdelmonem AM, Behzadi S, Clement JH, Dutz S, Ejtehadi MR, Hartmann R, Kantner K, Linne U, Maffre P, Metzler S, Moghadam MK, Pfeiffer C, Rezaei M, Ruiz-Lozano P, Serpooshan V, Shokrgozar MA, Nienhaus GU, Parak WJ. Temperature: The “Ignored” Factor at the NanoBio Interface. ACS Nano. 2013;7:6555–6562. doi: 10.1021/nn305337c. [DOI] [PubMed] [Google Scholar]
- 19.Arvizo RR, Miranda OR, Moyano DF, Walden CA, Giri K, Bhattacharya R, Robertson JD, Rotello VM, Reid JM, Mukherjee P. Modulating Pharmacokinetics, Tumor Uptake and Biodistribution by Engineered Nanoparticles. PLoS One. 2011;6:e24374. doi: 10.1371/journal.pone.0024374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakulkhu U, Maurizi L, Mahmoudi M, Motazacker M, Vries M, Gramoun A, Ollivier Beuzelin M-G, Vallee J-P, Rezaee F, Hofmann H. Ex Situ Evaluation of the Composition of Protein Corona Of Intravenously Injected Superparamagnetic Nanoparticles in Rats. Nanoscale. 2014;6:11439–11450. doi: 10.1039/c4nr02793k. [DOI] [PubMed] [Google Scholar]
- 21.Mahmoudi M, Sant S, Wang B, Laurent S, Sen T. Superparamagnetic Iron Oxide Nanoparticles (SPIONS): Development, Surface Modification and Applications in Chemotherapy. Adv Drug Delivery Rev. 2011;63:24–46. doi: 10.1016/j.addr.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW. Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J Am Chem Soc. 2012;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 23.Owens DE, III, Peppas NA. Opsonization, Biodistribution, and Pharmacokinetics of Polymeric Nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Beduneau A, Ma Z, Grotepas CB, Kabanov A, Rabinow BE, Gong N, Mosley RL, Dou H, Boska MD, Gendelman HE. Facilitated Monocyte-Macrophage Uptake and Tissue Distribution of Superparmagnetic Iron-Oxide Nanoparticles. PLoS One. 2009;4:e4343. doi: 10.1371/journal.pone.0004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walkey CD, Chan WCW. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 26.Pelaz B, Charron G, Pfeiffer C, Zhao Y, de la Fuente JM, Liang X-J, Parak WJ, del Pino P. Interfacing Engineered Nanoparticles with Biological Systems: Anticipating Adverse Nano–Bio Interactions. Small. 2013;9:1573–1584. doi: 10.1002/smll.201201229. [DOI] [PubMed] [Google Scholar]
- 27.Brust M, Fink J, Bethell D, Schiffrin DJ, Kiely C. Synthesis and Reactions of Functionalised Gold Nanoparticles. J Chem Soc, Chem Commun. 1995;801:1655–1656. [Google Scholar]
- 28.Hostetler MJ, Wingate JE, Zhong C-J, Harris JE, Vachet RW, Clark MR, Londono JD, Green SJ, Stokes JJ, Wignall GD, Glish GL, Porter MD, Evans ND, Murray RW. Alkanethiolate Gold Cluster Molecules with Core Diameters from 1.5 to 5.2 nm: Core and Monolayer Properties as a Function of Core Size. Langmuir. 1998;14:17–30. [Google Scholar]
- 29.Moyano DF, Rotello VM. Nano Meets Biology: Structure and Function at the Nanoparticle Interface. Langmuir. 2011;27:10376–10385. doi: 10.1021/la2004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z-J, Posati T, Moyano DF, Tang R, Yan B, Vachet RW, Rotello VM. The Interplay of Monolayer Structure and Serum Protein Interactions on the Cellular Uptake of Gold Nanoparticles. Small. 2012;8:2659–2663. doi: 10.1002/smll.201200794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha K, Moyano DF, Rotello VM. Protein Coronas Suppress the Hemolytic Activity of Hydrophilic and Hydrophobic Nanoparticles. Mater Horiz. 