Abstract
Pathogens are exposed to different temperatures during an infection cycle and must regulate gene expression accordingly. However, the extent to which virulent bacteria alter gene expression in response to temperatures encountered in the host is unknown. Group A Streptococcus (GAS) is a human-specific pathogen that is responsible for illnesses ranging from superficial skin infections and pharyngitis to severe invasive infections such as necrotizing fasciitis and streptococcal toxic shock syndrome. GAS survives and multiplies at different temperatures during human infection. DNA microarray analysis was used to investigate the influence of temperature on global gene expression in a serotype M1 strain grown to exponential phase at 29°C and 37°C. Approximately 9% of genes were differentially expressed by at least 1.5-fold at 29°C relative to 37°C, including genes encoding transporter proteins, proteins involved in iron homeostasis, transcriptional regulators, phage-associated proteins, and proteins with no known homologue. Relatively few known virulence genes were differentially expressed at this threshold. However, transcription of 28 genes encoding proteins with predicted secretion signal sequences was altered, indicating that growth temperature substantially influences the extracellular proteome. TaqMan real-time reverse transcription–PCR assays confirmed the microarray data. We also discovered that transcription of genes encoding hemolysins, and proteins with inferred roles in iron regulation, transport, and homeostasis, was influenced by growth at 40°C. Thus, GAS profoundly alters gene expression in response to temperature. The data delineate the spectrum of temperature-regulated gene expression in an important human pathogen and provide many unforeseen lines of pathogenesis investigation.
Keywords: temperature regulation, microarray, real-time TaqMan reverse transcription–PCR, iron
One of the major themes identified in prokaryotic research is that pathogenic bacteria coordinately control gene expression and virulence factor production by complex regulatory circuits in response to alterations in the host milieu. For example, virulence factor synthesis is influenced by nutrient availability, pH, osmolarity, growth phase, oxygen tension, iron levels, and temperature (1). Although knowledge has accumulated about the molecular mechanisms used by pathogenic bacteria to regulate gene expression in response to changing environments, study of global changes in gene transcription induced by temperature has not been conducted.
Group A Streptococcus (GAS) is a Gram-positive pathogen that causes many infections (pharyngitis, septicemia, toxic shock, and necrotizing fasciitis) and postinfectious sequelae (rheumatic heart disease and glomerulonephritis). Humans are the natural host and sole reservoir of GAS. The organism can survive and replicate in diverse anatomic sites such as the skin, throat, female urogenital tract, lower gastrointestinal tract, and blood (2). Hence, GAS must alter gene expression to survive and grow in these environments. At the extremes of superficial skin and deep tissue infections, GAS must differentially express virulence and other genes at temperatures ranging from roughly 25°C to 40°C.
Relatively little is known about gene regulatory circuits used by GAS in the course of human infection. Studies of differential gene expression in response to environmental changes have been conducted by analysis of relatively few target loci that encode proven or putative extracellular virulence factors, and hence have been preselected on the basis of presumed involvement in host-pathogen interactions (3–6). With the exception of limited data available from analysis of the genes encoding the antiphagocytic M protein (emm) (4) and the superantigen streptococcal pyrogenic exotoxin A (speA) (6), very little is known about the spectrum of temperature-regulated gene expression in GAS. To gain insight into the strategies used by a bacterial pathogen to interact with the host, we used DNA microarray technology to identify global changes in gene expression in response to physiologically relevant temperature changes.
Materials and Methods
Bacterial Growth Conditions.
GAS strain SF370 (serotype M1) was studied because its genome has been sequenced (7). Liquid cultures were grown in Todd Hewitt broth supplemented with 0.2% yeast extract (THY) in 5% CO2 at 29°C, 37°C, and 40°C. Cells were harvested in exponential growth (A600 of 0.3–0.4), and viable counts for organisms grown at 29°C, 37°C, and 40°C were similar (data not shown). Iron-limiting conditions were achieved by deferrating THY with the chelating resin Chelex 100 and supplementing it with 1 mM each of MgCl2, MnCl2, ZnCl2, and CaCl2 (DTHY) (8).
