Abstract
The FEZ1/LZTS1 gene maps to chromosome 8p22, a region that is frequently deleted in human tumors. Alterations in FEZ1/LZTS1 expression have been observed in esophageal, breast, and prostate cancers. Here, we show that introduction of FEZ1/LZTS1 into Fez1/Lzts1-negative cancer cells results in suppression of tumorigenicity and reduced cell growth with accumulation of cells at late S–G2/M stage of the cell cycle. Fez1/Lzts1 protein is hyperphosphorylated by cAMP-dependent kinase during cell-cycle progression. We found that Fez1/Lzts1 is associated with microtubule components and interacts with p34cdc2 at late S–G2/M stage in vivo. Present data show that FEZ1/LZTS1 inhibits cancer cell growth through regulation of mitosis, and that its alterations result in abnormal cell growth.
Human cancers result from the accumulation of genetic alterations at specific chromosomal regions involving a multistep process (1–3). The short arm of human chromosome 8 (8p) is frequently deleted in human cancer (4–7). We have positionally cloned and characterized the FEZ1/LZTS1 (leucine zipper, putative tumor suppressor 1) gene at 8p22 (8), a region that is lost in many tumors, including prostate, breast, head-and-neck, esophageal, and urinary bladder carcinomas. FEZ1 expression is altered in cancers, including >50% of tumor cells tested, and somatic mutations were observed in several cancers (8, 9). Immunohistochemical analysis shows that Fez1 expression is absent or reduced in 44% of gastric cancer with significant correlation to diffuse histotype (P < 0.001); its inactivation is caused by several factors, including genomic deletion and methylation (9). The data show that the FEZ1 gene is prone to inactivation in human cancer, and that the FEZ1 is inactivated by “two-hit” events that include allelic loss, point mutations, and, possibly, allelic loss plus shut-down transcription. These events suggest alterations of FEZ1 contribute to cancer development.
Although Fez1 has a region of homology with protein kinase A (PKA)-activated leucine-zipper protein, little is known about its biological function. Leucine-rich repeats have been found in peptide sequences of over 70 proteins with diverse functions and origins (10). Leucine-rich repeat superfamily proteins are located in the nucleus, cytoplasm, plasma membrane, and extracellular matrix (10). The present analysis shows that Fez1 localizes predominantly in the cytoplasm. Yeast two-hybrid screening has allowed the identification of Fez1-binding partners, including elongation factor (EF) 1γ, which has been shown to associate with microtubules (11–14), and p34cdc2 kinase (14, 15). Investigation of the interaction of Fez1/Lzts1 indicates Fez1/Lzts1 is associated with microtubule components and interacts with p34cdc2 in vivo. The present data show that Fez1 protein plays a role in mitosis and is involved in the stabilization of active p34cdc2, and that alterations of Fez1 lead to early exit from mitosis.
Materials and Methods
Cell Culture and Cell-Cycle Synchronization.
Cell lines were obtained from the American Type Culture Collection and were maintained as recommended. Synchronization of cells at G1/S was performed by double-thymidine block, as described (16). PKA inhibitor N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (Sigma) was used in culture medium (17).
Expression Vectors and Transfection.
Full-length and mutated FEZ1 cDNAs were cloned by reverse transcription–PCR amplification from human normal brain cDNA (CLONTECH) or from cancer cDNAs as templates (8). FEZ1 cDNAs were ligated in the sense direction into pcDNA vectors (Invitrogen), tetracycline (Tet)-Off-inducible vector (CLONTECH), glutathione S-transferase (GST)-fusion pGEX vector (Amersham Pharmacia), or cytomegalovirus-driven green fluorescent protein (GFP)-fusion plasmid-enhanced-GFP vector (CLONTECH). Full-length FEZ1 cDNA was ligated in an antisense direction into ecdysone-inducible vector (Invitrogen). Nucleotide sequences of inserted cDNAs were verified by sequencing. For transfection, cells were grown and incubated with plasmid DNAs and GenePorter reagent (Gene Therapy Systems, San Diego, CA). To obtain stable transfectants, cells were cultured in selection medium containing 300 μg/ml neomycin/G418 or 200 μg/ml hygromycin.
Cell Growth Assay.
MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] assay was performed with the CellTiter 96 Aqueous nonradioactive cell-proliferation assay kit (Promega). A reading of OD at 490 nm in the MTS assay in a 96-well plate showed linear compatibility with the number of cells in a range between 2 × 102 and 1 × 105 cells per well, counted by trypan-blue staining to exclude dead cells. Four independent assays were performed for each experimental group. For the tumorigenicity study, cells were inoculated s.c. into the left dorsal subclavicular region of 6-week-old female BALB/c nude mice. The tumor volume for each mouse was determined by measuring in two directions and was calculated as follows: tumor volume = [length × (width)2]/2.
Cellular Protein Fractionation.
Cytoplasmic and nuclear protein was extracted as described (18). Briefly, 293 cells (107) were suspended in hypotonic buffer and centrifuged, and the supernatant was collected as cytoplasmic extract C1. The pellet was broken by homogenization and centrifuged, and the supernatant was collected as intermediate extract C2. The pellet was resuspended in nuclear sample buffer.
Protein–Protein Interaction.
