Abstract
Objective:
MicroRNAs (miRs) are class of small non-coding regulatory RNA aberrantly expressed in various types of malignancies including prostate cancer and serves as potential targets to develop new diagnostic and therapeutic strategies. In this quiet we investigated miRNAs expression profile in benign prostatic hyperplasia (BPH) and prostate cancer (PCa) tissue samples and correlated their expression with clinicopathological parameters.
Methodology:
The miRNAs expression profile as well as their validation has been done by using Microarray and RT-PCR, respectively. Additionally, we also tried to speculate microRNA-mRNA regulatory module through computational target predictions by using Targetscan, Miranda and MirWalk and obtained results were analysed through DAVID software.
Result:
We observed that miR-711 is significantly deregulated in BPH and PCa, compared to controls. The lower expression of miR-711 was found to be significantly associated with high Gleason score and metastatic disease. Furthermore, the computational target prediction analysis explored miR-711 association to various cancer cells signalling cascade key molecules associated with cancer cell survival.
Conclusion:
From our observations we suggest that miR-711 may play a critical role in PCa progression, regulation of various cancer cell survival signalling cascades and that it may be a valuable biomarker for prediction of metastatic disease and poor prognosis in PCa.
Keywords: MicroRNA, Prostate cancer, BPH and Biomarkers
Introduction
Prostate cancer (PCa) is second most prevalent and fifth leading cause of cancer related death among men worldwide (Stuopelytė et al., 2016) and it continues to be second leading cause of motility after lung cancer in western world (Siegel et al., 2016; Bray et al., 2008). However, according to population based cancer registries of India, PCa is second leading site of incidence among males belonging to metro cities (Jain S et al., 2014). Discovery of serum based prostate specific antigen (PSA) revolutionized prostate cancer diagnosis, although the success was not far reaching and evidenced several shortcomings. Major clinical issues addressed with PSA includes inaccuracy of this serum marker to distinguish between patients with and without PCa leading to over diagnosis and most importantly its levels not exactly corresponding to disease severity. Other diagnostic methods are Digital rectal examination (DRE), and trans-rectal ultrasound-guided biopsy (on basis of Gleason score) all of which are appropriate for early diagnosis but again fail to discriminate between pathological stages of PCa (Hugosson et al., 2010; Schröder et al., 2009). Therefore it is imperative to identify novel diagnostic biomarkers with high sensitivity and specificity and above all more precision for stage specific diagnosis. MicroRNAs (miRNAs) are small non-coding RNAs of approximately 22 nucleotides in length and are considered as powerful regulators of cellular gene expression (Jackson et al., 2007). Since their initial discovery, their involvement has been proved in almost all of the biological processes including cellular growth, development and proliferation, metabolism, differentiation and apoptosis (Xu L et al., 2014). MicroRNAs predominantly bind to 3´-untranlated regions (3´-UTR) and sometimes to 5´UTR, codons of the transcribed mRNAs through specific seed sequences. This binding eventually results in either translation repression of target mRNAs to protein or induces their exonuclease-mediated cleavage, thereby regulating the expression of target genes (Inui et al., 2010; Deng et al., 2008). Moreover, their deregulated expression is also linked to the onset and progression of many diseases including prostate cancer (Ambs et al., 2008). Misexpression of much microRNA has been reported to be associated with cancer progression, metastasis as well as chemoresistance and is therefore classified as tumor suppressor miRNAs or oncogeneic miRNAs (oncomiRs) (Ding et al., 2015). Depending upon their relative expression and biological importance, miRs are very much presumed to be valuable diagnostic, predictive and prognostic biomarkers in all cancer types (Trang et al., 2008; Lu et al., 2005). Recent studies have identified many novel miRs in human tissues which have uncertain their biological functions (Londin et al., 2015). On the basis of this it may be presumed that there are still many unidentified miRNAs which may have significant role in cancer etiology. Identification of these miRNAs and their targetome will help in characterizing their role in cancer initiation as well as progression from low grade tumors to high grade tumor.