RNA Isolation.
Bacterial cell lysis and RNA isolation were conducted with the FastRNA Kit (Qbiogene, Carlsbad, CA) as described in Supplemental Materials and Methods, which is published on the PNAS web site, www.pnas.org. RNA quality was assessed spectrophotometrically and with gel electrophoresis. The absence of contaminating DNA was confirmed by PCR and TaqMan real-time reverse transcription (RT-PCR).
Microarray Hybridization.
ORF-specific primers were designed with the PRIMER3 program (www.genome.wi.mit.edu) based on the SF370 draft genome sequence (wit.mcs.anl.gov/WIT2). ORFs are designated with SPy numbers in accordance with the completed SF370 genome sequence (7). Preparation of DNA microarrays, slide processing, cDNA probe synthesis, hybridization, and data analysis are described in Supplemental Materials and Methods.
Differentially transcribed genes were classified into functional groups as described (9) except that genes encoding hypothetical and phage-associated proteins were categorized separately (Table 1, which is published as supplemental data on the PNAS web site). Genes encoding hypothetical proteins were analyzed by blast (www.ncbi.nlm.nih.gov/BLAST) to determine whether homologues were present in other bacteria. signalp software (www.cbs.dtu.dk/services) was used to identify potential secretion signal sequences for all proteins encoded by differentially regulated genes (Table 1).
Real-Time RT-PCR.
Primers and probes (Table 2, which is published as supplemental data on the PNAS web site) were designed with primerexpress software (Applied Biosystems) and purchased from Applied Biosystems and MegaBases (Evanston, IL). TaqMan assays were done as described (10). Triplicate assays were performed with RNA samples isolated from at least two independent exponential phase cultures of strain SF370, and standard curves were made with SF370 genomic DNA. Triplicate assays using three independent RNAs confirmed that transcript levels of gyrase subunit A (gyrA) were not significantly different (P > 0.05) at 29°C compared with 37°C or at 40°C compared with 37°C (data not shown). Therefore, gyrA (spy1152) was used to normalize TaqMan data. TaqMan assays also were performed to determine whether the iron content of the culture medium influenced transcription of specific GAS genes. For these studies, the fold difference of cDNA molecules present in GAS grown at 40°C in DTHY relative to THY was determined.
Enzyme Assays.
A600 measurements of GAS grown at 29°C and 37°C were taken over time (data not shown). Aliquots were centrifuged, and supernatants were filter-sterilized. Glucose utilization was assessed by measuring the glucose concentration in culture supernatants with a commercial kit (Sigma). Lactate accumulation was determined with a commercial kit (R-Biopharm, Marshall, MI). Superoxide dismutase activity was measured as described (11).
Results and Discussion
DNA Microarray Analysis and Verification.
A DNA microarray was used to study global gene expression differences in GAS grown in vitro at biomedically relevant temperatures. Although a 2-fold difference in relative transcript level is a threshold commonly used for analysis and interpretation of microarray data (e.g., ref. 12), Hughes et al. (13) recently reported that less than a 2-fold change in relative transcript abundance can be biologically significant. Hence, a 1.5-fold threshold was used in our study.
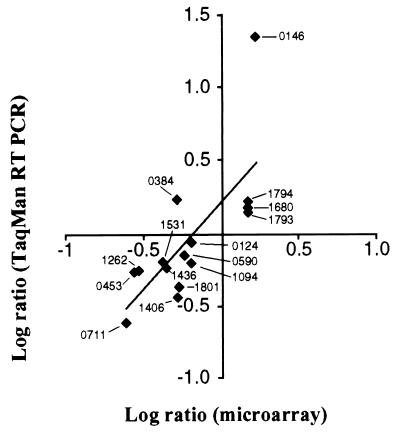
TaqMan RT-PCR analysis of a sample of 15 genes, representing six functional categories, was conducted to verify the microarray transcription profiling data. The average fold difference in quantity of cDNA molecules present at 29°C relative to 37°C was determined with RNA isolated from two independent cultures. There was a strong positive correlation (r = 0.74) between the data obtained by the two techniques (Fig. 1).