Yeast two-hybrid screening was performed with MATCHMAKER system 2 (CLONTECH). For in vitro binding assay, peptide EF family proteins α, β, and δ cDNAs were cloned by reverse transcription–PCR amplification from human liver cDNA (CLONTECH) and ligated into vectors. In vitro transcription/translation was performed with a rabbit reticulocyte system (Amersham Pharmacia) by labeling with [35S]methionine (Amersham Pharmacia). Proteins were incubated in two binding buffers: buffer A (100 mM NaCl/0.5% Nonidet P-40/0.75 mg/ml BSA/20 mM Tris⋅HCl, pH 8.0/1 mM EDTA), or buffer B [150 mM NaCl/0.1% Tween 20/0.75 mg/ml BSA/50 mM Tris⋅HCl, pH 8.0/5 mM EDTA/10% (vol/vol) glycerol] in 100 μl. After adding glutathione–agarose beads preincubated with 10% (wt/vol) BSA, samples were washed five times with each binding buffer and were centrifuged to pull down binding proteins. Samples were boiled and subjected to SDS/PAGE, and gels were exposed to x-ray film.
Microtubule Assembly.
Tubulin purified by two heat-dependent assemble/disassemble cycles from bovine brain (Sigma) containing 15% microtubule-associated proteins were disassembled at 4°C and subjected to in vitro assembly as described (19). Forty micrograms of assembled or disassembled tubulin was subjected to 4–20% gradient SDS/PAGE followed by Coomassie blue staining or by immunoblot analysis. In vitro tubulin polymerization assay was performed as described (20).
Immunoassay.
Immunoblotting and immunoprecipitation followed by detection of signals with the enhanced chemiluminescence (ECL) system (Amersham Pharmacia) were performed as described (21). Mouse monoclonal antibodies used were anti-V5 tag (Invitrogen), anti-Xpress (EXP) tag (Invitrogen), anti-α tubulin (Santa Cruz Biotechnology), anti-p34cdk1/cdc2 (Santa Cruz Biotechnology), anti-cyclin B1 (Santa Cruz Biotechnology), anti-phosphoserine (Sigma), and anti-β-actin (ICN). Rabbit polyclonal antibodies used were anti-Fez1 (9) and anti-EF1γ, kindly given by W. Moller and G. M. C. Janssen (Univ. of Leiden, The Netherlands). Immunofluorescence staining was performed by culturing cells on chambered coverglass, followed by methanol fixation, 0.05% Triton X-100 treatment, and staining with first and secondary antibodies (Zymed) as indicated. Confocal analysis was performed on a Bio-Rad MRC-600 laser scanning confocal microscope.
Phosphorylation Assay.
The PKA catalytic subunit (Sigma) was used for the PKA phosphorylation assay in reaction buffer, as described (20). The p34cdk1/cdc2 kinase assay was performed with the kit (Upstate Biotechnology, Lake Placid, NY) by using [γ-32P]ATP with or without the addition of cyclin B. Samples were separated by SDS/PAGE, and gels were exposed to x-ray film.
Results and Discussion
Fez1 Protein Expression in Cancer Cells Results in Reduced Cell Growth in Vitro and in Vivo.
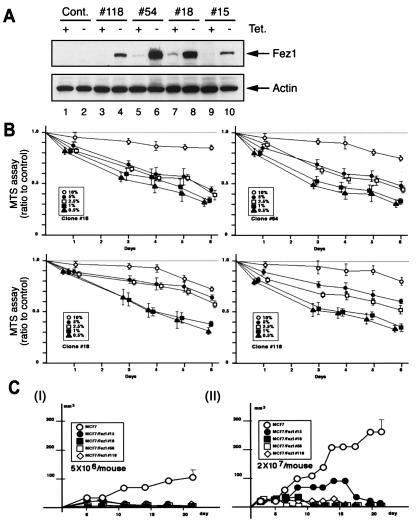
By using a Tet-inducible Fez1 expression system, we constructed FEZ1 transfectants of Fez1-negative breast cancer MCF7 cells. In this system, the transgene expression is suppressed by Tet, whereas transfectants express the transgene in the absence of Tet (22). Transfectants were grown and maintained in medium with Tet to repress Fez1 expression. We picked up 120 well isolated, independent clones after 5 weeks of selection in medium with hygromycin B and Tet. Immunoblot analysis showed that four well isolated clones showed substantial induction of Fez1 expression when cultured without Tet, whereas Fez1 expression either was barely detectable or was not detectable when cultured with Tet. We repeated immunoblot analysis in medium with or without Tet by using different serum concentrations (0.5–10%) to confirm the reproducibility of Fez1 induction [representative data in 5% (vol/vol) serum culture are shown in Fig. 1A]. In vitro cell growth of inducible MCF7 FEZ1 transfectants was analyzed by MTS assay (Promega) to analyze viable cell numbers (Fig. 1B). Results show that cell growth was reduced after Fez1 induction, compared with control transfectants. This reduction was more pronounced when cells were cultured in 0.5–5% serum (Fig. 1B) and when cells were grown at subconfluent densities (40–80%; data not shown). In vivo tumorigenicity of MCF7 FEZ1 transfectants was reduced, whereas control cells formed tumors (Fig. 1C). These data indicate that Fez1 expression inhibited cell growth in vitro and tumor formation in vivo.
Figure 1.
