Abstract
Human pluripotent stem-cell derived cardiomyocytes (hPSC-CMs) hold a great promise for applications in human disease modeling, drug discovery, cardiotoxicity screening, and ultimately regenerative medicine. The ability to study multiple parameters of hPSC-CM function, such as contractile and electrical activity, calcium cycling and force generation, is therefore of paramount importance. hPSC-CMs cultured on stiff substrates like glass or polystyrene do not have the ability to shorten during contraction making them less suitable for the study of hPSC-CM contractile function. Other approaches require highly specialized hardware and are difficult to reproduce. Here we describe a protocol for the preparation of hPSC-CMs on soft substrates that enable shortening and subsequently the simultaneous quantitative analysis of their contractile and electrical activity, calcium cycling and force generation at single cell resolution. This protocol only requires affordable and readily available materials and works with standard imaging hardware.
Introduction
Herein we describe a protocol for the preparation of patterned hPSC-CMs for functional analysis. Firstly, we describe the preparation of flexible poly-di-methyl-siloxane (PDMS) substrates on glass-bottomed dishes. Next, we report our protocol to coat the PDMS layer with a protein micropattern using microcontact printing. These substrates are then plated with hPSC-CMs that bind to the protein micropattern and assume its anisotropic shape. This allows for a high yield of single anisotropic hPSC-CMs that show fractional shortening up to 20% along the longitudinal axis for robust contractility and force generation measurements. In addition, we outline the use of fluorescent probes to quantify the calcium cycling and electrical activity of hPSC-CMs.
1. Preparation of PDMS substrates
This protocol describes the preparation of the flexible PDMS substrates. When combined with a protein coating, these substrates allow for long-term culture of hPSC-CMs (up to at least 6 months) that are able to shorten during contraction by deforming the underlying soft PDMS substrate. Alternatively, Sylgard 527 can be mixed with Sylgard 184 to obtain PDMS substrates with a tunable stiffness >5 kPa (Palchesko et al., 2012).
Materials
Fluorodish or other 35 mm glass-bottom dishes with a glass bottom well of at least 23 mm in diameter (World Precision Instruments, cat. no. FD35-100)
150 mm Petri dish
PDMS (Dow Corning, Sylgard 527 A&B Silicone Dielectric Gel)
50 ml Falcon tube
Vacuum chamber that can hold at least one 50 ml Falcon tube
10 ml serological pipet
Vortex mixer
PBS without Mg2+ or Ca2+
Prepare PDMS substrates
In a 50 ml Falcon tube, add equal parts of component A and B of the Sylgard 527 (at least 10 ml per preparation recommended).
Mix the components with a 10 ml serological pipet by inserting one end in the Falcon tube and pushing the other end on the vortex mixer for 10 seconds.
Degas the PDMS mixture by leaving it in a vacuum chamber for 30 min.
To maintain optimal aseptic technique, proceed to a cell culture flow hood for the next step.
- Place 8 Fluorodishes in a 150 mm Petri dish, pipet 100 μl of the degassed PDMS mixture in the middle of each glass bottom.Due to the viscous nature of PDMS, pipetting exactly 100 μl can be challenging. This exact amount is not essential— the goal is to achieve a thin layer of PDMS that allows for microscopic imaging through the PDMS.
- Place the dishes in an oven at 60° C for 6 hrs.If the dishes are not used directly, they can be stored at room temperature for at least 3 months.If these dishes are to be subsequently plated with hPSC-CMs on a regular protein coating (i.e. not from microcontact printing), they must then be soaked in regular PBS without Mg2+ or Ca2+overnight before protein coating and cell plating.
2. Microcontact printing on soft PDMS substrates
Here we describe a protocol for microcontact printing on soft and sticky substrates such as PDMS (Sylgard 527). The protocol is based on the original method published by Yu et al. (2012) with several modifications. The authors used a micropattern of 100 × 20 μm to obtain a high yield of anisotropic single hPSC-CMs (Figure 1).
Figure 1.