Figure 1.
Correlation of microarray and TaqMan assays. The fold difference in the number of cDNA molecules present at 29°C relative to 37°C for 15 genes influenced by temperature was log-transformed, and values were plotted. Points on the graph represent genes that were analyzed by both methods.
Global Response of GAS to Temperature.
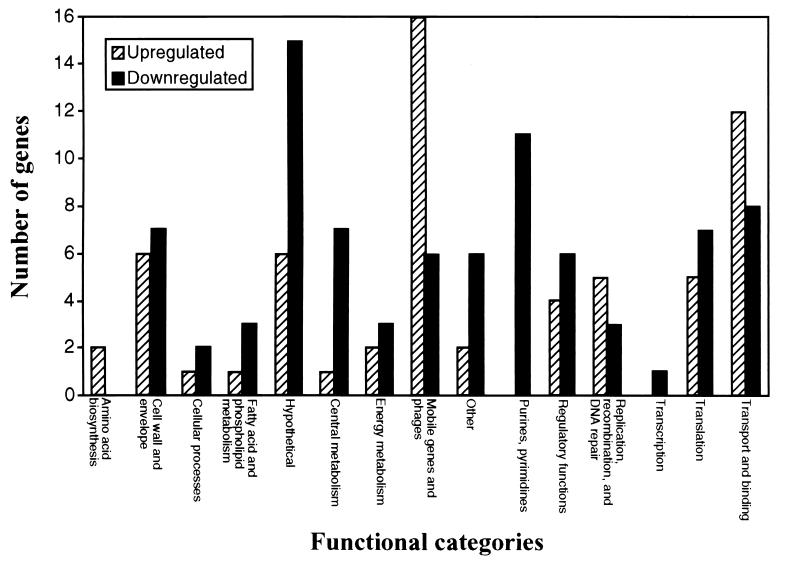
Globally, 9% of the genes represented on the microarray (n = 1,605) were differentially transcribed by organisms grown at 29°C compared with 37°C (Table 1). Forty-three percent (n = 63) of the differentially transcribed genes were up-regulated, and 57% (n = 85) were down-regulated at 29°C relative to 37°C (Fig. 2). Many of the differentially expressed genes encode extracellular proteins that may be critical in GAS-host interactions. In addition to known or putative virulence factors (e.g., streptococcal collagen-like protein 1 and streptococcal pyrogenic exotoxin C encoded by spy1893 and spy0711, respectively), 26 genes encoding proteins with a predicted secretion signal sequence were differentially transcribed (Table 1). These genes encode inferred proteases, cell wall-associated proteins, transporters, phage-associated proteins, and uncharacterized hypothetical proteins. Taken together, the data suggest that the composition of the GAS extracellular proteome is substantially influenced by temperature.
Figure 2.
Differentially expressed GAS genes at 29°C relative to 37°C. The number of up-regulated (filled bars) and down-regulated (stippled bars) genes in each functional group is shown.
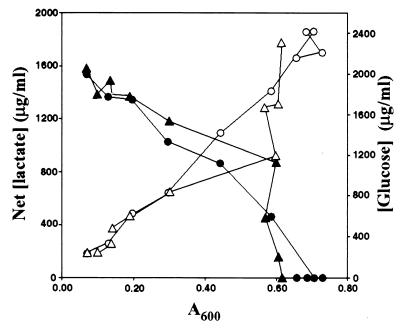
Global expression of genes encoding proteins involved in fundamental processes such as purine and pyrimidine metabolism, fatty acid metabolism, and glycolysis were primarily down-regulated at 29°C relative to 37°C (Fig. 2 and Table 1). Although the doubling times of GAS cultured at 29°C and 37°C were similar, bacteria grown at 29°C entered exponential phase later than GAS grown at 37°C. Bacteria grown at 29°C required 5 h longer to achieve the equivalent growth yield as bacteria grown at 37°C (data not shown). The decreased transcription of genes encoding glycolytic enzymes at 29°C compared with 37°C correlated with a 2-fold reduction in the rate of glucose utilization and lactate formation (Fig. 3).