Cell growth and tumorigenicity of Tet-inducible MCF7 FEZ1 transfectants. (A) Immunoblot analysis with anti-Fez1 antibody of MCF7 FEZ1 transfectants. Four Tet-inducible MCF7 FEZ1 transfectants (clones 15, 18, 54, and 118) were cultured in Tet-free 5%-serum medium to induce Fez1 expression. Proteins were extracted and analyzed before (lanes 1, 3, 5, 7, and 9) and after (lanes 4, 6, 8, and 10) Fez1 induction. Lanes 1 and 2, control vector transfectants. (B) In vitro cell-growth analysis. After Fez1 induction, four clones were seeded in 10, 5, 2.5, 1, or 0.5% serum medium without Tet. Cell growth of Tet-inducible MCF7 FEZ1 transfectants were analyzed by MTS assay. Data from transfectants were divided by data from control vector transfectants for the ratio. (C) Tumorigenicity of MCF7 FEZ1 transfectants. Cells of four MCF7 FEZ1 transfectants precultured in Tet-free medium for 72 hr [5 × 106 (I) or 2 × 107 (II)] were inoculated into four nude mice for each. ●, clone 15; ■, clone 18; □, clone 54; ⋄, clone 118; ○, control vector transfectant.
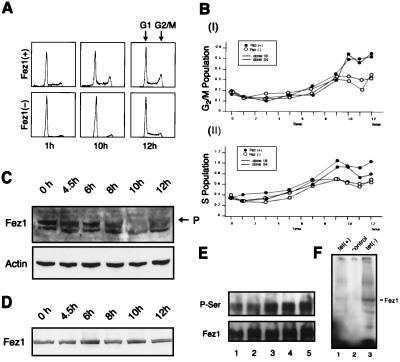
MCF7 FEZ1 transfectants were subjected to flow-cytometry analysis to assess the effect of Fez1 expression on cell-cycle progression (Fig. 2 A and B). FEZ1 transfectants in medium with or without Tet were synchronized at G1/S with a double-thymidine block. After the removal of thymidine, cell populations corresponding to G2/M (Fig. 2BI) and S (Fig. 2BII) were measured at indicated times. Data show that Fez1-positive cells accumulated in S phase at 7–12 hr and in G2/M phase at 9–12 hr. Taken together with the analysis shown in Fig. 1B, these data indicate that Fez1 expression exerts an inhibitory effect on cell growth by affecting cell-cycle progression.
Figure 2.
Cell-cycle analysis and protein modification of Fez1. (A and B) Cell-cycle analysis of synchronized MCF7 FEZ1 transfectants. FEZ1 transfectants were cultured in Tet-free medium and treated with double-thymidine block. After medium was exchanged for fresh medium, cells were fixed at indicated times and subjected to flow-cytometry analysis. Representative data for clone 54 is shown in A. Data in B are shown as ratios of G2/M (Upper) or S (Lower) to G1 population of clone 18 (solid line) or clone 54 (dotted line). ●, with induced Fez1; ○, without Fez1. (C and D) Endogenous Fez1 protein analysis in synchronized 293 cells in medium without (C) or with (D) 20 μM PKA inhibitor. After medium was exchanged for fresh medium, cellular proteins were extracted at indicated times and subjected to immunoblot analysis with anti-Fez1 or anti-actin antibody. P in figure denotes Fez1 with lower mobility. (E) Serine phosphorylation of Fez1 protein in MCF7 Tet-inducible FEZ1 transfectants (clone 54). After synchronizing, cells were incubated in fresh medium for indicated times. Lysates were immunoprecipitated with anti-Fez1 antibody, followed by immunoblot analysis with anti-phosphoserine (P-Ser) or anti-Fez1 antibody (Fez1). Cells were harvested at 0 (lane 1), 1 (lane 2), 2.5 (lane 3), 6 (lane 4), or 8 hr (lane 5) after relief from G1/S block. (F) In vivo phosphorylation of Tet-inducible MCF7 FEZ1 transfectants (clone 54) with [32P]orthophosphate. Cells were cultured in medium containing [32P]orthophosphate with Tet (lane 1) and without Tet (lane 3). Lane 2, control vector transfectant. Lysates were immunoprecipitated with anti-Fez1 antibody, subjected to SDS/PAGE, dried, and exposed to film.
Fez1 Protein Is Phosphorylated During Cell-Cycle Progression.
To assess whether Fez1 protein is modified during cell-cycle progression, we synchronized endogenous Fez1-positive 293 cells or Tet-inducible MCF7 FEZ1 transfectants by double-thymidine block, followed by culture in fresh medium to allow progression through the cell cycle. Immunoblot analysis of 293 cell lysates with anti-Fez1 antibody showed Fez1 protein with lower mobility at 8–12 hr after relief from G1/S block, when cells are at late S–G2/M phase (Fig. 2C). The presence of Fez1 protein with a different mobility suggests SDS-resistant modification of Fez1, or different charges of the protein. Proteins were subjected to a denaturing condition of 6 M urea–SDS/PAGE, followed by immunoblot analysis with anti-Fez1 antibody, which showed the disappearance of the lower mobility Fez1 protein (H.I. and C.M.C., unpublished data), suggesting that SDS-resistant modification(s) of Fez1 occurred during cell-cycle progression. Database searches indicated that Fez1 has a region homologous to PKA-responsive activated protein (8), and it has been shown that PKA phosphorylates serine and threonine residues of various proteins (23). Immunoblot analysis with anti-phosphoserine antibody showed that Fez1 protein immunoprecipitated from FEZ1 transfectants was hyperphosphorylated on serines during cell-cycle progression (Fig. 2E). When we treated synchronized 293 cells with PKA inhibitor in medium, immunoblot analysis of 293 cell lysate with anti-Fez1 antibody showed an absence of lower mobility Fez1 protein during cell-cycle progression (Fig. 2D). In addition, Tet-inducible MCF7 FEZ1 transfectants were radiolabeled to analyze phosphorylation in vivo. Results showed that, after Fez1 induction, a 67-kDa radiolabeled protein was immunoprecipitated with anti-Fez1 antibody after a 2-hr culture with [32P]orthophosphate (Fig. 2F). These data demonstrate protein modifications of Fez1, including serine phosphorylation by PKA during cell-cycle progression.
