Abstract
Background
BCoV is identified in both healthy and diarrheic calves, complicating its assessment as a primary pathogen.
Objectives
To investigate the detection rates of bovine coronavirus (BCoV) in feces of healthy and diarrheic calves and to describe the usefulness of a pancoronavirus reverse transcriptase (RT) PCR (PanCoV‐RT‐PCR) assay to identify BCoV in samples of diarrheic calves.
Animals
Two hundred and eighty‐six calves <21 days. Calves with liquid or semiliquid feces, temperature >39.5°C, and inappetence were considered as cases, and those that had pasty or firm feces and normal physical examination were designated as controls.
Methods
Prospective case–control study. A specific BCoV‐RT‐PCR assay was used to detect BCoV in fecal samples. Association between BCoV and health status was evaluated by exact and random effect logistic regression. Fecal (n = 28) and nasal (n = 8) samples from diarrheic calves were tested for the presence of BCoV by both the PanCoV‐RT‐PCR and a specific BCoV‐RT‐PCR assays. A Kappa coefficient test was used to assess the level of agreement of both assays.
Results
BCoV was detected in 55% (157/286) of calves; 46% (66/143), and 64% (91/143) of healthy and diarrheic calves, respectively. Diarrheic calves had higher odds of BCoV presence than healthy calves (OR: 2.16, 95% CI: 1.26 to 3.83, P = 0.004). A good agreement between PanCoV‐RT‐PCR and BCoV‐RT‐PCR to detect BCoV was identified (κ = 0.68, 95% CI: 0.392 to 0.967; P < 0.001).
Conclusions and Clinical Importance
BCoV was more likely to be detected in diarrheic than healthy calves. The PanCoV‐RT‐PCR assay can be a useful tool to detect CoV samples from diarrheic calves.
Keywords: Emerging, Epidemiology, Infectious diseases, Pancoronavirus, Prevalence, Re‐emerging
Abbreviations
- BCoV
bovine coronavirus
- BRoV
bovine rotavirus
- ELISA
enzyme‐linked immunosorbent assay
- PanCoV
pancoronavirus
- PCR
polymerase chain reaction
- RT
reverse transcription
Neonatal diarrhea is the most important cause of disease in calves worldwide,1, 2 leading to large economic losses to cattle herds.3 Common etiologic agents involved in calf diarrhea include Escherichia coli, Cryptosporidium spp, bovine rotavirus (BRoV), and bovine coronavirus (BCoV). Most of the studies investigating the etiology of calf diarrhea have focused on sick calves.4, 5, 6, 7, 8 However, some agents, especially viruses, could be frequently detected in healthy calves.9, 10, 11 For instance, BCoV has been identified in both healthy and diarrheic calves,10, 11 complicating assessment of its role as a primary pathogen. BCoV is the causative agent of diarrhea in newborn calves,12 winter dysentery in adult cows,13 and respiratory tract illness in calves and adults.14, 15 However, some studies have identified numerical but not statistical association between BCoV in fecal or nasal samples and clinical signs.16, 17, 18 Cases of BCoV enteritis occur in both dairy and beef herds, with clinical signs developing between 5 and 30 days of life and a peak of incidence between days 7 and 10.19 Transmission from calf‐to‐calf is mainly by the fecal–oral route but respiratory transmission can be possible.16, 19 Infected calves can excrete high levels of virus within 48 h of experimental infection, and this can continue up to 14 days.19 The mechanisms of diarrhea due to BCoV infection are still unclear, but malabsorption and hypersecretion appear to play important roles.20 Clinically recovered calves can continue to shed low levels of virus for weeks.21, 22
Diagnosis of BCoV enteritis can be achieved by viral culture, antigen‐capture ELISA, hemagglutination assay using mouse erythrocytes, and PCR.19 Recently, a pancoronavirus reverse transcription (RT) PCR assay (PanCoV‐RT‐PCR) was described to identify human CoV from samples of people with respiratory diseases,23 with primers that should amplify a 251‐bp fragment of the polymerase gene of all known coronaviruses.24 The utility of PanCoV‐RT‐PCR to detect BCoV in samples of animals with clinical diseases is yet to be determined.
The objectives of this study were to investigate the detection rates of BCoV in feces of healthy and diarrheic dairy calves and to describe the usefulness of a PanCoV‐RT‐PCR assay to identify BCoV in nasal and fecal samples of a group of calves from a dairy farm suffering an outbreak of diarrhea.
Material and Methods
Detection of BCoV in Healthy and Diarrheic Dairy Calves from Southern Ontario
Sample Size Calculation
A prospective case–control study approach was used, and the sample size was calculated based on an estimated 30% BCoV detection rate in healthy calves. The set parameters to detect an odds ratio of 2, including equal number of cases and controls, with a 95% confidence and a power of 0.8, yielded a total of a 138 calves in each group.