In the present study we therefore, attempted to identify novel microRNAs in PCa through comprehensive profiling of benign prostate hyperplasia (BPH) and PCa tissue samples. Through this study we aimed to find out the putative miRNAs which may be responsible for transitions between early stage of PCa to more invasive stage and that may serve as stage specific biomarker for PCa progression.
Materials and Methods
Human prostate tissue samples(n=144, including PCa, BPH and adjacent Control) from BPH and PCa patients were collected from Department of Urology, King George’s Medical University, Lucknow with informed written consent. The adjacent tissue samples were also obtained from the same patient who served as control. The BPH tissue samples (N=35) were obtained from Transurethral resection of prostate (TURP) method, whereas PCa tissues (N=74) were recovered through transrectal ultrasound (TRUS) biopsy and channel TURP and adjacent control tissues (N=35). The tissue samples were collected in RNA later and stored at - 80°C for further analysis. Histopathological reports of the respective tissues were obtained from Department of Pathology, King George’s Medical University and Lucknow. In case of prostate cancer patients prostate specific antigen (PSA) levels in serum were evaluated. The histopathological grading was done according to Gleason score grading methods. On the basis of Gleason score, patients were divided in two groups. The patients with Gleason score (GS) 7 or below were considered as low prostate cancer. However, the patients with Gleason score greater than 7 were considered as high prostate cancer. For the evaluation of metastatic disease, patients went under for bone scan. Clinical information of the patients were also collected and maintained for statistical evaluation. The clinical characteristics for these patients are described in Table 1. The present study was approved by Ethics Committee, King George’s Medical University, Lucknow.
Table 1.
Clinicopathological Characteristics of Prostate Cancer Patients
| Clinical Characteristic’s | MicroArray Group (n=11) | RT-PCR Group (n=74) |
|---|---|---|
| Age | ||
| <60 | 3(27.27%) | 13 (17.56%) |
| ≥60 | 8(72.73%) | 61 (82.44%) |
| Smoking habits | ||
| Smokers | 7 (63.63) | 49 (66.21) |
| Non-smokers | 4 (36.57) | 25 (43.03) |
| Nodal status | ||
| N0 | 6(54.55%) | 41(55.41%) |
| N+ | 5(45.45%) | 33(44.59%) |
| Metastatic Disease | ||
| M0 | 6(54.55%) | 30(40.55%) |
| M1 | 5(45.45%) | 44(59.45%) |
| PSA Level (ng/mL) | ||
| Median | 3(27.27%) | 23 (31.09%) |
| High | 8(72.73%) | 51 (68.91%) |
| Gleason Score | ||
| 7 | 3(27.27%) | 31(41.89%) |
| ≥7 | 8(72.73%) | 43(58.11%) |
MicroArray analysis
MicroRNA expression profiling through microarray was carried out in 27 prostate tissue samples from out of 144 tissue samples. The tissue samples were randomly selected for miR expression profiling through microarray analysis. The demographic profile of patients is mentioned in Table 1. We used one pooled RNA sample in each group of human sample set for microarray analysis, [Control sample (n=3); 2 individuals and 1 pooled sample of 6 individual), BPH sample (n=3); 2 individuals and 1 pooled of 6 individual, PCa sample (n=6); 5 individuals and 1 pooled of 6 individual)].
For microarray profile, the total RNA from tissues samples were isolated and enriched by using mirVana miRNA Isolation kit from Ambion according to manufacturer’s recommendations with slight modification. One microgram of total RNA from each sample was labelled with the FlashTag Biotin RNA Labelling Kit for Affymetrix microRNA 4.0 platforms and array type. Hybridization, washing, and scanning of the slides were done according to Affymetrix’s recommendations. The data analysis of miRs expression profiling was extracted from the images, quantile-normalized, summarized (median polish) and log2-transformed with the miR QC tool software from Affymetrix. We shortlisted up and down regulated miRNAs (miR-363-3p, miR-329-5p, miR-711, miR-132-3p, miR-193b-3p, miR-183-5p, miR-423-3p, miR-502-3p, miR-1246 and miR-548x-3p) on the basis of Fold change values ≥ 2.0.
