Abstract
Background:
Overexpression of proangiogenic vascular endothelial growth factor A family VEGFAxxx is associated with tumor growth and metastasis. The role of the alternatively spliced antiangiogenic family VEGFAxxxb is poorly investigated in head and neck squamous cell carcinomas (HNSCCs). The antiangiogenic isoform binds to bevacizumab and its expression level could influence the treatment response and progression-free survival. In this study, the relative expression of VEGFAxxx and VEGFA165b isoforms and splicing regulatory factors genes was investigated in a series of HNSCCs.
Methods:
VEGFAxxx, VEGFA165b, SRSF6, SRSF5, SRSF1 and SRPK1 gene expression was quantified by quantitative real time PCR in 53 tissue samples obtained by surgery from HNSCC patients. Protein expression was evaluated by immunohistochemistry.
Results:
VEGFAxxx and VEGFA165b were overexpressed in HNSCCs. Elevated protein expression was also confirmed. However, VEGFA isoforms demonstrated differential expression according to anatomical sites. VEGFAxxx was overexpressed in pharyngeal tumors while the VEGFA165b isoform was up-regulated in oral tumors. The VEGFA165b isoform was also positively correlated with expression of the splicing regulatory genes SRSF1, SRSF6 and SRSF5.
Conclusions:
We concluded that VEGFAxxx and VEGFA165b isoforms are overexpressed in HNSCCs and the splicing regulatory factors SRSF1, SRSF6, SRSF5 and SRPK1 may contribute to alternative splicing of the VEGFA gene. The findings for the differential expression of the antiangiogenic isoform in HNSCCs could facilitate effective therapeutic strategies for the management of these tumors.
Keywords: Head and neck cancer, VEGFA, alternative splicing, pathologic angiogenesis, gene expression
Introduction
Angiogenesis is an essential mechanism for tumor growth and results from genetic and/or environmental alterations. The formation of new vessels is a complex process that involves receptors, cytokines, enzymes and growth factors (Carmeliet et al., 2002), such as the vascular endothelial growth factor A (VEGFA). In vitro and in vivo experiments have shown that increased expression of VEGFA is associated with tumor growth and metastasis (Harper et al., 2008).
VEGFA gene consists of eight exons and seven introns with approximately 14 kilobases in length (Houck et al., 1991, Arcondéguy et al., 2013). Alternative splicing of exon 8 from pre-mRNA originates two families of protein: VEGFAxxx and VEGFAxxxb, where xxx is the number of amino acids encoded. VEGFA isoforms are generally co-expressed in all tissues and VEGF165 and VEGFA165b are the main isoforms. VEGFA165b protein has 96% homology with VEGFA165, but it presents different C-terminus amino acid sequence (Eswarappa and Fox, 2015). Studies have shown that VEGFA165b binds to VEGFR2 with the same affinity as VEGF165, but does not activate it completely resulting in alteration of the downstream signaling (Cébe-Suarez et al., 2006, Biselli-Chicote et al., 2012). This alternative isoform was associated with inhibition of endothelial proliferation, migration and vasodilation (Bates et al., 2002), and can reduce the physiological angiogenesis and tumor growth (Qiu et al., 2007), because the isoforms compete for the receptor ligation (Woolard et al., 2004). Tumors expressing VEGF165b grow significantly slowly than tumors expressing VEGF165 (Woolard et al., 2004), suggesting that a switch in splicing from VEGF165 to VEGF165b can inhibit the tumor growth.
The regulation of VEGFAxxx and VEGFAxxxb splicing remains unclear. However, it is known that exon splicing depends on the balance of the activity of serine-rich (SR) proteins, such as SRSF1, SRSF5 and SRSF6, in determining the C-terminus region. The splicing factors SRSF1 and SRSF5 support the proximal splicing site selection of the VEGFA pre-mRNA, leading to VEGFAxxx expression, while SRSF6 supports the distal splicing site selection promoting VEGFAxxxb expression (Nowak et al., 2008). Other important regulators of splicing include some SR protein kinases, such as SRPK1, that phosphorylate serine-arginine domains present in splicing factors (Manley et al., 1996). SR proteins can be regulated both directly by SR kinase proteins, such as Clk1 and SRPKs, and indirectly by mitogen-activated protein kinase (MAPK) and protein kinase C (PKC). SRPKs phosphorylate SRSF1 favoring the proximal splicing and Clk1 results in phosphorylation of SRSF1, SRSF6 and SRSF5 (Prasad et al., 1999, Lai et al., 2003).
Since most previous studies on HNSCC have not distinguished between proangiogenic and antiangiogenic isoforms and there are insufficient data for understanding the role of VEGF165b isoform in HNSCC, mainly in oral tumor, we aimed to evaluate the expression of VEGFAxxx and VEGFA165b isoforms in HNSCC, and investigate alternative splicing of VEGFA in this tumor type.