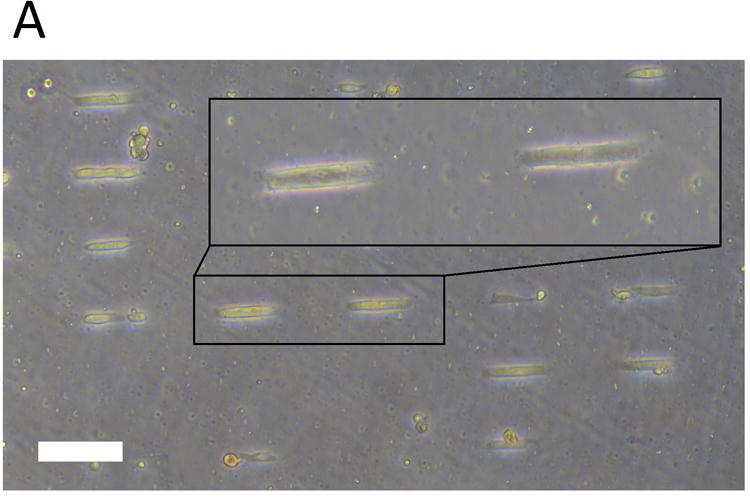
Microcontact printing of hPSC-CMs on soft substrates. (A) Using micropatterned PDMS stamps and dissolvable PVA films, Matrigel is microcontact printed on soft PDMS substrates. Subsequently hPSC-CMs are plated on these substrates. As shown here, upon attachment to the substrate, the hPSC-CMs conform geometrically to the Matrigel micropattern of 100 × 20 μm. Scale bar is 100 μm.
Materials
Erlenmeyer flask
Magnetic hotplate stirrer
Magnetic rod
Pair of scissors
Scalpel
Aluminum foil
Tweezer
Cell Strainer 100 μm Nylon or similar filter (Falcon cat. no. 352360)
Polyvinyl alcohol (Sigma, cat. no. P8136)
Polystyrene 150 mm Petri dish
50 g rod weight (stack of coins wrapped in Parafilm)
Gas duster can (Sigma, cat. no. Z379522, or similar product)
Matrigel Growth Factor Reduced Basement Membrane Matrix (Corning, cat. no. 354230)
Micropatterned PDMS stamps of 1 × 1 cm (protocol for stamp fabrication not provided here, see Théry et al., 2009)
DMEM:F-12, 1:1 Mixture (Lonza, cat. no. BE12-719F)
PBS without Mg2+ or Ca2+
Prepare PVA films
In an Erlenmeyer flask, slowly pour PVA into deionized water for a final concentration of 5% weight, then leave the solution covered with aluminum foil at room temperature overnight.
Use a magnetic hot plate with a magnet to stir the solution at 90° C for 3 hr to further dissolve the PVA. Cover the flask with aluminum foil to prevent evaporation of the water and stick a thermometer through the foil to monitor the temperature.
Let the solution cool down at room temperature for 30 min, then filter it through the 100 μm cell strainer.
- Pour 20 ml of the solution in a 150 mm Petri dish and let it dry in a flow hood at room temperature overnight with the lid half open to form a thin film.To store these films, wrap the Petri dishes in Parafilm. They can be stored up to 6 months at room temperature.
Microcontact printing
NOTE: The rest of this protocol must be performed in a cell culture flow hood with proper aseptic technique.
Wipe down a pair of scissors, a pair of tweezers and a scalpel with 70% ethanol.
Place the PDMS-coated dishes, the PDMS stamps, and the PVA film 10 cm under the UV-light in a cell culture flow hood. In addition, place the rod weights, the pair of scissors, the tweezers, and the scalpel in the flow hood. To sterilize them, turn on the UV light for 15 min.
- Release the PVA film from the Petri dish.Wedge a scalpel in between the film and the side of the Petri dish and move it around the full perimeter of the dish using the blunt side of the scalpel while gently lifting up the film. Next, use the tweezers to gently pull the center of the film off of the Petri dish (Figure 2A).
Mix Matrigel with DMEM:F-12 in a 1:25 ratio and add 250 ul of this solution on each PDMS stamp. Leave for 1 hr at room temperature.
- Cut the PVA film into 1 by 1 cm pieces using scissors (Figure 2B).Often the pieces will curve slightly, forming an arch. Make sure the middle of the arch is pointing up before step 7.
Aspirate the Matrigel from the PDMS stamps and dry the stamp surface using the gas duster.
- Put each PDMS stamp with the Matrigel-coated side facing down on a piece of PVA film and add a 50 g rod weight on top of the stamp for pressure (Figure 2C). Leave for 20 min at room temperature.The PVA film should still be dry after step 7. After the PDMS stamp is lifted up, the PVA film should adhere to the PDMS stamp and not the polystyrene surface underneath. If it sticks to the surface underneath and appears very sticky, the PDMS stamp was not dry enough before placing it on the PVA film.
