Abstract
About 50% of subjects infected with HIV present deficits in cognitive domains, which are known collectively as HIV associated neurocognitive disorder (HAND). The underlying synaptodendritic damage can be captured using resting state functional MRI, as has been demonstrated by a few earlier studies. Such damage may induce topological changes of brain connectivity networks. We test this hypothesis by capturing the functional interdependence of 90 brain network nodes using a Mutual Connectivity Analysis (MCA) framework with non-linear time series modeling based on Generalized Radial Basis function (GRBF) neural networks. The network nodes are selected based on the regions defined in the Automated Anatomic Labeling (AAL) atlas. Each node is represented by the average time series of the voxels of that region. The resulting networks are then characterized using graph-theoretic measures that quantify various network topology properties at a global as well as at a local level. We tested for differences in these properties in network graphs obtained for 10 subjects (6 male and 4 female, 5 HIV+ and 5 HIV−). Global network properties captured some differences between these subject cohorts, though significant differences were seen only with the clustering coefficient measure. Local network properties, such as local efficiency and the degree of connections, captured significant differences in regions of the frontal lobe, precentral and cingulate cortex amongst a few others. These results suggest that our method can be used to effectively capture differences occurring in brain network connectivity properties revealed by resting-state functional MRI in neurological disease states, such as HAND.
Keywords: resting state fMRI, HIV, cognitive impairment, non-linear analysis, time series analysis, causality, network topology, graph measures
1. INTRODUCTION
The Human Immunodeficiency Virus (HIV) has been shown to invade the brain causing irreversible damage to synaptic connections, which can gradually lead to neuronal dysfunction. Such changes are reflected in the cognitive performance of HIV-infected individuals, which can be assessed using neuropsychological performance tests, however such tests are often inconclusive or cannot be administered easily. Newer methods in functional MRI (fMRI) analysis provide interesting insights into detecting global and local functional changes in the brain network performance, which may indirectly lead to a better understanding of the underlying pathophysiology of the disease. Specific brain regions belonging to extended resting state networks have been shown to exhibit altered connectivity patterns in HIV-infected subjects with cognitive impairment [1,2]. However, more work is needed to develop a better understanding of the pathophysiology of infection, to detect related early degenerative changes, and to develop imaging biomarkers for disease progression and therapy management in HIV-related cognitive impairment.
We present a global and a regional brain network analysis using our novel Mutual Connectivity Analysis (MCA) [3] framework to acquire brain network graphs. We hypothesize that this approach may be better suited to capturing changes in connectivity patterns better than conventionally used linear cross-correlation methods, because it enables a non-linear analysis that also avoids a priori assumptions on the underlying connection patterns frequently implied by model-based approaches, such as Dynamic Causal Modeling (DCM) [4] or Structural Equation Modeling (SEM) [5]. The connecitivty profiles obtained by MCA capture information regarding the interactions between the different brain network nodes. A wide range of literature in neuroscience has analyzed brain networks from the perspective of graph theory, using which such profiles can be quantitatively analyzed using a wide range of simple yet ‘neuro-biologically’ meaningful graph-theoretic measures [6,7]. This work is embedded in our group’s endeavor to expedite ‘big data’ analysis in biomedical imaging by means of advanced pattern recognition and machine learning methods for computational radiology and radiomics, e.g. [17–41].
In this study, we have considered some of these measures while focusing on a few metrics characterizing global properties, as well as measures that can provide local/regional information from the graphs. We evaluate these measures regarding their ability to identify differences between the two subject groups with the aim of better quantifying neurological damage in HIV infection.
2. DATA
Functional MRI scans from 5 healthy controls and 5 HIV+ subjects, aged 32–53 years, 4 females and 6 males, were acquired using a 3.0 Tesla Siemens Magnetom TrioTim scanner at the Rochester Center for Brain Imaging (Rochester, NY, US). A resting state sequence, where the subject stayed still with their eyes closed, was acquired. The following parameters were used — TR = 1650 ms, TE = 23 ms, 96 × 96 (2.5 mm × 2.5 mm) acquisition matrix, flip angle = 84°. Acquisition lasted 6 minutes and 40 seconds, during which 250 volumes from 25 slices, separated at 5 mm, were acquired. Additional high-resolution structural imaging was performed using a T1-weighted magnetization-prepared rapid gradient echo sequence (MPRAGE; TE = 3.44 ms, TR = 2530 ms, isotropic voxel size = 1mm, flip angle = 7°). The latter was used for registration of the functional MRI data to the standard MNI152 template [11].
