Abstract
Hepatic flavin-containing mono-oxygenase 3 (FMO3) metabolizes a broad array of nucleophilic heteroatom (e.g., N or S)-containing xenobiotics (e.g., amphetamine, sulindac, benzydamine, ranitidine, tamoxifen, nicotine, and ethionamide), as well as endogenous compounds (e.g., catecholamine and trimethylamine). To predict the effect of genetic and nongenetic factors on the hepatic metabolism of FMO3 substrates, we quantified FMO3 protein abundance in human liver microsomes (HLMs; n = 445) by liquid chromatography-tandem mass chromatography proteomics. Genotyping/gene resequencing, mRNA expression, and functional activity (with benzydamine as probe substrate) of FMO3 were also evaluated. FMO3 abundance increased 2.2-fold (13.0 ± 11.4 pmol/mg protein vs. 28.0 ± 11.8 pmol/mg protein) from neonates to adults. After 6 years of age, no significant difference in FMO3 abundance was found between children and adults. Female donors exhibited modestly higher mRNA fragments per kilobase per million reads values (139.9 ± 76.9 vs. 105.1 ± 73.1; P < 0.001) and protein FMO3 abundance (26.7 ± 12.0 pmol/mg protein vs. 24.1 ± 12.1 pmol/mg protein; P < 0.05) compared with males. Six single nucleotide polymorphisms (SNPs), including rs2064074, rs28363536, rs2266782 (E158K), rs909530 (N285N), rs2266780 (E308G), and rs909531, were associated with significantly decreased protein abundance. FMO3 abundance in individuals homozygous and heterozygous for haplotype 3 (H3), representing variant alleles for all these SNPs (except rs2066534), were 50.8% (P < 0.001) and 79.5% (P < 0.01), respectively, of those with the reference homozygous haplotype (H1, representing wild-type). In summary, FMO3 protein abundance is significantly associated with age, gender, and genotype. These data are important in predicting FMO3-mediated heteroatom-oxidation of xenobiotics and endogenous biomolecules in the human liver.
Introduction
The flavin-containing mono-oxygenases (FMOs, EC 1.14.13.8) belong to a family of NADPH-, flavine-adenine dinucleotide-, and O2-dependent microsomal enzymes that catalyze the oxidation of a wide variety of nucleophilic heteroatom compounds containing a nitrogen, sulfur, phosphorous, or selenium atom (Krueger and Williams, 2005). Eleven human FMO genes (FMO1–FMO11P) have been identified to date, but human FMO1–FMO5 are the only protein-coding genes that have been functionally characterized (Yamazaki and Shimizu, 2013). Members of the FMO gene family display distinct temporal-, tissue-, and species-specific expression patterns (Klick and Hines, 2007). Typical FMO substrates include trimethylamine, catecholamines (Türkanoğlu Özçelik et al., 2013), nicotine (Bloom et al., 2013), imipramine (Wagmann et al., 2016), prothipendyl (Krueger and Williams, 2005), amitriptyline (Krueger and Williams, 2005), phospho-sulindac amide (Xie et al., 2014), benzydamine (BZD) (Taniguchi-Takizawa et al., 2015), ranitidine (Overby et al., 1997), tamoxifen (Parte and Kupfer, 2005), clozapine (Tugnait et al., 1997), diphenhydramine (Cruciani et al., 2014), and amphetamine and methamphetamine (Cashman et al., 1999).
FMO3, a protein with 532 amino acids, is the predominant form of the FMO enzyme family expressed in the adult human liver (Yamazaki and Shimizu, 2013). Although the FMO3 protein content in the liver is high (Haining et al., 1997; Overby et al., 1997), comparable to cytochrome P450 enzymes (P450s) (Koukouritaki et al., 2007), the role of FMO3 in xenobiotic or endobiotic metabolism has been often overlooked. The overlapping substrate specificity between FMO3 and P450s, the thermal instability of the enzyme during in vitro incubation (Cashman, 2008), and the unavailability of selective FMO inhibitors could be some of the reasons why contributions of FMO3 to drug biotransformation have not been studied more extensively. Further, many oxidation reactions previously found to be catalyzed by P450s were later determined to be catalyzed solely or predominately by FMO3 (e.g., itopride, clozapine, and cimetidine) (Cashman et al., 1993; Tugnait et al., 1997; Mushiroda et al., 2000). It has recently been concluded that FMOs are responsible for about 2.2% of all metabolic reactions and about 6% of all the phase 1 metabolic reactions (Testa et al., 2012). FMO3 is considered primarily responsible for almost all FMO-mediated reactions of pharmaceutical interest.
Unlike the P450s, FMO3 is not generally induced or inhibited by xenobiotics (Klick and Hines, 2007) and therefore poses limited drug-drug interaction potential (Cashman, 2008); however, interindividual differences in FMO3 activity (Overby et al., 1997) could result in significant differences in FMO3-dependent biotransformation of exogenous and endogenous compounds, leading to unexpected drug reactions or pathologic consequences (Koukouritaki et al., 2005). Many single nucleotide polymorphisms (SNPs) are associated with FMO3 function (Mitchell and Smith, 2001; Zhou and Shephard, 2006; Phillips and Shephard, 2008); however, the mechanism is unclear as to whether these SNPs affect substrate affinity (Km) or FMO3 protein levels. Moreover, some of the FMO3 SNPs exhibit linkage disequilibrium (LD) (Cashman and Zhang, 2002), but a systematic classification of haplotypes and diplotypes of FMO3 and their associations with protein activity or abundance has not been established. Although the association of the nongenetic factors, such as age and gender, with FMO3 abundance has been studied previously (Koukouritaki et al., 2002; Shimizu et al., 2011), investigations with multiple samples and with a more precise method for protein quantification are needed.
Therefore, the current study investigated the effect of genetic variations and nongenetic factors, including age, gender, and ethnicity, on FMO3 hepatic abundance using liquid chromatography-tandem mass spectrometry (LC-MS/MS). The investigation was conducted in a large cohort of well characterized pediatric and adult human liver samples (n = 445, age range, 0.00-87 Years).
Materials and Methods
Chemicals and Reagents.
Iodoacetamide, dithiothreitol, and Pierce trypsin protease (MS grade) were purchased from Thermo Fisher Scientific (Rockford, IL). Ammonium bicarbonate buffer (ABB, 98% purity) was purchased from Acros Organics (Geel, Belgium). Chloroform, MS-grade acetonitrile (99.9% purity), methanol, and formic acid (≥99.5% purity) were purchased from Fisher Scientific (Fair Lawn, NJ). Human serum albumin and bovine serum albumin (BSA) were obtained from Calbiochem (Billerica, MA) and Thermo Fisher Scientific, respectively. Synthetic isotopically pure heavy stable isotope-labeled peptides were produced by Thermo Fisher Scientific. The purified recombinant FMO3 protein was obtained from Abnova (H00002328-P01; Walnut, CA). NADPH and BZD were purchased from Sigma-Aldrich (St. Louis, MO). BZD N-oxide and dazidamine were obtained as previously described (Yeung et al., 2007).
Human Liver Microsome Samples.