Materials and Methods
Patients and tissue samples
The study protocol was approved by Institutional Ethics Committee. Informed consent was obtained from the participants of the study. Fresh tissue from 52 HNSCC and 26 adjacent non-tumor tissues were collected from 1998 to 2000 at the Head and Neck Surgery Service, Arnaldo Vieira de Carvalho Cancer Institute, Sao Paulo, Brazil, and from 2007 to 2012 at the Otolaryngology and Head and Neck Surgery Service, Hospital de Base / FAMERP, Sao Jose do Rio Preto, Brazil. Samples were immediately frozen in liquid nitrogen and stored at -80ºC until processing. Microdissection of the samples was performed in Pathology Laboratory at Hospital de Base. Representative formalin-fixed block from 26 tumor samples and 15 adjacent non-tumor tissues were selected for immunohistochemical staining.
Median age of the patients was 58±11.56 years. Eighty three percent of the patients were smokers, and 68% were alcohol consumers. The study cohort consisted mostly of male patients (72.1%) and the most frequent primary tumor site was the oral cavity (51%) followed by larynx (26%) and pharynx (23%). Twenty-two percent of the patients performed radio and/or chemotherapy. Despite the relationship of Human Papillomavirus (HPV) infection with HNSCC development in some populations, HPV status was not evaluated in our study because the low prevalence of HPV infection in HNSCC in Brazilian population (Ribeiro et al., 2011). Tumor staging was performed according to the 7th edition of the TNM staging system (Sobin et al., 2010). T1N0 tumors were classified as stage I; T1N1 and T2N0-1 tumors as stage II; T3N0-1 and T1-3N2 tumors as stage III; and T4N0-3, T1-3N3 and T1-4N0-3M1 as stage IV (Fleming et al., 1997). For statistical analyses stage I and II tumors were grouped and classified as non-advanced tumors and stage III and IV tumors were classified as advanced tumors. Based on histopathological examination of the surgical specimen, 33% presented nodal metastasis and 56% of the patients had advanced primary tumors.
Quantitative real time PCR
Total RNA was extracted with TRIzol Reagent (Ambion, TX) following manufacturer’s instructions. Two micrograms of total RNA were reverse-transcribed using RT-PCR kit (Applied Biosystems, CA). VEGFAxxx and VEGFA165b expression in HNSCC and adjacent non-tumor tissues was evaluated using primers that distinguish both families of VEGFA isoform. Primers and probe were designed using Primer Express v.3.0 software (Applied Biosystems, CA) using the VEGFA complementary DNA (cDNA) sequence (GenBank: NM_001171623.1). Primer set and probe used for detection of VEGFAxxx family amplify VEGFA148, VEGFA165, VEGFA183, VEGFA189 and VEGFA206 isoforms: forward primer 5’ AACACAGACTCGCGTTGCAA 3’, reverse primer 5’ CGCCTCGGCTTGTCACAT 3’ and TaqMan MGB 6-FAM probe 5’ AGCTTGAGTTAAACGAAC 3’. Reactions were performed in triplicate in 96 wells plate using 100ng of cDNA, 100 nM of forward primer, 300 nM of reverse primer and 250 nM of probe (Applied Biosystems, CA). The reactions were performed on StepOne Plus Real-Time PCR System (Applied Biosystems, CA) and cycled following manufacturer’s instructions.
Foward primer and probe used for VEGFAxxx were also used for VEGFA165b (GenBank:NM_001171629.1); however a specific reverse primer to the end of the exon 7 and beginning of the exon 8b was used to detect only the VEGFA165b isoform (5’ TTCCTGGTGAGAGATCTGCAAGTA 3’).
The reactions for VEGFA165b quantification were performed using 100 ng of cDNA, 900 nM of forward and reverse primers and 250 nM of probe (Applied Biosystems, CA). Primers and probes sequences for SRSF6, SRSF5, SRSF1 and SRPK1 genes were analyzed using TaqMan Gene Expression Assay (Applied Biosystems, CA).
Raw qPCR data were calculated by StepOne software version 2.0 (Applied Biosystems, CA) after manual adjustment of the basal fluorescence signal and the threshold. Relative gene expression from tumor samples was analyzed using non-tumor samples as calibrator group and TBP and RPLPO as reference genes (Applied Biosystems, CA). Fold change (FC) was calculated by ddCt algorithm.