Real- time quantitative: For Specific MicroRNA validation through RT- PCR, Total RNA was isolated from the clinical BPH, PCa tissue and adjacent control samples by using the Mirvana microRNA isolation kit. We selected three miRNAs (miR-711 miR-1246 and miR-548x-3p) from microarray data which were differentially expressed gradually in decreasing order in prostate tissue. The quantitative real time PCR (qRT-PCR) expression analysis were done by using the SYBR Advantage qPCR premix kit (Cat no.639676) The sequence of reverse and forward primers for Real-Time PCR were listed in Table 2. First, 250 ng of total RNA was reverse transcribed by using the Mir-X miRNA first-stand synthesis kit. Subsequently, the reverse-transcribed miRNAs were used as templates in the qRT-PCR analysis by using the fast real-time PCR System (7500HT, ABI, Applied Biosystems). The reaction volume is 25µl per reaction where RT-PCR condition where denaturation 95ºC- 10 sec qPCR x40 cycles 95ºC-5 sec, 60ºC- 20 sec and dissociation curve 95ºC- 60 sec, 55ºC-30 sec and 95ºC-30 sec was performed at this standard condition. Both melting curve analysis and agarose gel run were used to confirm the specificity of the amplification reactions. For normalization, expression of the U6 was used as the internal control. The Cycle of threshold (Ct) value of each sample was calculated by using 2-dd CT method for fold change expression of miRs.
Table 2.
Real Time- PCR Primer Sequence Used for Expression Analysis
| S.No. | MicroRNA | Primer sequence for SYBR Green reactions | GC contained % | Tm oC |
|---|---|---|---|---|
| 1 | Mir-711 | GGA CCC AGG GAG AGA CGT AA | 60 | 62 |
| 2 | miR-1246 | CAU GGA UUU UUG GAG CAG G | 47 | 60 |
| 3 | miR-548x-3p | GTC AAA ACT GCA ATT ACT CGC | 43 | 60 |
Bioinformatics analysis of miR-711
Online target prediction software Targetscan, Mirwalk and Miranda were used to find specific target genes targeted by miR-711. To minimize the number of false positives result, we took only target genes identified by above software. The Gene Ontology (GO) analysis and Kyoto encyclopedia of genes and genomes (KEGG) Pathway enrichment analysis were done by using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov) by taking top 25% of targeted genes of miR-711 analysed by targetscan software. Predicted target genes were pasted in DAVID v6.7 web-based tool for annotation, visualization, integrated discovery and for functional analysis. We performed GO and KEGG pathway analysis of target genes with differential expression of miR-711. p-value< 0.05 was considered as significant.
Statistical Analysis:
All statistical analysis and graphs were performed with the SPSS 21 (SPSS, Inc., Chicago, IL, USA) and graph pad Prism 5 Software. For fold change expression >=2.0 has been taken as the cut off value. The summary measurements were expressed as mean ±SEM. Depending on the distributions of continuous variables the significance of associations was determined by one way ANOVA test and student t-test. All differences were considered positive if P<0.05.
Results
Demographics of the study
A total of 144 patients were enrolled in the present study and that included PCa, BPH and healthy adjacent controls. The pathological diagnosis of PCa is shown in (Table 1) Mean age of PCa patients was 63.9 years (range, 45-75). 49 (66.21%) patients were smokers while 25 (33.03%) of patients were found to be non-smokers. Low PSA level was present in 31 % of samples, whereas 69 % had high PSA level (Table 1).