Previously isolated human liver microsomes (HLMs) (Pearce et al., 2016; Shirasaka et al., 2016) were used in this study. The liver tissue samples for the microsomal preparation were originally received from three liver banks: 1) the University of Washington Human Liver Bank (Seattle, WA) (n = 46), 2) Children’s Mercy Kansas City (Kansas City, MO) (n = 128), and 3) the Liver Bank at the St. Jude Children’s Research Hospital (Memphis, TN) (n = 271). The samples from Children’s Mercy Kansas City were obtained from the University of Maryland Brain and Tissue Bank for Developmental Disorders and the Liver Tissue Cell Distribution System. Additional details on the selection, procurement, and storage of the livers and investigator blinding for sample analyses have been described previously (Prasad et al., 2014; Shirasaka et al., 2016; Boberg et al., 2017; Tanner et al., 2017). Age, gender, and ethnicity were known for >95%, 98%, and 87% of the liver donors, respectively (Supplemental Table 1). The age range for donors was from 0 to 87 years (mean ± S.D.: 30 ± 24 years). Of the donors with known gender, 61.2% were male. The panel of samples consisted of 79.1% Caucasian, 5.8% African American, 0.9% Hispanic, 0.2% Native American, 0.2% Pacific Islander, and 12.6% unknown ethnicity donors. Cause of death, medications used, and liver pathology were known for less than 50% of donors, and smoking status was unknown for >88% of donors; therefore, these factors were not assessed as a predictor of FMO3 phenotypes in the present study. The collection and use of these tissues for research purposes were approved by the human subjects Institutional Review Boards of the University of Washington (Seattle, WA) and the St. Jude Children’s Research Hospital (Memphis, TN) and the Pediatric Institutional Review Board of Children’s Mercy Kansas City (Kansas City, MO).
FMO3 Protein Quantification.
FMO3 protein quantification was carried out using a validated LC-MS/MS proteomics method. Two surrogate peptides were selected for FMO3 protein quantification, and corresponding heavy peptides containing terminal labeled [13C6, 15N4]-arginine and [13C6, 15N2]-lysine residues were used as internal standards. VAIIGAGVSGLASIR was used as the quantifier and NNLPTAISDWLYVK as the qualifier (Supplemental Fig. 1). HLM samples were diluted to 2 mg/ml, and 80 μl (160 µg) of microsomal protein (or purified FMO3 protein as calibrator) was digested as described previously (Boberg et al., 2017) with minor modifications. Briefly, microsomal protein and 10 μl of human serum albumin (10 mg/ml) and/or 10 μl of BSA (0.2 mg/ml) were denatured and reduced with 10 μl of 250 mM dithiothreitol and 40 μl of ABB buffer (100 mM) at 95°C for 10 minutes with gentle shaking at 300g. After cooling to room temperature for 10 minutes, the denatured protein was alkylated by the addition of 20 μl of 500 mM iodoacetamide; the reaction was carried out in the dark for 30 minutes. Ice-cold methanol (500 µl), chloroform (100 µl), and water (400 µl) were added to each sample. After vortex-mixing and centrifugation at 16,000g (4°C) for 5 minutes, the upper and lower layers were removed using vacuum suction, and the pellets were dried at room temperature for 10 minutes. The pellets were then washed with 500 µl of ice-cold methanol and subjected to centrifugation at 8000g (4°C) for 5 minutes. After the supernatant was removed, the pellets were dried at room temperature for 30 minutes and resuspended in 60 µl of ABB buffer (50 mM, pH 7.8). Subsequently, the protein pellets were digested by adding 20 μl of trypsin (protein-trypsin ratio, approximately 80:1) and incubating at 37°C for 16 hours. The reaction was quenched by the addition of 20 μl of peptide internal standard cocktail (prepared in 80% acetonitrile in water containing 0.5% formic acid) and 10 μl of 80% acetonitrile in water containing 0.5% formic acid. The samples were vortex-mixed and subjected to centrifugation at 4000g for 5 minutes, and the supernatants were collected in LC-MS vials. The calibration curve standards ranged from 3.99 to 511 fmol (on-column amount based on the protein calibrator) and were generated by serial diluting of recombinant FMO3 protein standard in phosphate buffer (50 mM Kpi, 0.25 M sucrose, 10 mM EDTA, pH 7.4) and processed similarly to HLM sample. Quantification was performed using a triple-quadrupole MS instrument (SCIEX Triple Quad 6500; PE SCIEX, Concord, ON, Canada) in ESI positive ionization mode coupled to an Acquity UPLC, I-class (Waters Technologies, Milford, MA). Five microliters of the trypsin digest was injected onto the column (Acquity UPLC HSS T3 1.8 μm, C18 100A; 100 × 2.1 mm; Waters). Surrogate light and heavy (internal standards) peptides were monitored using instrument parameters provided in Supplemental Table 2. The LC-MS/MS data were processed using Analyst 1.6.2 version software (SCIEX).
FMO3 mRNA Quantification.
FMO3 mRNA expression was quantified only in a subset of unprocessed liver tissue samples (n = 214, Supplemental Table 1). Details of the RNA-seq procedures, including RNA isolation, truSeq stranded mRNA preparation, read processing, and analysis pipeline were described previously (Tanner et al., 2017). The transcript levels of mRNA are displayed in FPKM (fragments per kilobase per million reads) values.
FMO3 Enzyme Activity Assay.
FMO3 enzyme activity was assessed on a limited number of HLM samples prepared from fresh liver tissues (donor age range, 9–68 years; mean ± S.D., 40.1 ± 17.9 years, n = 37; Supplemental Table 1) owing to the potential instability of the protein to freeze-thaw cycles (Cashman, 2008). Activity was determined by quantifying the rate of BZD N-oxidation (Stormer et al., 2000). The assay conditions include the following: 0.2 mg/ml of HLM protein, 0.1 M phosphate buffer (pH 7.4), 50 μM BZD, 0.5 mM NADPH, and water to a final volume of 250 μl. All reactions were performed in triplicate. After a preincubation of 3 minutes at 37°C, the reaction was initiated with the addition of substrate and allowed to continue with gentle agitation (70 rpm) for 20 minutes. The reaction was quenched with an equal volume of acetonitrile containing 0.1 mg/ml of dazidamine (internal standard). Incubations were then placed on ice for 5 minutes to allow for full protein precipitation before centrifugation (14,000 rpm × 5 minutes). The supernatant was analyzed by an optimized high-performance liquid chromatography method as reported previously (Yeung and Rettie, 2006). Briefly, chromatographic separations of BZD, BZD N-oxide, and dazidamine were performed on an Agilent Hypersil ODS C18 column (4.0 × 250 mm, 5 μm). The mobile phase consisted of methanol:acetonitrile:water: 29% NH4OH (50:40:10:0.05, v/v/v/v) (A) and water (B). The mobile-phase components were mixed at a fixed ratio of 92% A + 8% B, and a flow rate of 1.0 ml/min was used. The effluent was monitored fluorometrically with an excitation wavelength of 307 nm and an emission wavelength of 377 nm. Approximate retention times for BZD N-oxide, dazidamine, and BZD were 4, 6, and 8 minutes, respectively.
FMO3 Gene Resequencing, Genotyping, and Haplotype Determination.