2014;1:102–105. doi: 10.1039/C3MH00075C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He C, Hu Y, Yin L, Tang C, Yin C. Effects of Particle Size and Surface Charge on Cellular Uptake and Biodistribution of Polymeric Nanoparticles. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 33.Moyano DF, Goldsmith M, Solfiell DJ, Landesman-Milo D, Miranda OR, Peer D, Rotello VM. Nanoparticle Hydrophobicity Dictates Immune Response. J Am Chem Soc. 2012;134:3965–3967. doi: 10.1021/ja2108905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen K, Xu Y, Rana S, Miranda OR, Dubin PL, Rotello VM, Sun L, Guo X. Electrostatic Selectivity in Protein–Nanoparticle Interactions. Biomacromolecules. 2011;12:2552–2561. doi: 10.1021/bm200374e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekmekci Z, Saha K, Moyano DF, Tonga GY, Wang H, Mout R, Rotello VM. Probing the Protein–Nanoparticle Interface: The Role of Aromatic Substitution Pattern on Affinity. Supramol Chem. 2014;27:123–126. doi: 10.1080/10610278.2014.914627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De M, Miranda OR, Rana S, Rotello VM. Size and Geometry Dependent Protein-Nanoparticle Self-Assembly. Chem Commun. 2009:2157–2159. doi: 10.1039/b900552h. [DOI] [PubMed] [Google Scholar]
- 37.Milani S, Bombelli FB, Pitek AS, Dawson KA, Rädler J. Reversible versus Irreversible Binding of Transferrin to Polystyrene Nanoparticles: Soft and Hard Corona. ACS Nano. 2012;6:2532–2541. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- 38.Casals E, Puntes VF. Inorganic Nanoparticle Biomolecular Corona: Formation, Evolution and Biological Impact. Nanomed. 2012;7:1917–1930. doi: 10.2217/nnm.12.169. [DOI] [PubMed] [Google Scholar]
- 39.Monopoli M, Pitek A, Lynch I, Dawson K. Formation and Characterization of the Nanoparticle–Protein Corona. Methods Mol Biol. 2013;1025:137–155. doi: 10.1007/978-1-62703-462-3_11. [DOI] [PubMed] [Google Scholar]
- 40.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc Natl Acad Sci USA. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dashty M, Akbarkhanzadeh V, Zeebregts CJ, Spek CA, Sijbrands EJ, Peppelenbosch MP, Rezaee F. Characterization of Coagulation Factor Synthesis in Nine Human Primary Cell Types. Sci Rep. 2012;2:787–796. doi: 10.1038/srep00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezaee F, Casetta B, Levels JHM, Speijer D, Meijers JCM. Proteomic Analysis of High-Density Lipoprotein. Proteomics. 2006;6:721–730. doi: 10.1002/pmic.200500191. [DOI] [PubMed] [Google Scholar]
- 43.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, Dawson KA. Physical−Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J Am Chem Soc. 2011;133:2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 44.Dobrovolskaia MA, Patri AK, Zheng J, Clogston JD, Ayub N, Aggarwal P, Neun BW, Hall JB, McNeil SE. Interaction of Colloidal Gold Nanoparticles with Human Blood: Effects on Particle Size and Analysis of Plasma Protein Binding Profiles. Nanomed Nanotech Biol Med. 2009;5:106–117. doi: 10.1016/j.nano.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. What the Cell “Sees” in Bionanoscience. J Am Chem Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 46.Lynch I, Salvati A, Dawson KA. Protein-Nanoparticle Interactions: What Does the Cell See? Nat Nanotechnol. 2009;4:546–547. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 47.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 48.Dashty M, Motazacker MM, Levels J, de Vries M, Mahmoudi M, Peppelenbosch MP, Rezaee F. Proteome of Human Plasma Very Low-Density Lipoprotein and Low-Density Lipoprotein Exhibits a Link with Coagulation and Lipid Metabolism. Thromb Haemost. 2014;111:518–530. doi: 10.1160/TH13-02-0178. [DOI] [PubMed] [Google Scholar]
