Abstract
Hair follicle stem cells (HFSCs) are noted for their relative quiescence and therefore can be distinguished from other cells by their differential history of cell division. Replicating cells can be labeled by pulsing the animals repeatedly with 5-bromo-2′-deoxyuridine (BrdU) or tritiated thymidine ([3H]TdR), thymidine analogs that get incorporated into DNA during DNA synthesis. Because dividing cells dilute the label after each cell division, frequently dividing cells will lose the label over time while slow cycling cells will retain the label and thus are termed label retaining cells (LRCs). [3H]TdR can be visualized by autoradiography and BrdU can be detected by immunofluorescence with anti-BrdU antibodies. Alternatively, a well-established tet-regulatable transgenic mouse model can be used to express histone H2B-GFP in epithelial proliferative cells and their dilution and retention of the GFP signal can be followed. In this chapter, we detail the steps to perform BrdU pulse-chase and H2B-GFP pulse-chase experiments to identify quiescent cells in the hair follicle.
Keywords: Skin, Hair follicle epithelial stem cells, Epidermis, Quiescence, Pulse-chase, Label retaining cells, Hair cycle
1 Introduction
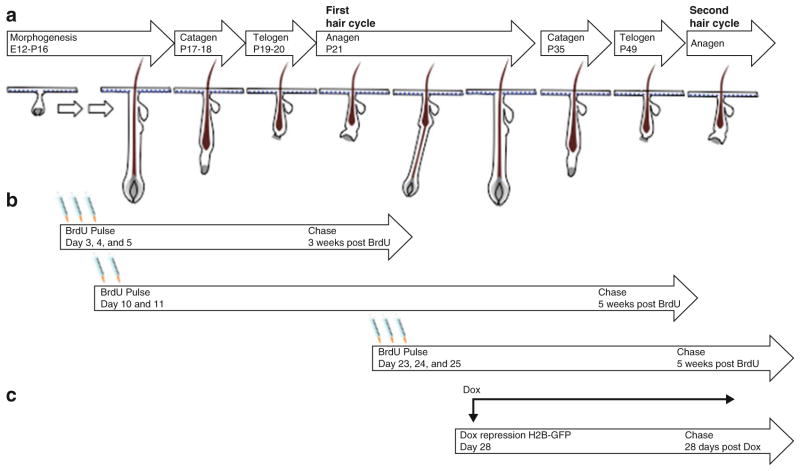
The skin is a self-renewing tissue made up of the stratified epidermis and its appendages: the hair follicle (HF) and sebaceous gland [1]. Each compartment contains designated stem cell populations that contribute to their homeostasis [2, 3]. Throughout life, HFs undergo cyclical stages of regression (catagen), rest (telogen), and growth (anagen) [4, 5] as illustrated in Fig. 1a. During catagen, cells in the lower two-third portion of the HF degenerate, leaving intact the upper region, the permanent structure of the HF. This region contains a specialized structure called the bulge, where hair follicle stem cells (HFSCs) are localized [6–11]. Through interaction with adjacent cells including the dermal papilla, a cluster of specialized mesenchymal cells at the base of the HF, HFSCs in the bulge transition from being quiescent in telogen to being activated in anagen. In anagen, progeny of HFSCs rapidly amplify before differentiating into mature progeny that occupy distinct layers within the hair follicle [12]. Since murine HFs enter specific phases of the hair cycle quite synchronously within an expected timeframe in the first two hair cycles [5, 13], the HF is an excellent model system to study the regulation of SCs, in particular the signals that govern their quiescence and activation. The HFSCs can be distinguished from other cells based on their high level of quiescence [6, 11, 14–17].
Fig. 1.