MicroRNA expression profiling in prostate tissue using microarray
To identify differentially expressed miRs, we performed comprehensive miRNA microarray analysis in prostate tissue samples. Among the unique 667 human miRNAs, we found 212 miRNAs to be differentially expressed in BPH and PCa as compared to adjacent control. The Affymetrix TAC software was used for data analysis by using default settings and one way ANOVA method was employed for assessing statistical significance between two groups. The p-value less than 0.05 was considered statistically significant and fold change >=2.0 was taken as the cut off value. We found ten novel microRNAs (miR-363-3p, miR-329-5p, miR-711, miR-132-3p, miR-193b-3p, miR-183-5p, miR-423-3p, miR-502-3p, miR-1246 and miR-548x-3p) differentially expressed in PCa tissue samples as compared to BPH and control (Figure 1). To the best of our knowledge these miRNAs have not been previously described in PCa and therefore gained our attention for further detailed analysis. Out of these ten we selected three miRNAs viz, miR-711, miR-1246 and miR-548x-3p with most significant fold change for validation through qRT-PCR.
Figure 1.
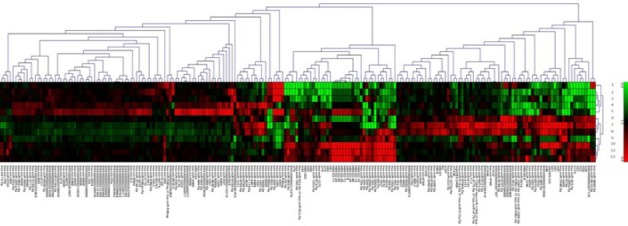
Hierarchical Clustering (HCL) Analysis. The above HCL expression image indicates that the experiment from the same group has a similar expression profile. Red colour shows over-expressed miRNAs(>0) &green colour shows under-expressed miRNAs(<0). (The HCL heat map image has been generated on the basis of log2 normalized intensity value). HCL analysis was performed in control, BPH, Prostate cancer (PCa) where, 1 (BPH pooled), 2,3 (BPH individual), 4, 5,9,10,11(PCa Individual),12 (PCa pooled), 6,8 (Control Individual), 7(Control pooled).
Validation of selected miRs by Real Time PCR
To validate our microarray findings we evaluated prostate tissue samples of BPH (n= 35), PCa (n=74) and control (n=35) for validation of miR-711, miR-1246 and miR-548x-3p expression through qRT-PCR. Among the three microRNAs we found only miR-711 to follow the trends of our microarray data. We observed that miR-711 was significantly down regulated in BPH and PCa tissue as compared to adjacent normal tissue (p= 0.01 and 0.0001 respectively, Figure 2A). The overall miR-711 expression level was approximately -1.777 ± 0.3783 fold in BPH and -6.042 ± 0.3497 fold in PCa as compared with control which shows consistency to our microarray analysed data. However, we did not observe any significant change in expression pattern of miRNA miR-1246 and miR-548x-3p as compared to control (p=0.7 and 0.15 respectively).
Figure 2.
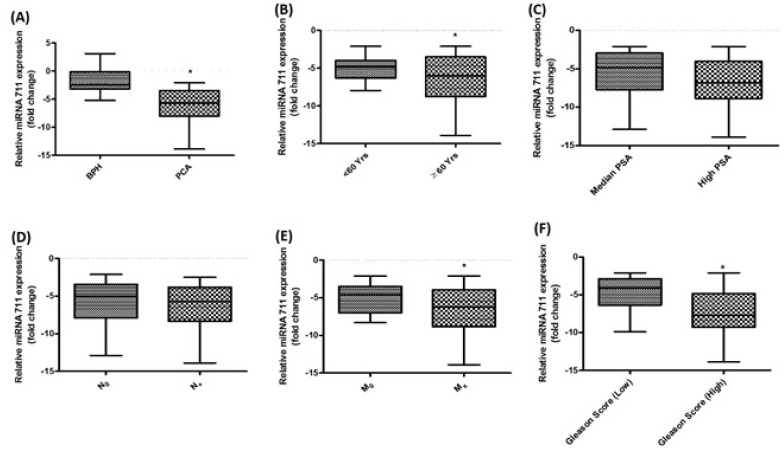
Relative Expression Levels of miR-711 in Prostate Cancer. (A) Differential expression in BPH and PCa (B) Differential expression in <60 years and >60years of PCa (C) Differential expression in Median and high PSA (D) Differential expression in Lymph Node –ve (N0) and in Lymph Node +ve (E) Differential expression in metastasis and non- metastasis(F) Differential expression in low Gleason score vs. high Gleason in PCa.
