Abstract
Background
The purpose of this study was to investigate the prognostic significance of methylation of RAS association domain family protein 1 (RASSF1A) in the promoter region for patients with stage II and III colorectal cancer (CRC) receiving oxaliplatin-based chemotherapy.
Material/Methods
There were 108 eligible CRC patients and 78 healthy controls included in this study. Methylation-specific polymerase chain reaction (MSP) was applied to detect the methylation status of RASSF1A in patients before and after chemotherapy. The effects of RASSF1A methylation on chemotherapy-sensitivity and prognosis for patients were also evaluated in the present study.
Results
The frequency of RASSF1A methylation was higher in CRC patients than in the healthy controls (48.44% versus 5.13%, p<0.001). After two cycles of chemotherapy, methylation ratio was significantly decreased (21.30%, p<0.001). Promoter methylation of RASSF1A was significantly correlated with tumor stage and pathological differentiation (p=0.008 and p=0.007, respectively). Patients without methylation had a favorable objective response (OR), compared with those with methylation (53.33% versus 25%, p=0.014). Methylation status of RASSF1A could influence progression-free survival and overall survival (log rank test, p<0.05). Cox regression analysis indicated that RASSF1A methylation (HR=2.471, 95% CI=1.125–5.428, p=0.024) and OR (HR=2.678, 95% CI=1.085–6.610, p 0.033) were independently correlated with prognosis for patients treated with oxaliplatin-based chemotherapy.
Conclusions
Promoter methylation of RASSF1A can influence sensitivity to oxaliplatin-based chemotherapy, which can be used to predict outcomes for patients with stage II and III CRC. In addition, the aberrant methylation may be a promising target for improving chemotherapy efficacy.
MeSH Keywords: Chemoradiotherapy, Adjuvant; Colorectal Neoplasms; Methylation; Prognosis
Background
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers that has contributed to a great deal of cancer-deaths in the world [1,2]. Treatments for CRC include surgery, chemotherapy, radiotherapy, targeted therapy, or combinations thereof [3]. Surgery is the optimal method to localize CRC lesions, but outcomes for the patients are usually poor due to metastasis [4]. For metastatic CRC, the accepted first-line therapeutic regimen is 5-fluorouracil (5-FU)/oxaliplatin [5]. However, patients with similar clinical characteristics may react differently to the treatments and have various outcomes [6]. Therefore, to exploit novel markers which can further stratify CRC beyond tumor node metastasis (TNM) staging may significantly improve outcomes for patients with advanced CRC treated with oxaliplatin-based chemotherapy.
Methylation of specific genes might play essential roles in chemotherapy resistance. HOTAIR for HOX transcript antisense RNA) has been proved to significantly affect carboplatin resistance in patients with ovarian cancer [7]. Methylation of HYAL2 (hyaluronoglucosaminidase 2) has also been reported to influence progress-free survival (PFS) and overall survival of patients who were with colon cancer under 5-FU therapy [8]. RAS association domain family protein 1 (RASSF1A), located on chromosome 3p21.3, is a known tumor suppressor gene which could regulate cell proliferation and apoptosis [9,10]. Aberrant methylation of RASSF1A is associated with several tumors such as lung cancer and esophageal squamous carcinoma [11,12]. The effects of RASSF1A methylation on outcomes for patients with ovarian cancer treated with platinum-based chemotherapy have been previously reported [13]. However, the effects of RASSF1A methylation on prognosis for patients with CRC receiving oxaliplatin-based chemotherapy have been rarely explored.
In this study, we are aimed to investigate the effects of RASSF1A promoter methylation on sensitivity to oxaliplatin-based chemotherapy in patients with CRC. The relationship between RASSF1A methylation and tumor response, as well as the long-term effects on PFS and overall survival in patients with stage II and III CRC treated with oxaliplatin-based chemotherapy were evaluated.
Material and Methods
Study subjects
The present study was carried out between December 2009 and February 2015 in Weifang People’s Hospital. The patients collected in the study accorded with the following criterion: 1) pathologically diagnosed with stage II and III CRC; 2) aged 18–75 years; 3) not diagnosed with serious body diseases; 4) firstly diagnosed with CRC, not with recurrent CRC; and 5) no radiotherapy or chemotherapy treatment before the specimens collected.