Mouse hair cycle in relation to pulse-chase schemes. (a) After completion of hair morphogenesis, hair follicles enter the destructive phase (catagen) on postnatal day 16–19. During catagen cells in the lower portion of the hair follicle degenerate, leaving intact the upper part, which contains the bulge region. HFSCs in the bulge enter a quiescent state (telogen) on postnatal day 19 lasting 2–3 days. Hair follicles enter the growth phase (anagen) of the first hair cycle when HFSCs in the bulge are activated and a new hair follicle downgrowth begins. Subsequently the hair follicles enter catagen at day 35 and then telogen at day 49 before reentering anagen of the second hair cycle weeks later. (b) BrdU pulse-chase scheme. Multiple injections of BrdU are delivered to mice from postnatal day 3–5, 10–11, or 23–25. After a chase period for an indicated amount of time post-BrdU injection, frequently dividing cells will dilute the amount of incorporated BrdU while the infrequently dividing cells retain BrdU. (c) Pulse-chase scheme for using the tet-off system to label and chase K5 positive cells with H2B-GFP. K5-tTA;TRE-H2B-GFP mice are placed on a diet containing doxycycline at postnatal day 28 to turn off transcription of H2B-GFP. After 4 weeks, H2B-GFP expression is diluted in frequently dividing cells and is retained in infrequently cycling cells
The BrdU pulse-chase method is most commonly used to label dividing cells and to follow their cell division history based on their differential dilution of label [17, 18]. Because BrdU, a pyrimidine analog of thymidine, is incorporated into DNA during DNA synthesis, multiple injections of BrdU into mice in the pulse period will label all cells that replicate within the pulse period. Subsequently in the “chase” period, proliferative cells will lose half of their incorporated BrdU after each division, while slow-cycling cells retain BrdU label. BrdU can then be detected by immunostaining skin sections with an antibody against BrdU. Depending on the end point of interest (either the first or second hair cycle), BrdU can be pulsed during morphogenesis from postnatal day 3–5 or day 10–11, or in early anagen from day 23–25 (Fig. 1b). When pulsed at day 3–5, LRCs can be visualized in the bulge at the first hair cycle when chased for a period of a minimum of 3 weeks. Pulsing in early anagen of the first hair cycle requires a chase period of a minimum of 5 weeks to localize the LRCs in the second hair cycle.
Alternatively, the well-established tet-regulatable transgenic mice expressing GFP-tagged histone H2B can be used to label and chase epithelial cells. This transgenic mouse model carries two transgenes: one expressing a tetracycline repressor fused to a transactivator VP16 under the control of a keratin 5 promoter (K5-tTA) [19], and another expressing histone H2B fused to green fluorescent protein under the control of tetracycline response element (TRE-H2B-GFP) [11]. These double transgenic mice constitutively express H2B-GFP in K5 positive cells throughout the epithelium, but the transcription of H2B-GFP will cease when the mice are placed on a diet containing a tetracycline analog, doxycycline. As with the BrdU pulse chase procedure, proliferative cells will dilute the H2B-GFP signal after each round of division, while cells that divide less frequently will retain GFP signal. After a 4-week period on doxycycline, H2B-GFP signal will be diluted in frequently dividing cells but will still be substantial in quiescent cells. One caveat of the BrdU pulse chase approach is that the labeling of quiescent cells may be incomplete, since the quiescent cells that are not proliferating during the 2-day period of BrdU pulse will not incorporate BrdU and hence cannot be labeled and chased. The advantage of the tet-regulatable histone H2B-GFP transgenic mouse model is that it allows more complete labeling of quiescent cells, due to the earlier initiation of induced expression of H2B-GFP and a longer duration of pulse. Because of the GFP signal retained in LRCs, this mouse model can also be used to isolate LRCs from the hair follicle bulge through fluorescence activated cell sorting.
2 Materials
2.1 BrdU Pulse-Chase
Postnatal day 3, 10, or 22 mice (see Note 1). Mice can be wild type or a genetically engineered mouse model of interest.
BrdU (Sigma, B5002) at 10 mg/mL in phosphate buffered saline (PBS).
1 mL insulin syringe (or 1 mL syringe and 26G needle).
2.2 H2B-GFP Labeling and Chase
K5-tTA transgenic mice [19] expressing tetracycline repressor fused to transactivator VP16 under the control of the Keratin 5 promoter.
TRE-H2B-GFP transgenic mice [11] expressing H2B-GFP under the control of tetracycline response element-driven (TRE), which are currently available from Jackson Laboratories (stock number 005104).
Doxycycline containing mouse feed 1 mg/g (Bioserv).
2.3 Backskin Harvesting and Embedding in O.C.T
Electric shavers or clippers.
4-in. fine point dissection scissors.
