Abstract
Chronobiological studies of prokaryotic organisms have generally lagged far behind the study of endogenous circadian clocks in eukaryotes, in which such systems are essentially ubiquitous. However, despite only being studied during the past 25 years, cyanobacteria have become important model organisms for the study of circadian rhythms and, presently, their timekeeping mechanism is the best understood of any system in terms of biochemistry structural biology, biophysics and adaptive importance. Nevertheless, intrinsic daily rhythmicity among bacteria other than cyanobacteria is essentially unknown; some tantalizing information suggests widespread daily timekeeping among Eubacteria and Archaea through mechanisms that share common elements with the cyanobacterial clock but are distinct. Moreover, the recent surge of information about microbiome–host interactions has largely neglected the temporal dimension and yet daily cycles control important aspects of their relationship.
Many bacteria experience substantial environmental changes during the day and night. Most free-living bacteria are exposed to daily cycles of light and darkness, and/or temperature. The gut microbiota is also exposed to daily changes in the intestinal environment, as most animal hosts eat on a daily schedule — usually during the day for diurnal animals and during the night for nocturnal animals — which creates a daily rhythm of feast and fast in the digestive system. Although endogenous circadian rhythms were once thought to be an exclusive characteristic of eukaryotic organisms, all defining properties of circadian clocks (BOX 1) were also found in cyanobacteria1–3. Many processes are circa-dian in cyanobacteria4–12; for example, nitrogen-fixing unicellular cyanobacteria rhythmically fix nitrogen during the night, approximately twelve hours out of phase from photosynthesis, which peaks at midday13. This is adaptive biochemistry, as nitrogenase, a nitrogen-fixing enzyme, in these cyanobacteria is inactivated by even tiny amounts of oxygen, and its activity is thus incompatible with photosynthesis, which releases oxygen. This is one simple example of why bacteria may have evolved timekeeping systems to coordinate metabolic events during the day to optimize performance and/or to temporally separate potentially conflicting metabolic processes. Whereas the timekeeper of cyanobacteria is well established as a prototypical circadian clock, other bacteria may have evolved alternative timekeeping systems, such as an hourglass timer, which confers a fitness advantage14.
Box 1. What constitutes a circadian clock?
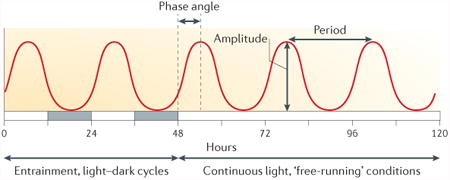
Circadian rhythms are defined by three well-established phenomenological criteria and not by their molecular mechanism98 (see the figure). The first criterion is that they persist with a self-sustained oscillation of approximately 24 h under constant conditions (that is, under constant temperature and either constant light or constant darkness). The expression of the endogenous clock under constant conditions is termed a ‘free-running’ rhythm. The second criterion is that these endogenous rhythms of approximately 24 h can be entrained to exactly 24 h by daily cyclic cues in the environment, such as light and darkness, temperature, and feeding and fasting. Finally these rhythms are temperature compensated, so that they proceed at almost the same rate and period in a physiological range of constant ambient temperatures. Another common property, but not a defining characteristic, is conditionality; that is, endogenous rhythmicity is robustly expressed under some environmental conditions but not others99,100. The fascination of this phenomenon is to explain how its biochemical mechanism can robustly sustain a long period of oscillation (∼24 h) with high precision, and how it enhances fitness in the natural environment.
What might other functions of a daily timekeeper in bacteria be? The most common hypothesis is that having a ∼24 h timekeeper enables the cell to anticipate and/or prepare for the daily transformation of environmental conditions that affect many bacteria. Anticipation might be most easily discerned in the case of photoautotrophic bacteria, such as cyanobacteria (FIG. 1), owing to the fitness disadvantage of maintaining energetically expensive photosynthetic pathways during the night. Thus, bacteria that anticipate dawn by activating quiescent photosynthetic processes late in the night could have an advantage over bacteria that merely react to dark-to-light transitions. Anticipating daily changes might also facilitate protective responses in bacteria that are exposed to the damaging effects of sunlight (especially UV light; FIG. 1). Even visible sunlight can be absorbed by cytochromes in the electron-transport chain and affect metabolism15. The effects of sunlight inspired the ‘escape from light’ hypothesis, which proposes that circadian clocks evolved owing to the selective pressure of daily cycles of light and darkness, in which light impaired growth16,17.
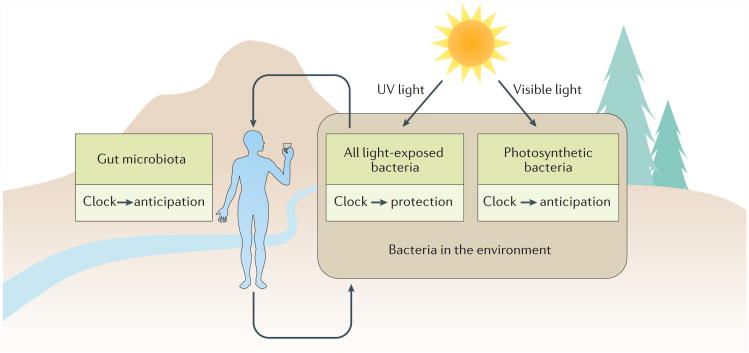
Figure 1. Many bacteria are exposed to daily selective pressures.
Free-living bacteria can be exposed to daily cycles of light and temperature that affect viability (for example, exposure to UV light) and/or provide energy (for example, through photosynthesis). Even the gut microbiota is often exposed to daily cycles of nutrients, owing to the rhythmic eating habits of the host, and temperature, as most animal hosts have daily rhythms of body temperature that are metabolically controlled in endotherms and behaviourally controlled in ectotherms. These same bacteria may be exposed to daily environmental cycles of light and temperature following excretion from the gut.
A gut bacterium that anticipates when fresh nutrients are entering the digestive tract could have an advantage in the competition for resources. Furthermore, gut bacteria may also benefit from daily timing when they attempt to survive outside of the host following excretion, in a similar manner to bacteria that are always exposed to the external world (FIG. 1). Therefore, bacteria that have a daily timekeeping mechanism, even if it is not a complete circadian system such as those found in cyanobacteria, probably have adaptive advantages. In this Review, we highlight recent mechanistic insights into circadian oscillators in cyanobacteria and timekeepers in other bacteria14,18,19. Moreover, we consider the potential adaptive value of both oscillating clocks and hourglass timers. We also examine recent research on the evolution of proto-circadian rhythms and how the relationship between a host and its microbiota is influenced by biological timekeeping14,20–25.