- Using the tweezers, gently peel the PVA film off the stamp and place the patterned side in contact with the PDMS substrate (Figure 2D). Leave for 30 min at room temperature.Once initial contact between part of the film and the PDMS-coating is established, the film will adhere automatically automatically. If it does not so fully, gently guide the adherence of the film by pushing on the sides of the film.
- Add 4 ml of PBS to each dish and leave it on for 5 min before aspirating, repeat this twice.This will completely dissolve and wash away the PVA film and leave the micropattern of Matrigel on the PDMS substrate.The dishes are now ready for cell plating. Alternatively, they can be stored at 4° C up to at least 2 months.
Figure 2.
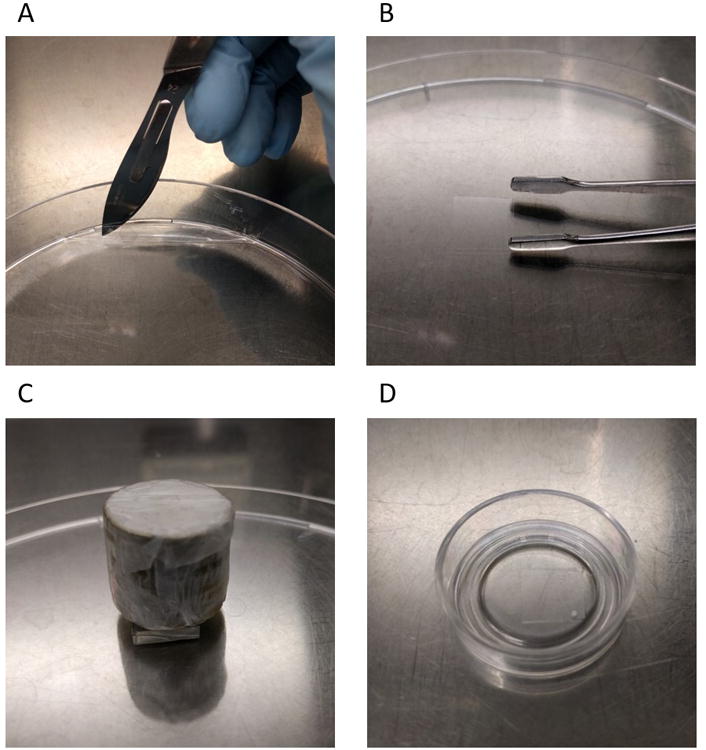
Microcontact printing using a PVA film. (A) Using a scalpel, the prepared PVA film is lifted from the Petri dish. (B) The PVA film is cut to 1 by 1 cm pieces using scissors, shown here is a PVA film piece between the tips of a tweezer. (C) Protein transfer from the PDMS stamp (middle) to the PVA film (bottom) is established through microcontact printing using a rod weight (top). (D) Protein transfer from the PVA film to the flexible PDMS substrate coating the glass-bottom dish is established through adherence.
3. Dissociation of hPSC-CMs into single cell suspension
The authors cultured several human embryonic stem cell lines. They were differentiated into hPSC-CMs using the protocol described by Lian et al. (2012).
NOTE: This protocol must be performed in a cell culture flow hood with proper aseptic technique. Solutions and equipment coming into contact with live cells must be sterile.
Materials
Collagenase A and B dissolved in cell medium (5mg/ml each), for a total collagenase concentration of 10mg/ml
Trypsin-EDTA 0.05% (Thermo Fisher, cat. no. 25300054)
G21 NeuroPlex Serum-Free Supplement dissolved in DMEM:F12 to 1× (Gemini Bioproducts, cat. no. 400-160)
PBS without Mg2+ or Ca2+
15 ml Falcon tube
Serological 10 ml pipet
Dissociate hPSC-CMs
NOTE: This protocol is based on the dissociation of hPSC-CMs in 1 well of a 6-well plate. Adjust volumes accordingly.
Aspirate cell medium.
Add 2 ml collagenase and incubate at 37° C for 10 min.
Using a 1 ml pipet, pipet up and down 5 times, breaking apart the cell sheet. Incubate for another 5 min at 37° C.
Put 5 ml PBS in a 15 ml Falcon tube, add the 2 ml collagenase with the broken cell sheet.
Rinse the well with 7 ml of PBS and add this to the tube.
Centrifuge cell solution at 1000 rpm for 5 min.