Fez1 Protein Binds to EF1γ Protein.
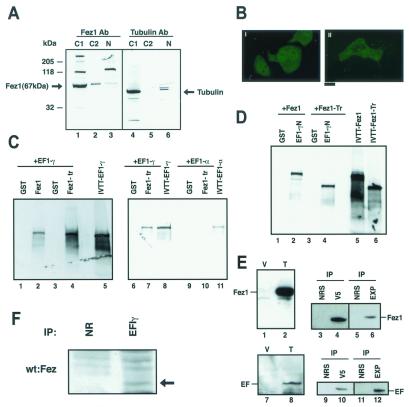
Cytoplasmic and nuclear proteins were extracted separately from 293 fetal kidney cells to assess cellular localization of Fez1 protein. Immunoblot analysis showed that Fez1 protein was localized predominantly in the cytoplasm with only a small fraction in the nucleus (Fig. 3A). An expression vector with GFP-fused FEZ1 cDNA was transfected to analyze cellular localization in living cells, showing GFP-Fez1 predominantly in the cytoplasm, whereas a control GFP vector transfectant showed GFP expression in both the cytoplasm and nucleus (Fig. 3B), suggesting that Fez1 protein caused localization of fusion protein preferentially in the cytoplasm in vivo.
Figure 3.
Intracellular localization of Fez1 protein and characterization of Fez1-interacting proteins. (A and B) Cytoplasmic localization of Fez1 protein. (A) Proteins were extracted from the cytoplasm (C1) and nucleus (N) of 293 cells, electrophoresed, and immunoblotted with anti-Fez1 (lanes 1–3) or anti-tubulin antibody (lanes 4–6) by using 40 μg of protein per lane. C2, intermediate extract. (B) GFP-fusion Fez1 expression vector was transfected into Fez1-negative HeLaS3 cells (II), and GFP expression was observed under a fluorescent microscope. (I) GFP vector control transfectant. (Bar = 5 μm.) (C) In vitro binding of EF1γ to Fez1. The interaction was shown by binding assay with [35S]methionine-labeled in vitro-translated EF1γ protein (lanes 1–8), which was incubated with GST-Fez1 (67 kDa; lane 2) or GST-C-terminal truncated Fez1 (40 kDa, corresponding to nucleotides 1–1128; lanes 4 and 7). Lanes 1, 3, and 6, addition of only GST protein as negative control; lanes 5 and 8, translated EF1γ protein alone as positive control (showing input). Reaction was shown in two different buffers, buffer A (lanes 1–4) and buffer B (lanes 6 and 7), as described in text. Binding of in vitro-translated EF1α (lanes 9 and 10) to GST-Fez1 (lane 10) was analyzed with buffer B. Lane 9, addition of only GST; lane 11, in vitro-translated protein alone, showing input. (D) In vitro binding of Fez1 proteins to N-terminal EF1γ protein. Similar to C, binding assay in buffer B was performed with in vitro-translated full-length 67-kDa Fez1 (lanes 1 and 2) or truncated 40-kDa Fez1 (lanes 3 and 4), incubated with GST-N-terminal EF1γ protein (lanes 2 and 4). Lanes 1 and 3, addition of only GST. In vitro-translated full-length 67-kDa (lane 5) or truncated 40-kDa Fez1 (lane 6) were shown as positive controls. (E) Interaction of Fez1 with EF1γ in transfectants. Full-length FEZ1 or EF1γ cDNA was ligated to pcDNA4V5 or pcDNA3His vector to express V5 tag- or EXP tag-fused protein, respectively. HeLaS3 cells were cotransfected with the vectors. Immunoblot analysis with antitag antibodies shows transfectants express ≈70-kDa Fez1 protein (lane 2) or ≈50-kDa EFγ protein (lane 8). Lanes 1 and 7, vector control transfectants. Immunoprecipitation (IP) with antitag antibodies (V5 and EXP) or control normal serum (NRS) was performed from lysates, followed by immunoblot analysis with anti-V5 (lanes 3–6) or anti-EXP antibody (lanes 9–12). (F) Immunoprecipitation of Fez1 and EF1γ in synchronized 293 cells. Similar to Fig. 5 A–C, 293 cells were synchronized and analyzed after a 6-hr culture plus a 2-hr treatment with 40 μg/ml nocodazole (late S–G2/M). Cellular proteins (1.5 mg) were subjected to immunoprecipitation with anti-EF1γ antibody (EF1γ) or normal rabbit serum (NR), followed by immunoblotting with anti-Fez1 antibody (wt:Fez, indicated by arrow).