Timekeeping mechanisms in cyanobacteria
In cyanobacteria, the clock system controls multiple output processes, including the transcriptome4–6, the proteome7, the timing of cell division8,9, nitrogen fixation1,13, chromosomal topology6,10,11 and metabolism12. In photoautotrophic cyanobacteria, there are changes in metabolite composition during the day and night, and, owing to the self-sustained function of the clock, these changes persist in continuous light12. Even the redox status of Synechococcus elongatus cells oscillates in continuous light and may influence the oxidation status of peroxiredoxins26,27. Interestingly, changes in metabolism that depend on the light and dark cycle, in particular redox status and/or energy levels, can directly entrain the cyanobacterial clockwork28–31. Metabolic feedback might even provide mechanisms to compensate for temperature and other changes to enable the clock to maintain accurate timekeeping32.
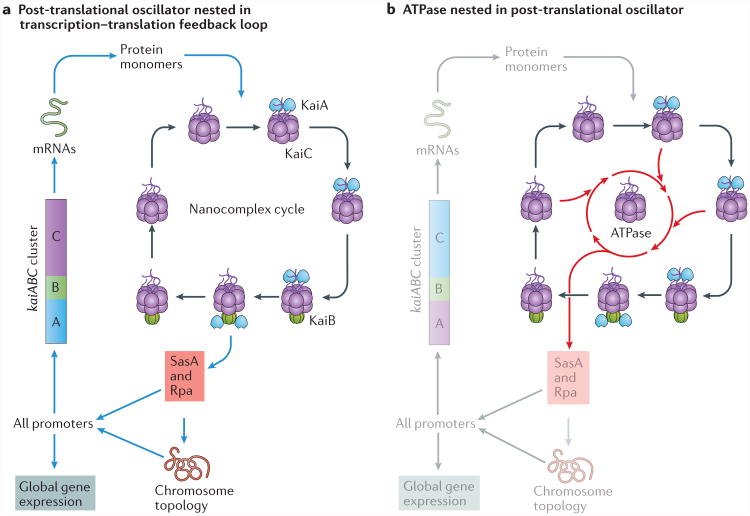
The core mechanism of the circadian clock in eukaryotic cells is widely thought to be based on a transcription–translation feedback loop (TTFL)33,34, although there is evidence that this model may be incomplete or inaccurate26,35–37. In cyanobacteria, initial experiments also supported a TTFL model38. However, the remarkable discovery that three cyanobacterial clock proteins (KaiA, KaiB and KaiC) could reconstitute a circadian oscillator in vitro for many days39 led to our current understanding of the clock system in cyanobacteria; a biochemical oscillator is the core pacemaker, which operates in (and regulates) a larger TTFL that, in turn, controls outputs and replenishes the essential proteins of the oscillator40–43 (FIG. 2a). At the present time, the cyanobacterial clock is the best understood of any circadian mechanism from a biochemical and biophysical perspective.
Figure 2. Circadian molecular and genetic oscillators in cyanobacteria.
a | A self-sustained post-translational oscillator (PTO) is embedded in a transcription–translation feedback loop (TTFL). The synthesis of non-phosphorylated KaiC monomers feeds into the molecular oscillator, which undergoes a cycle of association into, and disassociation from, the KaiA–KaiB–KaiC nanocomplex71. KaiC interacts with competing KaiB and SasA proteins to mediate the activity of transcriptional factors (such as RpaA) and rhythmic DNA torsion to control global transcription levels, including those that drive the expression of the essential clock genes kaiA, kaiB and kaiC. b | The core clock reaction that is embedded in the PTO is ATP hydrolysis, which is primarily catalysed by the ATPase in the CI domain of KaiC. In this view, the phosphorylation and nanocomplex cycles enhance and regulate the ATPase activity to promote robustness and temperature compensation. Part a is adapted with permission from REF. 42, AAAS.
The key component of the cyanobacterial clock is KaiC38,44. This protein has an astonishing range of biochemical activities, including autokinase, autophosphatase, phosphotransferase and ATPase, and is the core of a rhythmically associating and dissociating nanocomplex (FIG. 3). KaiC is a double hexamer that consists of two similarly sized rings, CI and CII, and a carboxy-terminal region in CII that interacts with KaiA through protruding peptide ‘tentacles’ (REF. 44). Both rings catalyse the hydrolysis of ATP45. As described below, the ATPase activity of the CI ring provides the rate-limiting reaction of the timing circuit19,45, whereas the ATP that is hydrolysed by the CII ring catalyses sequential phosphorylation and dephosphorylation reactions of serine and threonine residues44,46–50. The level of KaiC phosphorylation is a convenient marker of circadian phase in vivo and in vitro, including when transcription and/or translation are suppressed51,52. As neither transcription nor translation is required for KaiC phosphorylation to oscillate, it is a post-translational oscillator (PTO; FIGS 2a,3).
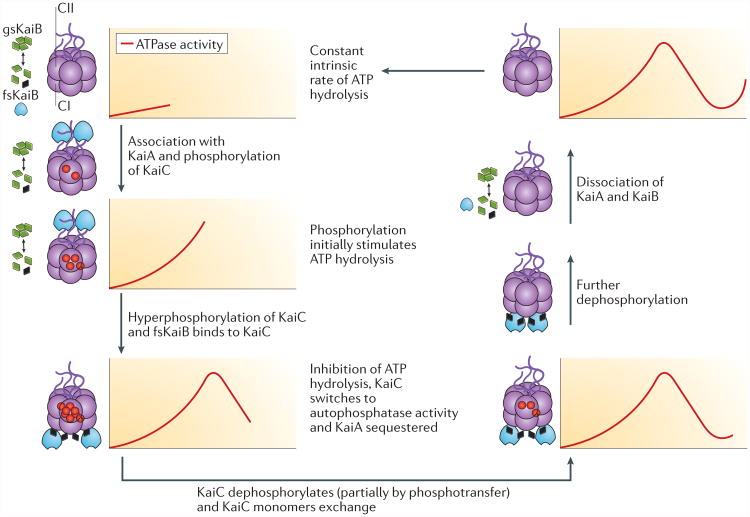
Figure 3. The post-translational oscillator (PTO).
The post-translational oscillator (PTO) is composed of rhythmic associations between KaiC (purple), KaiA (blue) and KaiB (green or black) to form a nanocomaplex that ATPase and phosphorylation activities of KaiC. KaiC forms two hexameric rings, CI and CII. KaiB monomers undergo a slow transition from ground-state KaiB (gsKaiB; green diamonds) to the fold-shifted state (fsKaiB; black diamonds), which binds to the CI domain of KaiC hexamers. KaiA associates with hypophosphorylated KaiC and stimulates the autophosphorylation of KaiC (red circles indicate the added phosphates), which accelerates the rate of ATP hydrolysis. The association and dissociation of the nanocomplex coincide with changes in the phosphorylation state of KaiC; residue T432 is phosphorylated first, followed by S431, and dephosphorylation occurs in the opposite order. Adapted with permission from REF. 37, Elsevier.