Study Design, Definitions, and Sample Collection
Calves from six dairy farms selected from a convenience sample of commercial dairy farms within a 120‐km radius from the University of Guelph (Guelph, ON) were sampled between the fall of 2014 and the fall of 2016. The number of diarrheic calves enrolled from each farm (approximately 5% of the total number of calves raised at each farm per year), the year and season in which the samples were collected, the management practices as well as the total duration the farm was enrolled in the study are presented in Table 1. Cases (diarrheic) and control (healthy) calves were farm‐, season‐, year‐, and age‐matched. Cases and controls were defined on the basis of the physical examination and fecal consistency (liquid, semiliquid, pasty, or firm). Calves that had liquid or semiliquid feces, fever (temperature >39.5°C) and decreased appetite were considered as diarrheic (i.e., cases), and those that had pasty or firm feces and normal physical examination were designated as controls. Once a diarrheic calf was identified an age‐matched (±2 days), control calf was enrolled on each farm. Calves were not included in the study if they had a previous incident of diarrhea since birth (only the first case of diarrhea was included in the study). All samples were screened for the presence of BCoV by the BCoV‐RT‐PCR assay.
Table 1.
Farms characteristics, number, and demographic data of the calves tested for the presence of BCoV, year, and season of sampling
| Farm 1 | Farm 2 | Farm 3 | Farm 4 | Farm 5 | Farm 6 | |
|---|---|---|---|---|---|---|
| Farms characteristics | ||||||
| Breed | Holstein | Holstein | Holstein | Holstein | Holstein | Jersey |
| Calves born per year | 800 | 1000 | 350 | 330 | 150 | 80 |
| Housing | Group pen | Individual | Group pen | Hutches | Hutches | Group pen |
| Bedding | Saw dust | Shavings | Straw | Straw | Straw | Straw |
| Colostrum (pooled) | 4L first 6 h | 6L first 6 h | 6L first 4 h | 5L first 6 h | 5L first 6 h | 5L first 6 h |
| Diet (up to 12 wks.) | Pasteurized milk | Milk replacer | Pasteurized milk | Pasteurized milk | Pasteurized milk | Pasteurized milk |
| Feeding method | Robot machine | Individual bucket | Robot machine | Individual bucket | Individual bucket/Robot machine | Bottle |
| Vaccination of pregnant cowsa | Yes | Yes | Yes | Yes | Yes | Yes |
| Sampled calves | ||||||
| Calves enrolled | 84 | 94 | 48 | 30 | 20 | 10 |
| Sex (n,%) | ||||||
| Female | 60 (71%) | 65 (69%) | 39 (81%) | 30 (100%) | 20 (100%) | 8 (80%) |
| Male | 24 (29%) | 30 (31%) | 9 (19) | 0 (0%) | 0 (0%) | 2 (20%) |
| Age (mean ± SD) | 10 ± 4.7 | 8.9 ± 2.2 | 7.7 ± 3.4 | 8.5 ± 2.1 | 10.5 ± 1.7 | 9.5 ± 2.7 |
| Sampling year and season | F‐2015 | F‐2014 | F‐2014 | W‐2015 | Sp‐2016 | Su‐2016 |
| W‐2016 | W‐2015 | W‐2015 | Sp‐2015 | Su‐2016 | F‐2016 | |
| Sp‐2016 | Sp‐2015 | Sp‐2015 | Su‐2015 | |||
| Su‐2016 | Su‐2015 | |||||
wks, weeks; F, fall; W, winter; Sp, spring; Su, summer.
In all farms, cows were vaccinated against bovine rotavirus and coronavirus 8 and 4 weeks before calving using a vaccine containing attenuated strains of bovine rotavirus and coronavirus.
Bovine Coronavirus RT‐PCR Assay
Briefly, RNA was extracted with a commercial kit1 according to the manufacturer's instructions. Amplification of an 81 bp fragment of the BCoV nucleocapsid protein was performed with the following primer set: Cor‐FW (5′‐CTAGTAACCAGGCTGATGTCAATACC‐3′) and Cor‐RV (5′‐GGCGGAAACCTAGTCGGAATA‐3′). These one‐step RT‐PCR assays (One‐Step RT‐PCR kit; QIAGEN) were undertaken in a 50 μL reaction volume containing 10 μL RNA‐extract, 10 μL 5× QIAGEN One‐Step RT‐PCR Buffer, 2 μL dNTP mix (final concentration of 400 μM of each dNTP), 1.8 μL One‐Step RT‐PCR Enzyme Mix (a combination of Omniscript and Sensiscript reverse transcriptase and Hot‐ StarTaq DNA polymerase)2, 4 μM of each primer, and RNase‐free water to 50 μL. The reaction was carried out with an initial reverse transcription step at 50°C for 30 minutes, followed by PCR activation at 95°C for 10 minutes, 40 cycles of amplification (30 seconds at 94°C; 45 seconds at 60°C; 1 minute at 72°C), and a final extension step at 72°C for 10 min in a GeneAmp PCR system 9600 thermal cycler.3 A BCoV isolate recovered from a cow suffering from winter dysentery was used as a positive control. Specific methods to assess for the presence of nonspecific inhibitors of PCR were not performed.