4-in. dissection forceps.
Razor blade.
PBS.
Brown paper towel.
Cryomold 25 × 20 mm.
O.C.T. compound.
Dry ice block.
2.4 Tissue Sectioning
Cryostat.
Microscope slides.
Hematoxylin 2.
Staining jar.
Light microscope.
2.5 Immunostaining
Humidified chamber (see Note 2).
Glass staining jar.
37 °C water bath.
PAP pen.
PBS.
1 N Hydrochloric acid.
4% Paraformaldehyde in PBS.
10% Triton X-100 solution.
20% Bovine serum albumin (BSA).
Cold water fish skin gelatin (Sigma).
Normal Donkey Serum (NDS).
Normal Goat Serum (NGS).
PBS-GT: 2% fish gelatin, 0.2% Triton X-100, in PBS.
Blocking buffer: 10% NDS and 2% BSA in PBS-GT.
Staining buffer: 1% BSA and 5% NDS in PBS-GT.
BrdU gelatin blocking buffer: 2.5% NDS, 2.5% NGS, 1% BSA, 2% fish skin gelatin, 0.1% Triton X-100 in PBS.
Hoechst 33342 (Sigma).
ProLong Diamond antifade mounting solution (LifeTech, P36961).
Microscope slide glass coverslips.
Fluorescent microscope.
2.6 Primary and Secondary Antibodies
Chicken α mouse K5 (BioLegend, Sig-3475-100, dilution 1:1000).
Rat α mouse CD34 (eBioscience, dilution 1:50).
Rat α BrdU clone BU1/75 (Abcam, ab6326, dilution 1:200).
Alexa Fluor 488-conjugated donkey α chicken (Jackson Lab, dilution 1:100).
RRX-conjugated donkey α rat (Jackson Lab, dilution 1:150).
3 Methods
We describe here two methods for in vivo labeling dividing cells and identifying the quiescent cells in the hair follicles using pulse-chase techniques. We detail the steps to harvest and embed skin sections to best obtain intact hair follicles. We also include a co-immunostaining protocol to visualize BrdU or H2B-GFP with other epithelial/hair follicle markers.
3.1 BrdU Pulse-Chase
Administer injections of BrdU (50 mg/kg) every 12 h for a total of six injections subcutaneously into 3-day-old mice, or intraperitoneally into 23-day-old mice. Alternatively, administer BrdU every 12 h for a total of four injections subcutaneously into 10-day-old mice (see Note 1).
Allow a chase period of a minimum of 21 days (pulse at day 3–5) or 5 weeks (pulse at day 10–13 or day 23–25) to detect LRCs before continuing to Subheading 3.3.
3.2 H2B-GFP Labeling and Chase in Skin Epithelial Cells
Mice are maintained as single transgenic parental lines K5-tTA and TRE-H2B-GFP. Generate double transgenic mice by crossing K5-tTA single transgenic and TRE-H2B-GFP single transgenic lines.
Keep double transgenic pups on a normal diet until 4 weeks of age to allow H2B-GFP expression in all K5 positive cells. To turn off transcription of H2B-GFP, feed the double transgenic mice doxycycline containing chow and maintain them on this dox diet for a minimum of 4 weeks. Continue to Subheading 3.3 at the desired time point after chase.
3.3 Harvesting and Embedding Back Skin
Euthanize mice according to institution approved protocols and guidelines (see Note 3).
Shave off dorsal hair closely with an electric shaver.
Mark the midline of the mouse by drawing a line from head to tail with a marker.