Phosphorylation and the nanocomplex cycle
The status of KaiC phosphorylation is not merely a marker of the circadian phase, it also regulates the association and dissociation of the KaiA–KaiB–KaiC nanocomplex, thereby switching KaiC between its autokinase and autophosphatase states (FIG. 3). The rhythm begins with unphosphorylated KaiC. KaiA stimulates the intrinsic autokinase activity of KaiC through interaction with the CII-terminal tentacles. This leads to the phosphorylation of two key residues in the CII domain, T432 followed by S431 (REFS 46,47,49,53–56). Dephosphorylation proceeds in the same order, phosphorylated T432 (pT432) followed by pS431, such that the overall process from the hypophosphorylated to the hyperphosphorylated and back to the hypophosphorylated state of KaiC follows the sequence: (T432/S431) to (pT432/S431) to (pT432/pS431) to (T432/pS431) to (T432/S431)46,47 (FIG. 3). The dephosphorylation phase proceeds, at least partially, by regenerating ATP from CII-bound ADP and the phosphates that are bound to T432 and S431 (REFS 57,58). This remarkable phosphotransferase activity limits the overall consumption of ATP, which possibly preserves clock function when cellular ATP concentrations fluctuate; the cyanobacterial clock accurately tracks time during extended exposure to the dark, when cells are not able to produce ATP by photosynthesis28,32,51,52,57.
Concomitant with the phosphorylation and dephosphorylation cycle is a rhythm of association and dissociation of the KaiA–KaiB–KaiC nanocomplex (FIG. 3). At the beginning of the nanocomplex cycle, KaiA rapidly interacts intermittently with the CII tentacles of KaiC59–62. This transient interaction stimulates the phosphorylation of KaiC (or inhibits dephosphorylation, thereby shifting the reaction towards phosphorylation)57,58,63, and when S431 of KaiC achieves the (T432/pS431) state in the phosphorylation sequence, KaiB binds to KaiC. A recent study reported that KaiB exists in two diametrically different conformations: a ground state (gsKaiB) that does not interact readily with KaiC, and a substantially fold-shifted state (fsKaiB) that binds to the CI domain18 (FIG. 3). The fsKaiB state is conformationally similar to a structural motif of SasA, which is a histidine kinase that interacts with KaiC and transmits its oscillatory information. fsKaiB competes with SasA for binding to KaiC and thus regulates the downstream transcriptional cascade of SasA and its response regulator RpaA30,64–69 (FIG. 2a). In addition to displacing SasA from CI of KaiC, KaiB forms a complex with CI domains that also stably sequesters and inactivates KaiA, so that KaiA can no longer stimulate the phosphorylation of KaiC, and KaiC switches to its autophosphatase mode18,41,47,70 (FIG. 3). When KaiC fully dephosphorylates, KaiB and KaiA are released, which restarts the nanocomplex and phosphorylation cycles (FIG. 3).
The slow pace of ATPase activity
How can this biochemical oscillator maintain such a long time constant (for example, a period of 24 h)? Are the phosphorylation and nanocomplex cycles the core pacemakers of the cyanobacterial circadian clock? Similar to the TTFL, there is nothing intrinsic about these cycles that makes them oscillate with a 24 h period. Certainly, phosphorylation events and protein associations and dissociations can occur very rapidly; these biochemical reactions do not dictate a slow 24 h timekeeping. Kondo and co-workers have championed the hypothesis that the ATPase activity of KaiC is the core rate-limiting reaction19,45 (FIG. 2b). This reaction is very slow (only 15–16 ATP molecules are hydrolysed by one KaiC monomer per day) and is temperature compensated45. Moreover, an analysis of KaiC mutants in vivo and in vitro showed a strict correlation between the ATPase activity of KaiC and circadian frequency, which implies that the circadian period depends directly on the energy that is provided by ATP hydrolysis19,45. There is even evidence for a highly damped oscillation of the ATPase activity of KaiC in the absence of KaiA and KaiB19.
However, is the ATPase hypothesis as the core pacemaker just an ‘infinite regress’, with another process underlying the slow rate? Given that many ATP-hydrolysing reactions have very fast rates, can we explain the sedate tempo of the KaiC ATPase? A structural study identified features in the CI domain of KaiC that might explain its slow ATPase activity19. The lytic water molecule that is involved in nucleophilic attack on the γ-phosphate of ATP is positioned away from the optimal position, so that hydrolysis is less favourable than in the catalytic sites of more typical ATPases, such as kinesin and F1-ATPase. A second source of the slow activity is slow cis–trans isomerization of a crucial peptide bond, which increases the activation energy barrier of the ATPase reaction. These two features decrease the probability of ATP hydrolysis and thereby establish a remarkably slow but stable ATPase cycle in the CI domains of the KaiC hexamer19. This landmark study provided the first biochemical and biophysical explanations of how a circadian oscillator could have such a long time constant.
Cycles within cycles: robustness and synchrony
The aforementioned studies reveal TTFL, phosphorylation– nanocomplex and ATPase cycles in the cyanobacterial clockwork (FIGS 2b,3). Why are there so many cycles? An analysis of the relationship between the PTO and the TTFL found that they have a definite hierarchical interdependency; the PTO is the core pacemaker, whereas the TTFL is a ‘slave oscillator’, which damps when the PTO stops71 and is no longer temperature-compensated under some metabolic conditions. The TTFL feeds back into the PTO through the synthesis of new clock proteins, which can reinforce and/or fine-tune the core PTO pacemaker. This coupled oscillator system is more resilient to intracellular noise71. Moreover, an analysis of cells in a microfluidic device found that the clocks of individual cells were less stable and synchrony was not maintained among cells in the population in the absence of the TTFL72. The advantage of coupled oscillators can not only be demonstrated mathematically71,73, it also makes sense biologically. The TTFL controls transcription and translation, whereas the clock-specific nanocomplex, which is composed of KaiA–KaiB–KaiC, is a mass-action biochemical oscillator; these two types of oscillator are likely to be sensitive to different kinds of intrinsic and extrinsic cellular noise (cell division, transcriptional noise, fluctuating metabolites, and so on). Moreover, it is very likely that the core ATPase reaction19,45 is stabilized and temperature-compensated through its interactions with the phosphorylation–nanocomplex and TTFL cycles. Therefore, the three nested and interacting cycles (that is, the TTFL, phosphorylation–nanocomplex and ATPase cycles; FIG. 2b) provide a resilient circadian system71,72,74.
A separate but related issue regarding robustness is synchrony among the KaiC hexamers in the PTO. Remarkably, in vitro, KaiA–KaiB–KaiC reactions can oscillate for up to ten days without detectable dampening75. Why is the period of each KaiC hexamer not sufficiently different from every other hexamer in the reaction so that the phasing in the population of hexamers becomes asynchronous after a few cycles? There must be a mechanism, or mechanisms, to keep the hexamers synchronized; indeed, at least two such mechanisms are known. First, a KaiC hexamer can exchange monomeric subunits with neighbouring hexamers59,60,76, and this monomer exchange maintains synchrony of the phosphorylation status among the KaiC hexamers in the population60,75. Another important factor for maintaining synchrony among KaiC hexamers is the sequestration and inactivation of KaiA activity during the dephosphorylation phase41,47. Therefore, KaiC monomer exchange and KaiA sequestration ensure that KaiC hexamers remain synchronized in the PTO.