3. METHODS
3.1 Preprocessing and Parcellation
Standard preprocessing steps were applied to the data. The first 10 volumes were deleted to remove initial saturation effects. The volumes were then motion-corrected and the brain was extracted. High-pass filtering (0.01 Hz) was performed to remove the effects of signal drifts. Subsequently, the slices were registered to the standard MNI152 template [11]. In addition, the ventricle mask based on the standard MNI152 template was used to eliminate time series in the corresponding regions. All these steps were carried out using the FMRI Expert Analysis Tool (FEAT) of FSL [8]. Also, the time series were normalized to zero mean and unit standard deviation to focus on signal dynamics rather than amplitude [9]. For each dataset, 90 regional time-series were computed based on the average time series of each of these 90 regions, excluding regions located in the cerebellum and the brain stem according to the Automated Anatomic Labeling (AAL) template [12].
3.2 Mutual Connectivity Analysis (MCA) Using a Generalized Radial Basis Function (GRBF) Neural Network
Our first step is to build a pair-wise affinity/similarity matrix A for all time series of the n = 90 brain regions using Mutual Connectivity Analysis (MCA) [3]. The pair-wise affinity between two regional time series X and Y (where X, Y ∈ {Xk, k = 1,…,n}) describes the degree of their dynamic coupling as a measure of their cross-prediction performance. For example, to compute matrix element (A)X,Y, we break down time series X of length l into a set of vectors xt, t ∈ {1,2,…,l-d+1} of dimension d, which can be interpreted as a sliding window of length d moving along X. The corresponding prediction target vectors for xt are vectors yt of dimension e. In this study, the parameters d and e were chosen d = 10 and e = 1. Here, xt is mapped to future yt, e.g. the vector xt that comprises of the first 10 time points of X is mapped to yt, which corresponds to the 11th time point of Y.
The set of and their corresponding are split into a training (Tr) and test (Te) set. The training set is then used to create a non-linear mapping f, i.e.,
For defining the approximating function f, we use a GRBF neural network with three layers, i.e. the input, hidden and output layers. The activation pattern of the input layer with d neurons is represented by d-dimensional vector . In the training phase, this activity ( ) is propagated to k neurons of the hidden layer through directed connections with prototypical weight vectors . These weight vectors are representations of the training set and are computed using an unsupervised clustering approach. The activity of neurons in the hidden layer is defined as
where the ρ parameter controls the width of the radial basis function kernel and defines the neighborhood of vectors that contributes to the computation of f [10]. In the final step of the training phase, the activity of the output layer is computed as a weighted sum of the hidden layer activations , i.e.,
where are the output weights obtained through minimization of the cost function . After the training phase is completed, f is subsequently used to process the test set and is constructed from target vector estimates . Further details concerning our GRBF approach can be found in [3]. We haven chosen the value of ρ as 0.5 and used 20 hidden layer neurons based on initial experiments.
3.3 Graph Theoretic Measures
The network graph induced by the affinity matrix A was symmetrized to average out the directional influence between the nodes. An undirected binary graph a was obtained from the symmetrized matrix A by thresholding. The appropriate choice of the threshold for binarization of network graphs obtained from fMRI data analysis is subject to ongoing discussions in the pertinent literature [5]. Based on earlier studies [10], we chose a threshold value that retained 45% of the connections. At such a threshold, the networks are not fragmented, and they still preserve their ‘small-world’ topology [10]. Measures to characterize local and global network properties (summarized in Table 1) were computed for the thresholded graphs obtained for each subject.
Table 1.