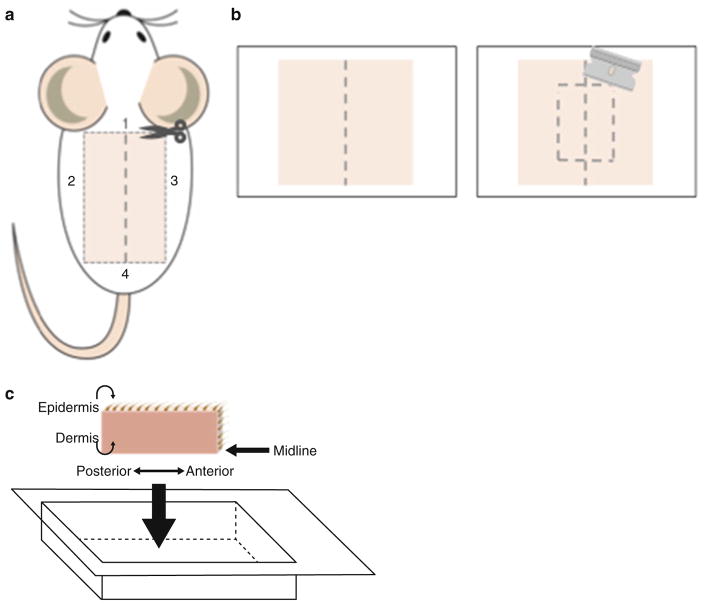
As illustrated in Fig. 2a, use fine forceps to lift the skin at the anterior end and generate an incision perpendicularly to the midline (step 1). Generate two more incisions that run down both lateral sides of the mouse (steps 2 and 3). Finally, generate an incision at the posterior end that runs perpendicular to the midline to create a rectangle (step 4).
Dampen a paper towel with PBS and flatten skin on paper towel with the dermis facing down on the paper towel (see Note 4) (Fig. 2b).
With a new razor blade cut the skin along the midline. Then generate two parallel cuts about 0.5 cm away from either side of the midline. Lastly, create two cuts perpendicular to the midline to generate two 0.5 cm × 2.5 cm strips (Fig. 2b).
Fill the Cryomold with O.C.T.
Use forceps to submerge 0.5 cm × 2.5 cm skin strips in O.C.T. (see Note 5), keeping the skin straight and its midline flush with the bottom of the Cryomold. Place the Cryomold on dry ice (see Note 6) and allow the O.C.T. to completely freeze (Fig. 2c).
Move the O.C.T. tissue blocks to a prechilled cryostat for sectioning or store at −80 °C.
Fig. 2.
Illustration of back skin harvesting and embedding. (a) After removing hair from the back skin, draw a midline from neck to tail. Create an incision at the anterior portion of the back skin (step 1) and two incisions parallel to the midline (steps 2–3). Pull the skin from the anterior end then cut across at the posterior side to remove the skin from the animal (step 4). (b) Place the skin on a PBS dampened paper towel with the dermis face down. Use a razor blade to create 0.5 cm × 2.5 cm strips. (c) Embed the skin in O.C.T. filled Cryomold, with the midline against the bottom of the Cryomold
3.4 Cryosectioning
Set up the cryostat for skin sectioning according to manufacturer instructions (see Note 7). We typically section skin at 8 μm thickness (6–12 μm) and include two sections per slide.
Cut a few sections and air-dry them for a few minutes, then counterstain with hematoxylin for 1–2 min before rinsing briefly in tap water. View slide under light microscope to determine if multiple intact hair follicles can be seen in the skin section (see Note 8). Adjust the angle of the block accordingly, and recut sections until full hair follicles can be visualized.
Collect sections and allow slides to dry for approximately 20 min (see Note 9). Slides can be used immediately for staining or can be kept at −80 °C for long-term storage.
3.5 Immunostaining
3.5.1 Co-immunostaining for Skin Epithelial Markers (or Other Proteins of Interest) in Chased Skin
If slides were previously frozen, allow slides to thaw and dry for 10–15 min at room temperature.
Use a PAP pen to draw a circle around the skin section on the slide. The hydrophobic PAP pen mark surrounding the skin will retain blocking/staining solution inside the marked area and prevent drying out of the skin section.
Fix slides in 4% PFA/PBS at room temperature for 10 min.
Wash slides with PBS in a staining jar three times for 5 min for each wash. Aspirate excess liquid from the slides after the final wash, add 200 μL of blocking buffer directly on the skin section encircled by the PAP pen mark, and incubate for 1 h at room temperature.
Prepare primary antibody solution by diluting primary antibody of interest in staining solution. Use primary antibody anti-K5 (diluted 1:1000) to mark the basal layer of the stratified epidermis and the outer root sheath of the hair follicle or anti-CD34 to mark bulge cells (see Note 10).
Aspirate the blocking buffer and add 150 μL of primary antibody solution. Incubate overnight in a humidified chamber at 4°C or 1 h at room temperature.