The sun also rises for other bacteria
In cyanobacteria, all three Kai proteins are required for circadian oscillations in vivo and in vitro38,39. Homologues of kaiB and kaiC are distributed widely among both photosynthetic and non-photosynthetic members of the Eubacteria and the Archaea, but kaiA is found only in cyanobacteria77,78. Even among cyanobacteria, the kaiA gene was lost during evolution in the important cyanobacterial genus Prochlorococcus, which retained kaiB and kaiC and has daily rhythms, but not a sustained circadian rhythm, under constant conditions79,80. The loss of the kaiA gene from Prochlorococcus may have ‘de-evolved’ an original KaiA–KaiB–KaiC circadian clock to a simpler timer that is based on only KaiB–KaiC and keeps track of time similarly to an hourglass81,82. Another example of a potential hourglass timer is found in the halophilic archaeal genus Halobacterium, which has a single-domain kaiC but neither kaiA nor kaiB; Halobacterium spp. exhibit light–dark-entrained daily transcription but not sustained oscillations under constant conditions83. Does this mean that all three Kai proteins are required for self-sustained circadian oscillations, or is it possible that a circadian clock could be composed of KaiB and KaiC, or even just KaiC alone?
In this context, the example of purple bacteria may be instructive. Purple bacteria are globally distributed members of the Proteobacteria that frequently inhabit soil and water environments together with cyanobacteria. Tests on circadian rhythmicity have been reported for two species of purple bacteria, Rhodobacter sphaeroides84 and Rhodopseudomonas palustris14, which both have homologues of kaiB and kaiC. R. palustris does not sustain rhythmicity, as monitored by nitrogen fixation and the phosphorylation of KaiC, in constant conditions14 (FIG. 4), which is a defining characteristic of circadian rhythms (BOX 1). In R. sphaeroides, rhythms of gene expression were detected, 21 h under aerobic conditions and 11 h under anaerobic conditions, but other defining circadian characteristics, such as temperature compensation and entrainment, were not reported84.
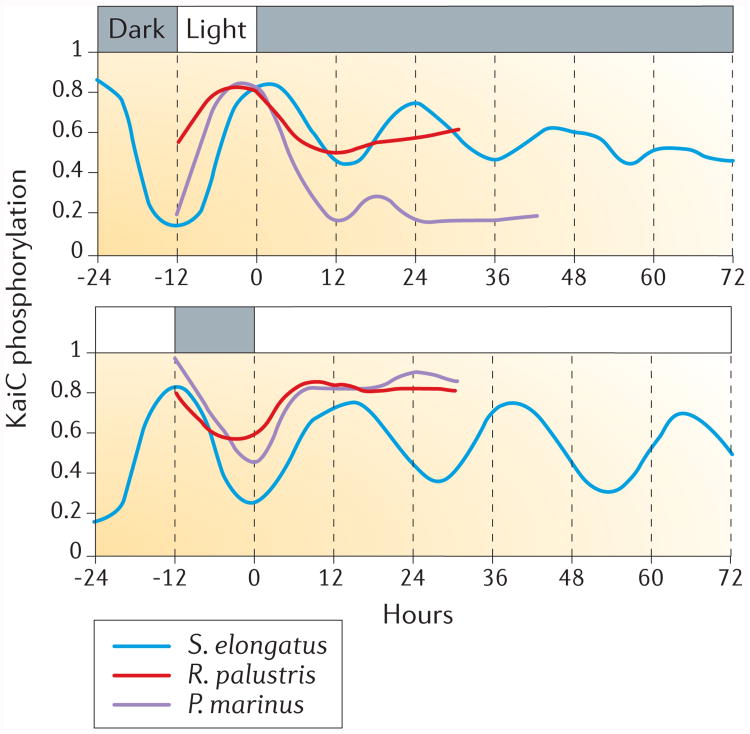
Figure 4. KaiC phosphorylation temporal patterns differ between complete circadian clocks that consist of KaiA–KaiB–KaiC and proto-circadian KaiB–KaiC systems.
The phosphorylation of KaiC in light and dark cycles, constant darkness and constant light is shown for three different species of bacteria: the two cyanobacteria Synechococcus elongatus and Prochlorococcus marinus and the purple bacterium Rhodopseudomonas palustris. S. elongatus harbours kaiABC and has a complete circadian system, whereas P. marinus and R. palustris only harbour kaiBC.
Recently, the daily profiles of KaiC phosphoryla-tion during light–dark cycles, continuous darkness and continuous light were compared among S. elongatus, Prochlorococcus marinus and R. palustris14,51,71 (FIG. 4). S. elongatus, which contains kaiA, kaiB and kaiC, exhibits daily rhythms during light–dark cycles that are sustained in both continuous darkness and continuous light. By contrast, P. marinus and R. palustris, which contain kaiB and kaiC, exhibit daily rhythms of KaiC phosphorylation during light–dark cycles that dampen rapidly under constant conditions (FIG. 4). We previously proposed that the timekeeping system in R. palustris is a ‘proto-circadian’ oscillator that does not persist under constant conditions14. This hypothesis is consistent with the observation that P. marinus — which ‘lost’ kaiA77–79 — also does not sustain robust rhythms of gene expression or KaiC phosphorylation under constant conditions79–82 (FIG. 4). Taken together, the data support the hypothesis that all three Kai proteins are necessary for sustained circadian oscillations in bacteria, and that kaiBC-based and kaiC-based systems might be capable of hourglass timekeeping but not a persistent circadian oscillation81,82. Given that the sequestration of KaiA promotes synchrony in S. elongatus41,47, KaiA was perhaps an evolutionary innovation of cyanobacteria to sustain oscillations, which resulted in their robust persistent rhythms2.
Timekeeper evolution and new criteria for testing adaptive fitness
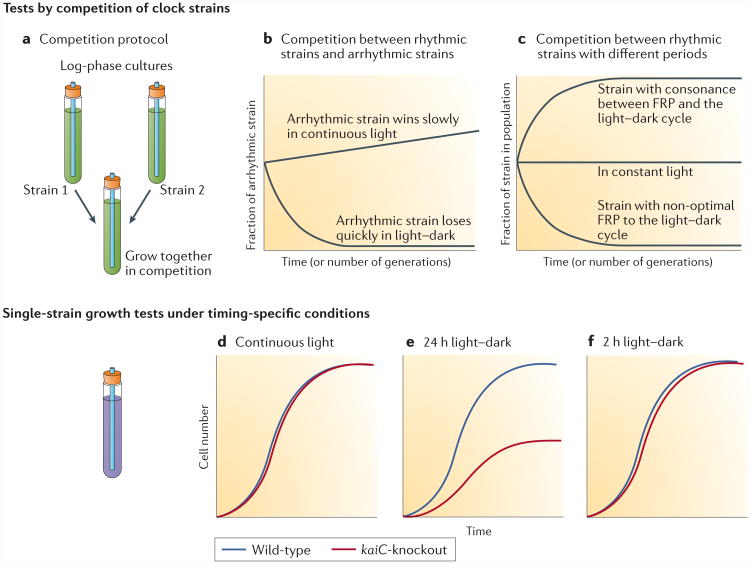
Speculations about the potential adaptive values of timekeeping in bacteria are thought-provoking, but have there been any rigorous tests of a fitness advantage? In cyanobacteria, strains that have a functioning bio logical clock out-competed arrhythmic strains in rhythmic environments (for example, light–dark cycles), whereas in constant environments (for example, continuous light), rhythmic and arrhythmic strains grew at comparable rates85 (FIG. 5a,b). Moreover, among rhythmic strains, those that had endogenous free-running periods that closely matched the period of the environmental cycle had the greatest fitness advantage86 (FIG. 5c). Competition experiments are a sensitive test of fitness and they clearly indicated that the cyanobacterial circadian system interacts with the environment to mediate an adaptive advantage, especially as clock mutants could out-compete wild-type cells under appropriate conditions86.