List of the graph-theoretic measures used in this study. For a detailed description, please refer to [7].
| Measure | Undirected definition | Description | |
|---|---|---|---|
| Degree |
|
Defined for each node i; Represents the total number of links connected to a node. It is a marker of network development and resilience. | |
| Characteristic Path Length |
where, Li is the distance between node i, and all other nodes |
Characteristic Path Length is a measure of functional integration and is primarily influenced by long paths. The inverse (global efficiency) measure is considered a superior measure and is primarily influenced by short paths. | |
| Global Efficiency |
|
||
| Local Efficiency |
where, Eloc,,i is the local efficiency of node i, and djh (Ni) is the length of the shortest path between j and h, that contains only neighbors of i. |
Characterizes the efficiency of node connections at a smaller scale. | |
| Clustering Coefficient |
where Ci is the clustering coefficient of node i (Ci = 0 for ki < 2). |
The mean clustering coefficient represents the presence of clustered connectivity around individual nodes |
All procedures, excluding the pre-processing steps discussed in section 3.1, were implemented using MATLAB (MathWorks Inc., Natick, MA, 2013). The network measures definitions are taken from [7]. Significance testing was performed using a Wilcoxon signed rank test.
4. RESULTS
The results obtained using global graph measures are represented in Figure 1. We can see differences in the distribution of the various properties between the subject groups. Trending differences (p<0.1) are seen in characteristic path length, assortativity and degree variance, where as significant difference is observed for the clustering coefficient (p < 0.05). This suggests low local efficiency of the network in subjects affected with HIV, implying a loss of more local connections than connections across different regions. This has been seen in the case of other diseases such as Schizophrenia [15] and Alzheimer’s [16]. Transitivity is a variant of the clustering coefficient, it is less sensitive to the degree of the nodes that are connecting an edge. This characteristic also shows similar reduction although the differences seen are not significant.
Figure 1.
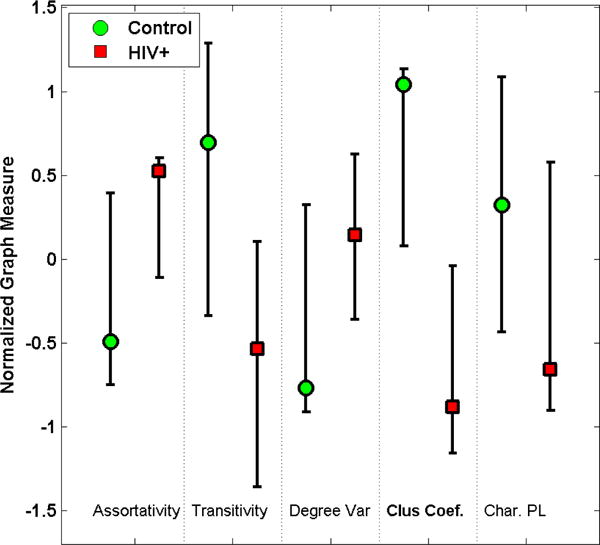
The distribution of global graph measures across the two subject groups. Certain trends are seen, though significant difference (p < 0.05) is obtained only when using the Clustering Coefficient (highlighted in boldface). Transitivity is also comparatively reduced in HIV+ subjects, though the difference was not found to be significant (p = 0.08). Note that the measures have been z-score normalized for the plot here.
We have also explored the possibility of regional characteristics being different amongst the subject groups. The different regional properties as listed in Table 1, were used for comparison across each node. There are no specific trends seen in this for example: some regions show an increase in degree whereas in other regions this is reduced, however, significant differences are noted in degree distribution are found for the frontal pole, precuneus region of the lateral occipital cortex. This indicates a loss of connections to or from these regions as a possible result of neuronal damage. Clustering coefficient of most nodes is reduced. Significant reduction is seen in the regions of of the cingulate cortex and the basal ganglia. The regions of the posterior cingulate gyrus, the basal ganglia and the lateral occipital cortex are known to be affected in HIV infection in earlier studies [1,10,11]. The regions of the brain highlighting differences using the local properties are shown in Figure 2.
Figure 2.
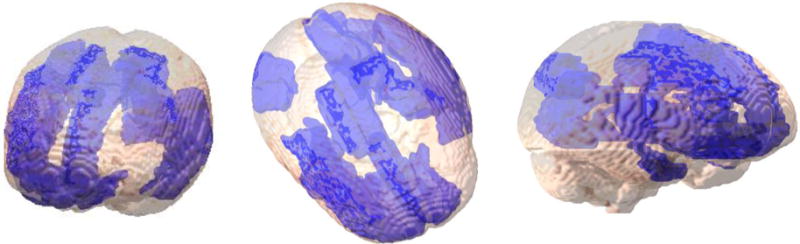
A simplified view of the brain representing the regions highlighting differences (seen in red) in local properties. These regions are consistent with some differences identified in other studies examining HIV-related cognitive impairment [1,10,11]. The regions shown here represent the differences using all the features and mapped over the standard atlas. Regions are specifically described in the text.