There were 108 eligible patients with CRC included in this study as the test group. In addition, 78 healthy volunteers were recruited for the study as controls. All the participators signed an informed consent at the beginning of the study. This study was approved by the Ethical Committee of our hospital.
All the patients were enrolled in a three-year investigation and the clinical characteristics and survival status were collected in order to evaluate the long-term effects of RASSF1A methylation on patients with CRC undergoing platinum-based chemotherapy.
Therapy
The patients in the test group were treated with oxaliplatin-based chemotherapy. A cycle of chemotherapy lasted for five days. On the first day of treatment, patients received 130 mg/m2 of oxaliplatin intravenously for two hours; 130 mg/m2 of leucovorin was intravenously injected into the patients for two hours every day during the chemotherapy duration. In addition, 300 mg/m2 of 5-fluoropyrimidine was given to the patients through intravenous injection for four hours every day during the treatment. This treatment was repeated every three weeks.
Response to chemotherapy
Tumor response to oxaliplatin-based chemotherapy was assessed according to Response Evaluation Criteria in Solid Tumors (RECIST). Complete response (CR), partial response (PR), no change (NC), and objective response (OR) were respectively defined as complete tumor disappearance, tumor reduction of at least 50%, tumor reduction less than 50% or tumor enlargement, and the sum of CR and PR percentages.
Specimen collection
At the beginning of the study, 5 mL blood samples were collected from all the participators after six to eight hours of fasting. After two cycles of chemotherapy, 5 mL blood samples were collected from the patients in test group again. Ethylene diamine tetra-acetic acid (EDTA) was used for anticoagulation.
DNA isolation and methylation specific polymerase chain reaction (MSP)
DNA samples were isolated from the collected blood samples using genomic DNA Extraction Kit (Tiangen Biotech, China) according to the manufacturer’s instructions. The quality and concentration of the obtained DNA samples were measured by 1% agarose gel electrophoresis and ultraviolet spectrophotometer, respectively. After sodium bisulfite modification, the DNA samples were purified and prepared for templates [14].
Promoter methylation of RASSF1A was detected by MSP. The specific primer pairs included methylated-RASSF1A primers and unmethylated-RASSF1A primers. The primer sequences were as follows: methylated-RASSF1A sense primer: 5′ GTGTTAACGCGTTGCGTATC 3′, antisense primer: 5′ AACCCCGCGAACTAAAAACGA3′; and unmethylated-RASSF1A sense primer: 5′ TTTGGTTGGAGTGTGTTAATGTG 3′, antisense primer: 5′ CAAACCCCACAAACTAAAAACAA 3′ [15]. A 20 μL reaction system was used in this study, which included 2×Tag PCR Master Mix 10 μL, 7 μL ddH2O, 2 μL purified DNA samples, and 0.5 μL primers. The reaction condition for unmethylated amplification was as follows: 94°C (5 minutes); 94°C (30 seconds), 60°C (30 seconds), 72°C (30 seconds), 35 cycles; 72°C (10 minutes). For methylated primers, the annealing temperature was 55°C.
Statistical analysis
The statistical analysis was performed using SPSS 18.0 software. The continuous variables were presented as average ± standard deviation (SD) and analyzed by student t-test, while discontinuous variables analysis was performed using chi-square analysis. Kaplan-Meier method with log rank test was applied to evaluate PFS and overall survival for patients in the test group. The prognostic significance of RASSF1A methylation was assessed by Cox regression analysis. A value of p<0.05 was considered statistically significant.
Results
Demographic characteristics of the study groups
There were 108 patients with CRC and 78 eligible volunteers included in this study. The average age for the patients and healthy controls were 46.79±11.95 and 45.68±14.17 years, respectively. The gender ratio was similar among the two groups (p=0.323). Clinical information including tumor size, site, differentiation, and stages are listed in Table 1.
Table 1.