Two different approaches, gene resequencing and genotyping, were used for genetic characterization of the liver donors because these data were generated in two different institutes (Supplemental Table 1). FMO3 gene resequencing was performed using the Pharmacogenomics Research Network (PGRN)-Seq platform, a targeted sequencing approach, for the University of Washington and St. Jude Liver Bank samples, as discussed elsewhere (Gordon et al., 2016). DNA of the tissues provided by Children’s Mercy were genotyped on the PharmacoScan (Affymetrix, Santa Clara, CA). LD analyses of FMO3 variants and inferred haplotypes were determined using Haploview 4.2 (Cambridge, MA).
Data Analysis.
We used a robust strategy to ensure optimum reproducibility when quantifying FMO3 proteins. For example, ion suppression was addressed by using heavy peptide internal standards. BSA was used as exogenous protein internal standards, which was added to each sample as a fixed quantity before methanol-chloroform-water extraction and trypsin digestion. Addition of BSA addresses the variability introduced during the predigestion processing steps, such as 1) protein loss during methanol-chloroform-water extraction and 2) sample-to-sample trypsin digestion artifacts. To address interbatch variability, we processed three sets of pooled representative HLM samples each day, which served as quality controls across the entire study. In total, a three-step data normalization approach was used; first, the average of detectable light peak areas for specific peptide daughter fragments was divided by the average of detectable heavy peak areas. For example, we used a ratio of the average of three fragments of VAIIGAGVSGLASIR (light) and the average of two fragments of VAIIGAGVSGLASI[13C6,15N4]R (heavy) for FMO3 quantification. More fragments of the light peptide were used in this case because the dynamic range for analyte peptide (i.e., light) was unpredictable compared with the heavy internal standard peptide. Next, this ratio was further divided by the BSA light/heavy area ratio. Finally, for each day, these data were further normalized by mean values of the quality control values run with individual batch to adjust for any interday variability.
The samples were classified based on the following age categories: neonatal plus infancy (0–364 days), toddler/early childhood (1 to <6 years), middle childhood (6 to <12 years), adolescence (12–18 years), and adulthood (>18 years).
Statistical analyses were performed using GraphPad Prism 5 (La Jolla, CA) and Microsoft Excel (Version 14; Redmond, WA). The protein abundance, mRNA expression levels and activity data were non-normally distributed, and therefore nonparametric tests were used to test age, gender, ethnicity, or genotype dependence. To compare two groups (e.g., male vs. female), the Mann-Whitney test was used. The Kruskal-Wallis test followed by Dunn’s multiple comparison test were used to perform the age-dependent data analysis and determine the associations between FMO3 genotype and mRNA expression levels, FMO3 protein abundance, and enzyme activity. For correlation analysis, the nonparametric Spearman regression test was used:
 |
(1) |
A nonlinear regression equation (eq. 1) was used to fit the ontogeny data, as described previously (Boberg et al., 2017), where Adultmax is the maximum average relative protein abundance, Age is the age in years of the subject at the time of sample collection, Age50 is the age in years at which half-maximum adult protein abundance is obtained, F is the fractional protein abundance in adult samples, Fbirth is the fractional protein abundance (of adult) at birth, and n is the exponential factor.
The number of variants was directly counted. Hardy-Weinberg equilibrium was determined by comparing the variant frequencies with the expected values using a contingency table χ2 statistic with the Yates correction. The numbers of haplotypes, statistics D, D’, and LD were estimated by Haploview 4.2 software. A P value below 0.05 was considered statistically significant.
Results
Absolute Quantification of Human Hepatic FMO3 Based on LC-MS/MS and Correlation to mRNA and Activity Data.
The calibration curves for FMO3 (3.99–511 fmol, on-column) demonstrated good linearity (r2 > 0.99, Supplemental Fig. 2). The lower limit of quantification for the FMO3 protein standard in the trypsin digestion buffer was 3.99 fmol (on-column). The intraday precision (% coefficient of variance) of the analytical method, based on pooled quality control samples, was within 25%.
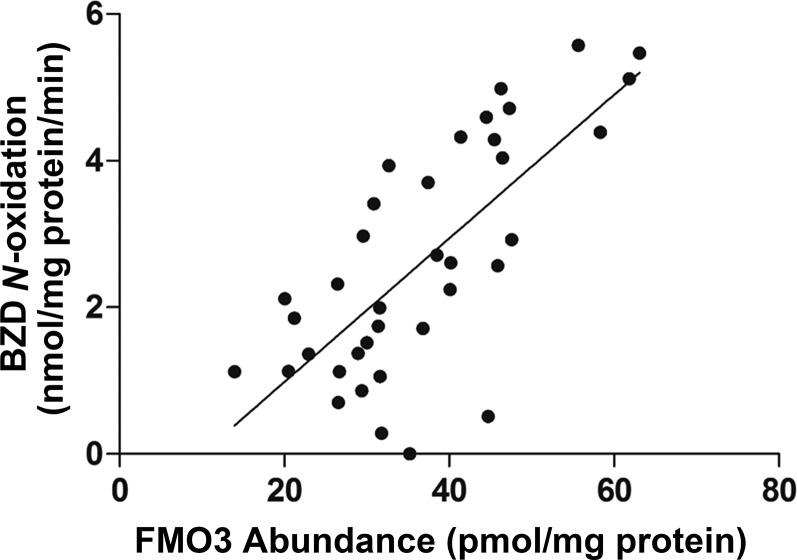
The average human liver FMO3 protein concentration in HLM samples (n = 445) was 25.1  12.0 pmol/mg microsomal protein with 124-fold interindividual variation (0.76–94.5 pmol/mg microsomal protein). Furthermore, FMO3 protein abundance was positively correlated with BZD N-oxidation activity (r = 0.73, P < 0.001, Fig. 1); however, the correlation between FMO3 mRNA and protein abundance was poor (r = 0.19, P > 0.05) (data not shown).
12.0 pmol/mg microsomal protein with 124-fold interindividual variation (0.76–94.5 pmol/mg microsomal protein). Furthermore, FMO3 protein abundance was positively correlated with BZD N-oxidation activity (r = 0.73, P < 0.001, Fig. 1); however, the correlation between FMO3 mRNA and protein abundance was poor (r = 0.19, P > 0.05) (data not shown).
Fig. 1.
Association between FMO3 protein abundance (pmol/mg protein) and BZD N-oxidation activity in human liver microsomes (n = 37).
Association of Nongenetic Factors with Human Hepatic FMO3 Abundance.
Age was positively associated with FMO3 protein abundance. Stratifying by age as a categorical variable, the FMO3 protein abundance was 1.9-fold (P < 0.001) higher in adults compared with neonates (0–1 year) (Fig. 2A). Similarly, FMO3 abundance in neonatal liver was significantly lower compared with that observed in middle childhood (6–12 years, P < 0.01) and adolescence (12–18 years, P < 0.001). FMO3 protein levels were lower in infants (1–6 years, P < 0.001) compared with adults. Age versus FMO3 abundance with age as a continuous variable is shown in Fig. 2B. Nonlinear regression of the HLM protein abundance data revealed that the FMO3 protein was 50% of the values observed in adults at the age of approximately 10 months. The effect of age on FMO3 protein abundance was independent of the genotype (Fig. 2, C and D) as discussed to follow.