Yeast two-hybrid screening was performed in the Y190 yeast strain to identify binding partner(s) of the Fez1 protein. The Y190 was transformed with pAS2–1 full-length FEZ1 vector as a “bait,” to screen 3 × 106 clones of pACT2 cDNA expression library, followed by β-galactosidase assay. We isolated clones and performed DNA sequencing. Positive clones were redundant and classified into 24 sequences (>100 bp), including six unknown fragments. Of the 18 known gene sequences, 8 corresponded to EF1γ (12, 13), whereas 6, 2, and 2 corresponded to other genes. To confirm the result, β-galactosidase assay was performed with four isolated cDNA clones, demonstrating that the candidate clone EF1γ showed strong interaction with Fez1 (the reaction time was <15 min, whereas positive control reaction was 15–20 min, and negative control was >48 hr). The other three clones took >1 hr to express the β-galactosidase reaction.
EF1γ is a member of the protein family composed of EF1α, β, γ, and δ proteins (12, 13). To analyze binding of EFs to Fez1 further, we performed an in vitro binding assay with GST-fused Fez1 proteins and in vitro-translated EF1α, β, γ, and δ proteins. We found that Fez1 bound to EF1γ (Fig. 3C), whereas binding of Fez1 to EF1α (Fig. 3C) or δ (data not shown) was not detected. Binding of Fez1 to EF1β was not informative, because EF1β bound to GST (data not shown). We made three deletion mutants of in vitro-translated EF1γ, retaining the N-terminal 153 aa, the C-terminal 126 aa, or the mid-portion 149 aa, and analyzed binding with GST-Fez1. Fez1 bound to N-terminal EF1γ but not to the other two parts (data not shown). To confirm this fact, we performed another binding assay with GST-fused N-terminal 153-aa protein of EF1γ and in vitro-translated Fez1, which showed truncated Fez1 protein (N-terminal 2/3 part of Fez1 protein, which lacks a C-terminal region downstream of the leucine-zipper repeat) bound to the N-terminal 153-aa protein of EF1γ (Fig. 3D). HeLaS3 cells were cotransfected by using two expression vectors with full-length FEZ1 and EF1γ cDNAs to assess in vivo interaction, showing Fez1 was coimmunoprecipitated with EF1γ protein (Fig. 3E). We also detected in vivo association of endogenous Fez1 and EF1γ proteins in synchronized 293 cells (Fig. 3F). It is shown that EF1γ protein is involved in microtubule association (11–14) as well as in peptide synthesis (12, 13). Our current study shows that induction of Fez1 results in the accumulation of cells in S–G2/M, when microtubule dynamics are developed (24). We then examined Fez1 involvement in microtubule function.
Fez1 Is Associated with Assembled Microtubules.
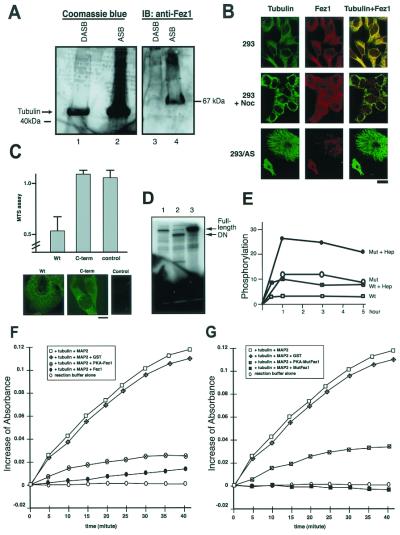
Tubulin- and microtubule-associated proteins extracted from brain by heat-dependent assemble/disassemble treatment were used to analyze the association of Fez1 with tubulin. Coomassie blue staining after SDS/PAGE showed assembled tubulin contained proteins of various sizes, including 65- to 70-kDa proteins (Fig. 4A). Immunoblot analysis with anti-Fez1 antibody detected a 67-kDa band in assembled tubulin but not in disassembled tubulin (Fig. 4A). To investigate the effect of tubulin assembly on Fez1 association, cytoplasmic fractions were extracted from synchronized 293 cells and incubated with paclitaxel to inhibit tubulin depolymerization in vitro (25). Results showed that Fez1 is associated with assembled tubulin precipitates from the extracted cytoplasmic protein of cells in the G1/S, S, and, to a lesser extent, the G2/M phase, but not with disassembled precipitates induced by colchicine (data not shown).
Figure 4.