Demographic characteristics of the study groups.
| Characteristics | Test group (n=108) | Control group (n=78) | P |
|---|---|---|---|
| Age (year) | 46.79±11.95 | 45.68±14.17 | 0.565 |
| Gender | 0.323 | ||
| Male | 55 | 34 | |
| Female | 53 | 44 | |
| Tumor size | – | ||
| ≥5 cm | 58 | – | |
| <5 cm | 50 | – | |
| Tumor stage | – | ||
| II | 67 | – | |
| III | 41 | – | |
| Tumor site | – | ||
| Rectum | 57 | – | |
| Colon | 51 | – | |
| Pathological differentiation | – | ||
| Well + moderate | 58 | – | |
| Poor | 50 | – |
’–’ – indicated no available data.
Incidences of RASSF1A methylation
MSP was used to analyze the methylation status in the collected specimens. The PCR products were 125 bp and the results are shown in Figure 1. Before chemotherapy, promoter methylation of RASSF1A was detected in 48 (44.44%) patients in the test group, while there were only 4 (5.13%) persons with RASSF1A methylation in the control group (Table 2). After two cycles of chemotherapy, the proportion of methylation was significantly decreased in test group, compared with methylation before treatment (21.30% versus 44.44%, p<0.001). However, the methylation ratio was still obviously higher than in the control group (p=0.002).
Figure 1.
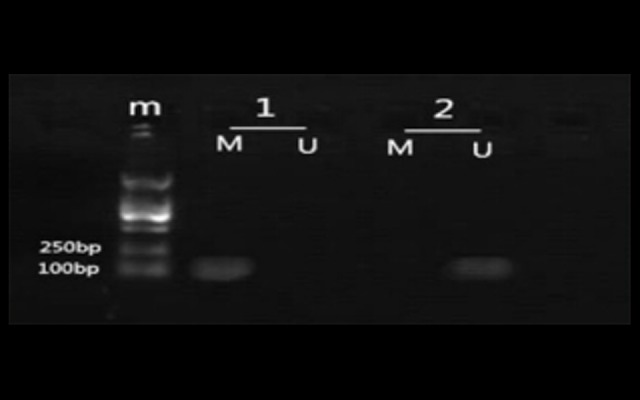
Electrophoresis gel appearance of RASSF1A methylation status. m – marker; M – methylated products; U – unmethylated products. Sample 1 shows methylation and sample 2 shows unmethylation.
Table 2.
The incidence rate of RASSF1A methylation.
| Group | Time | Methylation | Unmethylation | Proportion of methylation |
|---|---|---|---|---|
| Test group (n=108) | Before chemotherapy | 48 | 60 | 44.44%*** |
| After chemotherapy | 23 | 85 | 21.30%***,## | |
| Control group (n=78) | At the beginning of the study | 4 | 74 | 5.13% |
* Indicated a significant difference between test group and control group, * P<0.05, ** P<0.01, *** P<0.001. # Predicted significant differences, compared with before chemotherapy, # P<0.05, ## P<0.01, ### P<0.001.
Association between RASSF1A methylation and clinical characteristics
The patients in the test group were divided into a methylation group (n=48) and an unmethylation group (n=68). The relationship between RASSF1A methylation and clinical characteristics was evaluated (Table 3). It was demonstrated that RASSF1A methylation was associated with pathological differentiation and tumor stage (p=0.008 and p=0.007, respectively). However, methylation status of RASSF1A was not shown to be correlated with age, gender, tumor site or tumor size (p>0.05).
Table 3.
Relationship between RASSF1A methylation and clinical characteristics.
| Characteristics | Methylation group (n=48) | Unmethylation group (n=60) | P |
|---|---|---|---|
| Age (year) | 46.96±11.28 | 45.85±12.47 | 0.365 |
| Gender | 0.547 | ||
| Male | 26 | 29 | |
| Female | 22 | 31 | |
| Tumor size | 0.635 | ||
| ≥5 cm | 27 | 31 | |
| <5 cm | 21 | 29 | |
| Tumor stage | 0.007 | ||
| II | 23 | 44 | |
| III | 25 | 16 | |
| Tumor site | 0.605 | ||
| Rectum | 24 | 33 | |
| Colon | 24 | 27 | |
| Pathological differentiation | 0.008 | ||
| Well + moderate | 19 | 39 | |
| Poor | 29 | 21 |
Relationship between RASSF1A methylation and objective response (OR)
Objective response (OR) was used to evaluate the effects of platinum-based chemotherapy. The OR for patients without RASSF1A methylation (53.33%) was significantly higher than that for patients with methylation (25%) p=0.014, Table 4)
Table 4.