Figure 5. Criteria for testing the adaptive fitness of daily timekeeping mechanisms.
a | The fitness advantages of timekeeping can be assessed by competition between strains in mixed cultures under different environmental conditions. b | In competition between rhythmic and arrhythmic strains of Synechococcus elongatus, the arrhythmic strain is rapidly out-competed by the rhythmic wild-type strain in light–dark cycles, but slowly outgrows the wild-type strain in constant light, which provides no selective pressure for circadian timekeeping. c | In competition among strains that are rhythmic, a strain that has an endogenous rhythm (free-running period (FRP)) that closely matches the environmental light–dark cycle out-competes strains with non-optimal free-running periods. Under continuous light, all strains are maintained in the population. d | Fitness can also be inferred from single-strain growth rates of cultures of wild-type or kaiC-knockout cells in the purple bacterium Rhodopseudomonas palustris growing phototrophically. Under continuous light, KaiC-dependent timekeeping confers no advantage to the growth of R. palustris. e | A cycle of 12 h of light and 12 h of darkness, which matches the environmental conditions to which R. palustris has adapted, provides wild-type cells with a fitness advantage over the kaiC-knockout strain. f | Under a cycle of 1 h of light and 1 h of darkness, both the wild-type and kaiC-knockout cells grow at the same rate. Parts a–c are adapted from REF. 101, Elsevier. Parts d–f are adapted from REF. 14.
More recently, the fitness benefits of a timekeeping programme have been assessed in R. palustris14. As mentioned above, R. palustris does not exhibit sustained oscillations under constant conditions14 (FIG. 4). Nonetheless, does KaiC carry out an adaptive timekeeping function in this purple bacterium? To address this question, the growth rates of wild-type R. palustris and a kaiC-knockout strain were compared in single-strain cultures14. Both strains grew at the same rate under non-selective continuous light conditions, but in the 24 h cycle (12 h of light and 12 h of darkness), the kaiC-knockout cells grew slower than wild-type cells14 (FIG. 5d,e). Therefore, KaiC enhanced growth in a 24 h light–dark cycle, but not under continuous conditions. Importantly, additional experiments with a 1 h light and 1 h dark cycle (FIG. 5f) ruled out the reduced amount of light exposure during cycles of 12 h of light and 12 h of darkness as a confounding factor. Therefore, the kaiC gene of R. palustris has an important effect on growth and fitness in 24 h cyclic environments, but not in non-24 h environments (continuous conditions or cycles of 1 h of light and 1 h of darkness); this result strongly implies that the fitness advantage of kaiC in R. palustris is related to adaptation to environments that have a 24 h periodicity14. Growth experiments similar to the ones described above can be considered as new criteria to assess whether KaiC is involved in an adaptive 24 h timekeeping mechanism in any bacterium14.
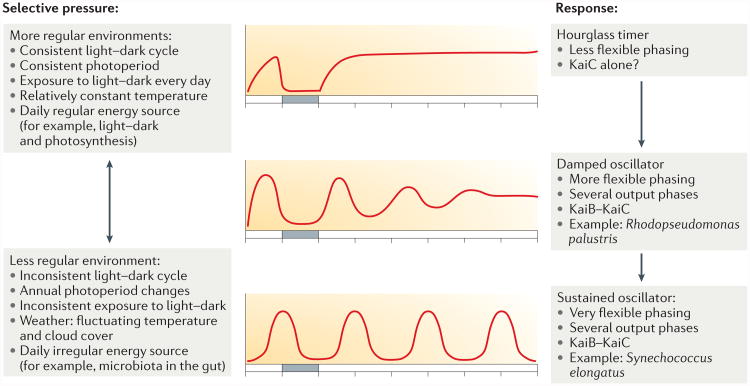
What might be the selective pressures that led to the evolution of timers versus oscillators? In a sufficiently regular environment, a damped oscillator or even an hourglass timer might enable adequate anticipation of daily changes in the environment (FIG. 6). Some chrono biologists have questioned the necessity of a sustained circadian rhythm; as circadian clocks evolved in a cyclic world (not continuous conditions), it is the mechanism of entrainment that evolved and not self-sustained oscillations87. Nevertheless, among eukaryotes, daily biological clocks are almost universally self-sustained circadian systems. How and why is this the case? Perhaps studying the evolution of daily timekeepers in bacteria can help to address these questions, especially in bacteria that have kai homologues that can be identified and targeted for fitness tests, as shown in FIG. 5. For example, a bacterial species that is always exposed to a regular light–dark cycle with a consistent photoperiod and consistent temperature might effectively adapt to the cyclic world with an hourglass timer (FIG. 6). However, an oscillator enables several outputs and/or flexible phasing, even if the oscillator is damped and requires a relatively consistent light–dark cycle, such as the kaiBC-based system of R. palustris (FIG. 6). Finally, in bacteria that inhabit temporally irregular environments (for example, environments with unreliable exposure to light and darkness, inconsistent light intensities, annually varying photoperiods or irregularly available energy sources), a fully developed circadian oscillator provides the maximum temporal buffering to accurately track the solar day88 (FIG. 6). As an example, seasonality may create very different selective pressures for free-living bacteria compared with gut bacteria. For a free-living phototroph, the differences in average temperature and photoperiod between winter and summer affect growth rates. Gut bacteria may enjoy a more consistent annual environment, but the diet of the host can differ substantially between winter and summer, which leads to drastic annual nutrition cycles that are known to affect the composition of the microbiome89. Currently, the model shown in FIG. 6 is speculative. However, the widespread distribution of kai homologues in various bacteria, which inhabit vastly different environmental niches, presents a unique set of test-cases with which to analyse the evolution of daily biological timekeeping.
Figure 6. A hypothesis for the evolution of Kai-based timekeepers in bacteria.
The type of time keeper (that is, hourglass timer, damped oscillator or self-sustained circadian oscillator) that evolves is primarily determined by the regularity of the environmental daily cycle of the bacterium, as this is the dominant selective pressure. In very regular environments, an hourglass timer that is based on KaiC alone is sufficient. In fluctuating environments, which require more flexibility of the timer, a sustained oscillator based on KaiA–KaiB–KaiC is advantageous. In intermediate environments, a damped oscillator that is based on KaiB–KaiC is sufficient.