Fig. 2.
Association between age and FMO3 protein levels in human liver without (A and B) and with (C and D) considering interplay with diplotype E158K-E308G. (A and C) represent categorical data, and (B and D) show continuous data. Dots indicate individual protein abundance; the mean and S.D. are presented as the horizontal line and error bar. The Kruskal-Wallis test followed by Dunn’s multiple comparison test were used to compare protein levels; ** and *** indicate P values of < 0.01 and < 0.001, respectively.
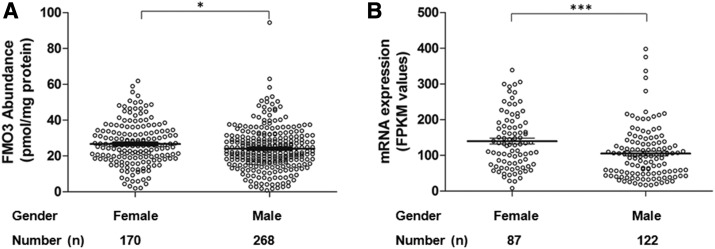
Female liver donors exhibited modestly higher FMO3 protein (26.7  12.0 pmol/mg protein vs. 24.1 ± 12.1 pmol/mg protein, P < 0.05, Fig. 3A) and mRNA levels (FPKM values, 139.9 ± 76.9 vs. 105.1 ± 73.1, P < 0.001, Fig. 3B) compared with males. The gender differences in both protein and mRNA levels were consistent even when pediatric samples from subjects younger than 6 years of age were excluded from the analysis (Supplemental Fig. 3); however, no significant difference in FMO3 activity was observed between female and male samples, perhaps because of the small number of samples (data not shown).
12.0 pmol/mg protein vs. 24.1 ± 12.1 pmol/mg protein, P < 0.05, Fig. 3A) and mRNA levels (FPKM values, 139.9 ± 76.9 vs. 105.1 ± 73.1, P < 0.001, Fig. 3B) compared with males. The gender differences in both protein and mRNA levels were consistent even when pediatric samples from subjects younger than 6 years of age were excluded from the analysis (Supplemental Fig. 3); however, no significant difference in FMO3 activity was observed between female and male samples, perhaps because of the small number of samples (data not shown).
Fig. 3.
Association between gender and FMO3 protein (A) and mRNA (B) levels in human liver. Dot plots are displayed with mean abundance as the horizontal line, and the error bar displays S.D. with individual maximum and minimum values shown in the dot plot. The Mann-Whitney test was used to compare FMO3 levels; * and *** indicate P values of < 0.05 and < 0.001, respectively.
No significant difference in FMO3 protein abundance was observed among Caucasians (25.4 ± 12.4 pmol/mg protein, n = 352) and African Americans (22.1 ± 14.1 pmol/mg protein, n = 30).
Association of Genetic Factors with Human Hepatic FMO3 Abundance.
Gene resequencing of the 285 tissue samples (263 Caucasians, two African Americans, one Asian, and 19 unknown) from the University of Washington and St. Jude Children’s Research Hospital identified 21 variants in FMO3. Eleven of these variants were within the coding sequence, and 10 were intronic variants (Table 1). Of these 21 variants identified, 11 variants (rs2064074, rs1800822, rs72549326, rs2266782, rs2066534, rs1736557, rs909530, rs61753344, rs2266780, rs909531, and rs72549334) have been reported previously in the literature, and an additional seven variants (rs4140653, rs28363536, rs143661234, rs75904274, rs79952472, rs373775407, and rs373403267) were found in the Single Nucleotide Polymorphism Database (dbSNP, https://www.ncbi.nlm.nih.gov/snp/); however, no reports for three of the variants could be found. The minor allele frequencies (MAF) of these variants are shown in Table 1.
TABLE 1.
FMO3 variants identified in a subset of samples of this study (n = 285)
| Variant No. | Chromosome Position | Nucleotide Change | Location | Amino Acid Change | rs No. | HW P Value | MAF | FMO3 Function | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 171061989 | C > T | Intron 2 | rs4140653 | 0.6354 | 0.037 | |||
| 2 | 171061990 | A > G | Intron 2 | rs2064074 | 0.1809 | 0.47 | Koukouritaki et al. (2005) | ||
| 3 | 171072862 | G > T | Intron 2 | rs28363536 | 0.3712 | 0.198 | |||
| 4 | 171076813 | T > G | Intron 3 | 1 | 0.002 | ||||
| 5 | 171076935 | C > T | Exon 4 | S147S | rs1800822 | 1 | 0.058 | Koukouritaki et al. (2005), Yamazaki and Shimizu (2013) | |
| 6 | 171076952 | C > T | Exon 4 | P153L | rs72549326 | 1 | 0.002 | Inactive | Treacy et al. (1998), Yeung et al. (2007) |
| 7 | 171076966 | G > A | Exon 4 | E158K | rs2266782 | 0.1308 | 0.412 | Decreased | Furnes et al. (2003), Koukouritaki et al. (2005), Yeung et al. (2007), Yamazaki and Shimizu (2013) |
| 8 | 171077038 | GTCT > G | Intron 4 | rs143661234 | 1 | 0.058 | |||
| 9 | 171077274 | G > T | Exon 5 | rs75904274 | 1 | 0.016 | |||
| 10 | 171077372 | C > G | Intron 5 | rs2066534 | 0.9376 | 0.209 | Koukouritaki et al. (2005) | ||
| 11 | 171077393 | C > A | Intron 5 | rs79952472 | 1 | 0.002 | |||
| 12 | 171079971 | G > A | Exon 6 | V220V | rs373775407 | 1 | 0.002 | ||
| 13 | 171080080 | G > A | Exon 6 | V257M | rs1736557 | 1 | 0.081 | No change | Furnes et al. (2003), Koukouritaki et al. (2005), Yeung et al. (2007), Yamazaki and Shimizu (2013) |
| 14 | 171083128 | GTC > G | Intron 6 | 1 | 0.002 | ||||
| 15 | 171083174 | C > T | Exon 7 | N285N | rs909530 | 1 | 0.261 | Furnes et al. (2003), Koukouritaki et al. (2005) | |
| 16 | 171083208 | G > T | Exon 7 | 1 | 0.002 | ||||
| 17 | 171083232 | G > T | Exon 7 | E305 | rs61753344 | 1 | 0.002 | Inactive | Treacy et al. (1998) |
| 18 | 171083242 | A > G | Exon 7 | E308G | rs2266780 | 0.3712 | 0.198 | Decreased | Koukouritaki et al. (2005), Yeung et al. (2007), Yamazaki and Shimizu (2013) |
| 19 | 171083537 | T > C | Intron 7 | rs909531 | 0.6644 | 0.209 | Koukouritaki et al. (2005) | ||
| 20 | 171083540 | A > G | Intron 7 | rs373403267 | 1 | 0.002 | |||
| 21 | 171086457 | C > T | Exon 9 | R492W | rs72549334 | 1 | 0.002 | Inactive | Yeung et al. (2007) |
HLM samples (with donor age over age 6 years) were grouped according to the results of FMO3 gene-resequencing data. Six SNPs were associated with decreased FMO3 protein abundance (Fig. 4). These SNPs include three missense (rs2266782, E158K; rs909530, N285N; and rs2266780, E308G) and three intronic (rs2064074, rs28363536, and rs909531) SNPs; however, there was no association between FMO3 mRNA expression and BZD N-oxidase activity among those SNPs that showed a correlation with protein abundance (Supplemental Figs. 4 and 5).