Use of Pancoronavirus Assay to Detect CoV in Fecal and Nasal Samples of Diarrheic Calves from an Outbreak
Sampling
A large dairy farm located in Southern Ontario, Canada, experienced a sudden increase in the incidence of diarrhea and death attributable to diarrhea in calves younger <30 days of age that was sustained over a 2‐month period. At the time of the outbreak, approximately 200 preweaned calves were maintained in the calf barn. The calculated incidence of diarrhea increased from (calculated from the month before the outbreak) 36% to 57% (54/94) during the outbreak period in calves <30 days. The case fatality increased from 5% to 17% (9/54). Eight severely affected diarrheic calves were referred to the Ontario Veterinary College Health Sciences Centre (OVC‐HSC) for diagnosis and treatment of diarrhea. Rectal swabs or fecal samples and nasal swabs were collected at admission. Fecal samples were tested by culture for Salmonella spp using Rappaport‐Vassiliadis and tetrathionate broths and inoculation onto brilliant green sulpha and Hektoen agars. Escherichia coli was isolated on blood and MacConkey's agars, and isolates were serotyped using antiserum against F4 (K88) and F5 (K99) fimbriae for detection of ETEC. Sucrose wet mount assay was used for detection of Cryptosporidium spp and Giardia Spp, and real‐time polymerase chain reaction was performed to detect ruminant rotavirus A and B. PanCoV‐RT‐PCR and BCoV‐RT‐PCR assays were used to screen fecal samples and nasal swabs for the presence of BCoV. Additionally, a farm visit took place 3 days after presentation of the eight affected calves. Fecal samples were collected from 20 diarrheic calves for PanCoV‐RT‐PCR and BCoV‐RT‐PCR.
Pancoronavirus RT‐PCR Assay
Amplification of a 251‐bp fragment of the BCoV polymerase gene was performed with the following primer set: Cor‐FW (5′‐ACWCARHTVAAYYTNAARTAYGC‐3′) and Cor‐RV (5′‐TCRCAYTTDGGRTARTCCCA‐3′). These one‐step RT‐PCR assays were undertaken similarly than those described for the BCoV‐RT assay. The reaction was carried out with an initial reverse transcription step at 50°C for 30 minutes, followed by PCR activation at 95°C for 15 minutes, 50 cycles of amplification (30 seconds at 94°C; 30 seconds at 48°C; 1 minutes at 72°C), and a final extension step at 72°C for 10 minutes in a GeneAmp PCR system 9600 thermal cycler.3 Specific methods to asses for the presence of nonspecific inhibitors of PCR were not performed. A BCoV isolate recovered from a cow suffering from winter dysentery was used as a positive control. All samples were further tested with specific BCoV primers.
Statistical Analysis
The primary exposure of interest for development of diarrhea in this study was the presence of BCoV, and sex of animals was considered as a possible confounder. The matching variables were as follows: age, farm, and time of diarrhea cases. We, therefore, summarized primary exposure in case and control animals by simple cross‐tabulations, and additionally examined frequency of primary exposure in case and control animals across variables representing the matched variables including age, farm, calendar year, and season. Statistical analysis was conducted by two approaches: conditional exact logistic regression accounting for the matched pair as strata and by random intercept logistic regression where the matched pair designation represented a random effect on the intercept. After the univariable analysis, difference in the effect of the BCoV across each of the matched factors was evaluated by evaluating statistical significance of interaction between BCoV with each of the matching variables. The final models (Exact logistic regression and Random effect logistic regression) considered inclusion of all variables that were statistically significant at liberal P‐value of <0.20. Interaction between exposure of interest and sex was also tested. Choice of the final model was based on the combination of the achieved level of statistical significance and the AIC obtained in the random effect model. Analysis was performed by a statistical software.4
To test whether the PanCoV‐RT‐PCR and BCoV‐RT‐PCR assays were equally likely in detecting CoV in samples from sick calves, the McNemar's test was used. The level of agreement between both assays in detecting BCoV was assessed by Kappa coefficient test. Analysis was performed by the statistical software JMP.5
Results
Detection of BCoV in Healthy and Diarrheic calves from Farms of Ontario
A total of 143 diarrheic calves between 3 and 21 days of age and 143 healthy farm‐ and age‐matched control calves were enrolled (Table 1). None of the healthy calves developed diarrhea within one week after sampling. Cases were distributed across six herds (Fig 1), all four seasons (Fig 2), and over a period of 3 years (Fig 3). BCoV was detected in 157/286 (55%) of calves; 46% (66/143) of healthy calves, and 64% (91/143) of diarrheic calves (Table 2). Overall, diarrhea cases had higher odds of BCoV presence than the age‐matched control animals, although the numerical estimates varied slightly between the estimates based on exact logistic regression and random intercept logistic regression (Table 3). Sex was also associated with the presence of BCoV, with male calves at increased risk (Table 3). Based on the nonsignificant interaction between BCoV and matching factors (age, farm, season, and year), the association of BCoV with diarrhea did not appear to vary according to age of the animals, season, and farm (Tables 3 and 5, Figs 2 and 3). In exact logistic regression, interaction of exposure with year of sampling was close to statistical significance and indicated that in year 2015, the effect of BCoV could have been different than in the other 2 years (Table 3). The effect of year in 2015 could not be identified as statistically significant in the random effect model for the year of 2015 (Fig 3). The final model (exact logistic regression and random effect logistic regression) contained covariates for BCoV and sex. Their interaction was not statistically significant, and the coefficients for both BCoV and sex changed by very small quantity once both variables were in the model. This suggests that sex failed to act as confounder for the association between BCoV and diarrhea in this dataset (Table 3).
Figure 1.