Daily timing and the microbiome
The relationship between the host and its microbiome has become a central topic in biology and medicine. The temporal dimension of this relationship warrants serious investigation. At the very least, because most animal hosts eat on a daily schedule, the environment that is experienced by the gut microbiota undergoes a ∼24 h transformation in nutrient availability. A bacterium that can anticipate the arrival of fresh nutrients might readily metabolize these resources and thus gain an advantage over time-ignorant competitors. Our knowledge that at least some free-living bacteria have daily timekeepers1,2,14 opens the possibility that bacteria of the microbiota have endogenous timekeepers that interact with the biological clock of the host. This blossoming area of enquiry has already identified a gut bacterium, Enterobacter aerogenes, that seems to express circadian rhythms when isolated outside of the host20. Interestingly, the robustness of its rhythm is enhanced if melatonin is included in the culture medium. Melatonin is a circulating hormone that is rhythmically controlled by the circadian clock in vertebrates, and these results imply that melatonin could stimulate rhythmicity in members of the microbiota in vivo. In addition to melatonin, other candidates for daily time cues include diurnal cycles of nutrients and circadian body temperature rhythms (core body temperature oscillates diurnally in most endothermic animals).
Moreover, if members of the gut microbiota have timekeeping capability, might they release signals that influence the circadian system of the host? The micro-biota secretes a vast range of compounds, many of which are bioactive90. For example, some bacteria that inhabit or infect mammals can release cytokine-like fac-tors91, which, if released rhythmically, could influence the circadian clock of the host. For example, microbial metabolites (in particular, short-chain fatty acids, such as butyrate) can modulate circadian gene expression in hepatic organoids22. Conversely, perturbations of the biological clock of the host affect the composition of the gut microbiome. Several studies have reported diurnal changes in the relative abundance of bacterial phyla in the gut microbiome21–25. Disruption of the biological clock of the host either by jet-lag or by mutations in the clock genes of the host alters the proportion of these phyla that exhibit daily changes23–25. Moreover, alterations to diet, such as time-restricted feeding and transition between a regular and a high-fat diet, are known to affect circadian clocks in mice and they also modify the composition of the gut microbiome21–24.
At this time, it seems clear that the composition of the mammalian gut microbiome is temporally changing, and that changes to the circadian clock of the host affect the viability and relative growth of some members of the microbiota. Whether there is feedback from the microbiota to the circadian clock of the host is less well characterized, but this is a fascinating possibility to investigate22. Along these lines, Thaiss and co-workers reported a remarkable interplay between host and microbiota92. Antibiotic treatment of the host affects rhythmic localization and adherence of bacteria to the intestinal epithelium and this, in turn, reprogrammes chromatin and transcription oscillations in the host. Remarkably, these effects occur not only in host intestinal cells but also extend to the liver, which modulates hepatic circadian gene expression and detoxification92. Finally, at least some members of the gut microbiota may have endogenous timekeepers20 that promote their adaptation to the intestinal environment and to the external environment after they are excreted (FIG. 1).
Beyond the gut environment, there are other fascinating examples of host–bacteria relationships that have temporal dimensions, such as the symbiosis between the squid Euprymna scolopes and the luminous bacterium Aliivibrio fischeri (formerly known as Vibrio fischeri)93. E. scolopes has a specialized light organ that is colonized by the bacteria. The luminescence that is produced by A. fischeri is used by the squid as a camouflage strategy, in which the animal emits light from its ventral surface to mimic the nocturnal moonlight and starlight that shine from above. The luminescence varies during the day–night cycle, with the highest levels of luminescence in the early evening when the animal begins to forage94,95. An examination of the transcriptome of the resident A. fischeri revealed day–night oscillations of metabolism between anaerobic respiration and fermentation95. This adaptive temporal interaction between host and symbiont illustrates diurnal reciprocity between hosts and microbial partners93.
Conclusions and future perspectives
Although cyanobacteria are an archetypal model system for endogenous self-sustained timekeeping mechanisms and fitness, we have barely glimpsed the panoply of likely timing mechanisms that have evolved among bacteria in response to various environments and selective pressures (FIGS 1,6). Homologues of the kai genes are widely distributed among Eubacteria and Archaea. Although the cyanobacterial ‘cycles within cycles’ design (FIG. 2) is optimized for resilience to internal and external noise, other bacteria might use Kai proteins in different configurations to adjust to the daily patterns of their particular environment (FIG. 6). There is every reason to believe that new types of timekeeper other than circadian clocks exist and are ready to be discovered. As we expand this research to other bacterial species, new tools will be required to monitor daily rhythms. Luminescence reporters have been crucial for optimally monitoring rhythmic gene expression in aerobic eukaryotes and cyanobacteria2,96,97, but luciferase reporters require oxygen. Therefore, to monitor rhythms from bacteria that operate in various metabolic modes, the development of automated technologies for micro-aerobic and anaerobic environments will facilitate the assessment of the generality of timekeeping mechanisms among Eubacteria and Archaea.
Understanding that host–microbiota relationships have a temporal dimension is an exciting new research area. It is clear that the feeding rhythms and diet of the host affect which bacterial phyla thrive in the gut. Signals that emanate from the microbiota reciprocally modulating the circadian clock of the host is a tantalizing possibility, and, if found to be true, the implications for chronobiology are profound. As many members of the microbiota are not restricted to the gut but can also exist in a larger ecosystem that is also rhythmic, a daily timekeeper is potentially useful in both environments (FIG. 1). Bacteria that alternate between internal and external environments might adjust the timekeeping mechanism if they relocate; for example, after excretion. For a timekeeper in the regular 24 h external environment, the precision of cadence may be essential, whereas in the more variable environment of a host gut, flexibility of tempo may be more valuable.
From the broader perspective of chronobiology as an example of evolutionary adaptation, testing simple hypotheses about fitness can be challenging in higher organisms, in which many processes become interdependent over evolutionary time so that the adaptive value of a specific trait independent of the background is difficult to ascertain. Bacteria may provide clearer insights into the relative roles of various environmental factors in the natural selection of biological timekeepers. This is not to say that bacteria are ‘simple’, but that their small and generally streamlined genomes may enable more precise manipulation and measurement of the fitness consequences of variation in traits that underlie daily timekeepers. These factors lead to greater confidence about the functional basis of interactions between trait variation and fitness because of greater experimental control over both the genomic background and the environmental conditions. Therefore, the adaptive role of daily timekeepers, and the selective pressures that led to the evolution of temporal precision, temperature compensation and entrainment, may be best studied in bacteria. It is time (and timely) for microbiology and chronobiology to contribute more to each other.