5. NEW AND BREAKTHROUGH WORK
We present a clinical application of our computational framework for non-linear Mutual Connectivity Analysis (MCA) in the human brain from resting state fMRI data. Based on the evaluation of pair-wise cross-prediction quality we construct connectivity profiles from a parcellated human brain. Subsequently, quantitative analysis of the resulting network graphs is carried out using graph theoretic approaches of characterizing network properties. We have applied this approach to identify differences in network connectivity patterns between the network profiles of healthy and diseased subjects with HIV infection. We are able to identify significant differences in these profiles at a global as well as a regional level. We show that there are certain changes in the overall topological organization of the brain in HIV infection. The results obtained here using our novel methodology are in line with earlier studies on HAND. We hypothesize that our non-linear approach has the potential to capture more accurate information on brain connectivity patterns than more traditional linear or model-based approaches. This combined with graph theoretical approach has be useful to develop biomarkers for disease progression and therapy management in patients with HIV-associated neurological disease.
6. CONCLUSION
In conclusion, the results presented in this work indicate that our non-linear time-series modelling approach (MCA) in combination with subsequent graph-theoretic network characterization can be applied to detecting changes in brain network connectivity patterns observed in patients with HAND. Hence, MCA can serve as an alternate method for fMRI-based functional connectivity analysis in the human brain, as it avoids certain inherent limitations of traditional approaches. The initial results suggest that our method may contribute to biomedical research endeavors aiming at a better understanding of underlying disease mechanisms in HAND and can potentially be useful in clinical applications to quantitatively monitor its progression.
Acknowledgments
This research was funded by the National Institutes of Health (NIH) Award R01-DA-034977. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. This work was conducted as a Practice Quality Improvement (PQI) project related to American Board of Radiology (ABR) Maintenance of Certificate (MOC) for Prof. Dr. Axel Wismüller. The authors would like to thank Prof. Dr. Dorothee Auer at the Institute of Neuroscience, University of Nottingham, UK, for her assistance with the fMRI data acquisition process. The authors would also like to thank Dr. Lutz Leistritz and Prof. Dr. Herbert Witte of Bernstein Group for Computational Neuroscience and the Institute of Medical Statistics, Computer Sciences, and Documentation, Jena University Hospital, Friedrich Schiller University Jena, Germany, Dr. Oliver Lange and Prof. Dr. Maximilian F. Reiser, Institute of Clinical Radiology, Ludwig Maximilian University, Munich, Germany for their support.
Footnotes
This work is not being and has not been submitted for publication or presentation elsewhere.
References
- 1.Thomas JB, Brier MR, Snyder AZ, Vaida FF, Ances BM. Pathways to neurodegeneration Effects of HIV and aging on resting-state functional connectivity. Neurology. 2013;80(13):1186–1193. doi: 10.1212/WNL.0b013e318288792b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Foryt P, Ochs R, Chung JH, Wu Y, Parrish T, Ragin AB. Abnormalities in resting-state functional connectivity in early human immunodeficiency virus infection. Brain Connectivity. 2011;1(3):207–217. doi: 10.1089/brain.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wismüller A, Wang X, DSouza AM, Nagarajan MB. A Framework for Exploring Non-Linear Functional Connectivity and Causality in the Human Brain: Mutual Connectivity Analysis (MCA) of Resting-State Functional MRI with Convergent Cross-Mapping and Non-Metric Clustering. 2014 arXiv:1407.3809 [cs.NE] [Google Scholar]