Immunohistochemistry
Three-micrometer thick sections of formalin-fixed paraffin-embedded tissues were cut and mounted onto silanized glass slides. Sections were dewaxed, rehydrated, washed in distilled water and the antigenic retrieval was performed in microwave with sodium citrate (pH 6.0) heated at 97 ºC for 20 minutes. Reactions were performed with REVEAL Biotin-Free Detection System (Spring Bioscience, CA) following manufacturer’s instructions. Incubation with pan-VEGF antibody (MAB293, R and D Systems, MN) diluted 1:50 or VEGFA165b antibody (MAB3045, R and D Systems, MN) diluted 1:50 was performed overnight at 4ºC. Subsequently, the slides were washed with phosphate-buffered saline for 5 minutes and incubated with HRP conjugate and diaminobenzidine (DAB) as chromogen. The development time in DAB solution was 10 minutes. The slides were counterstained with Harris’s hematoxylin for 40 seconds. Sections from mammary tumor tissue were used as positive control, and slides without primary antibody treatment were used as negative control. Images were obtained with a Camera Retiga 4000R (QImaging, CA) attached to an Olympus microscope (Model BX53, NY), and captured, averaged, and digitized using Image-Pro Plus 7.01 software (Media Cybernetics, USA). Illumination exposure was uniformly maintained and regularly checked in order to prevent any distortion of measurements (immunopositive area, gray level) among the samples. After capture, the image was analyzed using the software ImageJ, version 1.43m (Bethesda, USA) (Jensen et al., 2013). After selection of the squamous cell region, the software calculated the immunopositive area by counting all pixels with gray intensity equal or superior to the threshold of staining. The threshold was defined for each protein evaluated, based on the mean immunopositivity of all control cases and taking into account the nuclear hematoxylin staining. Results were shown as a percentage of immunopositive area/total area. Three areas from each image were evaluated and the mean of percentage of immunopositivity was obtained for statistical analyses.
Statistical analyses
Statistical analyses were performed using the GraphPad Prism software version 5.01 and StatsDirect software version 3.0.171. Wilcoxon Signed Rank Test was performed to analyze gene expression in tumor samples. Analyses of gene expression according the tumor progression was performed by Mann-Whitney Test. Comparison of gene expression among the primary tumor sites was evaluated by Kruskal-Wallis Test followed by Dunn’s Multiple Comparison Post Test. Spearman correlation was used to evaluate the correlation between VEGFA isoforms and splicing factor expression in tumors. Immunohistochemistry data were evaluated by Mann-Whitney Test or Kruskal-Wallis Test. Survival analysis was performed using the Kaplan-Meier curve and Log-rank test. Results with a p value < 0.05 were considered statistically significant.
Results
Overexpression of VEGFAxxx and VEGF165b in HNSCC
VEGFxxx transcripts was detected in 51 samples and VEGF165b transcripts was detected in 49 samples. Overexpression of VEGFAxxx (p < 0.0001) and VEGFA165b (p < 0.0001) was observed in tumor samples as compared to adjacent non-tumor tissues (Table 1 and Figure 1). Radio and/or chemotherapy did not change VEGFAxxx and VEGFA165b expression in the analyzed casuistic (data not shown). Corroborating gene expression data, immunohistochemistry analyses showed that VEGFAxxx and VEGFA165b proteins are significantly (p <0.0001) overexpressed in tumor samples (75.11% and 63.89% of the stained area, respectively) as compared to non-tumor tissues (33.64% and 18.16%) (Figure 2).
Table 1.
Association Between Gene Expression and Clinicopathologic Features of the HNSCC Patients
| Variables | VEGFAxxx | VEGFA165b | SRSF1 | SRSF6 | SRSF5 | SRPK1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | FC* | p Value | N | FC | p Value | N | FC | p Value | N | FC | p Value | N | FC | p Value | N | FC | p Value | |
| Total Tumors | 51 | 1.50 | <0.0001 | 49 | 3.06 | <0.0001 | 52 | 0.86 | 0.293 | 52 | 0.68 | 0.305 | 52 | 0.75 | 0.545 | 52 | 1.02 | 0.3171 |
| Primary site | ||||||||||||||||||
| Oral | 25 | 1.30 | 0.038 | 24 | 2.9 | 0.02 | 26 | 0.31 | ** | 26 | 0.61 | 0.169 | 27 | 0.79 | 0.033 | 27 | 0.65 | <0.0001 |
| Pharynx | 12 | 4.93 | 12 | 1.55 | - | - | 11 | 0.93 | 11 | 1.45 | 13 | 12.05 | ||||||
| Larynx | 14 | 2.30 | 14 | 1.39 | 14 | 0.20 | 13 | 0.44 | 13 | 2.0 | 14 | 2.96 | ||||||
| TNM stage | ||||||||||||||||||
| I-II (non-advanced) | 14 | 1.47 | 0.214 | 15 | 1.45 | 0.153 | 16 | 0.97 | 0.412 | 16 | 0.75 | 0.034 | 16 | 0.86 | 0.439 | 16 | 0.81 | 0.745 |
| III-IV (advanced) | 31 | 2.40 | 30 | 4.50 | 30 | 0.82 | 30 | 0.62 | 30 | 0.55 | 30 | 1.12 | ||||||
| Tumor extent | ||||||||||||||||||
| T1-T2 | 15 | 1.48 | 0.800 | 16 | 1.55 | 0.188 | 17 | 0.82 | 0.601 | 17 | 0.75 | 0.108 | 17 | 0.58 | 0.543 | 17 | 0.81 | 0.406 |
| T3-T4 | 29 | 1.49 | 29 | 4.50 | 29 | 0.91 | 29 | 0.62 | 29 | 0.85 | 29 | 1.13 | ||||||
| Nodal metastasis | ||||||||||||||||||
| N0 | 26 | 1.49 | 0.765 | 27 | 1.76 | 0.492 | 28 | 0.90 | 0.220 | 18 | 0.73 | 0.442 | 28 | 1.0 | 0.229 | 28 | 0.95 | 0.721 |
| N+ | 19 | 1.50 | 18 | 3.88 | 18 | 0.5 | 28 | 0.57 | 18 | 0.51 | 18 | 1.12 | ||||||
FC, fold change.