Interaction of Fez1 with Microtubules. (A) Association of Fez1 with assembled tubulin. Forty μg each of disassembled (lanes 1 and 3) or assembled (lanes 2 and 4) tubulin, which was extracted from brain, was electrophoresed and stained with Coomassie blue (lanes 1 and 2) or immunoblotted with anti-Fez1 antibody (lanes 3 and 4). (B) Double immunofluorescence with mouse anti-tubulin and fluorescein-labeled anti-mouse antibody (Tubulin), and with rabbit anti-Fez1 and rhodamine-labeled anti-rabbit antibody (Fez1). Yellow areas depict colocalization of the two fluorochromes (Tubulin + Fez1). Fetal kidney 293 cells were cultured in medium without (293) or with (293 + Noc) nocodazole (40 μg/ml) before staining. 293/AS (similar to Fig. 5 A–C), 293 cell transfectants expressing ecdysone-inducible antisense FEZ1, cultured with ecdysone derivative ponasterone A to inhibit endogenous Fez1 expression. (Bar = 5 μm.) (C) Fez1-negative cervical cancer HeLaS3 cells transfected with full-length (Wt) or N-terminal truncated (C-term, lacking N-terminal 388-aa portion) FEZ1 cDNA in pcDNA4V5 vector. MTS cell-growth assay (Upper) was performed, similar to Fig. 1B. Immunofluorescence (Lower) was performed with anti-V5 tag and fluorescein-labeled anti-mouse antibody. wt, wild-type FEZ1; C-term, truncated FEZ1; Control, without primary antibody. (Bar = 5 μm.) (D) In vitro kinase assay of Fez1 proteins. Purified recombinant GST-Fez1 proteins (wt, S29P, and Q501Stop) were subjected to the analysis. PKA was incubated with wt (lane 1), mutated (Q501Stop) (lane 2), or mutated (S29P) protein (lane 3) at 30°C for 45 min for [γ-32P]ATP labeling. Samples were subjected to SDS/PAGE, dried and exposed to film. (E) In vitro kinetics of Fez1 phosphorylation by PKA. One μg each of purified GST-wt or S29P mutated Fez1 protein (Mut) was subjected to kinase assay using [γ-32P]ATP, without or with (+ Hep) heparin at 20 μg/ml. Samples were separated by SDS/PAGE, dried, and exposed to film for quantitative analysis with the PhosphorImager (Molecular Dynamics). y axis, mol of phosphate per mol of Fez1. (F and G) Involvement of Fez1 with tubulin polymerization in vitro. Purified tubulin and purified MAP2 were incubated at 37°C for indicated times with GST-Fez1 (F, ●), PKA-phosphorylated GST-Fez1 (F, ⊗), GST-mutated Fez1 (S29P) (G, ■), and PKA-phosphorylated GST-mutated Fez1 (S29P) (G, square with cross). Controls in F and G include reactions with tubulin and MAP2 (□), with tubulin, MAP, and GST (diamonds with cross), or reaction buffer alone (○). Absorbance at 350 nm was measured with spectrophotometer, and the increase is shown.
We performed double immunofluorescence to assess colocalization of Fez1 protein with microtubules in vivo. The results demonstrated the filamentous nature of Fez1 throughout the cytoplasm (Fig. 4B). Tubulin and Fez1 filaments were colocalized in 293 cells (Fig. 4B) and esophageal cancer TE1 FEZ1 transfectants (data not shown). When cells were treated with nocodazole to inhibit microtubule polymerization, both tubulin and Fez1 staining were diffusely cytoplasmic, suggesting that the filamentous pattern of Fez1 protein depends on polymerized microtubules. To elucidate Fez1 function further, we analyzed microtubule association and cell-growth inhibition. When we transfected Fez1-negative cells with FEZ1 cDNA vectors, the wt Fez1 protein was associated with microtubules and cell growth was inhibited, whereas the truncated Fez1 lacking the N-terminal segment neither associated with microtubules nor inhibited cell growth (Fig. 4C). The data show that Fez1 protein, but not its C-terminal segment, plays a role in association with microtubules and inhibition of cell growth.
Previous analysis of human cancer cells has shown altered expression and somatic mutations of FEZ1, including missense mutation (S29P) and nonsense mutation (Q501Stop; ref. 8). cDNAs were synthesized from mRNAs of these cancer cells and ligated into expression vector to make recombinant, mutated Fez1 proteins. Five micrograms of purified GST-fused mutated Fez1 proteins, S29P, and Q501Stop, and wt Fez1 protein were incubated with [γ-32P]ATP and PKA catalytic subunit in vitro. Results showed that wt protein was phosphorylated to a level similar to that of mutated Q501Stop protein, whereas mutated S29P protein was hyperphosphorylated compared with wt Fez1 protein (Fig. 4D). The data suggest that the phosphorylation sites are N-terminal to Gln-501. The S29P protein seems to be more susceptible to PKA phosphorylation. To confirm this possibility, phosphorylation kinetics were analyzed by incubating purified GST-fused wt and mutated S29P proteins with [γ-32P]ATP and PKA. Results showed that mutated S29P protein was hyperphosphorylated with a faster reaction and at a higher intensity than wt Fez1 protein, and that heparin enhanced phosphorylation (Fig. 4E). Purified GST-fused Fez1 proteins, wt, and S29P mutant, with and without PKA phosphorylation, were incubated with purified tubulin in the presence of purified mitogen-activated protein (MAP)2, which had been shown to induce tubulin polymerization (26). Results showed that both wt and mutant (S29P) Fez1 proteins without PKA phosphorylation inhibited (by 90% and >95%) MAP2-induced tubulin polymerization in vitro, whereas GST did not (Fig. 4 F and G), suggesting that Fez1 protein is involved in MAP2-induced assembly of microtubules in vitro. The inhibition of tubulin polymerization was reduced by 70–75% when wt and mutant (S29P) Fez1 proteins had been phosphorylated by PKA (Fig. 4 F and G). The release was more evident in mutant Fez1 (S29P) than in wt Fez1 protein, suggesting that hyperphosphorylation of mutant Fez1 (S29P) might result in the loss of normal Fez1 function. The present data demonstrate that Fez1 protein is involved in microtubule assembly, which is regulated, at least in part, by Fez1 phosphorylation by PKA.
p34cdc2 Mitotic Kinase Interacts with Fez1 Protein.