- 4.Friston Karl J, Harrison Lee, Penny Will. Dynamic causal modelling. Neuroimage. 2003;19(4):1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- 5.Mclntosh AR, Gonzalez-Lima F. Structural equation modeling and its application to network analysis in functional brain imaging. Human Brain Mapping. 1994;2(1–2):2–22. [Google Scholar]
- 6.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature Reviews Neuroscience. 2009;10(3):186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 8.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Niazy RK. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 9.Wismüller A, Lange O, Dersch DR, Leinsinger GL, Hahn K, Pütz B, Auer D. Cluster analysis of biomedical image time-series. International Journal of Computer Vision. 2002;46(2):103–128. [Google Scholar]
- 10.Lynall ME, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, Bullmore E. Functional connectivity and brain networks in schizophrenia. The Journal of Neuroscience. 2010;30(28):9477–9487. doi: 10.1523/JNEUROSCI.0333-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Mazoyer B. A four-dimensional probabilistic atlas of the human brain. Journal of the American Medical Informatics Association. 2001;8(5):401–430. doi: 10.1136/jamia.2001.0080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cysique LA, Moffat K, Moore DM, Lane TA, Davies NW, Carr A, Rae C. HIV, Vascular and Aging Injuries in the Brain of Clinically Stable HIV-Infected Adults: A 1 H MRS Study. PloS one. 2013;8(4):e61738. doi: 10.1371/journal.pone.0061738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aylward EH, Henderer JD, McArthur JC, Brettschneider PD, Harris GJ, Barta PE, Pearlson GD. Reduced basal ganglia volume in HIV‐1‐associated dementia Results from quantitative neuroimaging. Neurology. 1993;43(10):2099–2099. doi: 10.1212/wnl.43.10.2099. [DOI] [PubMed] [Google Scholar]
- 14.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Liang M, Zhou Y, He Y, Hao Y, Song M, Jiang T. Disrupted small-world networks in schizophrenia. Brain. 2008;131:945–61. doi: 10.1093/brain/awn018. [DOI] [PubMed] [Google Scholar]
- 16.Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens Ph. Small-world networks and functional connectivity in Alzheimer’s disease. Cerebral cortex. 2007;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- 17.Hoole P, Wismüller A, Leinsinger G, Kroos C, Geumann A, Inoue M. Analysis of tongue configuration in multi-speaker, multi-volume MRI data. Proc 5th Semin Speech Prod Model Data CREST Work. Model Speech Prod Mot Plan Articul Model. 2000:157–160. [Google Scholar]
- 18.Wismüller A. Thesis PhD. Technical University of Munich, Department of Electrical and Computer Engineering; 2006. Exploratory Morphogenesis (XOM): a novel computational framework for self-organization. [Google Scholar]
- 19.Wismüller A, Dersch DR, Lipinski B, Hahn K, Auer D. ICANN 98. Springer; London: 1998. A neural network approach to functional MRI pattern analysis—clustering of time-series by hierarchical vector quantization; pp. 857–862. [Google Scholar]
- 20.Wismüller A, Vietze F, Dersch DR, Behrends J, Hahn K, Ritter H. Neurocomputing. 1. Vol. 48. Elsevier; 2002. The deformable feature map-a novel neurocomputing algorithm for adaptive plasticity in pattern analysis; pp. 107–139. [Google Scholar]
- 21.Behrends J, Hoole P, Leinsinger GL, Tillmann HG, Hahn K, Reiser M, Wismüller A. Bildverarbeitung für die Medizin 2003. Springer; Berlin, Heidelberg: 2003. A segmentation and analysis method for MRI data of the human vocal tract; pp. 186–190. [Google Scholar]
- 22.Wismüller A, Dersch DR. Neural network computation in biomedical research: chances for conceptual cross-fertilization. Theory in Biosciences. 1997;116(3):229–240. [Google Scholar]
- 23.Bunte K, Hammer B, Villmann T, Biehl M, Wismüller A. Exploratory Observation Machine (XOM) with Kullback-Leibler Divergence for Dimensionality Reduction and Visualization. ESANN. 2010;10:87–92. [Google Scholar]
- 24.Wismüller A, Vietze F, Dersch DR, Hahn K, Ritter H. ICANN 98. Springer; London: 1998. The deformable feature map—adaptive plasticity for function approximation; pp. 123–128. [Google Scholar]