Figure 3.
Differential utilization of glucose and concomitant lactate accumulation in GAS grown at 29°C and 37°C. The concentration of glucose (closed symbols) and lactate (open symbols) present in culture supernatants of GAS grown at 29°C (triangles) and 37°C (circles) was measured.
Genes encoding regulatory proteins are of special interest because changes in their transcription may have pleiotropic effects, and hence may significantly influence host-pathogen interaction and disease outcome. Ten presumed regulatory genes were differentially transcribed (Table 1 and Fig. 2). For example, genes (spy0124 and spy0245) encoding the characterized transcriptional regulators RofA and FasA, respectively, were down-regulated at 29°C relative to 37°C (Table 1). RofA negatively regulates several virulence genes and positively activates transcription of the gene encoding extracellular protein F (14). FasA is a response regulator that negatively regulates expression of genes encoding virulence factors and positively regulates expression of fasX, a putative regulatory RNA molecule (15). Our results indicate that fasA and rofA transcription is influenced by temperature, implying the existence of a broader gene regulatory network than heretofore suggested in GAS.
Microarray analysis also demonstrated that several genes (spy0146, spy0533, and spy1680) encoding transcriptional regulator homologues were up-regulated at 29°C relative to 37°C (Table 1). For example, spy0146, which encodes a protein that is 59% identical to the Clostridium perfringens perfringolysin O activator PfoR, was up-regulated 1.7-fold. It is unknown whether Spy0146 regulates toxin production in GAS. Similarly, genes controlled by the transcriptional activator homologues Spy0533 and Spy1680 are also unknown.
Transcription of Genes Encoding Iron Homeostasis Proteins Is Influenced by Growth Temperature.
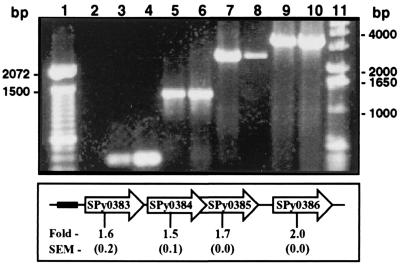
A central theme identified by the microarray analysis was that growth temperature influenced the level of transcription of many genes encoding proteins presumed to be involved in iron homeostasis, including genes encoding homologues of iron transport proteins, considered to be virulence factors for many bacteria (16, 17). For example, transcription of the four-gene operon containing spy0386 (ferrichrome transport ATP-binding protein), spy0385 (ferrichrome iron-binding protein), spy0384 (ferrichrome transport permease), and spy0383 (ferric-dicitrate transport permease) was up-regulated at 29°C relative to 37°C (Fig. 4). Several other genes (e.g., spy1406 and spy1531) encoding proteins with a putative role in iron homeostasis also were differentially expressed at 29°C relative to 37°C (Table 1).
Figure 4.
RT-PCR analysis of a polycistronic mRNA encoding a putative iron transporter. RNA from GAS grown at 29°C was reverse-transcribed into cDNA, and PCR was done with ORF-specific primers. The templates for PCR were SF370 genomic DNA (lanes 3, 5, 7, and 9), cDNA (lanes 4, 6, 8, and 10), and RNA (lane 2). Primers for PCR were 0383–5′ and 0383–3′ (lanes 2–4); 0383–5′ and 0384–3′ (lanes 5 and 6); 0383–5′ and 0385–3′ (lanes 7 and 8); and 0383–5′ and 0386–3′ (lanes 9 and 10). The schematic shows the arrangement of the genes in the chromosome, including intergenic spaces, and the black box represents the putative promoter. The average fold difference in transcription of each ORF at 29°C relative to 37°C was determined with TaqMan assays. The SEM is shown in parentheses. Dashes denote DNA sizes in the 100-bp (lane 1) and 1-kb (lane 11) ladders.