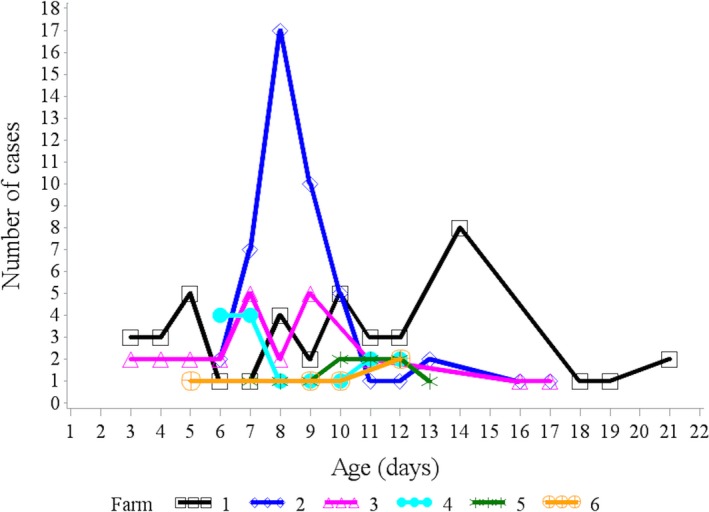
Age distribution of diarrheic cases from 6 different farms that tested positive for bovine coronavirus in feces.
Figure 2.
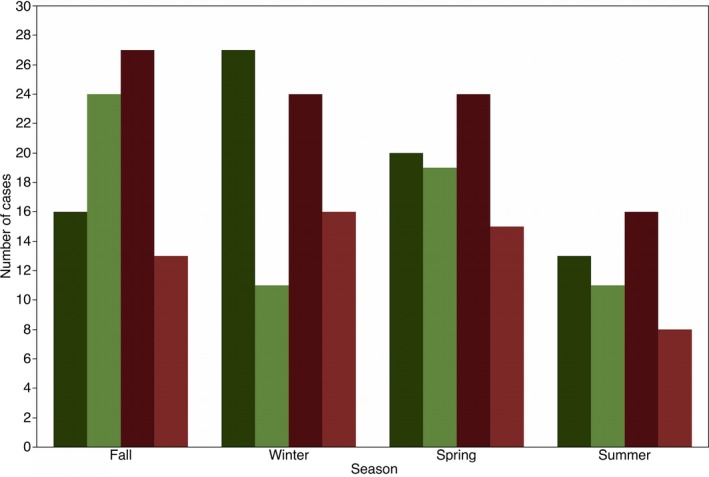
Frequency distribution of healthy (green) and diarrheic (red) calves that tested positive (dark) and negative (light) for bovine coronavirus (BCoV) in feces during the different sampling seasons. (%) represents the percentage of the total number of cases.
Figure 3.
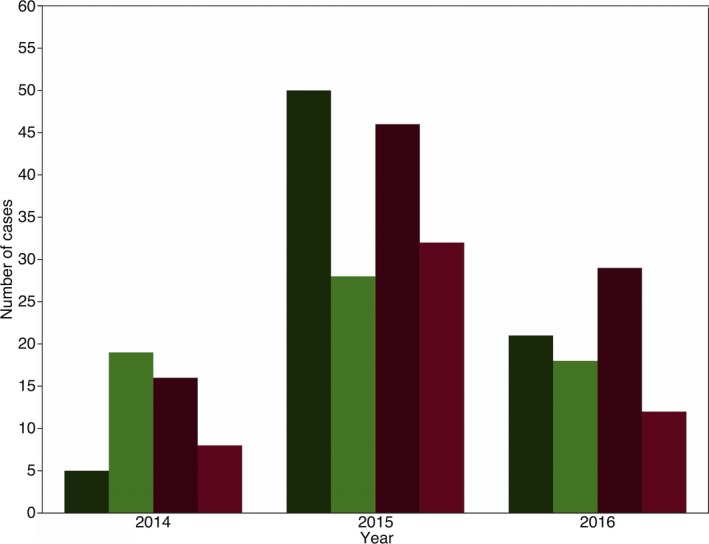
Frequency distribution of healthy (green) and diarrheic (red) calves that tested positive (dark) and negative (light) for bovine coronavirus (BCoV) in feces during the different sampling years. (%) represents the percentage of the total number of cases.
Table 2.
Frequency distribution of bovine coronavirus positivity status stratified by clinical status and sex of 286 calves from six dairy farms
| BCoV | Farm | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | All farms | Sex | |||||||||
| H | D | H | D | H | D | H | D | H | D | H | D | H | D | M | F | |
| − | 14 (33%) | 10 (24%) | 24 (51%) | 15 (32%) | 19 (79%) | 3 (60%) | 9 (60%) | 8 (53%) | 7 (70%) | 4 (40%) | 4 (80%) | 3 (60%) | 77 (54%) | 52 (36%) | 25 (27%) | 118 (82%) |
| + | 28 (67%) | 32 (76%) | 23 (49%) | 32 (68%) | 5 (21%) | 2 (40%) | 6 (40%) | 7 (47%) | 3 (30%) | 6 (60%) | 1 (20%) | 2 (40%) | 66 (46%) | 91 (64%) | 39 (18%) | 104 (73%) |
| Total | 42 | 42 | 47 | 47 | 24 | 5 | 15 | 15 | 10 | 10 | 5 | 5 | 143 | 143 | 64 | 222 |
BCoV, Bovine Coronavirus; +, positive; −, negative; H, healthy (controls); D, diarrhea (cases); M, male; F, female.
Table 3.