Median
No amplification of the calibrator sample;. Significant p Value in bold.
Figure 1.
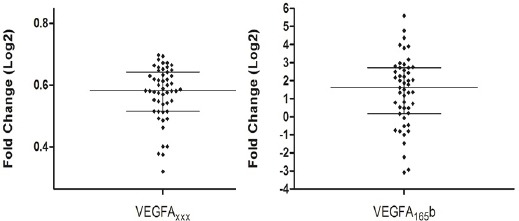
VEGFAxxx and VEGFA165b Expression in HNSCC as Compared to Non-Tumor Tissues. Fold change were Log2 transformed (y-axis). VEGFAxxx and VEGFA165b were overexpressed in tumors (Wilcoxon Signed Rank Test: p < 0.0001). The bars represent median with interquartile variation (25th percentile and 75th percentile). Calibrator (non-tumor tissues) log RQ = 0.
Figure 2.
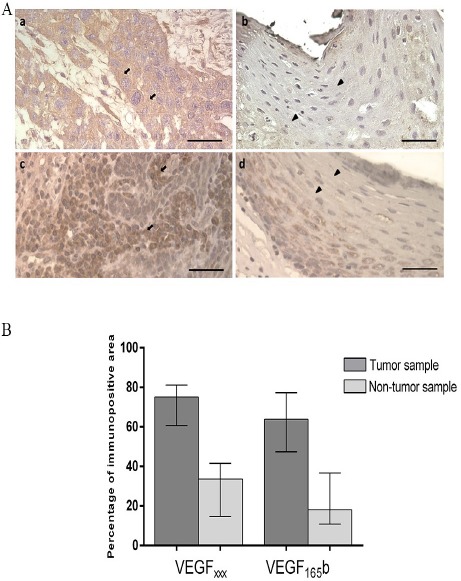
A. Representative Graphic of Immunolocalization of VEGFAxxxand VEGFA165b proteins in HNSCC and non-tumor tissue. A. Cytoplasmic immunostaining for VEGFAxxx and VEGFA165b in formalin-fixed paraffin-embedded sections of oral tumors (a and c, respectively) and non-tumor tissues (b and d, respectively). Arrows indicate strong staining in the cytoplasm of tumor samples and arrows head indicate weak staining in the cytoplasm of non-tumor samples. Bar = 1000µM. B. Comparison of VEGFA165b and VEGFAxxx immunopositive area in HNSSC and non-tumor tissue. Tumors presented increased VEGFAxxx and VEGFA165b immunoreactivity compared to non-tumor tissue (Mann-Whitney test, p <0.0001). The bars represent median with interquartile variation (25th percentile and 75th percentile).
Differential expression of VEGFA isoforms according to the anatomical sites
Analysis of expression in tumors compared to non-tumor tissues showed that VEGFAxxx was significantly overexpressed in pharynx tumors (p = 0.001). Overexpression of VEGFAxxx was also observed in larynx tumors (p = .035). VEGFAxxx expression differed significantly among the anatomical sites (p = 0.038), and the Post Test showed higher expression in the pharynx than oral tumors (Table 1 and Figure 3). VEGFA165b showed significantly higher expression only in oral tumors as compared to non-tumor tissues (p = 0.0005) and presented differential expression among the anatomical sites (p = 0.02); the Post Test showed higher expression in oral tumor than in larynx tumor (Table 1 and Figure 4). Regarding protein expression, there was no significant difference in expression of VEGFAxxx and VEGFA165b among the anatomical sites (p = 0.8473).