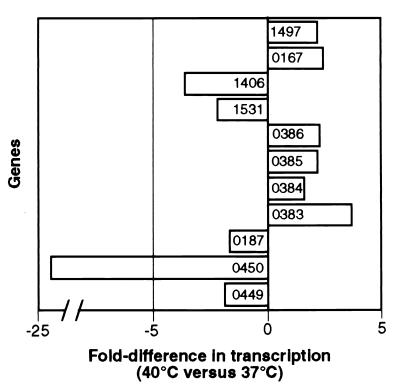
Patients with severe invasive disease caused by GAS usually have fever, frequently exceeding 40°C. It is well known that one of the initial host responses to microbial infection is to restrict the amount of free iron available to the pathogen by sequestering it with iron-binding proteins (18–20). This process is especially important in infections characterized by high fever and is thought to be an innate adaptive response that inhibits bacterial growth. Hence, we hypothesized that genes encoding iron transport and acquisition proteins are up-regulated at elevated temperatures, whereas genes encoding proteins that repress iron-regulated gene expression are down-regulated. TaqMan assays were used to test this hypothesis. Transcription of iron-regulated genes in bacteria is often controlled by the ferric uptake repressor (Fur) (16, 17), a protein that binds as a dimer to consensus sequences (Fur boxes) located in the promoter region of iron-regulated genes (21). spy0187, which encodes a protein with 43% identity to a Staphylococcus aureus Fur homologue, is present in the genome of strain SF370. Transcription of spy0187 was down-regulated 1.6-fold at 29°C relative to 37°C and 1.7-fold at 40°C relative to 37°C (Fig. 5), indicating that expression is influenced by growth temperature.
Figure 5.
TaqMan analysis of GAS genes influenced by growth at 40°C. TaqMan assays were done with RNA isolated from independent cultures of GAS grown at 40°C and 37°C. The average fold difference in transcription at 40°C relative to 37°C is shown.
The draft genome sequence of strain SF370 also contained two contiguous genes encoding proteins with homology to the amino and carboxyl terminus, respectively, of an inferred iron-dependent repressor in Streptococcus pneumoniae and the iron-regulated diphtheria toxin repressor DtxR, a global regulator of gene expression in Corynebacterium diphtheriae (22). Analysis of the recently completed SF370 genome (7) actually revealed that these coding sequences, which are annotated as spy0450 and spy0449, are one ORF (spy0450). Our data show that the 5′ (spy0450) and 3′ (spy0449) portions of this gene were down-regulated at 40°C relative to 37°C (Fig. 5), a result supporting our hypothesis.
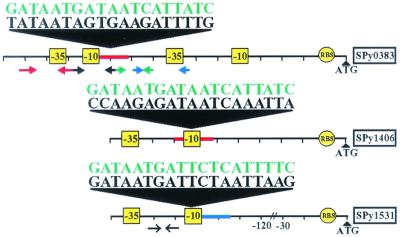
We next tested whether down-regulation of transcriptional repressor genes spy0187 and spy0450 at 40°C coincided with increased transcription of genes in the operon encoding Spy0386, Spy0385, Spy0384, and Spy0383. Transcription of genes in this operon was up-regulated 2.4-fold at 40°C relative to 37°C (Fig. 5). Putative transcriptional regulatory elements (i.e., four imperfect dyad repeats) are located in the promoter region of this operon (Fig. 6). This region also has a DNA sequence with 11 of 19 nucleotides identical to the Bacillus subtilis dhb Fur box consensus sequence (23). Of note, Ochsner and Vasil (24) demonstrated that sequences with 10 of 19 identical nucleotides to the Fur box consensus sequence bound Pseudomonas aeruginosa Fur and were functional.
Figure 6.