Acknowledgments
The authors dedicate this paper to the memory of their colleague and friend D. McCauley, a population geneticist who encouraged and facilitated their tests on the adaptive fitness of circadian clocks in microorganisms. The authors thank T. Kondo, S. Golden and M. Ishiura (and past and present members of their laboratories), as well as past and present members of the laboratory of C.H.J., for an exhilarating scientific journey into the circadiana of cyanobacteria. This research in the laboratory of C.H.J. is currently supported by grants from the US National Institutes of Health (GM 067152 and GM107434).
Glossary
- Diurnal
Active during the day
- Nocturnal
Active during the night
- Hourglass timer
A simple non-oscillatory timer that is set in motion and keeps track of time linearly, and thus does not self-sustain a cycle
- Photoautotrophic bacteria
Bacteria that derive their energy exclusively from light to drive photosynthetic carbon fixation and the synthesis of organic compounds
- Proto-circadian
A broad term that includes hourglass timers, damped oscillators and other potential timekeeping mechanisms that may be ancestral systems that include some, but not necessarily all, of the canonical properties of circadian clocks. Proto-circadian systems may be a step along an evolutionary trajectory that might ultimately lead to a bona fide circadian system
- Peroxiredoxins
A class of antioxidant enzymes that control peroxide levels
- Entrain
The process whereby the period of a biological rhythm becomes equal to that of an environmental cycle (for example, of light and dark). Entrainment also establishes a stable phase relationship between the entraining cycle and the biological rhythm
- Ectotherms
‘Cold-blooded’ animals in which the body temperature depends on the environmental temperature but is often modulated behaviourally
- Halophilic
Adapted to high concentrations of salt
- Endothermic
‘Warm-blooded’, that is, the maintenance of body temperature metabolically so that it is independent of the environment and behaviour
Footnotes
Competing interests statement: The authors declare no competing interests.
References
- 1.Grobbelaar N, Huang T, Lin H, Chow T. Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol Lett. 1986;37:173–177. This is the first persuasive report of a circadian rhythm expressed by a bacterium. [Google Scholar]
- 2.Kondo T, et al. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. This paper establishes the cyanobacterial circadian system that has been so productive in understanding the mechanism and adaptive importance of circadian rhythms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. doi: 10.1046/j.1365-2958.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 5.Ito H, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijayan V, Zuzow R, O'Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerreiro AC, et al. Daily rhythms in the cyanobacterium Synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol Cell Proteomics. 2014;13:2042–2055. doi: 10.1074/mcp.M113.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140:529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci USA. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond S, Jun D, Rubin BE, Golden SS. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc Natl Acad Sci USA. 2015;112:E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsui A, et al. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature. 1986;323:720–722. [Google Scholar]
- 14.Ma P, Mori T, Zhao C, Thiel T, Johnson CH. Evolution of KaiC-dependent timekeepers: a proto-circadian timing mechanism confers adaptive fitness in the purple bacterium Rhodopseudomonas palustris. PLoS Genet. 2016;12:e1005922. doi: 10.1371/journal.pgen.1005922. This paper reports daily timekeepers that might not be circadian and also establishes an alternative fitness tests for biological timers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson JB, Davis CR, Johnson CH. Visible light alters yeast metabolic rhythms by inhibiting respiration. Proc Natl Acad Sci USA. 2013;110:21130–21135. doi: 10.1073/pnas.1313369110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:17–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido SS, Johnson CH. Daily and circadian variation in survival from ultraviolet radiation in Chlamydomonas reinhardtii. Photochem Photobiol. 2000;71:758–765. doi: 10.1562/0031-8655(2000)071<0758:dacvis>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Chang YG, et al. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science. 2015;349:324–328. doi: 10.1126/science.1260031. This study of the biochemical oscillator of the cyanobacterial clock unveils a substantial conformational change in the KaiB protein prior to its binding to KaiC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe J, et al. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science. 2015;349:312–316. doi: 10.1126/science.1261040. This study provides the first biochemical explanation of how circadian clockwork can have such a long (slow) time constant of approximately 24 hours. [DOI] [PubMed] [Google Scholar]
- 20.Paulose JK, Wright JM, Patel AG, Cassone VM. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS ONE. 2016;11:e0146643. doi: 10.1371/journal.pone.0146643. A new study that reports a gut bacterium that seems to exhibit daily rhythms when isolated outside of the host. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leone V, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voigt RM, et al. Circadian disorganization alters intestinal microbiota. PLoS ONE. 2014;9:e97500. doi: 10.1371/journal.pone.0097500. This study shows that disruption of the host circadian system alters the composition of the gut microbiome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 25.Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. 2015;112:10479–10484. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, et al. Regulation of the cyanobacterial circadian clock by electrochemically controlled extracellular electron transfer. Angew Chem Int Ed Engl. 2014;53:2208–2211. doi: 10.1002/anie.201309560. [DOI] [PubMed] [Google Scholar]
- 28.Rust MJ, Golden SS, O'Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YI, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci USA. 2012;109:17765–17769. doi: 10.1073/pnas.1216401109. References 28 and 29 show that environmental resetting of the cyanobacterial clock seems to be mediated by daily light-driven and dark-driven driven changes in intracellular ATP and/or redox levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen SE, Golden SS. Circadian rhythms in cyanobacteria. Microbiol Mol Biol Rev. 2015;79:373–385. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattanayak GK, Phong C, Rust MJ. Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr Biol. 2014;24:1934–1938. doi: 10.1016/j.cub.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson CH, Egli M. Metabolic compensation and circadian resilience in prokaryotic cyanobacteria. Annu Rev Biochem. 2014;83:221. doi: 10.1146/annurev-biochem-060713-035632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 34.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 35.Dibner C, et al. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson CH. Circadian clocks and cell division: what's the pacemaker? Cell Cycle. 2010;9:3864–3873. doi: 10.4161/cc.9.19.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egli M, Johnson CH. A circadian clock nanomachine that runs without transcription or translation. Curr Opin Neurobiol. 2013;23:732–740. doi: 10.1016/j.conb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. This study details a unique reconstitution of a circadian biochemical oscillator in vitro with three purified clock proteins from cyanobacteria. [DOI] [PubMed] [Google Scholar]
- 40.Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin X, et al. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA. 2010;107:14805–14810. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson CH, Egli M, Stewart PL. Structural insights into a circadian oscillator. Science. 2008;322:697–701. doi: 10.1126/science.1150451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosokawa N, Kushige H, Iwasaki H. Attenuation of the posttranslational oscillator via transcription–translation feedback enhances circadian-phase shifts in Synechococcus. Proc Natl Acad Sci USA. 2013;110:14486–14491. doi: 10.1073/pnas.1302243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattanayek R, et al. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell. 2004;15:375–388. doi: 10.1016/j.molcel.2004.07.013. This study reports the 3D structure of the core cyanobacterial clock protein, as well as the identification of the first phosphorylation site that regulates its function. [DOI] [PubMed] [Google Scholar]