Evaluation of the association between BCoV, diarrhea, and matched variables (age, farm, season, and year)
| Analysis of single factors of interests | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Exact logistic regression | Random effect logistic regression | ||||||||
| OR | 95% CI | P‐value | OR | 95% CI | P‐value | ||||
| BCoV (+ve) | +ve | 2.19 | 1.28 | 3.86 | 0.031 | 2.04 | 1.27 | 3.29 | 0.0037 |
| Sex (Male) | Male | 1.87 | 0.99 | 3.68 | 0.054 | 1.77 | 0.99 | 3.14 | 0.050 |
| BCov*age | – | – | – | – | 0.92 | – | – | – | 0.97 |
| BCov*season | – | – | – | – | 0.47 | – | – | – | 0.86 |
| BCov*year | – | – | – | – | 0.03 | – | – | – | 0.17 |
| 2014*BCoV | 2.51 | 1.13 | +inf | 4.74 | 1.62 | 13.9 | |||
| 2015*BCoV | 0.43 | −inf | 0.83 | 1.67 | 0.95 | 2.91 | |||
| 2016*BCoV | 3.36 | 1.85 | +inf | 2.11 | 1.09 | 4.11 | |||
| BCoV*farm | – | – | – | – | 0.76 | – | – | – | 0.84 |
| Final model | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Exact logistic regression | Random effect logistic regression | ||||||||
| OR | 95% CI | P‐value | OR | 95% CI | P‐value | ||||
| BCoV | +ve | 2.16 | 1.26 | 3.83 | 0.004 | 1.96 | 1.21 | 3.17 | 0.006 |
| Sex | Male | 1.84 | 0.95 | 2.95 | 0.072 | 1.63 | 0.91 | 2.92 | 0.097 |
BCoV, Bovine coronavirus; +ve, positive; OR, odds ratio, CI, confidence interval.
PanCoV‐RT‐PCR for Detection of CoV in Fecal and Nasal Samples
By PanCoV‐RT‐PCR, the CoV RNA was detected in 8/8 fecal samples and 6/8 nasal swabs from the calves presented to OVC‐HSC (Table 4). All but one sample that was positive for CoV by the PanCoV‐RT‐PCR were also positive on the BCoV‐RT‐PCR. Thirteen out of 20 (65%) diarrheic calves from the same farm tested positive for CoV by PanCoV‐RT‐PCR assay. All of the samples that tested positive in the PanCoV‐RT‐PCR assay were also positive when screening by BCoV‐RT‐PCR assay, but three samples positive for BCoV in the BCoV‐RT‐PCR assay were negative on the PanCoV‐RT‐PCR assay (Table 5). McNemar's test demonstrated that both tests were equally likely to detect CoV in samples from calves (P = 0.67). Kappa agreement test revealed a good agreement between both tests for detection of BCoV in fecal and nasal samples from diarrheic calves (κ = 0.68, 95% CI: 0.392 to 0.967; P < 0.001). The results of the bacterial, viral, and protozoal analyses performed in feces of the calves presented to the OVC‐HSC are presented in Table 5. All 20 fecal samples were negative for Salmonella.
Table 4.
Test results for bacterial, viral, and protozoal analyses performed in feces of eight hospitalized diarrheic calves
| Pathogen/Calf | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| E. coli (F5/K99) | − | − | − | − | − | − | − | − |
| Cryptosporidium | + | + | − | + | − | − | − | + |
| BRoV‐A | − | + | − | − | − | − | − | − |
| BRoV‐B | − | + | − | − | − | − | − | − |
| PanCoV‐RT‐PCR | + | + | + | + | + | + | + | + |
| BCoV‐RT‐PCR | + | + | + | + | + | + | + | + |
| Salmonella spp. | − | − | − | − | − | − | − | − |
| Giardia spp. | − | + | − | − | − | + | − | − |
F5/K99, E. coli F5 (K99) fimbriae attachment factor; BRoV, bovine rotavirus; PanCoV‐RT‐PCR assay, pancoronavirus reverse transcriptase PCR assay; BCoV‐RT‐PCR assay, bovine coronavirus reverse transcriptase PCR assay.
Table 5.
Comparison of Coronavirus detection results by Pancoronavirus reverse transcriptase (PanCoV) and BCoV reverse transcriptase (BCoV) assays
| PanCoV | |||
|---|---|---|---|
| + | − | ||
| BCoV | |||
| + | 26 | 3 | 29 |
| − | 1 | 6 | 7 |
| Total | 27 | 9 | 36 |
McNemar's test P‐value = 0.67. Kappa agreement test, κ = 0.68, 95% CI: 0.392 to 0.967; P < 0.001.