Schematic of putative promoters of temperature-regulated iron metabolism-related genes. Potential promoters (yellow boxes noting −10 and −35 regions) and ribosomal binding sites (RBS) are shown. The nucleotide sequences with similarity to the B. subtilis dhb Fur box (red line) and B. subtilis fhuD Fur box 2 (blue line) consensus sequences are shown. The Fur box consensus sequences are shown in green. Dyad repeats are denoted below the line with red, black, green, and blue arrows. An indirect repeat is denoted below the line by black arrows lacking a closed arrowhead.
Decreased Expression of Homologues of Iron Homeostasis and Oxidative Stress Proteins.
The hypothesis that elevated temperature influences transcription of genes encoding inferred iron homeostasis proteins was tested by analysis of additional genes. spy1406 encodes superoxide dismutase (SOD), an enzyme that protects the cell from oxidative stress by scavenging superoxide radicals. sod expression is controlled by iron in some bacteria (25, 26). When intracellular iron levels are low, fewer superoxide radicals are expected to be produced by the Fenton reaction (27), suggesting a decreased need for oxidative stress protection. Consistent with this idea, spy1406 transcription was down-regulated at 29°C relative to 37°C (Fig. 1 and Table 1) and at 40°C relative to 37°C (Fig. 5). In addition, less SOD activity was present in cell lysates of organisms grown at 29°C and 40°C compared with 37°C (data not shown). The putative −10 region of sod has a sequence with 10 of 19 nucleotides identical to the B. subtilis dhb Fur box (Fig. 6). Although the influence of temperature on sod transcription has not been previously evaluated in GAS, sod expression in the human mucosal pathogen Porphyromonas gingivalis is temperature-regulated (28).
Spy1531 is homologous to the iron storage protein bacterioferritin (Bfr) and a Dps (DNA-binding protein from starved cells)-like peroxide resistance protein (Dpr) in Streptococcus mutans (29). Yamamoto et al. (29) reported that Dpr binds iron and forms multiple subunits similar to ferritin and enhances aerotolerance by sequestering iron and limiting oxidative damage. The microarray and TaqMan data indicated that spy1531 transcription also was decreased at 29°C relative to 37°C (Fig. 1 and Table 1) and at 40°C relative to 37°C (Fig. 5). Potential regulatory elements that may respond to the level of available iron are located upstream of the ATG translation start codon in spy1531. A 13-nt indirect repeat and a sequence with 15 of 19 nt identical to the fhuD Fur box 2 consensus sequence of B. subtilis (30) are present in the putative promoter region (Fig. 6). Although it has not been demonstrated that transcription of genes encoding Spy1531 homologues is influenced by temperature in other bacteria, Quail et al. (31) suggested that bfr in Escherichia coli is positively regulated by Fur.
Differential Transcription of GAS Genes Encoding Hemolysins.
Hemolysins, which are virulence factors for many pathogens (16), also are used by microbes to acquire iron (16, 27). Therefore, we tested whether transcription of genes encoding inferred hemolysins (Spy0167, Spy1497, Spy0378, Spy0370, and Spy0738) was up-regulated at 40°C relative to 37°C. TaqMan assays showed that transcription of spy0167 encoding the virulence factor streptolysin O (SLO) was increased 2.4-fold at 40°C relative to 37°C (Fig. 5). In this regard, we note that transcription of the gene encoding S. aureus α-hemolysin was induced by growth at 42°C (32). The mechanism of slo regulation is not well understood, but transcription is not controlled by regulators such as Mga, Rgg, or CovR (33–35). spy1497 transcription was increased 2.1-fold at 40°C relative to 37°C (Fig. 5). Transcription of spy0378, spy0370, and spy0738 was not influenced by growth temperature, suggesting that spy0167 and spy1497 may be up-regulated in humans with infections accompanied by high fever.
In the aggregate, the results demonstrate that transcription of genes encoding iron transporters and iron-dependent regulators are similarly affected at decreased (29°C versus 37°C) and increased (40°C versus 37°C) temperatures. One explanation for this pattern of gene regulation is that growth at 29°C and 40°C is suboptimal for GAS, resulting in differential transcription of genes encoding proteins needed to enhance survival. Furthermore, the results suggest that many of the same proteins are important for GAS survival and pathogenesis at anatomic sites characterized by different temperatures, such as skin, throat, and blood.