Correlation of deregulated miR-711 and clinical characteristics
We further evaluated the expression level of miR-711 and compared with different clinic-pathological parameters. We observed that miR-711 level was significantly down regulated in metastatic disease as compared to non-metastatic disease (p=0.03, Figure 2E). Similarly, low expression was found to be significantly associated with high Gleason score in comparison with low score (p=0.0001, Figure 2F). On the other hand, expression level of miR-711 and its correlation with other clinicopathological parameters like age, PSA level and Nodal status were found to be non-significant (p= 0.15, 0.12 and 0.14 respectively, Figure 2B, C and D). In order to confirm statistical relevance of the miR-711 expression, a receiver operating characteristic (ROC) was established and Area under the curve (AUC) was determined to estimate the sensitivity and specificity of miR-711 expression (Figure 3). The AUC for the expression was 0.874 with 95% confidence interval (0.810 - 0.937). The result indicated that miR-711 may be used as a biomarker, with the ability to resolve PCa patients from the control.
Figure 3.
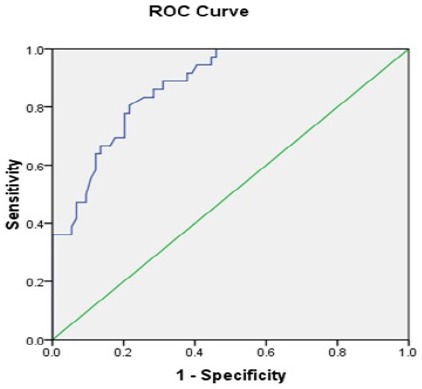
ROC Curve Analysis for Detection of Prostate Cancer. The ROC analysis showed that miRNA-711 could be distinguish PCa from Control (AUC=0.874).
Targetome analysis of miR-711
MiR-711 is a novel microRNA and to the best of our knowledge, very little is known about its precise role in cancer except for a few recent reports. So we next attempted to reveal the downstream target genes of miRNA-711 through target prediction analysis and its suggestive role in PCa. The potential targets of miR-711 were predicted by Target Scan database, Mirwalk and Miranda software (Table 3). We searched for target genes of miR-711 based on the seed sequence alignment.
Table 3.
Mir-711 Specific Targeted Gene Analysis Through Mirwalk, Miranda, Targetscan Software, Literature Reported
| MicroRNA | Gene Name |
|---|---|
| MicroRNA 711 | SP1, CDK4, Akt, IRS1, NKX3 - 1, XRCC3, BCAS1, CYP11B2, LEP, TLR4, C1R, ERBB2, skI, cyp2w1, CDK4, DNM2, ILK, ZNFR3, TXNIP, FADS1, VSIG2, CLCN5, ALDH9A1, BACE2, RHCG, RASA4, ACADSB, NCAN, SARDH, DUSP4, GNA11, LRPAP1, PCDHGC3, RXRA, NR2C2, BTG2, PCDHGB4, ABCG1, CELSR1, PCDHGA8, XPO7, TXN2, PCDHGA12, VPS4A, FAM203A, PCDHGC5, PCDHGC4, PCDHGB7, PCDHGB6, PCDHGB5, PCDHGB3, PCDHGB2, PCDHGB1, PCDHGA11. |
Gene Ontology bioinformatics analysis indicated significance role of miR-711 in cancer progression pathways.