Figure 3.
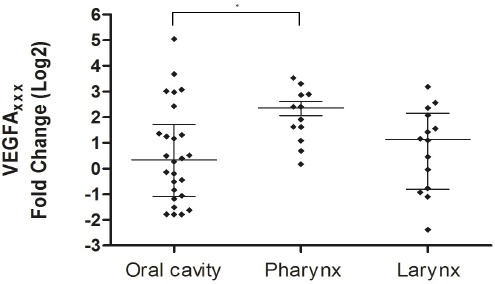
VEGFAxxx Expression According to the Anatomical Sites. Fold change were Log2 transformed (y-axis). The bars represent median with interquartile variation (25th percentile and 75th percentile). Calibrator (non-tumor tissues) log RQ = 0. *Statistically significant (Kruskal-Wallis test, p=0.038).
Figure 4.
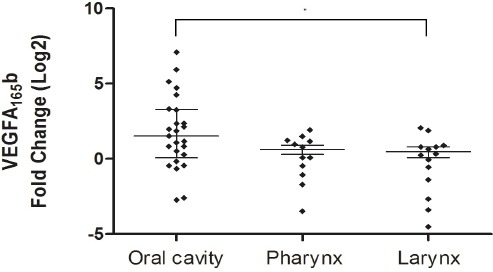
VEGFA165b expression according to the anatomical sites. Fold change were Log2 transformed (y-axis). The bars represent median with interquartile variation (25th percentile and 75th percentile). Calibrator (non-tumor tissues) log RQ = 0. *Statistically significant (Kruskal-Wallis test, p=0.02).
Expression of VEGFA isoforms was not associated with tumor progression
Information about TNM was possible for 47 patients. Comparison between tumors T1/T2 and T3/T4 showed no difference in VEGFAxxx and VEGFA165b expression (p = 0.800 and p = 0.188, respectively) (Table 1). VEGFAxxx and VEGFA165b (p = 0.765 and p = 0.492, respectively) did not differ between tumors grouped according to the presence of nodal metastasis (Table 1). Thirty one patients presented advanced tumors (stage III and IV) and 16 presented non-advanced tumors (stage I and II). There was no difference in gene (p = 0.214 for VEGFAxxx and p = 0.153 for VEGFA165b) and protein (p = 0.557 for VEGFAxxx and p = 0.103 for VEGFA165b) expression between advanced and non-advanced tumors.
Expression of SR splicing factors in HNSCC
SRSF1, SRSF5, SRSF6 and SRPK1 transcripts were detected in 52 tumor samples. SRSF5 presented higher expression in pharynx tumor as compared to non-tumor tissue (p = 0.003) and to the other anatomical sites (p = 0.033) (Table 1). SRPK1 was overexpressed in pharynx (p = 0.001) and larynx (p = 0.0009) tumors as compared to non-tumor tissue. SRPK1 presented differential expression among the anatomical sites (p < 0.0001), and the Post Test showed higher expression in pharynx and larynx tumors than in oral tumor (Table 1). SRSF6 expression was lower in advanced tumors as compared to non-advanced tumors (p = 0.0339) (Table 1).
Correlation between the expression of VEGFA isoforms and SR proteins
VEGF165b presented positive correlation with SRSF1, SRSF5 and SRSF6 expression in tumors (Table 2). No correlation was observed among SR factors and VEGFAxxx. Analysis of correlation between SR kinase SRPK1 and SR factors showed positive correlation with SRSF1 (r = 0.533, p < 0.0001), SRSF5 (r = 0.509, p = 0.002) and SRSF6 (r = 0.552, p < 0.0001) in tumor tissues.
Table 2.
Correlation among Splicing Factors and VEGFA Isoforms in HNSCC.
| Splicing factor | VEGFAxxx | VEGFA165b | ||
|---|---|---|---|---|
| ra | p Value | ra | p value | |
| SRSF1 | 0.081 | 0.566 | 0.387 | 0.005 |
| SRSF6 | -0.056 | 0.695 | 0.337 | 0.017 |
| SRSF5 | 0.193 | 0.17 | 0.444 | 0.001 |
| SRPK1 | -0.06 | 0.673 | 0.224 | 0.118 |
Survival Analysis
The prognostic role of VEGFAxxx and VEGFA165b was evaluated by Kaplan-Meier survival analysis. Disease-free survival was defined as the time from surgical resection of the primary tumor to the tumor recurrence. Overall survival was defined as the time from surgical resection to the death or last follow-up. Patients were followed for a period of 48 months (median) and a maximum of 101 months. Fold change (FC) above the median was considered high expression of VEGFAxxx (FC = 1.5) and VEGFA165b (FC = 3.06). The results showed that high expression of VEGFAxxx and VEGFA165b have no significant effect on disease-free survival or overall survival time (Table 3).