Effects of RASSF1A methylation on OR.
| Groups | CR (%) | PR (%) | SD (%) | PD (%) | CR+PR (%) | P |
|---|---|---|---|---|---|---|
| Methylation group (n=48) | 4 (8.33) | 8 (16.67) | 15 (31.25) | 21 (43.75) | 12 (25.00) | 0.014 |
| Unmethylation group (n=60) | 14 (23.33) | 18 (30.00) | 16 (26.67) | 12 (20.00) | 32 (53.33) |
CR – complete response; PR – partial response; SD – stable disease; PD – progress disease, OR – objective response.
PFS and overall survival analysis
Patients without methylation of RASSF1A had a longer PFS time than those with methylation (29.6 months versus 23.04 months), and the differences were significant (log rank test, p=0.004, Figure 2).
Figure 2.
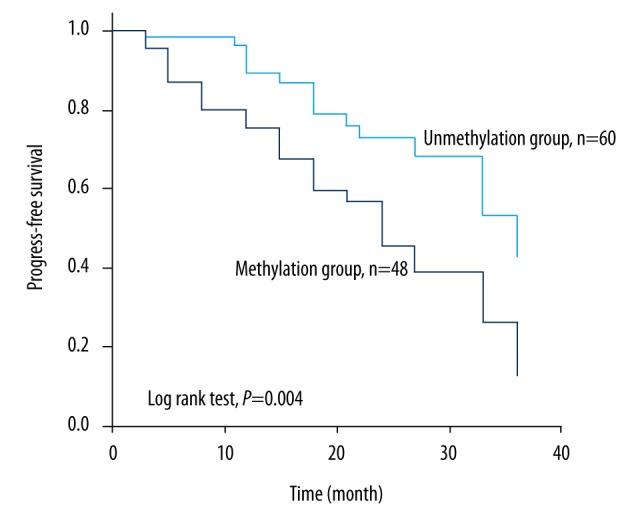
PFS curve for stage II and III CRC patients receiving oxaliplatin-based chemotherapy according to methylation status. Patients with RASSF1A methylation had a shorter PFS than those without methylation (23.04 months versus 29.6 months, log rank test, p=0.004).
Overall survival of patients with CRC was evaluated according to methylation status. The results, shown in Figure 3, indicated that patients in the methylation group had an obviously shorter overall survival time than those in the unmethylation group (25.90 months versus 31.66 months, log rank test, p=0.006).
Figure 3.
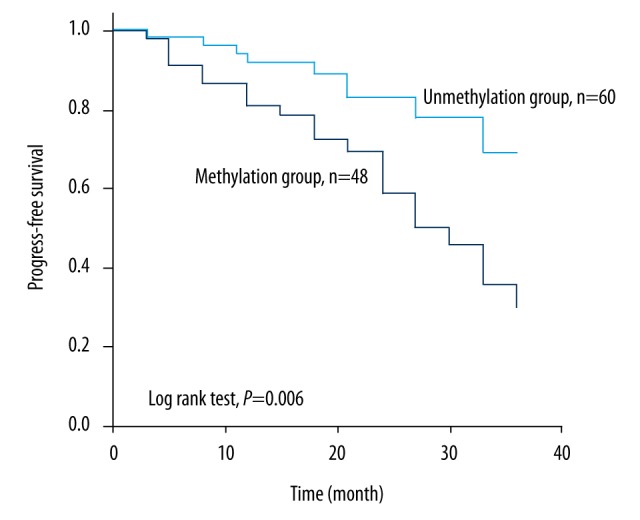
Overall survival analysis in stage II and III CRC patients treated with oxaliplatin-based chemotherapy according to methylation status of RASSF1A. Patients without RASSF1A methylation had a favorable prognosis, compared with those with methylation (31.66 months versus 25.90 months, log rank test, p=0.006).