To analyze the cell-growth regulatory function of Fez1 further, we studied its interaction with other cell-cycle regulators. Previous study showed that EF1γ is phosphorylated by the p34cdc2 kinase, which is an important factor in G2/M transition of cell cycle in vivo (14, 15). Infection of Fez1-negative SW780 cancer cells with adenoviral FEZ1 vector resulted in the inhibition of cell growth, and we detected Fez1 protein immunoprecipitated with p34cdc2 protein in the infected SW780 cancer cells (A.V. and C.M.C., unpublished data). In vitro binding data show that PKA phosphorylation of Fez1 results in a greater association with p34cdc2 (data not shown).
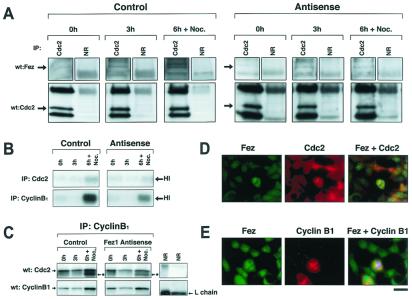
Because Fez1 is phosphorylated by PKA during cell-cycle progression, we investigated the association of endogenous Fez1 with p34cdc2 in 293 cells synchronized by double-thymidine block; for this experiment, we used synchronized 293 antisense FEZ1 transfectants. Data show that endogenous Fez1 was immunoprecipitated with p34cdc2 protein in synchronized 293 transfectants (Fig. 5A), which were cultured in the absence of ponasterone A to express endogenous Fez1. The association was greater for cells cultured in thymidine-free medium for 6 hr plus a 2-hr treatment with nocodazole (the late S–G2/M phase) than for cells cultured for 0 hr (G1/S phase) or for 3 hr (S phase). Less association was found in 293 antisense FEZ1 transfectants cultured with ponasterone A to inhibit endogenous Fez1 than in antisense noninduced transfectants. These data show that p34cdc2 associates with Fez1 predominantly in late S–G2/M phase.
Figure 5.
Association of Fez1 with p34cdc2. (A) Immunoprecipitation of p34cdc2 and Fez1 in synchronized 293 transfectants. 293 cell transfectants expressing antisense FEZ1 vector under control of ecdysone-inducible system were developed to inhibit endogenous Fez1 expression with the ecdysone derivative ponasterone A, as shown in Fig. 4B. 293 antisense FEZ1 transfectants (AS44) were cultured with (Right, Antisense) or without ponasterone A (Left, Control) in medium. 293 transfectants were synchronized by double-thymidine block. After thymidine medium was exchanged, cells were analyzed at 0, 3, or 6 hr plus an additional 2-hr treatment with 40 μg/ml nocodazole (6 h + Noc.). Cellular proteins (1.5 mg) were subjected to immunoprecipitation with anti-p34cdc2 antibody (Cdc2) or normal rabbit serum (NR), followed by immunoblot analysis with anti-Fez1 or with anti-p34cdc2 antibody. (B) Histone H1 phosphorylation after immunoprecipitation with anti-p34cdc2 or with anti-cyclin B1 antibody from synchronized 293 transfectants. Similar to A, samples were subjected to phosphorylation assay with [γ-32P]ATP and histone H1 after immunoprecipitation, separated by SDS/PAGE, and exposed to film. Arrows indicate phosphorylation of histone H1. (C) The p34cdc2 activation in synchronized 293 transfectants. Similar to A, samples were subjected to immunoblot analysis with anti-p34cdc2 or with anti-cyclin B1 antibody after immunoprecipitation with anti-cyclin B1 antibody. Asterisks with arrows indicate a dephosphorylated active form of p34cdc2. (D and E) Double immunofluorescence with mouse anti-p34cdc2 (D) or mouse anti-cyclin B1 antibody (E), followed by rhodamine-labeled anti-mouse antibody (Middle), or with rabbit anti-Fez1 antibody (D and E), followed by fluorescein-labeled anti-rabbit antibody (Left). Yellow areas depict colocalization of both fluorochromes (Right). Chromatin was stained with Hoechst 2495 to show in blue. (Bar = 10 μm.)
Studies have shown that the activation of p34cdc2 cyclin B1 induces entry into M phase (27), and that the inactivation of the kinase is indispensable for exit from M phase (28). Immunoprecipitation was performed with 293 antisense FEZ1 transfectants to assess histone H1 phosphorylation (Fig. 5B). Data show that, in antisense FEZ1 noninduced 293 transfectants, histone H1 phosphorylation was detected in late S–G2/M phase and was greater than in antisense-induced transfectants. Data were confirmed by immunoprecipitation with anti-cyclin B1 antibody, which showed that the active p34cdc2 form in late S–G2/M phase was reduced in 293 antisense FEZ1-induced transfectants (Fig. 5C), suggesting that Fez1 is involved in the stability of active p34cdc2-cyclin B1 complex in late S–G2/M phase in vivo, and that inactivation of Fez1 results in destabilization of the kinase, which leads to an early exit from M phase and uncontrolled cell proliferation.
We then performed double immunofluorescent staining to assess localization of Fez1, p34cdc2, and cyclin B1 in vivo. Results demonstrated that Fez1 colocalized with p34cdc2 and cyclin B1 in mitotic 293 cells (Fig. 5 D and E). It has been reported that cyclin B protein accumulates during interphase, reaches a peak level at metaphase, and is suddenly destroyed at the exit from M phase, whereas p34cdc2 remains present at a relatively constant level throughout cell cycle (29). The present study showed colocalization of Fez1 and cyclin B1 in mitotic cells, when cyclin B1 was detectable by immunofluorescence (Fig. 5E), indicating that the p34cdc2–cyclin B1 complex colocalizes with endogenous Fez1 in mitotic cells, and that Fez1 is involved in the regulation of mitosis, in which the p34cdc2–cyclin B1 complex plays a role.