Discussion
The results of this study demonstrated a positive association between BCoV and diarrhea in dairy calves as detection rates of this agent were higher in diarrheic calves than in farm‐, season‐, aged‐matched nondiarrheic calves. Similar results were found in one case–control study involving 380 calves.25 However, studies including small numbers of calves (n < 100) identified numerical but not statistical association between BCoV in fecal or nasal samples and clinical signs.16, 17, 18 Discrepancies between studies can be explained, at least in part, by differences in the source population, methodology, especially, the samples size used in the different investigations.16, 25
Detection of BCoV, either in diarrheic and in nondiarrheic calves, was higher in comparison with previous studies from the same geographic area,10, 26 other studies from different regions worldwide6, 7, 25 and more in line with recent studies from Europe.4 Earlier studies (1980 to 2007) investigating the presence of infectious agents in feces of healthy and diarrheic calves reported a prevalence of BCoV in healthy calves between 0 and 8.2% and in diarrheic calves between 3 and 40%.10, 26 More recent studies (2008 to 2012) reported prevalence of BCoV in healthy calves of 7.4% and in diarrheic calves between 3.4 and 40%.6, 7 Interestingly, a recent report from Europe identified a prevalence of BCoV of 79% of diarrheic calves;4 however, this study failed to investigate healthy controls. The prevalence of infectious agents causing diarrhea in calves was previously investigated in dairy farms from Ontario at three different periods by ELISA assays, 1982,26 1990–1991,27 and 1995–1997.10 The overall prevalence of BCoV in the 1982's study was 5% (3/56), but this study did not discriminate the prevalence of healthy and diarrheic calves.26 In the 1992 study, the prevalence of BCoV in diarrheic calves was 17% (54/312), whereas in 1998, the overall prevalence was 10% (16/161), and BCoV was detected in 13% (15/118) and 2.3% (1/43) of diarrheic and healthy calves, respectively. The reasons for these differences in the detection rates of BCoV in calves (healthy and diarrheic) between this study and previous investigations in the same geographic area (Ontario) are unclear. In fact, it is difficult to discern whether the increasing detection of BCoV reflects a true rise in prevalence in the course of time or whether it is due to simply to the better detection rate.28 The differences between studies could be explained, at least in part, by changes in farm management practices overtime (e.g., calf population density, antimicrobial drug use, and management/agricultural practices), differences in the type of assay used for detection of BCoV, and the emergence or re‐emergence of BCoV strains over the years.
Regarding the influence that farm management practices can have on the prevalence of infectious diseases, contradictory results have reported.1, 29, 30, 31 In one study, the prevalence of salmonellosis in preweaned calves was reported to be higher in group pen calves than those in individual pens.29 Contrary, some studies failed to identify an effect of management and infrastructure variables on the presence of enteropathogens in calves.30, 31 This study was not designed to evaluate farm‐level differences, something that would warrant further investigation.
Differences in the type of assay used for detection of BCoV (e.g., ELISA vs. RT‐PCR) also can explain differences in the detection rates of BCoV between studies. One study investigating the rates of BCoV fecal shedding in diarrheic calves found a detection rate of BCoV of 53% when using ELISA and 96% when using RT‐PCR16 suggesting that RT‐PCR was a more sensitive method. These results were further confirmed by one study reporting that the sensitivity and specificity of some ELISA assays were lower compared with quantitative RT‐PCR when both tests were used to identify the presence of BCoV in feces of diarrheic calves.32 Furthermore, more sensitive diagnostic assays also could contribute to detect lower levels of BCoV that cannot be detected by other methods.32
An alternative explanation for the changes in detection rates of BCoV in healthy and diarrheic calves over the years in dairy farms from Ontario could be the emerging and re‐emerging of BCoV strains. Similar to other RNA viruses, CoVs are believed to mutate frequently that could result in altered antigenicity and pathogenicity of new strains.33, 34 During the past decades, several CoVs have caused serious problems in humans and animals (e.g., severe acute respiratory syndrome coronavirus, SARS‐CoV; Middle East respiratory syndrome coronavirus, MERS‐CoV, and porcine epidemic diarrhea virus, PEDV).35 Emerging outbreaks associated with CoV also have been documented in horses worldwide,36 and emerging canine CoV variants have been associated with systemic infections in the European Union.37 In an Oklahoma study, a new CoV belonging to a BCoV clade 2 was isolated from nasal secretion and lung fluids of postweaned healthy beef calves and calves with respiratory disease.38 This new field isolate was antigenically and genetically different to previously isolated BCoV strains including those from neonatal diarrhea, reference strains, and the strain in the current modified live vaccine BCoV licence in United States that belong to CoV clade 1.38 Similar results have been reported in Europe4 and Asia.39 In 2012, the BCoV clade 2 strain was recovered from postweaned calves (6–7 months) suffering of severe enteritis with bloody diarrhea; the authors of that study suggested that this clinical syndrome caused by BCoV clade 2 should be added to the already known syndromes caused by BCoV.40 The results of these studies suggest that current BCoV strains circulating in worldwide are diverging from the prototype BCoV isolated in earlier studies and used in vaccines.38 This can result in the emergency of new clinical syndromes,40 outbreaks of enteric diseases with higher morbidity and case fatality (similar to the one described in this study), increased detection rates of BCoV in nasal and fecal samples of healthy and ill calves,4, 18, 41 and the lack of efficacy vaccination protocols.42 Future studies should focus on investigating the biological, antigenical, and genomical characteristic of BCoV circulating in Canada. It is important to highlight that in this study no other virus, parasites, and bacterial agents were investigated, and the role of BCoV in association with multiple pathogens was not determined which prevent us to clearly identify the role of BCoV as a primary cause of disease or as a agent associated with coinfection.