Wash the slides in PBS three times, 5 min for each wash.
Prepare secondary antibody solution by diluting the relevant fluorophore-conjugated secondary antibody in staining buffer at the dilution specified in material section.
Aspirate excess liquid after the last wash, then apply 150 μL of secondary antibody solution onto the washed skin sections. Incubate for 1 h at room temperature. Slides should be protected from light in all subsequent steps to minimize photobleaching.
Wash the slides in PBS three times, 5 min for each wash.
For H2B-GFP chased skin sections, proceed to Subheading 3.5.2. Continue to Subheading 3.5.3 for BrdU staining.
3.5.2 Visualizing H2B-GFP in Hair Follicle Cells
Dilute Hoechst 33342 in PBS (1:2000) and apply 150–200 μL to each tissue section. Incubate at room temperature for 2 min.
Wash the slides in PBS three times for a total of 10 min.
Aspirate to remove residual PBS, then mount a coverslip using ProLong Diamond antifade. Allow the slides to cure, protected from light, overnight to 24 h at room temperature.
After the slides have cured, seal the edges with nail polish. Allow the nail polish to air-dry for approximately 30 min prior to imaging.
Visualize and image with a fluorescent microscope with appropriate filters.
Slides can be stored at −20 °C for several months.
3.5.3 Detection of BrdU in Hair Follicle Cells
Incubate the slides in a glass Coplin jar that contains pre-warmed 1 N HCl in a 37 °C water bath for 40 min to denature DNA and unmask BrdU.
Wash in PBS once briefly, then four times for 5 min for each wash.
Aspirate any excess liquid after the last wash, then apply 200 μL of BrdU gelatin blocking buffer. Incubate for 1 h at room temperature.
Aspirate the blocking buffer then apply 150 μL of anti-BrdU antibody diluted (1:200) in gelatin blocking buffer. Incubate at room temperature for 1 h.
Wash the slides in PBS three times, 5 min for each wash.
Prepare secondary antibody (diluted as specified in material section) and Hoechst solution (1:2000) in gelatin blocking buffer.
Aspirate excess liquid after the last wash, then apply 150 μL of secondary antibody/Hoechst solution to each tissue section. Incubate at room temperature for 1 h (see Notes 11 and 12).
Wash the slides in PBS three times, 5 min for each wash.
Aspirate the residual PBS, then mount a coverslip using ProLong Diamond antifade. Allow the slides to cure, protected from light, overnight-24 h at room temperature.
After the slides have cured, seal the edges with nail polish. Allow the nail polish to air-dry for approximately 30 min prior to imaging.
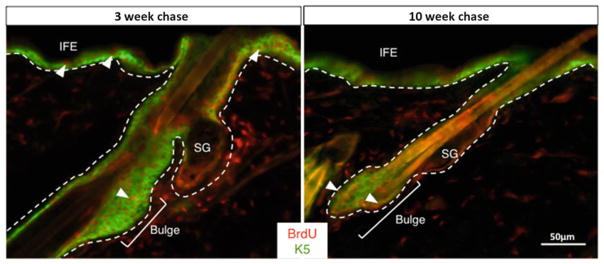
Visualize and image with a fluorescent microscope with appropriate filters (Fig. 3).
Slides can be stored at −20 °C for several months.
Fig. 3.
BrdU label retaining cells in hair follicle. Immunofluorescence images of BrdU in skin of wild-type mice pulsed at day 10–11 and chased for 3 weeks (left panel) or 10 weeks (right panel) after BrdU labeling. BrdU positive cells (red) are found throughout the skin 3 weeks after labeling and only in the bulge region of the hair follicle after 10 weeks. K5 (green) marks outer root sheath. (Scale bar = 50 μm)
Footnotes
The choice of the timing of BrdU pulse depends on the end point of interest. Pulsing at day 3–5 allows identification of LRCs at an earlier time point.
To create a humidified chamber, use an empty slide box where slides can lay on top of the slide storage slots during staining. Place dampened paper towels underneath to maintain moisture in the chamber and reduce evaporation during the incubation steps.