- 45.Terauchi K, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. This study details the discovery of the key rate-limiting reaction in KaiC that determines circadian period. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishiwaki T, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rust MJ, Markson JS, Lane WS, Fisher DS, O'Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, et al. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA. 2004;101:13933–13938. doi: 10.1073/pnas.0404768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pattanayek R, et al. Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS ONE. 2009;4:e7529. doi: 10.1371/journal.pone.0007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Y, et al. Intramolecular regulation of phosphorylation status of the circadian clock protein KaiC. PLoS ONE. 2009;4:e7509. doi: 10.1371/journal.pone.0007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription–translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. This study contains the first definitive evidence that indicates that the cyanobacterial clock system might not be based on a TTFL loop. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y, Mori T, Johnson CH. Circadian clock protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YI, Dong G, Carruthers CW, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105:12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pattanayek R, Egli M. Protein–protein interactions in the cyanobacterial circadian clock: structure of KaiA dimer in complex with C-terminal KaiC peptides at 2.8 Å resolution. Biochemistry. 2015;54:4575–4578. doi: 10.1021/acs.biochem.5b00694. [DOI] [PubMed] [Google Scholar]
- 57.Egli M, et al. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry. 2012;51:1547–1558. doi: 10.1021/bi201525n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 2012;287:18030–18035. doi: 10.1074/jbc.M112.350660. References 57 and 58 report the remarkable result that KaiC seems to regenerate ATP from ADP as it dephosphorylates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kageyama H, et al. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23:161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 60.Mori T, et al. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin J, Chew J, Chockanathan U, Rust MJ. Mixtures of opposing phosphorylations within hexamers precisely time feedback in the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2014;111:E3937–E3945. doi: 10.1073/pnas.1408692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vakonakis I, LiWang AC. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc Natl Acad Sci USA. 2004;101:10925–10930. doi: 10.1073/pnas.0403037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishiwaki-Ohkawa T, Kitayama Y, Ochiai E, Kondo T. Exchange of ADP with ATP in the CII ATPase domain promotes autophosphorylation of cyanobacterial clock protein KaiC. Proc Natl Acad Sci USA. 2014;111:4455–4460. doi: 10.1073/pnas.1319353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Espinosa J, et al. Cross-talk and regulatory interactions between the essential response regulator RpaB and cyanobacterial circadian clock output. Proc Natl Acad Sci USA. 2015;112:2198–2203. doi: 10.1073/pnas.1424632112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Markson JS, Piechura JR, Puszynska AM, O'Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takai N, et al. A KaiC-associating SasA–RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci USA. 2010;107:3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwasaki H, et al. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 69.Pattanayek R, et al. Combined SAXS/EM based models of the S. elongatus post-translational circadian oscillator and its interactions with the output His-kinase SasA. PLoS ONE. 2011;6:e23697. doi: 10.1371/journal.pone.0023697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iida T, et al. Importance of the monomer–dimer– tetramer interconversion of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. Genes Cells. 2015;20:173–190. doi: 10.1111/gtc.12211. [DOI] [PubMed] [Google Scholar]
- 71.Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8:e1000394. doi: 10.1371/journal.pbio.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teng SW, Mukherji S, Moffitt JR, De Buyl S, O'Shea EK. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science. 2013;340:737–740. doi: 10.1126/science.1230996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zwicker D, Lubensky DK, ten Wolde PR. Robust circadian clocks from coupled protein-modification and transcription–translation cycles. Proc Natl Acad Sci USA. 2010;107:22540–22545. doi: 10.1073/pnas.1007613107. References 71–73 show that the coupled PTO and TTFL oscillator systems in cyanobacteria promote an emergent robustness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mihalcescu I, Hsing W, Leibler S. Resilientcircadian oscillator revealed in individualcyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 75.Ito H, et al. Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat Struct Mol Biol. 2007;14:1084–1088. doi: 10.1038/nsmb1312. This paper reports the remarkable ability of the KaiA–KaiB–KaiC in vitro oscillator to maintain itself without damping for 10 or more days. [DOI] [PubMed] [Google Scholar]
- 76.Kitayama Y, Nishiwaki-Ohkawa T, Sugisawa Y, Kondo T. KaiC intersubunit communication facilitates robustness of circadian rhythms in cyanobacteria. Nat Commun. 2013;4:2897. doi: 10.1038/ncomms3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Loza-Correa M, Gomez-Valero L, Buchrieser C. Circadian clock proteins in prokaryotes: hidden rhythms? Front Microbiol. 2010;1:130. doi: 10.3389/fmicb.2010.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci USA. 2003;100:2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holtzendorff J, et al. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J Biol Rhythms. 2008;23:187–199. doi: 10.1177/0748730408316040. [DOI] [PubMed] [Google Scholar]
- 80.Zinser ER, et al. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE. 2009;4:e5135. doi: 10.1371/journal.pone.0005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Axmann IM, et al. Biochemical evidence for a timing mechanism in Prochlorococcus. J Bacteriol. 2009;191:5342–5347. doi: 10.1128/JB.00419-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mullineaux CW, Stanewsky R. The rolex and the hourglass: a simplified circadian clock in Prochlorococcus? J Bacteriol. 2009;191:5333–5335. doi: 10.1128/JB.00719-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Whitehead K, Pan M, Masumura K, Bonneau R, Baliga NS. Diurnally entrained anticipatory behavior in archaea. PLoS ONE. 2009;4:e5485. doi: 10.1371/journal.pone.0005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Min H, Guo H, Xiong J. Rhythmic gene expression in a purple photosynthetic bacterium, Rhodobacter sphaeroides. FEBS Lett. 2005;579:808–812. doi: 10.1016/j.febslet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 85.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 86.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. References 85 and 86 provide the first rigorous tests of the adaptive fitness that is conferred by circadian timing in any organism and establish the competition assay as a fitness test for circadian systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roenneberg T, Merrow M. Life before the clock: modeling circadian evolution. J Biol Rhythms. 2002;17:495–505. doi: 10.1177/0748730402238231. [DOI] [PubMed] [Google Scholar]
- 88.Troein C, Locke JC, Turner MS, Millar AJ. Weather and seasons together demand complex biological clocks. Curr Biol. 2009;19:1961–1964. doi: 10.1016/j.cub.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 89.Maurice CF, et al. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015;9:2423–2434. doi: 10.1038/ismej.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. A bacterial cytokine. Proc Natl Acad Sci USA. 1998;95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thaiss CA, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510. doi: 10.1016/j.cell.2016.11.003. This study finds that disruption of the microbiome by antibiotics feeds back on the host. [DOI] [PubMed] [Google Scholar]
- 93.McFall-Ngai M. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boettcher KJ, Ruby EG, McFall-Ngai MJ. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J Comp Physiol. 1996;179:65–73. [Google Scholar]
- 95.Wier AM, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Millar AJ, Short SR, Chua NH, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 98.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sinauer Associates; 2004. [Google Scholar]
- 99.Njus D, McMurry L, Hastings JW. Conditionality of circadian rhythmicity: synergistic action of light and temperature. J Comp Physiol. 1977;117:335–344. [Google Scholar]
- 100.Xu Y, et al. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013;495:116–120. doi: 10.1038/nature11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]