Analysis of the Effect of Iron-Restricted Growth Conditions on Temperature-Dependent Gene Expression.
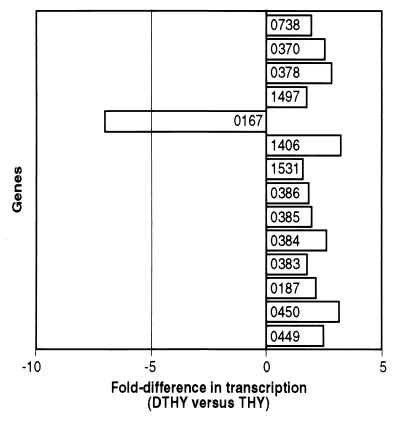
Importantly, temperature-induced differential transcription of genes encoding proteins involved in iron transport and homeostasis occurred during growth in THY, a rich medium. Based on the inferred function of the proteins and the increased sequestration of iron by iron-binding proteins during the human fever response, we investigated the influence on transcription of iron-restricted growth at 40°C. TaqMan assays revealed that all genes tested except spy0167 were up-regulated when grown in DTHY (Fig. 7). Of note, despite its role as a hemolysin, and up-regulation at 40°C (Fig. 5), spy0167 was down-regulated 7.0-fold under iron-limiting growth conditions (Fig. 7). In contrast, genes encoding three hemolysin homologues (Spy0378, Spy0370, and Spy0738) that were not differentially transcribed at 40°C in THY were up-regulated ≥2.0-fold in DTHY (Fig. 7). Genes encoding iron transport protein homologues (Spy0383, Spy0384, Spy0385, and Spy0386) were also up-regulated >2.0-fold in DTHY (Fig. 7). spy1406, spy1531, spy0187, spy0449, and spy0450 were down-regulated at 40°C in THY (Fig. 5) but up-regulated in DTHY, reinforcing the presence of a complex regulatory network in GAS that responds to multiple external stimuli. In summary, our results demonstrate that GAS genes encoding proteins involved in iron transport and homeostasis are influenced by both temperature and iron. Furthermore, the presence of secretion signal sequences in six of these proteins suggests that they may be expressed extracellularly in humans with infections accompanied by high fever and, hence, may also be therapeutic targets.
Figure 7.
Iron-limiting growth conditions influence GAS gene expression. TaqMan assays were done with RNA isolated from independent cultures of GAS grown at 40°C in DTHY and THY. The average fold difference in transcription in DTHY relative to THY is shown. Spy numbers of the relevant genes are shown.
Bacterial Pathogens and Temperature-Regulated Gene Expression.
The influence of temperature on bacterial gene expression has been widely studied, and many examples of temperature-regulated genes are known (36, 37). For example, arthropod-borne bacterial pathogens that infect mammals modulate gene expression in response to temperature changes encountered in their cold-blooded and warm-blooded hosts (36). Yersinia pestis, the cause of plague, differentially regulates genes encoding capsule, iron transporters, outer membrane proteins, and toxins in the flea vector and mammalian host (36). Similarly, Borrelia burgdorferi, the cause of Lyme disease, differentially regulates expression of genes encoding outer surface proteins required for successful transmission (38, 39). Pathogens such as Shigella and Listeria monocytogenes also have temperature-regulated genes, undoubtedly because their life cycles involve growth in a mammalian host and exposure to ambient environmental temperatures. Although temperature-regulated bacterial gene expression has been well described, our study delineates global changes in gene expression in response to alteration of this environmental condition.
Bacteria use multiple molecular strategies to alter gene expression in response to temperature change. For example, the level of DNA supercoiling in promoters of temperature-regulated genes influences transcription in E. coli and Shigella spp (40). Posttranscriptional temperature-sensing mechanisms that involve recognition of alterations in RNA secondary structure also have been described (41). In addition, a two-component signal transduction system that senses and responds to temperature in B. subtilis was described recently (42).