- 49.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors Affecting the Clearance and Biodistribution of Polymeric Nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahon E, Salvati A, Baldelli Bombelli F, Lynch I, Dawson KA. Designing the Nanoparticle–Biomolecule Interface for “Targeting and Therapeutic Delivery”. J Control Release. 2012;161:164–174. doi: 10.1016/j.jconrel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Ghavami M, Saffar S, Abd Emamy B, Peirovi A, Shokrgozar MA, Serpooshan V, Mahmoudi M. Plasma Concentration Gradient Influences the Protein Corona Decoration on Nanoparticles. RSC Adv. 2013;3:1119–1126. [Google Scholar]
- 52.Lescuyer P, Hochstrasser D, Rabilloud T. How Shall We Use the Proteomics Toolbox for Biomarker Discovery? J Proteome Res. 2007;6:3371–3376. doi: 10.1021/pr0702060. [DOI] [PubMed] [Google Scholar]
- 53.Mirshafiee V, Kim R, Park S, Mahmoudi M, Kraft ML. Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials. 2016;75:295–304. doi: 10.1016/j.biomaterials.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 54.Trouw LA, Bengtsson AA, Gelderman KA, Dahlbäck B, Sturfelt G, Blom AM. C4b-binding Protein and Factor H Compensate for the Loss of Membrane-bound Complement Inhibitors to Protect Apoptotic Cells against Excessive Complement Attack. J Biol Chem. 2007;282:28540–28548. doi: 10.1074/jbc.M704354200. [DOI] [PubMed] [Google Scholar]
- 55.Webb JH, Blom AM, Dahlbäck B. Vitamin K-Dependent Protein S Localizing Complement Regulator C4b-Binding Protein to the Surface of Apoptotic Cells. J Immunol. 2002;169:2580–2586. doi: 10.4049/jimmunol.169.5.2580. [DOI] [PubMed] [Google Scholar]
- 56.Berggård K, Johnsson E, Mooi FR, Lindahl G. Bordetella Pertussis Binds the Human Complement Regulator C4BP: Role of Filamentous Hemagglutinin. Infect Immun. 1997;65:3638–3643. doi: 10.1128/iai.65.9.3638-3643.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thern A, Stenberg L, Dahlbäck B, Lindahl G. Ig-Binding Surface Proteins of Streptococcus Pyogenes Also Bind Human C4b-Binding Protein (C4BP), a Regulatory Component of the Complement System. J Immunol. 1995;154:375–86. [PubMed] [Google Scholar]
- 58.Johnsson E, Thern A, Dahlbäck B, Hedén LO, Wikström M, Lindahl G. A Highly Variable Region in Members of the Streptococcal M Protein Family Binds the Human Complement Regulator C4BP. J Immunol. 1996;157:3021–9. [PubMed] [Google Scholar]
- 59.Moyano DF, Duncan B, Rotello V. Preparation of 2 nm Gold Nanoparticles for In Vitro and In Vivo Applications. In: Bergese P, Hamad-Schifferli K, editors. Nanomaterial Interfaces in Biology. Vol. 1025. Humana Press; 2013. pp. 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.You C-C, Miranda OR, Gider B, Ghosh PS, Kim I-B, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Detection and Identification of Proteins Using Nanoparticle-Fluorescent Polymer ‘Chemical Nose’ Sensors. Nat Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 61.Miller JN, Miller JC. Statistics and Chemometrics for Analytical Chemistry. 5th. Pearson Education Limited; Harlow, England: 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