Fig. 4.
Association between genetic variation and FMO3 protein levels in HLMs (with donor age over 6 years). (A–F) represent individual SNPs (i.e., rs2064074, rs28363536, rs2266782 (E158K), rs909530 (N285N), rs2266780 (E308G), and rs909531, respectively. Dots indicate individual protein abundance, and the mean and S.D. are presented as the horizontal line and error bar. The Kruskal-Wallis test followed by Dunn’s multiple comparison test were used to compare protein levels; *, ** and *** indicate P values of < 0.05, < 0.01, and < 0.001, respectively.
To avoid false-positive associations between these variants and FMO3 levels, LD analysis was subsequently performed. All SNPs within FMO3 were in Hardy-Weinberg equilibrium (P > 0.05, Table 1). Haplotype blocks and haplotypes were inferred based on the allele frequencies of the SNPs by using Haploview. In total, two haplotype blocks (Fig. 5) and six haplotypes, which occurred at a frequency greater than 1% accounting for more than 98.5% of the total chromosomes examined, were identified (Table 2). Six SNPs (rs2266782, rs909530, rs2266780, rs2064074, rs28363536, and rs909531), which were associated with FMO3 abundance, were found in strong LD (D’ > 0.975) with each other, except between rs2266782 and rs909530, where a median LD (D’ = 0.632) was observed. Regression coefficients (r2) of these pairwise SNPs indicated that rs28363536, rs2266780 (E308G), and rs909531 tag the same haplotype (r2 > 0.97); rs2266782 (E158K) was associated with rs22064074 (r2 = 0.78); and rs2266780 (E308G) was associated with rs909530 (N285N) (r2 = 0.71). Diplotype-dependent protein abundance of FMO3 was observed after excluding the effect of ontogeny. Samples (with donor age over 6 years) harboring the H3 haplotype, which consists of six SNPs, had up to 50% lower FMO3 protein levels compared with the reference haplotype H1 (Fig. 6). The association between H3 haplotype and FMO3 activity could not be investigated because of the smaller number of samples and higher variability (data not shown).
Fig. 5.
A schematic representation of human FMO3 gene structure and LD generated by Haploview 4.2. The HW P-value cutoff and minimum MAF were 0.01 and 0.02, respectively.
TABLE 2.
Haplotype analysis data of FMO3 in 285 samples
Haplotypes with frequencies 1%. Haplotype tag SNPs (htSNPs) are identified by an asterisk (*). The wild-type (reference) nucleotides are marked in italics.
1%. Haplotype tag SNPs (htSNPs) are identified by an asterisk (*). The wild-type (reference) nucleotides are marked in italics.
| Haplotype | Block 1 |
Block 2 |
Frequency | |||||
|---|---|---|---|---|---|---|---|---|
| rs2064074* (Intron 2) | rs28363536* (Intron 2) | rs2266782* (E158K) | rs2066534 (Intron 5) | rs909530* (N285N) | rs2266780* (E308G) | rs909531* (Intron 7) | ||
| H1 | A | G | G | C | C | A | T | 0.528 |
| H2 | G | G | A | G | C | A | T | 0.195 |
| H3 | G | T | A | C | T | G | C | 0.196 |
| H4 | G | G | G | C | T | A | T | 0.040 |
| H5 | G | G | G | C | C | A | T | 0.016 |
| H6 | G | G | A | G | T | A | T | 0.012 |
Fig. 6.
Association between FMO3 diplotypes and FMO3 protein levels in human liver (with donor age over 6 years). Only those diplotypes that show statistical significance in FMO3 protein abundance are presented. Dots indicate individual protein abundance, and the mean and S.D. are presented as the horizontal line and error bar. The Kruskal-Wallis test followed by Dunn’s multiple comparison test were used to compare protein levels; *, **, and *** indicate P values of < 0.05, < 0.01 and < 0.001, respectively.
Additionally, genotype analysis of the pediatric samples from Children’s Mercy Kansas City (n = 128) detected eight variants in FMO3 (seven exonic and one intronic; Supplemental Table 3). Since genotyping was performed on a different platform, the comprehensive Haploview analysis described already for the adult samples could not be performed on the pediatric samples. Consistent with the data described in the preceding section, two SNPs, rs2266782 (E158K) and rs2266780 (E308G), showed associations with protein abundance (Supplemental Fig. 6, A and B). No associations were detected for other variants, which is possibly due to the low variant frequencies and/or confounding age-related variability. Furthermore, the impact of haplotype E158K-E308G on FMO3 protein abundance is shown in Supplemental Fig. 6C. Considering the impact of allelic variants on FMO3 abundance, the association between age and human hepatic FMO3 protein was reanalyzed. Age-dependent protein abundance of FMO3 was still observed after excluding the samples harboring the homozygous E158K-G308G. The FMO3 protein abundance was 2.2-fold (P < 0.001) higher in adults compared with neonates (0–1 year) (Fig. 2C). After 6 years of age, no significant difference in FMO3 abundance was seen between children and adults. FMO3 protein was 50% of the values observed in adults by approximately 15 months based on the reanalysis of the data (Fig. 2D).
Discussion
In the present study, we identified several genetic and nongenetic factors that contribute to variation in human hepatic FMO3 in a large cohort of samples (n = 445). Although FMO quantification in the human livers has been accomplished by Western blotting and LC-MS/MS in previous studies (Overby et al., 1997; Chen et al., 2016), the sample sizes were small (n = 5–10) in these studies, and the associations of genetic factors, as well as the interplay of ontogeny and genotype, were not investigated. Consistent with the reported data based on the immunoblotting method (Koukouritaki et al., 2002; Shimizu et al., 2011), an age-dependent increase in FMO3 protein levels was observed in this study (Fig. 2). Koukouritaki et al. (2002) found that FMO3 expression increased 4.5-fold between the ages of 3 weeks and 10 months (1.1–4.7 pmol/mg protein), with another approximate 3-fold between 10 months and 11 years (4.7–12.7 pmol/mg protein). Between ages 11 and 18 years, there was a third significant increase in mean expression levels (12.7–26.9 pmol/mg protein) (Koukouritaki et al., 2002). In the present study, only a 2-fold increase in mean FMO3 abundance was observed between infancy (0–1 year) and middle childhood (6–12 years), and no significant increase was found in the donors over 6 years of age; however, the hepatic FMO3 levels found in both studies were comparable (24.2 pmol/mg protein vs. 26.9 pmol/mg protein) in adolescence. Differences in sensitivity at lower concentrations between Western blotting and LC-MS/MS methods, age classification criteria, quality of samples, and data analysis (3%–17% of samples in each age group were excluded in the study by Koukouritaki et al., 2002) may explain, at least in part, the differing observations between studies.