Prognostic significance of RASSF1A methylation
Cox regression analysis was used to assess the prognostic significance of RASSF1A methylation in patients with CRC treated with oxaliplatin-based chemotherapy. Univariate analyses indicated that RASSF1A methylation and OR were significantly correlated with outcomes of patients who were diagnosed with stage II and III CRC and who were treated with platinum-based chemotherapy (p<0.05). Multivariate analysis demonstrated that RASSF1A methylation (HR=2.471, 95% CI=1.125–5.428, p=0.024) and OR (HR=2.678, 95% CI=1.085–6.610, p=0.033) could be used to independently predict outcomes of patients with stage II and III CRC who were treated with oxaliplatin-based chemotherapy (Table 5).
Table 5.
Prognostic significance of RASSF1A methylation.
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| RASSF1A methylation (yes vs. no) | 2.782 | 1.273–6.080 | 0.010 | 2.471 | 1.125–5.428 | 0.024 |
| Gender (male vs. female) | 1.543 | 0.742–3.205 | 0.245 | – | – | – |
| Tumor size (≥5 cm vs. <5 cm) | 0.745 | 0.361–1.537 | 0.425 | – | – | – |
| Tumor site (rectum vs. colon) | 0.880 | 0.426–1.822 | 0.732 | – | – | – |
| Tumor stage (III vs. II) | 1.557 | 0.758–3.200 | 0.228/ | – | – | – |
| Pathological differentiation (poor vs. well+moderate) | 1.998 | 0.949–4.207 | 0.068 | – | – | – |
| OR (CR+PR vs. SD+PD) | 3.035 | 1.237–7.444 | 0.015 | 2.678 | 1.085–6.610 | 0.033 |
OR – objective response; ‘–’ – no available data.
Discussion
CRC is a frequently diagnosed malignancy worldwide. With the wide application of abdominal computerized tomography imaging (CTi) and colonoscopies, the detection rate of abdominal malignant diseases has an increasing trend [16]. Treatment still remains a great challenge for CRC patients in clinical setting, due to its unclear pathogenesis. With the development of molecular techniques, various disease-related specific genes have been identified in previous studies, which might be helpful for diagnosis and treatment. For instance, Isik et al. showed that MMP-1, -2, -9, and -13 expression levels were significantly associated with the formation of inguinal hernia, suggesting their potential as therapeutic targets [17]. Gene expression may be controlled by its methylation status in the promoter region. Growing evidence suggests that the methylation status of some specific genes plays a crucial role in tumor progression and treatment, such as tumor progression and treatment of colorectal tumors. Moreover, more frequent LPHN2 methylation has been detected in gastrointestinal cancer patients than in normal people, and the aberrant methylation was significantly correlated with sensitivity and cytotoxicity of cisplatin treatments [18]. In addition, epigenetic silencing of some key genes caused by aberrant methylation may influence prognosis of patients with glioblastoma [19]. In this study, we investigated the effects of RASSF1A methylation on outcomes of patients with stage II and III CRC treated with oxaliplatin-based chemotherapy.
In our study, RASSF1A methylation was more frequently detected in blood samples collected from CRC patients compared with healthy controls. In addition, In addition, the methylation status was obviously correlated with tumor stage and pathological differentiation, implying that promoter methylation of RASSF1A may affect tumor progression of CRC. This conclusion was in accordance with previous studies. A meta-analysis conducted by Wang et al. indicated that RASSF1A methylation was associated with clinical characteristics of patients with CRC among Asians. Sinha et al. reported that RASSF1A methylation could predict tumor stage and metastasis in adenocarcinomatous sporadic colorectal cancer in an Indian population [20]. Therefore, promoter methylation of RASSF1A may be an optimal indicator for tumor progression in CRC patients.
Previous studies have indicated that promoter methylation of RASSF1A could influence the efficacy of chemotherapy in various cancers. In a study by Gil et al., RASSF1A methylation was reported to be an important modulating factor for the efficacy of docetaxel-based chemotherapy in breast cancer [21]. Xie et al. suggested that patients with methylation in the promoter region of RASSF1A had a lower response rate to cisplatin-based neoadjuvant therapy than those without methylation [22]. A study carried out by Metei et al. proved that RASSF1A methylation could significantly influence PFS for ovarian cancer patients after decitabine treatment [13]. In the present study, we also detected effects of RASSF1A methylation on tumor response to oxaliplatin-based chemotherapy. Our results suggested that patients with methylation had a lower OR than those without methylation, and the methylation rate was significantly decreased after chemotherapy. These results could be explained by that aberrant methylation of RASSF1A, which might lead to suppression of protein expression, which could further influence the chemotherapy effects [22].