- 25.Wismüller A. Advances in Self-Organizing Maps. Springer; Berlin Heidelberg: 2009. The exploration machine–a novel method for data visualization; pp. 344–352. [Google Scholar]
- 26.Wismüller A. Method, data processing device and computer program product for processing data. US Patent 7,567,889. 2009
- 27.Meyer-Bäse A, Jancke K, Wismüller A, Foo S, Martinetz T. Eng Appl Artif Intell. 4. Vol. 18. Elsevier; 2005. Medical image compression using topology-preserving neural networks; pp. 383–392. [Google Scholar]
- 28.Huber MB, Nagarajan M, Leinsinger G, Ray LA, Wismüller A. SPIE Med Imaging 7624, 762410. International Society for Optics and Photonics; 2010. Classification of interstitial lung disease patterns with topological texture features. [Google Scholar]
- 29.Wismüller A. The exploration machine: a novel method for analyzing high-dimensional data in computer-aided diagnosis. SPIE Med Imaging, 72600G–72600G. 2009 [Google Scholar]
- 30.Bunte K, Hammer B, Villmann T, Biehl M, Wismüller A. Neurocomputing. 9. Vol. 74. Elsevier; 2011. Neighbor embedding XOM for dimension reduction and visualization; pp. 1340–1350. [Google Scholar]
- 31.Wismüller A. Advances in Self-Organizing Maps. Springer; Berlin Heidelberg: 2009. A computational framework for nonlinear dimensionality reduction and clustering; pp. 334–343. [Google Scholar]
- 32.Huber MB, Nagarajan MB, Leinsinger G, Eibel R, Ray LA, Wismüller A. Med Phys. 4. Vol. 38. American Association of Physicists in Medicine; 2011. Performance of topological texture features to classify fibrotic interstitial lung disease patterns; pp. 2035–2044. [DOI] [PubMed] [Google Scholar]
- 33.Wismüller A, Verleysen M, Aupetit M, Lee JA. Recent Advances in Nonlinear Dimensionality Reduction, Manifold and Topological Learning. ESANN. 2010 [Google Scholar]
- 34.Wismüller A, Meyer-Baese A, Lange O, Reiser MF, Leinsinger G. Cluster analysis of dynamic cerebral contrast-enhanced perfusion MRI time-series. Med Imaging, IEEE Trans. 2006;25(1):62–73. doi: 10.1109/TMI.2005.861002. IEEE. [DOI] [PubMed] [Google Scholar]
- 35.Otto TD, Meyer- Bäse A, Hurdal M, Sumners D, Auer D, Wismüller A. Model-free functional MRI analysis using cluster-based methods. AeroSense. 2003;2003:17–24. [Google Scholar]
- 36.Varini C, Nattkemper TW, Degenhard A, Wismüller A. Breast MRI data analysis by LLE. Neural Networks, 2004. Proceedings. 2004 IEEE Int Jt Conf. 2004;3:2449–2454. [Google Scholar]
- 37.Huber MB, Lancianese SL, Nagarajan MB, Ikpot IZ, Lerner AL, Wismüller A. Prediction of biomechanical properties of trabecular bone in MR images with geometric features and support vector regression. Biomed Eng IEEE Trans. 2011;58(6):1820–1826. doi: 10.1109/TBME.2011.2119484. IEEE. [DOI] [PubMed] [Google Scholar]
- 38.Meyer-Base A, Pilyugin SS, Wismüller A. Stability analysis of a self-organizing neural network with feedforward and feedback dynamics. Neural Networks, 2004. Proceedings. 2004 IEEE Int Jt Conf. 2004;2:1505–1509. [Google Scholar]
- 39.Wismüller A, Meyer- Bäse A, Lange O, Schlossbauer T, Kallergi M, Reiser MF, Leinsinger G. Segmentation and classification of dynamic breast magnetic resonance image data. J Electron Imaging. 2006;15(1) 13020-013020-13, International Society for Optics and Photonics. [Google Scholar]
- 40.Nagarajan MB, Huber MB, Schlossbauer T, Leinsinger G, Krol A, Wismüller A. Mach Vis Appl. 7. Vol. 24. Springer; Berlin Heidelberg: 2013. Classification of small lesions in dynamic breast MRI: eliminating the need for precise lesion segmentation through spatio-temporal analysis of contrast enhancement; pp. 1371–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagarajan MB, Huber MB, Schlossbauer T, Leinsinger G, Krol A, Wismüller A. Classification of Small Lesions in Breast MRI: Evaluating The Role of Dynamically Extracted Texture Features Through Feature Selection. J Med Biol Eng. 2013;33(1) doi: 10.5405/jmbe.1183. NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]