This study also demonstrated that a recently described PanCoV‐RT‐PCR assay can be as useful as the BCoV‐RT‐PCR assay to identify BCoV in fecal and nasal samples of diarrheic calves, with the added benefit of providing a broader test that would detect novel CoVs. Currently, viral culture is the gold standard for laboratory diagnosis of viral gastrointestinal disorders; however, detection of coronavirus by culture isolation or serological methods can be difficult.23 To overcome this issue and obtain rapid diagnostic results, several sensitive molecular assays for detection of CoV have been developed, including RT‐PCR, nested RT‐PCR, and real‐time RT‐PCR. Recently, a novel PanCoV‐RT‐PCR assay was designed to allow detection of all known CoV based on an alignment of conserved genome regions of several coronavirus.43 These results showed a good level of agreement between BCoV‐RT‐PCR and the novel PanCoV‐RT‐PCR assays indicating that PanCoV‐RT‐PCR can be a rapid tool to detect BCoV in samples from cattle. Future studies with a larger sample size are required to corroborate these findings.
Conclusion
Calves positive to BCoV were more likely to suffer from diarrhea than farm‐ and aged‐matched healthy calves. These results also showed an apparent higher prevalence of BCoV in fecal samples of healthy and diarrheic calves in Ontario, Canada, compared with previous years. A new PanCoV‐RT‐PCR was useful in identifying the presence of CoV in nasal and fecal samples from diarrheic calves during an outbreak of diarrhea.
Acknowledgments
We thank Dr. Davor Ojkic from the Animal Health Laboratory, University of Guelph, who kindly provided the BCoV isolate used as a positive control in these assays. We also thank Joyce Rousseau for her input during the process of carrying out the experiments described in this manuscript. This study was supported by a National Sciences Engineering and Research Council (NSERC) Discovery Grant.
Conflict of interest Declaration
Authors declare no conflict of interest.
Off‐label Antimicrobial Declaration
Authors declare no off‐label use of antimicrobials.
This work was presented as a research abstract at the 2016 ACVIM Forum Denver, Colorado, USA.
Footnotes
QIAamp Viral RNA Mini kit, QIAGEN, Toronto, ON
One‐Step RT‐PCR kit; QIAGEN, Toronto, ON
Applied Biosystems, Foster City, CA
SAS Institute Inc., Cary, NC
JMP 12, SAS Institute Inc., Cary, NC
Calf Guard, Zoetis, Parsippany, NJ
References
- 1. Svensson C, Lundborg K, Emanuelson U, Olsson SO. Morbidity in Swedish dairy calves from birth to 90 days of age and individual calf‐level risk factors for infectious diseases. Prev Vet Med 2003;58:179–197. [DOI] [PubMed] [Google Scholar]
- 2. USDA . Dairy 2007, Part I: Reference of Dairy Cattle Health and Management Practices in the United States. Fort Collins CO: USDA‐APHIS‐VS; 2007:2007. [Google Scholar]
- 3. Stanton AL, Kelton DF, LeBlanc SJ, et al. Effects of tulathromycin on incidence of various diseases and growth of young heifers. J Am Vet Med Assoc 2013;243:267–276. [DOI] [PubMed] [Google Scholar]
- 4. Lojkić I, Krešić N, Šimić I, Bedeković T. Detection and molecular characterisation of bovine corona and toroviruses from Croatian cattle. BMC Vet Res 2015;11:202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coura FM, Freitas MD, Ribeiro J, et al. Longitudinal study of Salmonella spp., diarrheagenic Escherichia coli, Rotavirus, and Coronavirus isolated from healthy and diarrheic calves in a Brazilian dairy herd. Trop Anim Health Prod 2015;47:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uhde FL, Kaufmann T, Sager H, et al. Prevalence of four enteropathogens in the faeces of young diarrhoeic dairy calves in Switzerland. Vet Rec 2008;163:362–366. [DOI] [PubMed] [Google Scholar]
- 7. Izzo MM, Kirkland PD, Mohler VL, et al. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust Vet J 2011;89:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gillhuber J, Rügamer D, Pfister K, Scheuerle MC. Giardiosis and other enteropathogenic infections: A study on diarrhoeic calves in Southern Germany. BMC Res Notes 2014;7:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koopmans M, van Wuijckhuise‐Sjouke L, Schukken YH, et al. Association of diarrhea in cattle with torovirus infections on farms. Am J Vet Res 1991;52:1769–1973. [PubMed] [Google Scholar]
- 10. Duckmanton L, Carman S, Nagy E, Petric M. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J Clin Microbiol 1998;36:1266–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snodgrass DR, Terzolo HR, Sherwood D, et al. Aetiology of diarrhoea in young calves. Vet Rec 1986;119:31–34. [DOI] [PubMed] [Google Scholar]
- 12. Saif LJ, Brock KV, Redman DR, Kohler EM. Winter dysentery in dairy herds: Electron microscopic and serological evidence for an association with coronavirus infection. Vet Rec 1991;128:447–449. [DOI] [PubMed] [Google Scholar]
- 13. Cho YI, Han JI, Wang C, et al. Case‐control study of microbiological etiology associated with calf diarrhea. Vet Microbiol 2013;166:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lathrop SL, Wittum TE, Loerch SC, et al. Antibody titers against bovine coronavirus and shedding of the virus via the respiratory tract in feedlot cattle. Am J Vet Res 2000;61:1057–1061. [DOI] [PubMed] [Google Scholar]
- 15. Storz J, Purdy CW, Lin X, et al. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp from cattle involved in two natural outbreaks of shipping fever. J Am Vet Med Assoc 2000;216:1599–1604. [DOI] [PubMed] [Google Scholar]