Furthermore, we assessed the effects of RASSF1A methylation on PFS of patients with stage II and III CRC treated with oxaliplatin-based chemotherapy. The results indicated that patients with methylation had a shorter progression-free time than those without methylation, and methylation status of RASSF1A in the promoter region was significantly correlated with overall survival. Cox regression analysis indicated that methylation status of RASSF1A was independently associated with prognosis of patients with stage II and III CRC treated with oxaliplatin-based chemotherapy. The prognostic significance of RASSF1A methylation for cancers has been reported in various cancers, such as hepatocellular carcinoma, prostate cancer, breast cancer, and Wilms tumor [23–26]. In addition, the prognostic value of RASSF1A methylation for cancer patients receiving chemotherapy has been proven. In a study by Fischer et al., RASSF1A promoter methylation was reported to significantly affect outcomes for patients with non-small cell lung cancer treated with gemcitabine [27]. A study carried out by Honda et al. indicated that RASSF1A methylation might be a promising biomarker for chemotherapeutic outcomes of patients with hepatoblastoma [29]. These conclusions were consistent with our results. However, no biologically relevant mechanism has yet been revealed.
Conclusions
Our results demonstrated that RASSF1A methylation frequency was higher in patients with stage II and III CRC than in the healthy controls, the methylation status was correlated with tumor stage and pathological differentiation, and RASSF1A methylation could significantly influence sensitivity to oxaliplatin-based chemotherapy for CRC patients. Therefore, we concluded that methylation of RASSF1A in the promoter region was independently associated with prognosis in CRC patients treated with oxaliplatin-based chemotherapy, and aberrant RASSF1A methylation might be a promising target for improving chemotherapeutic effects.
Footnotes
Source of support: Departmental sources
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Amirkhah R, Farazmand A, Irfan-Maqsood M, et al. The role of microRNAs in the resistance to colorectal cancer treatments. Cell Mol Biol (Noisy-le-grand) 2015;61(6):17–23. [PubMed] [Google Scholar]
- 4.Yu X, Li Z, Yu J, et al. MicroRNAs predict and modulate responses to chemotherapy in colorectal cancer. Cell Prolif. 2015;48(5):503–10. doi: 10.1111/cpr.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Cutsem E, Rivera F, Berry S, et al. Safety and efficacy of first-line bevacizumab with FOLFOX, XELOX, FOLFIRI and fluoropyrimidines in metastatic colorectal cancer: The BEAT study. Ann Oncol. 2009;20(11):1842–47. doi: 10.1093/annonc/mdp233. [DOI] [PubMed] [Google Scholar]
- 6.Deng Y. [Molecular phenotypes of colorectal cancer is critical in clinical individual treatment]. Zhonghua Wei Chang Wai Ke Za Zhi. 2015;18(10):994–97. [in Chinese] [PubMed] [Google Scholar]
- 7.Teschendorff AE, Lee SH, Jones A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015;7(1):108. doi: 10.1186/s13073-015-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfütze K, Benner A, Hoffmeister M, et al. Methylation status at HYAL2 predicts overall and progression-free survival of colon cancer patients under 5-FU chemotherapy. Genomics. 2015;106(6):348–54. doi: 10.1016/j.ygeno.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Pfeifer GP, Dammann R. Methylation of the tumor suppressor gene RASSF1A in human tumors. Biochemistry (Mosc) 2005;70(5):576–83. doi: 10.1007/s10541-005-0151-y. [DOI] [PubMed] [Google Scholar]