Acknowledgments
We thank Drs. W. Moller and G. M. C. Janssen, University of Leiden, The Netherlands, for kindly giving us anti-EF1γ antibody. We thank Dr. Kay Huebner for critical reading of the manuscript. This work was supported partially by U.S. Public Health Service Grants CA39860, CA51083, and CA56336, and Grant CA83698 from the National Cancer Institute.
Abbreviations
- EF
elongation factor
- PKA
protein kinase A
- GFP
green fluorescent protein
- Tet
tetracycline
- wt
wild type
- MAP
mitogen-activated protein
- GST
glutathione S-transferase
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nowell P C, Croce C M. Am J Pathol. 1986;125:8–15. [PMC free article] [PubMed] [Google Scholar]
- 2.Nowell P C. Adv Cancer Res. 1993;62:1–17. doi: 10.1016/s0065-230x(08)60313-9. [DOI] [PubMed] [Google Scholar]
- 3.Knudson A G. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagan J, Stein J, Babaian R J, Joe Y-S, Pisters L L, Glassman A B, von Eschenbach A C, Troncoso P. Oncogene. 1995;11:2121–2126. [PubMed] [Google Scholar]
- 5.Anbazhagan R, Fujii H, Gabrielson E. Am J Pathol. 1998;152:815–819. [PMC free article] [PubMed] [Google Scholar]
- 6.El-Naggar A K, Coombes M M, Batsakis J G, Hong W K, Goepfert H, Kagan J. Oncogene. 1998;16:2983–2987. doi: 10.1038/sj.onc.1201808. [DOI] [PubMed] [Google Scholar]
- 7.Wagner U, Bubendorf L, Gasser T C, Moch H, Gorog J-P, Richter J, Mihatsch M J, Waldman F M, Sauter G. Am J Pathol. 1997;151:753–759. [PMC free article] [PubMed] [Google Scholar]
- 8.Ishii H, Baffa R, Numata S, Murakumo Y, Rattan S, Inoue H, Mori M, Fidanza V, Alder H, Croce C M. Proc Natl Acad Sci USA. 1999;96:3928–3933. doi: 10.1073/pnas.96.7.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vecchione A, Ishii H, Shiao Y-H, Trapasso F, Rugge M, Tamburrino J F, Murakumo Y, Alder H, Croce C M, Baffa R. Clin Cancer Res. 2001;7:1546–1552. [PubMed] [Google Scholar]
- 10.Buchanan S G S C, Gay N J. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- 11.Janssen G M C, Moller W. Eur J Biochem. 1988;171:119–129. doi: 10.1111/j.1432-1033.1988.tb13766.x. [DOI] [PubMed] [Google Scholar]
- 12.Bec G, Kerjan P, Waller J-P. J Biol Chem. 1994;269:2086–2092. [PubMed] [Google Scholar]
- 13.Janssen G M C, van Damme H T F, Kriek J, Amons R, Moller W. J Biol Chem. 1994;269:31410–31417. [PubMed] [Google Scholar]
- 14.Belle R, Minella O, Cormier P, Morales J, Poulhe R, Mulner-Lorillon O. Prog Cell Cycle Res. 1995;1:265–270. doi: 10.1007/978-1-4615-1809-9_21. [DOI] [PubMed] [Google Scholar]
- 15.Moreno S, Nurse P. Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 16.Masson D, Kreis T E. J Cell Biol. 1995;131:1015–1024. doi: 10.1083/jcb.131.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. J Biol Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 18.Shapiro D J, Sharp P A, Wahli W W, Keller M J. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 19.Williams R C, Jr, Lee J C. Methods Enzymol. 1982;85:376–385. doi: 10.1016/0076-6879(82)85038-6. [DOI] [PubMed] [Google Scholar]
- 20.Tseng H-C, Lu Q, Henderson E, Graves D J. Proc Natl Acad Sci USA. 1999;96:9503–9508. doi: 10.1073/pnas.96.17.9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ausubel F M, Brent R, Kingston R E, Moor D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1989. [Google Scholar]
- 22.Gossen M, Freundieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 23.Cho-Chung Y S, Pepe S, Clair T, Budillon A, Nesterova M. Crit Rev Oncol Hematol. 1995;21:33–61. doi: 10.1016/1040-8428(94)00166-9. [DOI] [PubMed] [Google Scholar]
- 24.Nurse P. Nature (London) 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 25.Kumar N. J Biol Chem. 1981;256:10435–10441. [PubMed] [Google Scholar]
- 26.Ainsztein A M, Purich D L. J Biol Chem. 1994;269:28465–28471. [PubMed] [Google Scholar]
- 27.Berry L D, Gould K L. Prog Cell Cycle Res. 1996;2:99–105. doi: 10.1007/978-1-4615-5873-6_10. [DOI] [PubMed] [Google Scholar]
- 28.Nishiyama A, Tachibana K, Igarashi Y, Yasuda H, Tanahashi N, Tanaka K, Ohsumi K, Kishimoto T. Genes Dev. 2000;14:2344–2357. doi: 10.1101/gad.823200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King R W, Deshaies R J, Peters J M, Kirschner M W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]