Aspirate the supernatant and add 2 ml trypsin. Using a 1 ml pipet tip, pipet up and down 8 times to partly dissolve the cell pellet.
Bring the tube to a 37°C water bath and partly submerge it, now gently shake the tube every 10 seconds to a total of 1:45 min.
Pipet the cell solution up and down 8 times with a 10 ml serological pipet.
Add 3 ml of G21 and 9 ml PBS, pipet up and down 5 times with a 10 ml serological pipet.
Centrifuge the cell solution again for 5 min at 1000 rpm.
Dissolve the cell pellet in your cell medium of choice by pipetting up and 8 times with a 10 ml serological pipet.
Plate the cells on the micropatterned PDMS substrates in 250 ul of medium. After 2 hr aspirate the medium and add 2ml new cell medium to wash away unattached cells.
4. Live cell imaging including calcium and action potential imaging and data analysis
After the cells have been plated on micropatterned PDMS substrates they are ready for live-cell imaging and analysis of contractility, calcium cycling and electrical activity within several days. In the author's experience, the fractional shortening and force generation of hPSC-CMs increases during the first 5 to 7 days after plating. hPSC-CMs can be cultured on these PDMS substrates up to at least 3 months.
Materials
Fluovolt Membrane Potential Kit (Thermo Fisher, cat. no. F10488)
Fluo-4 AM (Thermo Fisher, cat. no. F14201)
Fiji image analysis software (http://imagej.net/Fiji/Downloads)
Visible video analysis software (Reify, available on request)
Contractility imaging
For analysis of contractility, the authors made videos of the hPSC-CMs at 50 or more frames per second. The imaging is performed at 37° C and 5% CO2. The acquired videos are then processed using Visible, a custom software program which is available on request. (Kijlstra et al., 2015)
In the author's experience, a frame rate of at least 50 frames per second is optimal for further analysis of the contractions of hPSC-CMs.
Calcium imaging
- Preheat and buffer 1 ml cell medium in an incubator at 37° C and 5% CO2 for 30 min.In the author's experience, hPSC-CMs functional properties are highly sensitive to changes in temperature and pH, therefore it is essential to preheat and buffer the cell medium prior to addition to the cells shortly before image acquisition.
- Dissolve Fluo-4 AM in the cell medium for a final concentration of 0.5 μM. Add this solution to the cells 10 min prior to image acquisition. Afterwards, wash once with cell medium.Caution must be taken that the Fluo-4 AM does not affect calcium cycling and hPSC-CM contractility.
- Capture the fluorescent signal of the Fluo-4 AM at 50 or more frames per second.Short term videos (preferably under 10 seconds) must be captured to minimize phototoxicity.
Use the plugin Time Series Analyzer V3 included in Fiji, to analyze the fluorescent signal of the intracellular calcium during contraction.
Action potential imaging
Preheat and buffer 1 ml cell medium in an incubator at 37° C and 5% CO2 for 30 min.
- Dissolve PowerLoad concentrate (Component B) and FluoVolt dye (Component A) in the cell medium at respectively a 1:100 and 1:1000 dilution. Add this solution to the cells 15 min prior to image acquisition.In the author's experience, FluoVolt gives a strong signal in hPSC-CMs of 10-15% increase in fluorescence during membrane depolarization.
- Use the plugin Time Series Analyzer V3 included in Fiji, to analyze the fluorescent signal of the intracellular calcium during contraction.In the author's experience, the signal of FluoVolt does not appear strictly limited to the cell membrane. Therefore, the fluorescent signal in the entire cell is analyzed.
Commentary
Background Information
Human pluripotent stem cells (hPSCs) can be obtained from blastocyst-stage embryos or through reprogramming pluripotency in adult human cells (Takahashi et al., 2007; Thomson et al., 1998). hPSCs have the ability to differentiate into multiple cell types from all three germ layers including hPSC-CMs. Adult human cardiomyocytes are very limited in their availability. Due to the inherent differences in murine and human cardiac physiology, adult mouse cardiomyocytes are limited in their usability to study human disease. Moreover, isolated adult cardiomyocytes rapidly dedifferentiate in culture, only allowing for short experiments. As such, hPSC-CMs have emerged as an attractive model to study human cardiac disease (Musunuru et al., 2010). In addition, hPSC-CMs are used for drug discovery, regenerative medicine and cardiotoxicity screening. hPSC-CMs can be cultured for at least 3 months whilst displaying contractile function, thus making them more suitable for medium- and long term in vitro experiments (Lundy et al., 2013).