Table 3.
Analysis of the Prognostic Role of VEGFAxxx and VEGFA165b Expression in HNSCC
| 12-month disease-free survival (%) | p value | 12-month overall survival (%) | p value | |
|---|---|---|---|---|
| VEGFAxxx | ||||
| Overexpression | 75 | 0.963 | 100 | 0.902 |
| Down expression | 81 | 90 | ||
| VEGFA165b | ||||
| Overexpression | 75 | 0.687 | 100 | 0.91 |
| Down expression | 79 | 100 | ||
Discussion
In the present study, VEGFA isoforms were overexpressed in HNSCC tumor samples as compared to non-tumor tissue. Overexpression of VEGFA is associated with tumor growth and results in increased angiogenesis (Das et al., 2007). Anti-VEGFA drugs, as bevacizumab and sunitinib, are often used in the treatment of patients with cancer (Prager et al., 2010). VEGFA overexpression have been found in solid tumors, such as breast cancer (Schneider et al., 2005), colorectal tumor (Ferroni et al., 2006) and head and neck squamous cell carcinoma (Uehara et al., 2004, Jaiswal et al., 2011). For oral tumors, VEGFA expression was significantly associated with a poor prognosis (Uehara et al., 2004). In bladder cancer, VEGFA protein was positively correlated with the tumor progression (Yang et al., 2015).
In 2002, Bates and colleagues identified the VEGFA165b isoform in renal tissue and proposed an antiangiogenic role for this variant. Analyses in vitro have shown that the recombinant VEGFA165b and VEGFA121b proteins induced human umbilical vein endothelial cell (HUVEC) proliferation and VEGFR-2 phosphorylation (Catena et al., 2010). However, HUVEC proliferation was approximately 50% less stimulated by VEGFA121/165b in comparison to VEGFA165. According to the authors, both VEGFAxxx and VEGFAxxxb isoforms equally compete for binding to the receptor, although VEGFAxxxb isoform induces less effectively the angiogenesis. Our findings show overexpression of the antiangiogenic isoform VEGF165b in HNSCC. Individuals with high relative levels of VEGFA165b could not benefit with antiangiogenic treatment, once this isoform also binds bevacizumab, preventing the binding with the proangiogenic isoforms and inhibition of angiogenesis.
The expression of VEGFA isoforms was not associated with head and neck tumor progression in this study. Overexpression of total VEGFA in oral and pharynx advanced tumors (T3 and T4) has been observed in the presence of nodal metastasis (Boonkitticharoen et al., 2008). High expression of total VEGFA was associated with larger tumor size, tumor progression and metastasis in larynx (Sullu et al., 2010). However, regarding the antiangiogenic isoform, recent data have shown that VEGFxxxb was overexpressed in 97.3% of the pharynx and larynx tumor, independent of HPV status, but the expression was not associated with lymph node metastasis and survival (Wilkie et al., 2016). These findings showed overexpression of VEGFxxxb in HNSCC, although it does not seem a reliable prognostic biomarker for tumors with presence of nodal metastasis, corroborating our results.
Analyzing the VEGFA isoforms expression data according to the tumor primary sites, we observed that pharynx tumors presented higher VEGFAxxx expression, and oral tumors presented higher VEGFA165b expression, reflecting the differential expression of these isoforms among head and neck tumor sites. In this study, the overexpression of SRSF5 in the pharynx tumor is in accordance with the high expression of VEGFAxxx isoforms in this anatomical site. SRSF5 and SRSF1 are associated with the selection of proximal splicing site favoring the synthesis of VEGFAxxx. Although the SRSF1 overexpression was not observed in tumor samples in the present study, it is possible that the high availability of SRPK1 results in more efficient activation of SRSF1 protein. SRPK1 is associated with the proximal splicing site selection, and presented high expression in pharynx and larynx tumors, as well as VEGFAxxx. SRSF6 was down expressed in advanced tumors as compared to non-advanced tumors. SRSF6 promotes VEGF165b expression; however, other factors, such as TGFβ1 and IGF-1, could also play a role in the regulation of the VEGFA alternative splicing (Nowak et al., 2008; Slomiany et al., 2004).
Concerning the splicing regulation of VEGFA isoforms by SR proteins, positive correlation was observed between gene expression of VEGFA165b isoform and all splicing factors, SRSF1, SRSF6 and SRSF5, in the present study. On the other hand, no correlation was found between the expression of SR proteins and VEGFAxxx. Although the effect of these splicing factors has been proposed by in vivo studies (Nowak et al., 2008; Nowak et al., 2010) the interaction between SR proteins and their target RNA at a specific site is influenced by several and multiple determining factors, such as competition with other SR proteins (Pandit et al., 2013) or other RNA binding proteins, including heterogeneous ribonucleoprotein proteins, and RNA secondary structure (Long et al., 2009).