- 10.Cao D, Chen Y, Tang Y, et al. Loss of RASSF1A expression in colorectal cancer and its association with K-ras status. Biomed Res Int. 2013;2013:976765. doi: 10.1155/2013/976765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei H, Fang N, Guo L, et al. [Meta-analysis of the association between RASSF1A gene promoter methylation and non-small cell lung cancer]. Zhongguo Fei Ai Za Zhi. 2015;18(7):443–50. doi: 10.3779/j.issn.1009-3419.2015.07.09. [in Chinese] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Z, Ma K, Sun X, et al. Methylation of RASSF1A gene promoter and the correlation with DNMT1 expression that may contribute to esophageal squamous cell carcinoma. World J Surg Oncol. 2015;13:141. doi: 10.1186/s12957-015-0557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matei D, Fang F, Shen C, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72(9):2197–205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinn RL, Pruitt K, Eguchi S, et al. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67(1):194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- 15.Lin Q, Geng J, Ma K, et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol. 2009;135(12):1675–84. doi: 10.1007/s00432-009-0614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isik A, Soyturk M, Süleyman S, et al. Correlation of bowel wall thickening seen using computerized tomography with colonoscopies: A preliminary study. Surg Laparosc Endosc Percutan Tech. :2017. doi: 10.1097/SLE.0000000000000389. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Isik A, Gursul C, Peker K, et al. Metalloproteinases and their inhibitors in patients with inguinal hernia. World J Surg. 2017;41(5):1259–66. doi: 10.1007/s00268-016-3858-6. [DOI] [PubMed] [Google Scholar]
- 18.Jeon MS, Song SH, Yun J, et al. Aberrant epigenetic modifications of LPHN2 function as a potential cisplatin-specific biomarker for human gastrointestinal cancer. Cancer Res Treat. 2016;48(2):676–86. doi: 10.4143/crt.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma J, Hou X, Li M, et al. Genome-wide methylation profiling reveals new biomarkers for prognosis prediction of glioblastoma. J Cancer Res Ther. 2015;11(Suppl 2):C212–15. doi: 10.4103/0973-1482.168188. [DOI] [PubMed] [Google Scholar]
- 20.Sinha R, Hussain S, Mehrotra R, et al. Kras gene mutation and RASSF1A, FHIT and MGMT gene promoter hypermethylation: Indicators of tumor staging and metastasis in adenocarcinomatous sporadic colorectal cancer in Indian population. PLoS One. 2013;8(4):e60142. doi: 10.1371/journal.pone.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gil EY, Jo UH, Jeong H, et al. Promoter methylation of RASSF1A modulates the effect of the microtubule-targeting agent docetaxel in breast cancer. Int J Oncol. 2012;41(2):611–20. doi: 10.3892/ijo.2012.1470. [DOI] [PubMed] [Google Scholar]
- 22.Xie G, Hu C, Huang M. [Methylation status of RASSF1A and clinical efficacy of neoadjuvant therapy in patients with advanced epithelial ovarian cancer]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011;36(7):631–33. doi: 10.3969/j.issn.1672-7347.2011.07.008. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 23.Lin JC, Wu YC, Wu CC, et al. DNA methylation markers and serum alpha-fetoprotein level are prognostic factors in hepatocellular carcinoma. Ann Hepatol. 2015;14(4):494–504. [PubMed] [Google Scholar]
- 24.Ge YZ, Xu LW, Jia RP, et al. The association between RASSF1A promoter methylation and prostate cancer: Evidence from 19 published studies. Tumour Biol. 2014;35(4):3881–90. doi: 10.1007/s13277-013-1515-3. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Cui L, Chen WD, et al. The prognostic role of RASSF1A promoter methylation in breast cancer: A meta-analysis of published data. PLoS One. 2012;7(5):e36780. doi: 10.1371/journal.pone.0036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohshima J, Haruta M, Fujiwara Y, et al. Methylation of the RASSF1A promoter is predictive of poor outcome among patients with Wilms tumor. Pediatr Blood Cancer. 2012;59(3):499–505. doi: 10.1002/pbc.24093. [DOI] [PubMed] [Google Scholar]
- 27.Fischer JR, Ohnmacht U, Rieger N, et al. Prognostic significance of RASSF1A promoter methylation on survival of non-small cell lung cancer patients treated with gemcitabine. Lung Cancer. 2007;56(1):115–23. doi: 10.1016/j.lungcan.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Honda S, Haruta M, Sugawara W, et al. The methylation status of RASSF1A promoter predicts responsiveness to chemotherapy and eventual cure in hepatoblastoma patients. Int J Cancer. 2008;123(5):1117–25. doi: 10.1002/ijc.23613. [DOI] [PubMed] [Google Scholar]