Plating of hPSC-CMs on protein micropatterns to obtain hPSC-CMs with specific anisotropic shapes have been described previously (Bray et al., 2008). However, hPSC-CMs were plated on stiffer substrates that did not allow for physiologic amounts of fractional shortening during contraction, making these approaches less suitable for the analysis of contractile function. An alternative to microcontact printing is plating hPSC-CMs on regular substrates and selecting anisotropic single cells. This yields a much lower percentage of usable hPSC-CMs and therefore is disadvantageous. An added benefit of microcontact printing is that hPSC-CM dimensions can also be strictly regulated.
Critical Parameters and Troubleshooting
Whilst working with hPSC-CMs, it is critical to plan ahead to assure a stable supply of hPSC-CMs for subsequent plating and experiments. As described below it is challenging to maintain hPSCs in a stable pluripotent state to subsequently generate hPSC-CMs with efficient differentiations. Therefore it is recommended to generate a large stock of frozen hPSCs at an early passage. The number of passages that hPSCs can be maintained and will generate high quality hPSC-CMs after differentiations varies between cell lines and cell culture protocols.
Anticipated Results
hPSCs and hPSC-CMs are now widely applied in research. Yet long-term culture of hPSCs whilst retaining pluripotency over a large number of passages remains challenging to most labs. Therefore, it remains somewhat challenging for most labs to generate a stable supply of hPSC-CMs.
Generation of the PDMS substrates and PVA films is rather straighforward. Performing the subsequent microcontact printing of protein on these substrates (as described above) is technically challenging. It is to be expected that several tries are necessary to perfect this technique.
Depending on the size of the PVA film, the efficiency of microcontact printing, and the efficiency of hPSC-CM adhesion to the substrate, several hundred to several thousand hPSC-CMs will be available for analysis per dish. If analysis is restricted to strictly single hPSC-CMs, it is to be anticipated that plating density must be optimized to obtain a high proportion of single hPSC-CMs adhering to the protein micropattern.
Time Considerations
Frozen hPSCs are cultured for 3-7 days depending on the plating density and are passaged every 5-7 days after this. Differentiation of hPSCs to hPSC-CMs can be started before the first passage but differentiation efficiency will usually increase during the first three passages. hPSC-CMs are ready to be plated on the PDMS substrates after day 18 of differentiation and in the author's experience are ideally plated between day 20 to day 30 of differentiation. After plating on PDMS substrates with a protein micropattern, hPSC-CM will demonstrate beating within 24 hr. The fractional shortening and force generation will increase during the first 7 days on the substrate. Thus, the minimum time required to generate stable beating hPSC-CMs on PDMS substrates from frozen hPSCs is about 30 days.
Acknowledgments
This work was supported by grants from the NIH/National Heart, Lung, and Blood Institute (U01HL100408-01 and 1K08 HL091209) and a grant from the Dutch Heart Foundation (2013SB013).
Literature Cited
- Bray MA, Sheehy SP, Parker KK. Sarcomere alignment is regulated by myocyte shape. Cell motility and the cytoskeleton. 2008;65:641–651. doi: 10.1002/cm.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijlstra JD, Hu D, Mittal N, Kausel E, van der Meer P, Garakani A, Domian IJ. Integrated Analysis of Contractile Kinetics, Force Generation, and Electrical Activity in Single Human Stem Cell-Derived Cardiomyocytes. Stem cell reports. 2015;5:1226–1238. doi: 10.1016/j.stemcr.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1848–1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem cells and development. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Domian IJ, Chien KR. Stem cell models of cardiac development and disease. Annual review of cell and developmental biology. 2010;26:667–687. doi: 10.1146/annurev-cellbio-100109-103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchesko RN, Zhang L, Sun Y, Feinberg AW. Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PloS one. 2012;7:e51499. doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thery M, Piel M. Adhesive micropatterns for cells: a microcontact printing protocol. Cold Spring Harbor protocols. 2009:pdb. prot5255. doi: 10.1101/pdb.prot5255. 2009. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Yu H, Xiong S, Tay CY, Leong WS, Tan LP. A novel and simple microcontact printing technique for tacky, soft substrates and/or complex surfaces in soft tissue engineering. Acta biomaterialia. 2012;8:1267–1272. doi: 10.1016/j.actbio.2011.09.006. [DOI] [PubMed] [Google Scholar]