The expression of the SR kinase SRPK1 presented positive correlation with SRSF1, SRSF6 and SRSF5 in tumor tissues. These results suggest the co-expression of these splicing factors and the SR kinase SRPK1 and their involvement in VEGFA splicing. To date, it is known that SRPK1 phosphorylates SRSF1, but it can also phosphorylate other factors, including SRSF6. It is also known that other protein kinases like Clk1 are associated with the phosphorylation of SRSF1, SRSF6 and SRSF5. Further, SR proteins can be indirectly regulated by PKC and MAPK (Prasad et al., 1999; Lai et al. 2003). Thus, other factors may contribute to the expression of these proteins in tumor samples.
In our point of view, the main finding was the overexpression of VEGFA165b in HNSCC as compared to non-tumor tissue. Importantly, there are no published studies investigating the gene expression pattern of VEGFA165b isoform in HNSCC. These results are important for the knowledge of the biology of head and neck cancer and may contribute to directing more effective therapeutic strategies in the treatment of these tumors. In addition, it is important to note that anti-VEGFA therapies currently used in cancer treatment target both the pro- and antiangiogenic isoforms, which could prevent the antiangiogenic activity of VEGFAxxxb isoforms in head and neck cancer, since this isoform is up regulated in this tumor type. Considering these issues, the use of therapy targeting only VEGFAxxx could improve the treatment outcomes in HNSCC (Carter et al., 2015).
In summary, the results showed that VEGFAxxx and VEGFA165b are overexpressed in HNSCC development with possible contribution of SRSF1, SRSF6, SRSF5 and SRPK1 regulatory factors in alternative splicing of VEGFA gene. The findings contribute to the understanding of the role of VEGFA165b in cancer angiogenesis and increase the knowledge about the mechanism related to carcinogenesis development in head and neck.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors thank Fundação de Amparo à Pesquisa do Estado de São Paulo (Grant 2009/07985-7), Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and Hospital de Base / Sao Jose do Rio Preto Medical School Foundation (FUNFARME) for support.
References
- Arcondéguy T, Lacazette E, Millevoi S, Prats H, Touriol C. VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. 2013;41:7997–8010. doi: 10.1093/nar/gkt539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DO, Cui TG, Doughty JM, et al. VEGF165b an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 2002;62:4123–31. [PubMed] [Google Scholar]
- Biselli-Chicote PM, Oliveira AR, Pavarino EC, Goloni-Bertollo EM. VEGF gene alternative splicing: pro- and antiangiogenic isoforms in cancer. J Cancer Res Clin Oncol. 2012;138:363–70. doi: 10.1007/s00432-011-1073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonkitticharoen V, Kulapaditharom B, Leopairut J, et al. Vascular endothelial growth factor A and proliferation marker in prediction of lymph node metastasis in oral and pharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2008;134:1305–11. doi: 10.1001/archotol.134.12.1305. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- Carter JG, Gammons MV, Damodaran G, et al. The carboxyl terminus of VEGF-A is a potential target for anti-angiogenic therapy. Angiogenesis. 2015;18:23–30. doi: 10.1007/s10456-014-9444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catena R, Larzabal L, Larrayoz M, et al. VEGFxxxb and VEGFxxxb are weakly angiogenic isoforms of VEGF-A. Mol Cancer. 2010;9:320. doi: 10.1186/1476-4598-9-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cébe-Suarez S, Zehnder-Fjällman A, Ballmer-Hofer K. The role of VEGF receptors in angiogenesis;complex partnerships. Cell Mol Life Sci. 2006;63:601–15. doi: 10.1007/s00018-005-5426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Zhao Y, Sugiono M, et al. Differential expression of vascular endothelial growth factor 165b in transitional cell carcinoma of the bladder. Urol Oncol. 2007;25:317–21. doi: 10.1016/j.urolonc.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Eswarappa SM, Fox PL. Antiangiogenic VEGF-Ax: A New Participant in Tumor Angiogenesis. Cancer Res. 2015;75:2765–9. doi: 10.1158/0008-5472.CAN-14-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferroni P, Spila A, Martini F, et al. Prognostic value of vascular endothelial growth factor tumor tissue content of colorectal cancer. Oncology. 2006;69:145–53. doi: 10.1159/000087838. [DOI] [PubMed] [Google Scholar]
- Fleming I, Cooper J, Henson D. Head and neck sites. In: Fleming I, Cooper J, Henson D, editors. In “AJCC cancer staging manual”. Philadelphia: Lippincott-Raven; 1997. pp. 21–46. [Google Scholar]