An association between gender and hepatic FMO3 levels has not been revealed in previous reports (Cherrington et al., 1998; Koukouritaki et al., 2002). In this study, we observed a modest, statistically significant (but perhaps not clinically relevant) gender difference in FMO3 mRNA and protein abundances (Fig. 3). Previous studies have shown that female mouse livers contain higher levels of FMO3 mRNA levels (80-fold) than those in male liver samples (Janmohamed et al., 2004), which may be due to the modulating effects of testosterone on FMO3 expression; testosterone, but not 17-estradiol, suppresses mouse FMO hepatic activity (dimethylaniline N-oxidation) (Duffel et al., 1981). After castration, hepatic FMO activity (methimazole oxidation) increased significantly, and serum testosterone levels decreased; however, administration of physiologic levels of testosterone to castrated animals returned FMO activity (Falls et al., 1997); however, there is little evidence for sex hormone regulation of the FMO3 abundance in human livers. Whereas gender difference was apparently observed for FMO3 mRNA, as well as protein expression, the regression analysis indicated a lack of correlation between FMO3 mRNA and protein expression, which is inconsistent with the findings of the previous study (Overby et al., 1997). The latter indicates that FMO3 mRNA is not a good surrogate of its protein abundance. This lack of association could be a result of different mechanisms of mRNA and protein regulation (Liu et al., 2016) or mRNA instability in the frozen-thawing procedure of the samples (Botling et al., 2009).
More than 300 SNPs have been reported for FMO3 (http://www.ncbi.nlm.nih.gov). Most are rare, and more than 40 of these polymorphisms have been linked to altered FMO3 function and symptomatic trimethylaminuria (also referred as “fish-like odor syndrome”) (Yeung et al., 2007). FMO3 variants have also been associated with higher risks of sudden infant death syndrome (Poetsch et al., 2010) and hypertension-related ischemic stroke (Türkanoğlu Özçelik et al., 2013), altered cigarette consumption (Bloom et al., 2013), and increased efficacy of sulindac to prevent familial adenomatous polyposis (Hisamuddin et al., 2004). In vitro FMO3 activity studies investigating the effects of nonsynonymous variants or some haplotypes, such as E158K, E132H, E308G, E132H-E158K, and E158K-E308G, on kinetic parameters revealed that Vmax values of the variant forms are lower than the reference allele (Lattard et al., 2003; Yamazaki and Shimizu, 2013); but changes in Km are substrate-dependent (Lattard et al., 2003; Yeung et al., 2007). These results indicate that the effects of nonsynonymous variants on FMO3 substrate affinity may be substrate-dependent, but their effects on FMO3 abundance are consistent; however, direct evidence of the association between genetic variation and FMO3 protein abundance has never been studied. In the present study, six SNPs were identified that are associated with decreased FMO3 protein expression. Individuals homozygous for the missense variant allele E158K (rs2266782) was associated with significantly lower FMO3 protein levels (Fig. 4), which was consistent with the 65% reduced FMO3 catalytic efficiency (Vmax/Km) in vitro toward multiple substrates such as methimazole, trimethylamine and 10-(N,N-dimethylaminopentyl)-2-(trifluoromethyl)-phenothiazine) (Lattard et al., 2003). Similarly, a positive association between protein abundance and activity was observed for another amino acid change, E308G, caused by rs2266780 (Lattard et al., 2003). Two other variants, P153L (rs72549326) and E305X (rs61753344), previously shown to have no functional activity (Treacy et al., 1998; Yeung et al., 2007), could not be tested in this study because of the lack of sufficient samples, although the FMO3 abundance was lower (11.8 and 7.8 pmol/mg protein vs. 26.4 pmol/mg protein for P153L and E305X vs. reference allele, respectively) in the single heterozygous sample present in our cohort. Similarly, another function-unchanging missense SNP, V257M (rs1736557) (Treacy et al., 1998), showed no relation to FMO3 protein levels in this study. Several mechanisms could potentially explain decreased protein levels resulting from inherited alterations in encoded amino acid sequence, but the mechanism has most often involved accelerated degradation of the variant allozyme, which is typically via a ubiquitin-proteasome–mediated pathway (Weinshilboum and Wang, 2004). Furthermore, one synonymous SNP (N285N, rs2266780) and three intronic variants (rs2064074, rs28363536, and rs909531) were also associated with decreased FMO3 protein abundance (Fig. 4). Although synonymous SNPs and variants in intronic regions may also contribute to changes in protein abundance by impacting splicing, translation fidelity, mRNA stability, and protein folding (Sauna and Kimchi-Sarfaty, 2011), no correlation between FMO3 function and these SNPs has been reported. Three samples showed relatively high FMO3 protein levels but very small BZD N-oxidation activity (Fig. 1). The potential mechanism of the disconnection between FMO3 protein levels and activity might be the instability of FMO3 protein or unique functional variant not investigated in the present study.
LD analysis data showed that SNP rs2266780 (N285N), rs2064074, rs28363536, and rs909531 are perhaps related to E308G and/or E158K, indicating that the suppressive impacts of these four SNPs on FMO3 protein abundance may be from one or both of the exonic SNPs. Moreover, consistent with previous studies reporting that the Vmax values of FMO3 were wild-type > E158K E308G > E158K-E308G (Lattard et al., 2003; Krueger and Williams, 2005), the FMO3 protein abundance of a donor homozygous for haplotype E158K-E308G was significantly lower than the reference allele in this study (Fig. 6; Supplemental Fig. 6). This is the first direct evidence that the decreased FMO3 protein abundance in liver tissues from donors harboring these SNPs (E158K and E308g variants) is one of the possible reasons for a significant reduction in FMO3 activity.
E308G > E158K-E308G (Lattard et al., 2003; Krueger and Williams, 2005), the FMO3 protein abundance of a donor homozygous for haplotype E158K-E308G was significantly lower than the reference allele in this study (Fig. 6; Supplemental Fig. 6). This is the first direct evidence that the decreased FMO3 protein abundance in liver tissues from donors harboring these SNPs (E158K and E308g variants) is one of the possible reasons for a significant reduction in FMO3 activity.
This study had some limitations. For instance, although we determined the protein levels of FMO3 in 445 samples, we were able to access only 37 fresh samples, and only one typical substrate was used to measure enzyme activity; this sample size was insufficient to draw definite conclusions regarding the associations of genotype with FMO3 activity using HLM samples. The samples used for mRNA expression (n = 214), gene resequencing (n = 285), and genotype determination (n = 129) were also limited by the lack of available liver tissue from all the donors. Thus, the interplay of ontogeny and genotype on FMO3 abundance could not be fully evaluated in this study. Finally, health conditions and smoking status were unknown for most of the donors, suggesting that the interindividual differences in FMO3 protein abundance and mRNA levels might also come from other potential factors.
In summary, the hepatic levels of FMO3 (25.1  12.0 pmol/mg protein) demonstrated marked interindividual variations (124-fold) in a large panel of HLM samples (n = 445). Age and genetic variations were the two main sources for this variation. Our results, combined with reported differences in intrinsic catalytic activity of the different FMO3 variants, may be useful in deriving scaling factors that may be applied to predict age- or genotype-dependent hepatic clearance of FMO3 substrates via PBPK modeling and simulations. Moreover, these data may serve to generate hypotheses for future clinical studies designed to predict the effects of FMO3 haplotype on the pharmacokinetics and pharmacodynamics of FMO3 substrates.