By using the database for annotation, DAVID network analysis, the molecular function of miR-711 and their possible targets signalling cascades key components were analysed according to gene ontology (GO) functional and annotation and categorise. GO and Pathway analysis of top 25% of genes sorted on the basis of level of significance was performed. GO analysis revealed that most of the target genes of miR-711 were involved in the regulation of cell activation, cell proliferation, MAPK signalling cascade, transcription regulator activity and protein homodimerization activity etc.
KEGG pathway enrichment analysis indicated most significant signalling cascade key molecules involved in cell cycle progression and proliferation were targeted by miR-711:
The KEGG pathway enrichment analysis indicated that cell cycle pathways, cytokine- cytokine receptor interaction, Jak-Stat signalling pathway, Hedgehog signalling pathway, basal cell carcinoma and hypoxia response pathway. Through David software we found that NKX3-1, XRCC3, BCAS1, CYP11B2, LEP, TLR4, MC1R and ERBB2 are targeted by miR-711 and also regulated in prostate cancer. These results indicated that miR-711 may have an important role in prostate cancer progression through targeting cellular genes involved in oncogenesis (Figure 4).
Figure 4.
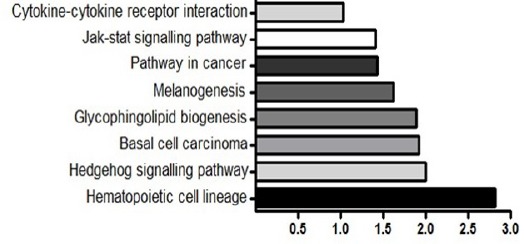
KEGG Pathway Analysis for Target Genes of the miR-711. Potential target genes of miR-711 were enriched in JAK-STAT signaling pathway, pathway in cancer, basal cell carcinoma. Each bar indicates the target genes involved in relevant pathway
Discussion
PCa load among populations is increasing due to extended life span as well as improper life style among men in both developed and developing countries (Maruthappuet al., 2010). Patients with advanced PCa still do not have adequate diagnostic tools as well as drugs for therapy. Many studies have underscored the importance of miRNAs in PCa as prospective biomarkers as well targets for therapy (Srivastava et al., 2014; Filella et al., 2015; Gao et al., 2016; Yin et al., 2016; Kachakova et al., 2015). However, miRNAs which can significantly distinguish between different stages of PCa are very less studied and their identification may help in improvising stage specific diagnosis with more accuracy (Nam et al., 2015). Furthermore, continuous increase in number of novel miRNAs in different human tissues including prostate is very much distracting and questions our present knowledge in this context. Ultimately it becomes very obvious to know the exact number of miRNAs that may have significant role in PCa progression and stage transitions and can improve our overall understanding.
In this report, we performed a comprehensive microRNA profiling in tissue samples from BPH and different pathological grades of PCa using an updated version of microRNA microarray chip. Bioinformatics analysis showed differential expression of hundreds of microRNAs. We found up regulation of miR-182, miR-183, miR-200a, miR-25, miR-26b and down regulation of miR-222, miR-205 and miR-1296 which were consistent with previous studies in PCa thereby validating our microarray results (Volinia et al., 2006; Schaefer et al., 2010; Xu et al., 2016; Srivastava et al., 2013; Majid et al., 2010). However, as a breakthrough we identified a set of ten novel microRs in PCa tissue samples amongst which we validated three most significant microRNAs, miR-711, miR-1246 and miR-548x-3p in independent set of tissue samples. We found miR-711 to be significantly down regulated in PCa tissues as compared to BPH and normal control consistently in microarray and qRT-PCR analysis. However, miR-1246 and miR-548x-3p did not correspond to our microarray data (p≤0.7). The expression of miR-711 inversely correlated (p≤0.5) with metastasis and advanced stage PCa implicating a tumor suppressor role of this novel miRNA in PCa progression. MiR-711 came out to be a good candidate biomarker with high specificity and sensitivity in ROC curve analysis. Previous reports on miR-711 