American Veterinary Medical Association (AVMA) guidelines require CO2 delivery that does not exceed 10–30% of the chamber volume per min to reduce distress. Deliver CO2 for 3 min at the appropriate flow rate for your chamber. Turn off CO2 delivery allowing mice to remain in the chamber for 2 min.
When embedding skins, we recommend working in batches of five mice maximum. If working with multiple mice, be sure to keep skins moist with PBS.
Multiple skin strips can be embedded in one block, but should be limited to no more than three strips per O.C.T. block. When embedding more than one skin strips, cover each strip with a thin layer of O.C.T then stack the pieces on top of one another. All pieces should be oriented epidermis face up with all mid-lines facing the same direction.
The dry ice block should be flat and level to ensure even freezing.
For skin sectioning, the chamber temperature (CT) is set to −24 °C while the object temperature (OT) is set to −27 °C.
In addition to proper embedding technique, proper set up of the tissue block on the cryostat is required to ensure that the sectioned skin contain intact hair follicles throughout the section. Hair follicles are considered intact when the entire hair follicle is seen contiguous to the interfollicular epidermis and is also in contact with the dermal papilla at its base.
Slides that are left to dry longer than 30 min will have increased background staining.
Alternative structural markers such as integrin α-6 (CD49f) or integrin β-4 (CD104) can be used to demark the boundary between epithelial cells and the dermis. Note that antibodies against CD34 and BrdU are both from rat and therefore cannot be used for co-immunostaining.
For BrdU staining, we find that acid treatment of the tissue can diminish the signal of some fluorophores. In our experience we find that Alexa Fluor 488 gives superior results and withstands the harsh acid treatment compared to FITC and RRX. (Ranked: AF488 > FITC> > RRX)
Acid treatment for BrdU staining tends to reduce Hoechst 33342 signal. We find it is beneficial to integrate Hoechst staining during the secondary antibody incubation.
References
- 1.Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3(3):199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 2.Schepeler T, Page ME, Jensen KB. Heterogeneity and plasticity of epidermal stem cells. Development. 2014;141(13):2559–2567. doi: 10.1242/dev.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodell MA, Nguyen H, Shroyer N. Somatic stem cell heterogeneity: diversity in the blood, skin and intestinal stem cell compartments. Nat Rev Mol Cell Biol. 2015;16(5):299–309. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy MH. The secret life of the hair follicle. Trends Genet. 1992;8(2):55–61. doi: 10.1016/0168-9525(92)90350-d. [DOI] [PubMed] [Google Scholar]
- 5.Stenn KS, Paus R. controls of hair follicle cycling. Physiol Rev. 2001;81(1):449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- 6.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61(7):1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 7.Rochat A, Kobayashi K, Barrandon Y. Location of stem cells of human hair follicles by clonal analysis. Cell. 1994;76(6):1063–1073. doi: 10.1016/0092-8674(94)90383-2. [DOI] [PubMed] [Google Scholar]
- 8.Oshima H, et al. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104(2):233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- 9.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22(4):411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 10.Blanpain C, et al. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118(5):635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider MR, Schmidt-Ullrich R, Paus R. The hair follicle as a dynamic miniorgan. Curr Biol. 2009;19(3):R132–R142. doi: 10.1016/j.cub.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Muller-Rover S, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117(1):3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 14.Morris RJ, Potten CS. Slowly cycling (label-retaining) epidermal cells behave like clonogenic stem cells in vitro. Cell Prolif. 1994;27(5):279–289. doi: 10.1111/j.1365-2184.1994.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 15.Morris RJ, Potten CS. Highly persistent label-retaining cells in the hair follicles of mice and their fate following induction of anagen. J Invest Dermatol. 1999;112(4):470–475. doi: 10.1046/j.1523-1747.1999.00537.x. [DOI] [PubMed] [Google Scholar]
- 16.Taylor G, et al. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102(4):451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 17.Braun KM, et al. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in whole-mounts of mouse epidermis. Development. 2003;130(21):5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 18.Bickenbach JR, Chism E. Selection and extended growth of murine epidermal stem cells in culture. Exp Cell Res. 1998;244(1):184–195. doi: 10.1006/excr.1998.4163. [DOI] [PubMed] [Google Scholar]
- 19.Diamond I, et al. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115(5):788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]