12.0 pmol/mg protein) demonstrated marked interindividual variations (124-fold) in a large panel of HLM samples (n = 445). Age and genetic variations were the two main sources for this variation. Our results, combined with reported differences in intrinsic catalytic activity of the different FMO3 variants, may be useful in deriving scaling factors that may be applied to predict age- or genotype-dependent hepatic clearance of FMO3 substrates via PBPK modeling and simulations. Moreover, these data may serve to generate hypotheses for future clinical studies designed to predict the effects of FMO3 haplotype on the pharmacokinetics and pharmacodynamics of FMO3 substrates.
Acknowledgments
We thank Prachi Jha and Dr. Aanchal Mehrotra for assistance with LC-MS/MS sample analysis.
Abbreviations
- ABB
ammonium bicarbonate buffer
- BSA
bovine serum albumin
- BZD
benzydamine
- FMO
flavin-containing monooxygenase
- FMO3
flavin-containing monooxygenase 3
- FPKM
fragments per kilobase per million reads
- HLM
human liver microsome
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LD
linkage disequilibrium
- MAF
minor allele frequency
- P450
cytochrome P450 enzyme
- PGRN
Pharmacogenomics Research Network
- SNPs
single nucleotide polymorphism
Authorship Contributions
Participated in research design: Xu, Bhatt, Yeung, Prasad.
Conducted experiments: Xu, Bhatt, Claw, Yeung, Broeckel, Gaedigk, Prasad.
Contributed new reagents or analytic tools: Thummel, Leeder, Rettie, Chaudhry, Gaedigk, Pearce, Nickerson, Schuetz.
Performed data analysis: Xu, Bhatt, Claw, Yeung, Chaudhry, Gaedigk, Prasad.
Wrote or contributed to the writing of the manuscript: Xu, Claw, Yeung, Bhatt, Chaudhry, Gaedigk, Pearce, Nickerson, Schuetz, Rettie, Leeder, Thummel, Prasad.
Footnotes
This work was primarily supported by the National Institutes of Health [Grant no. R01 HD081299-02]. Liver sample procurement, subcellular isolation, transcript, and genotyping data analysis for the University of Washington and St. Jude Children’s Research Hospital samples were supported by the NIH [Grants U01 GM092676 and P01 GM116691]. The National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland is funded by NIH contract HHSN275200900011C, reference no. N01-HD-9-0011, and the Liver Tissue Cell Distribution System is funded by NIH contract number N01-DK-7-0004/ HHSN267200700004C. M.X. was supported by the China Clinical Evaluation Research Institute [Grant no. KK11-1].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Bloom AJ, Murphy SE, Martinez M, von Weymarn LB, Bierut LJ, Goate A. (2013) Effects upon in-vivo nicotine metabolism reveal functional variation in FMO3 associated with cigarette consumption. Pharmacogenet Genomics 23:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boberg M, Vrana M, Mehrotra A, Pearce RE, Gaedigk A, Bhatt DK, Leeder JS, Prasad B. (2017) Age-dependent absolute abundance of hepatic carboxylesterases (CES1 and CES2) by LC-MS/MS proteomics: application to PBPK modeling of oseltamivir in vivo pharmacokinetics in infants. Drug Metab Dispos 45:216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botling J, Edlund K, Segersten U, Tahmasebpoor S, Engstrom M, Sundstrom M, Malmstrom PU, Micke P. (2009) Impact of thawing on RNA integrity and gene expression analysis in fresh frozen tissue. Diagn Mol Pathol 18:44–52. [DOI] [PubMed] [Google Scholar]
- Cashman JR. (2008) Role of flavin-containing monooxygenase in drug development. Expert Opin Drug Metab Toxicol 4:1507–1521. [DOI] [PubMed] [Google Scholar]
- Cashman JR, Park SB, Yang ZC, Washington CB, Gomez DY, Giacomini KM, Brett CM. (1993) Chemical, enzymatic, and human enantioselective S-oxygenation of cimetidine. Drug Metab Dispos 21:587–597. [PubMed] [Google Scholar]
- Cashman JR, Xiong YN, Xu L, Janowsky A. (1999) N-oxygenation of amphetamine and methamphetamine by the human flavin-containing monooxygenase (form 3): role in bioactivation and detoxication. J Pharmacol Exp Ther 288:1251–1260. [PubMed] [Google Scholar]
- Cashman JR, Zhang J. (2002) Interindividual differences of human flavin-containing monooxygenase 3: genetic polymorphisms and functional variation. Drug Metab Dispos 30:1043–1052. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zane NR, Thakker DR, Wang MZ. (2016) Quantification of flavin-containing monooxygenases 1, 3, and 5 in human liver microsomes by UPLC-MRM-based targeted quantitative proteomics and its application to the study of ontogeny. Drug Metab Dispos 44:975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington NJ, Cao Y, Cherrington JW, Rose RL, Hodgson E. (1998) Physiological factors affecting protein expression of flavin-containing monooxygenases 1, 3 and 5. Xenobiotica 28:673–682. [DOI] [PubMed] [Google Scholar]
- Cruciani G, Valeri A, Goracci L, Pellegrino RM, Buonerba F, Baroni M. (2014) Flavin monooxygenase metabolism: why medicinal chemists should matter. J Med Chem 57:6183–6196. [DOI] [PubMed] [Google Scholar]
- Duffel MW, Graham JM, Ziegler DM. (1981) Changes in dimethylaniline N-oxidase activity of mouse liver and kidney induced by steroid sex hormones. Mol Pharmacol 19:134–139. [PubMed] [Google Scholar]
- Falls JG, Ryu DY, Cao Y, Levi PE, Hodgson E. (1997) Regulation of mouse liver flavin-containing monooxygenases 1 and 3 by sex steroids. Arch Biochem Biophys 342:212–223. [DOI] [PubMed] [Google Scholar]
- Furnes B, Feng J, Sommer SS, Schlenk D. (2003) Identification of novel variants of the flavin-containing monooxygenase gene family in African Americans. Drug Metab Dispos 31:187–193. [DOI] [PubMed] [Google Scholar]
- Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. (2016) PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics [DOI: 10.1097/FPC.0000000000000202]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining RL, Hunter AP, Sadeque AJ, Philpot RM, Rettie AE. (1997) Baculovirus-mediated expression and purification of human FMO3: catalytic, immunochemical, and structural characterization. Drug Metab Dispos 25:790–797. [PubMed] [Google Scholar]
- Hisamuddin IM, Wehbi MA, Chao A, Wyre HW, Hylind LM, Giardiello FM, Yang VW. (2004) Genetic polymorphisms of human flavin monooxygenase 3 in sulindac-mediated primary chemoprevention of familial adenomatous polyposis. Clin Cancer Res 10:8357–8362. [DOI] [PubMed] [Google Scholar]
- Janmohamed A, Hernandez D, Phillips IR, Shephard EA. (2004) Cell-, tissue-, sex- and developmental stage-specific expression of mouse flavin-containing monooxygenases (Fmos). Biochem Pharmacol 68:73–83. [DOI] [PubMed] [Google Scholar]