- 16. Hasoksuz M, Hoet AE, Loerch SC, et al. Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in an Ohio feedlot. J Vet Diagn Invest 2002;14:308–313. [DOI] [PubMed] [Google Scholar]
- 17. Hasoksuz M, Kayar A, Dodurka T, Ilgaz A. Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in Northwestern Turkey. Acta Vet Hung 2005;53:137–146. [DOI] [PubMed] [Google Scholar]
- 18. Francoz D, Buczinski S, Bélanger AM, et al. Respiratory pathogens in Québec dairy calves and their relationship with clinical status, lung consolidation, and average daily gain. J Vet Intern Med 2015;29:381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boileau MJ, Kapil S. Bovine coronavirus associated syndromes. Vet Clin North Am Food Anim Pract 2010;26:123–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graham DY, Sackman JW, Estes MK. Pathogenesis of rotavirus‐induced diarrhea. Preliminary studies in miniature swine piglet. Dig Dis Sci 1984;29:1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mebus CA, Stair EL, Rhodes MB, Twiehaus MJ. Pathology of neonatal calf diarrhea induced by a coronavirus‐like agent. Vet Pathol 1973;10:45–64. [DOI] [PubMed] [Google Scholar]
- 22. Saif LJ, Redman DR, Moorhead PD, Theil KW. Experimentally induced coronavirus infections in calves: Viral replication in the respiratory and intestinal tracts. Am J Vet Res 1986;47:1426–1432. [PubMed] [Google Scholar]
- 23. Moës E, Vijgen L, Keyaerts E, et al. A novel pancoronavirus RT‐PCR assay: Frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis 2005;5:6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vijgen L, Moës E, Keyaerts E, et al. A pancoronavirus RT‐PCR assay for detection of all known coronaviruses. Methods Mol Biol 2008;454:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pérez E, Kummeling A, Janssen MM, et al. Infectious agents associated with diarrhoea of calves in the canton of Tilarán, Costa Rica. Prev Vet Med 1998;33:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waltner‐Toews D, Martin SW, Meek AH, et al. A field trial to evaluate the efficacy of a combined rotavirus‐coronavirus/Escherichia coli vaccine in dairy cattle. Can J Comp Med 1985;49:1–9. [PMC free article] [PubMed] [Google Scholar]
- 27. Carman PS, Hazlett MJ. Bovine coronavirus infection in Ontario 1990–1991. Can Vet J 1992;33:812–814. [PMC free article] [PubMed] [Google Scholar]
- 28. Jones KE, Patel NG, Levy MA, et al. Global trends in emerging infectious diseases. Nature 2008;451:990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Linton AH, Howe K, Pethiyagoda S, Osborne AD. Epidemiology of salmonella infection in calves (1): Its relation to their husbandry and management. Vet Rec 1974;94:581–585. [DOI] [PubMed] [Google Scholar]
- 30. Bartels CJ, Holzhauer M, Jorritsma R, et al. Prevalence, prediction and risk factors of enteropathogens in normal and non‐normal faeces of young Dutch dairy calves. Prev Vet Med 2010;93:162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Al Mawly J, Grinberg A, Prattley D, et al. Prevalence of endemic enteropathogens of calves in New Zealand dairy farms. N Z Vet J 2015;63:147–152. [DOI] [PubMed] [Google Scholar]
- 32. Izzo MM, Kirkland PD, Gu X, et al. Comparison of three diagnostic techniques for detection of rotavirus and coronavirus in calf faeces in Australia. Aust Vet J 2012;90:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steinhauer DA, Holland JJ. Direct method for quantitation of extreme polymerase error frequencies at selected single base sites in viral RNA. J Virol 1986;57:219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology 2001;279:371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lau SK, Chan JF. Coronaviruses: Emerging and re‐emerging pathogens in humans and animals. Virol J 2015;12:209–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pusterla N, Mapes S, Wademan C, et al. Emerging outbreaks associated with equine coronavirus in adult horses. Vet Microbiol 2013;162:228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buonavoglia C, Decaro N, Martella V, et al. Canine coronavirus highly pathogenic for dogs. Emerg Infect Dis 2006;12:492–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fulton RW, Ridpath JF, Burge LJ. Bovine coronaviruses from the respiratory tract: Antigenic and genetic diversity. Vaccine 2013;31:886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park SJ, Lim GK, Park SI, et al. Detection and molecular characterization of calf diarrhoea bovine coronaviruses circulating in South Korea during 2004–2005. Zoonoses Public Health 2007;54:223–230. [DOI] [PubMed] [Google Scholar]
- 40. Fulton RW, Herd HR, Sorensen NJ, et al. Enteric disease in postweaned beef calves associated with Bovine coronavirus clade 2. J Vet Diagn Invest 2015;27:97–101. [DOI] [PubMed] [Google Scholar]
- 41. O'Neill R, Mooney J, Connaghan E, et al. Patterns of detection of respiratory viruses in nasal swabs from calves in Ireland: A retrospective study. Vet Rec 2014;175:351–356. [DOI] [PubMed] [Google Scholar]
- 42. Meganck V, Hoflack G, Piepers S, Opsomer G. Evaluation of a protocol to reduce the incidence of neonatal calf diarrhoea on dairy herds. Prev Vet Med 2015;118:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stephensen CB, Casebolt DB, Gangopadhyay NN. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res 1999;60:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]