are very less but have shown its contrasting roles in various pathological conditions. According to recent studies in different cancer types, miR-711 has been shown to possess oncogenic trait in breast cancer and cutaneous T-cell lymphoma where as it was found to be tumor suppressor in gastric cancer (Sabirzhanov et al., 2016; Hu et al., 2016; Liao et al., 2016; Ralfkiaer et al., 2011). Likewise miR-711 was also reported to be identified as a novel miRNA in HPV positive cervical cancer cells (Sharma et al., 2016). Other than cancer, miR-711 was shown to possess important role in traumatic brain injury, myocardial infarction and insulin resistance (Sabirzhanov et al., 2016; Zhao et al., 2013). All these studies showed that miR-711 exhibited immense biological importance in carcinogenesis and proper understanding of its molecular function is pre-requisite for its clinical use. Our target prediction analysis revealed several cellular target genes of miR-711, including some of the validated targets like SP1, CDK4, Akt, IRS1 as well as large number of predicted targets like skI, cyp2w1, CDK4, DNM2, ILK, ZNFR3, TXNIP, FADS1 VSIG2 CLCN5 and ALDH9A1which are involved in the cancer progression.
Conclusively, in the present study we identified miR-711 as a novel microRNA in PCa. We also report its much less described tumor suppressor role in PCa progression. Selective deregulation of miR-711 in advanced and metastatic PCa patients advocates its proper assessment as a stage specific biomarker for tumor progression and metastasis. Our findings will provide data that can be used in future studies to investigate the possible role of miR-711 in PCa progression. Furthermore, we are currently focusing on its molecular mechanisms in vitro and its validation as non-invasive biomarker in serum samples of PCa patients. We anticipate that through this study and other ongoing studies in our laboratory may help in better understanding and functional characterization of miR-711 in PCa and ensure its significance as a diagnostic and prognostic biomarker.
Funding
Science and Engineering Research Board (SERB), Department of Science and Technology, New Delhi, India(SR/SO/HS/-219/2012).
Statement conflict of Interest
Authors have declared that no conflict of interests exists. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank all the members of Professor A A Mahdi laboratory for the helpful discussion. Mohammad Waseem is supported by Senior research fellowship (SRF) from Science and Engineering Research Board (SERB), Department of Science and Technology, New Delhi, India.
References
- Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–45. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- Deng S, Calin GA, Croce CM, et al. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–6. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- Ding HY, Qian WQ, Xu J. MicroRNA-146b acts as a potential tumor suppressor in human prostate cancer. J BUON. 2015;2:434–43. [PubMed] [Google Scholar]
- Filella X, Foj L. Emerging biomarkers in the detection and prognosis of prostate cancer. Clin Chem Lab Med. 2015;53:963–73. doi: 10.1515/cclm-2014-0988. [DOI] [PubMed] [Google Scholar]
- Gao Y, Guo Y, Wang Z, et al. Analysis of circulating miRNAs 21 and 375 as potential biomarkers for early diagnosis of prostate cancer. Neoplasma. 2016;63:623–8. doi: 10.4149/neo_2016_417. [DOI] [PubMed] [Google Scholar]
- Hu JY, Yi W, Zhang MY, et al. MicroRNA-711 is a prognostic factor for poor overall survival and has an oncogenic role in breast cancer. Oncol Lett. 2016;11:2155–63. doi: 10.3892/ol.2016.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugosson J, Carlsson S, Aus G, et al. Mortality results from the Göteborg randomised population-based prostate-cancer screening trial. Lancet Oncol. 2010;11:725–32. doi: 10.1016/S1470-2045(10)70146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–63. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Standart N. How do microRNAs regulate geneexpression. Sci Stke. 2007:367. doi: 10.1126/stke.3672007re1. DOI:10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Jain S, Saxena S, Kumar A. Epidemiology of prostate cancer in India. Meta Gene. 2014;29:596–605. doi: 10.1016/j.mgene.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachakova D, Mitkova A, Popov E, et al. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015;34:189–200. doi: 10.1089/dna.2014.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao A, Tan G, Chen L, et al. RASSF1A inhibits gastric cancer cell proliferation by miR-711-mediated down regulation of CDK4 expression. Oncotarget. 2016;7:5842–51. doi: 10.18632/oncotarget.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londin E, Loher P, Telonis AG, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate-and tissue-specific microRNAs. Proc Natl Acad Sci. 2015;112:1106–15. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Majid S, Dar AA, Saini S, et al. Regulation of minichromosome maintenance gene family by microRNA-1296 and genistein in prostate cancer. Cancer Res. 2010;70:2809–18. doi: 10.1158/0008-5472.CAN-09-4176. [DOI] [PubMed] [Google Scholar]
- Maruthappu M, Watkins J, Noor AM, et al. Universal health coverage and cancer mortality in high-income and middle-income countries 1990–2010: a longitudinal analysis. Lancet. 2016;388:684–95. doi: 10.1016/S0140-6736(16)00577-8. [DOI] [PubMed] [Google Scholar]
- Nam RK, Amemiya Y, Benatar T, et al. Identification and validation of a five microRNA signature predictive of prostate cancer recurrence and metastasis: a cohort study. J Cancer. 2015;6:1160–71. doi: 10.7150/jca.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralfkiaer U, Hagedorn PH, Bangsgaard N, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL) Blood. 2011;118:5891–900. doi: 10.1182/blood-2011-06-358382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirzhanov B, Stoica BA, Zhao Z, et al. miR-711 upregulation induces neuronal cell death after traumatic brain injury. Cell Death Differ. 2016;23:654–68. doi: 10.1038/cdd.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf HJ, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–76. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- Sharma S, Hussain S, Soni K, et al. Novel MicroRNA signatures in HPV-mediated cervical carcinogenesis in Indian women. Tumor Biol. 2016;37:4585–95. doi: 10.1007/s13277-015-4248-7. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Goldberger H, Dimtchev A, et al. MicroRNA profiling in prostate cancer –the diagnostic potential of urinary miR-205 and miR-214. PLoS One. 2013;8:e76994. doi: 10.1371/journal.pone.0076994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Goldberger H, Dimtchev A, et al. Circulatory miR-628-5p is down regulated in prostate cancer patients. Tumor Biol. 2014;35:4867–73. doi: 10.1007/s13277-014-1638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuopelytė K, Daniūnaitė K, Jankevičius F, et al. Detection of miRNAs in urine of prostate cancer patients. Medicina. 2016;52:116–24. doi: 10.1016/j.medici.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2009;27:52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CG, Yang MF, Fan JX, et al. MiR-30a and miR-205 are down regulated in hypoxia and modulate radiosensitivity of prostate cancer cells by inhibiting autophagy via TP53INP1. Eur Rev Med Pharmacol Sci. 2016;20:1501–8. [PubMed] [Google Scholar]
- Xu L, Qi X, Duan S, Xie Y, et al. MicroRNAs: potential biomarkers for disease diagnosis. Bio Med Mater Eng. 2014;24:3917–25. doi: 10.3233/BME-141223. [DOI] [PubMed] [Google Scholar]
- Yin C, Fang C, Weng H, et al. Circulating microRNAs as novel biomarkers in the diagnosis of prostate cancer: a systematic review and meta-analysis. Int Urol Nephro. 2016;48:1087–95. doi: 10.1007/s11255-016-1281-4. [DOI] [PubMed] [Google Scholar]
- Zhao N, Yu H, Yu H, et al. MiRNA-711-SP1-collagen-I pathway is involved in the anti-fibrotic effect of pioglitazone in myocardial infarction. Sci China Life Sci. 2013;56:431–9. doi: 10.1007/s11427-013-4477-1. [DOI] [PubMed] [Google Scholar]