- Klick DE, Hines RN. (2007) Mechanisms regulating human FMO3 transcription. Drug Metab Rev 39:419–442. [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Simpson P, Yeung CK, Rettie AE, Hines RN. (2002) Human hepatic flavin-containing monooxygenases 1 (FMO1) and 3 (FMO3) developmental expression. Pediatr Res 51:236–243. [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Poch MT, Cabacungan ET, McCarver DG, Hines RN. (2005) Discovery of novel flavin-containing monooxygenase 3 (FMO3) single nucleotide polymorphisms and functional analysis of upstream haplotype variants. Mol Pharmacol 68:383–392. [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Poch MT, Henderson MC, Siddens LK, Krueger SK, VanDyke JE, Williams DE, Pajewski NM, Wang T, Hines RN. (2007) Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants. J Pharmacol Exp Ther 320:266–273. [DOI] [PubMed] [Google Scholar]
- Krueger SK, Williams DE. (2005) Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 106:357–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattard V, Zhang J, Tran Q, Furnes B, Schlenk D, Cashman JR. (2003) Two new polymorphisms of the FMO3 gene in Caucasian and African-American populations: comparative genetic and functional studies. Drug Metab Dispos 31:854–860. [DOI] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550. [DOI] [PubMed] [Google Scholar]
- Mitchell SC, Smith RL. (2001) Trimethylaminuria: the fish malodor syndrome. Drug Metab Dispos 29:517–521. [PubMed] [Google Scholar]
- Mushiroda T, Douya R, Takahara E, Nagata O. (2000) The involvement of flavin-containing monooxygenase but not CYP3A4 in metabolism of itopride hydrochloride, a gastroprokinetic agent: comparison with cisapride and mosapride citrate. Drug Metab Dispos 28:1231–1237. [PubMed] [Google Scholar]
- Overby LH, Carver GC, Philpot RM. (1997) Quantitation and kinetic properties of hepatic microsomal and recombinant flavin-containing monooxygenases 3 and 5 from humans. Chem Biol Interact 106:29–45. [DOI] [PubMed] [Google Scholar]
- Parte P, Kupfer D. (2005) Oxidation of tamoxifen by human flavin-containing monooxygenase (FMO) 1 and FMO3 to tamoxifen-N-oxide and its novel reduction back to tamoxifen by human cytochromes P450 and hemoglobin. Drug Metab Dispos 33:1446–1452. [DOI] [PubMed] [Google Scholar]
- Pearce RE, Gaedigk R, Twist GP, Dai H, Riffel AK, Leeder JS, Gaedigk A. (2016) Developmental expression of CYP2B6: a comprehensive analysis of mRNA expression, protein content and bupropion hydroxylase activity and the impact of genetic variation. Drug Metab Dispos 44:948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips IR, Shephard EA. (2008) Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol Sci 29:294–301. [DOI] [PubMed] [Google Scholar]
- Poetsch M, Czerwinski M, Wingenfeld L, Vennemann M, Bajanowski T. (2010) A common FMO3 polymorphism may amplify the effect of nicotine exposure in sudden infant death syndrome (SIDS). Int J Legal Med 124:301–306. [DOI] [PubMed] [Google Scholar]
- Prasad B, Evers R, Gupta A, Hop CE, Salphati L, Shukla S, Ambudkar SV, Unadkat JD. (2014) Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab Dispos 42:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. (2011) Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12:683–691. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Denton T, Kozono M, Cashman JR, Leeder JS, Yamazaki H. (2011) Developmental variations in metabolic capacity of flavin-containing mono-oxygenase 3 in childhood. Br J Clin Pharmacol 71:585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, Fohner A, Thornton TA, Isoherranen N, Unadkat JD, et al. (2016) Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J 16:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Störmer E, Roots I, Brockmöller J. (2000) Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br J Clin Pharmacol 50:553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi-Takizawa T, Shimizu M, Kume T, Yamazaki H. (2015) Benzydamine N-oxygenation as an index for flavin-containing monooxygenase activity and benzydamine N-demethylation by cytochrome P450 enzymes in liver microsomes from rats, dogs, monkeys, and humans. Drug Metab Pharmacokinet 30:64–69. [DOI] [PubMed] [Google Scholar]
- Tanner JA, Prasad B, Claw KG, Stapleton P, Chaudhry A, Schuetz EG, Thummel KE, Tyndale RF. (2017) Predictors of variation in CYP2A6 mRNA, protein, and enzyme activity in a human liver bank: influence of genetic and nongenetic factors. J Pharmacol Exp Ther 360:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa B, Pedretti A, Vistoli G. (2012) Reactions and enzymes in the metabolism of drugs and other xenobiotics. Drug Discov Today 17:549–560. [DOI] [PubMed] [Google Scholar]
- Treacy EP, Akerman BR, Chow LM, Youil R, Bibeau C, Lin J, Bruce AG, Knight M, Danks DM, Cashman JR, et al. (1998) Mutations of the flavin-containing monooxygenase gene (FMO3) cause trimethylaminuria, a defect in detoxication. Hum Mol Genet 7:839–845. [DOI] [PubMed] [Google Scholar]
- Tugnait M, Hawes EM, McKay G, Rettie AE, Haining RL, Midha KK. (1997) N-oxygenation of clozapine by flavin-containing monooxygenase. Drug Metab Dispos 25:524–527. [PubMed] [Google Scholar]
- Türkanoğlu Özçelik A, Can Demirdöğen B, Demirkaya S, Adalı O. (2013) Flavin containing monooxygenase 3 genetic polymorphisms Glu158Lys and Glu308Gly and their relation to ischemic stroke. Gene 521:116–121. [DOI] [PubMed] [Google Scholar]
- Wagmann L, Meyer MR, Maurer HH. (2016) What is the contribution of human FMO3 in the N-oxygenation of selected therapeutic drugs and drugs of abuse? Toxicol Lett 258:55–70. [DOI] [PubMed] [Google Scholar]
- Weinshilboum R, Wang L. (2004) Pharmacogenetics: inherited variation in amino acid sequence and altered protein quantity. Clin Pharmacol Ther 75:253–258. [DOI] [PubMed] [Google Scholar]
- Xie G, Cheng KW, Huang L, Rigas B. (2014) The in vitro metabolism of phospho-sulindac amide, a novel potential anticancer agent. Biochem Pharmacol 91:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Shimizu M. (2013) Survey of variants of human flavin-containing monooxygenase 3 (FMO3) and their drug oxidation activities. Biochem Pharmacol 85:1588–1593. [DOI] [PubMed] [Google Scholar]
- Yeung CK, Adman ET, Rettie AE. (2007) Functional characterization of genetic variants of human FMO3 associated with trimethylaminuria. Arch Biochem Biophys 464:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung CK, Rettie AE. (2006) Benzydamine N-oxygenation as a measure of flavin-containing monooxygenase activity. Methods Mol Biol 320:157–162. [DOI] [PubMed] [Google Scholar]
- Zhou J, Shephard EA. (2006) Mutation, polymorphism and perspectives for the future of human flavin-containing monooxygenase 3. Mutat Res 612:165–171. [DOI] [PubMed] [Google Scholar]