- Harper SJ, Bates DO. VEGFA splicing: the key to antiangiogenic therapeutics? Nat Rev Cancer. 2008;8:880–7. doi: 10.1038/nrc2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck KA, Ferrara N, Winer J, et al. The vascular endothelial growth factor family: identification of a fourth molecular species and characterization of alternative splicing of RNA. Mol Endocrinol. 1991;5:1806–14. doi: 10.1210/mend-5-12-1806. [DOI] [PubMed] [Google Scholar]
- Jaiswal SG, Gadbail AR, Chaudhary MS, Jaiswal GR, Gawande M. Correlation of serum levels of vascular endothelial growth factor with TNM staging, histopathologic grading, and surgical therapy for oral squamous cell carcinoma. Quintessence Int. 2011;42:771–9. [PubMed] [Google Scholar]
- Jensen EC. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat Rec. 2013;296:378–81. doi: 10.1002/ar.22641. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Differential effects of hyperphosphorylation on splicing factor SRp55. Biochem J. 2003;371:937–45. doi: 10.1042/BJ20021827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–79. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Nowak DG, Amin EM, Rennel ES, et al. Regulation of vascular endothelial growth factor (VEGF) splicing from proangiogenic to antiangiogenic isoforms: a novel therapeutic strategy for angiogenesis. J Biol Chem. 2010;285:5532–40. doi: 10.1074/jbc.M109.074930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak DG, Woolard J, Amin EM, et al. Expression of pro- and antiangiogenic isoforms of VEGF is differentially regulated by splicing and growth factors. J Cell Sci. 2008;121:3487–95. doi: 10.1242/jcs.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, et al. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–35. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager GW, Lackner EM, Krauth MT, et al. Targeting of VEGF-dependent transendothelial migration of cancer cells by bevacizumab. Mol Oncol. 2010;4:150–60. doi: 10.1016/j.molonc.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Bevan H, Weeraperuma S, et al. Mammary alveolar development during lactation is inhibited by the endogenous antiangiogenic growth factor isoform, VEGF165b. FASEB J. 2007;22:1104–12. doi: 10.1096/fj.07-9718com. [DOI] [PubMed] [Google Scholar]
- Ribeiro KB, Levi JE, Pawlita M, et al. Low human papillomavirus prevalence in head and neck cancer: results from two large case-control studies in high-incidence regions. Int J Epidemiol. 2011;40:489–502. doi: 10.1093/ije/dyq249. [DOI] [PubMed] [Google Scholar]
- Schneider BP, Miller KD. Angiogenesis of breast cancer. J Clin Oncol. 2005;23:1782–90. doi: 10.1200/JCO.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Slomiany MG, Rosenzweig SA. IGF-1-induced VEGF and IGFBP-3 secretion correlates with increased HIF-1 alpha expression and activity in retinal pigment epithelial cell line D407. Invest Ophthalmol Vis Sci. 2004;45:2838–47. doi: 10.1167/iovs.03-0565. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Wittekind CH. Head and neck tumors. In: Sobin LH, Gospodarowicz MK, Wittekind Ch, editors. In “TNM classification of malignant tumors”. Berlin: Springer Verlag; 2010. pp. 22–45. [Google Scholar]
- Sullu Y, Gun S, Atmaca S, Karagoz F, Kandemir B. Poor prognostic clinicopathologic features correlate with VEGF expression but not with PTEN expression in squamous cell carcinoma of the larynx. Diagn Pathol. 2010;5:35. doi: 10.1186/1746-1596-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara M, Sano K, Ikeda H, et al. Expression of vascular endothelial growth factor and prognosis of oral squamous cell carcinoma. Oral Oncol. 2004;40:321–5. doi: 10.1016/j.oraloncology.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Wilkie MD, Emmett MS, Santosh S, et al. Relative expression of vascular endothelial growth factor isoforms in squamous cell carcinoma of the head and neck. Head Neck. 2016;38:775–81. doi: 10.1002/hed.23959. [DOI] [PubMed] [Google Scholar]
- Woolard J, Wang WY, Bevan HS, et al. VEGF165b an inhibitory vascular endothelial growth factor splice variant: mechanism of action, in vivo effect on angiogenesis and endogenous protein expression. Cancer Res. 2004;64:7822–35. doi: 10.1158/0008-5472.CAN-04-0934. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang Z, Guo Y, Wang Z. Correlation and significance of urinary soluble fas and vascular endothelial growth factor in bladder urothelial cancer. Dis Markers. 2015;2015:383509. doi: 10.1155/2015/383509. [DOI] [PMC free article] [PubMed] [Google Scholar]