One of the important themes that emerged from our work was that many genes encoding proteins involved in iron homeostasis were differentially regulated in response to growth temperature. Hence, a study evaluating the influence of temperature on expression of iron-regulated genes (tolQRA) encoding a toxin transporter in the human pathogen P. aeruginosa is pertinent to our study. Lafontaine and Sokol (43) reported that the tol genes were differentially expressed when bacteria were grown in iron-replete media at 25°C, 30°C, 37°C, and 40°C, and also were up-regulated in iron-limiting growth conditions. Maximal transcription occurred at 37°C (43), as we observed with many GAS genes involved in iron homeostasis. Growth temperature and iron concentration synergistically influenced tol expression but did not completely control transcription, indicating that multiple regulators participate. Additional evidence for an interactive regulatory process involving temperature and iron in P. aeruginosa was provided by studies of the synthesis of iron-regulated virulence factors pyoverdin and exotoxin A (44, 45).
Conclusions.
Microarray and TaqMan analyses demonstrated that GAS responds globally to physiologically important temperatures. Temperature-induced changes occurred at the level of transcription of genes encoding extracellular proteins, proven and potential virulence factors, regulatory proteins, transporters, phage-associated proteins, hypothetical proteins, and proteins involved in iron uptake and homeostasis. Although many environmental factors influence gene expression, our data demonstrate that GAS has the ability to alter gene transcription extensively in response to temperatures present at different infection sites in humans (e.g., pharynx, skin, and blood). Differential expression of 28 genes encoding proteins with predicted secretion signal sequences suggests that temperature substantially influences the GAS extracellular proteome and the array of pathogen proteins that may interact with the host. Importantly, many of the genes that are differentially regulated in response to growth temperature encode hypothetical proteins of unknown function, and thereby provide additional avenues for pathogenesis research. We will show elsewhere that humans with GAS infection make antibody to many of these proteins, indicating that they are expressed in the course of host-pathogen interaction.
Supplementary Material
Acknowledgments
We thank Drs. S. Falkow, V. Kapur, and D. Relman and members of their laboratories for assistance in the development of our microarray facility, N. Hoe for critical review of the manuscript, and A. Mora for assistance with graphics.
Abbreviations
- GAS
group A Streptococcus
- RT-PCR
reverse transcription–PCR
- THY
Todd Hewitt broth supplemented with 0.2% yeast extract
- DTHY
deferrated THY
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mekalanos J J. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham M W. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caparon M G, Geist R T, Perez-Casal J, Scott J R. J Bacteriol. 1992;174:5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIver K S, Heath A S, Scott J R. Infect Immun. 1995;63:4540–4542. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanHeyningen T, Fogg G, Yates D, Hanski E, Caparon M. Mol Microbiol. 1993;9:1213–1222. doi: 10.1111/j.1365-2958.1993.tb01250.x. [DOI] [PubMed] [Google Scholar]
- 6.Xu S, Collins C M. Infect Immun. 1996;64:5399–5402. doi: 10.1128/iai.64.12.5399-5402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti J J, McShan W M, Ajdic D, Savic D J, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov A N, Kenton S, et al. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichenbaum Z, Green B D, Scott J R. Infect Immun. 1996;64:1956–1960. doi: 10.1128/iai.64.6.1956-1960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Chaussee M S, Watson R O, Smoot J C, Musser J M. Infect Immun. 2001;69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marklund S, Marklund G. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 12.Ichikawa J K, Norris A, Bangera M G, Geiss G K, van 't Wout A B, Bumgarner R E, Lory S. Proc Natl Acad Sci USA. 2000;97:9659–9664. doi: 10.1073/pnas.160140297. . (First Published August 8, 2000; 10.1073/pnas.160140297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 14.Beckert S, Kreikemeyer B, Podbielski A. Infect Immun. 2001;69:534–537. doi: 10.1128/IAI.69.1.534-537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreikemeyer B, Boyle M D, Buttaro B A, Heinemann M, Podbielski A. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 16.Litwin C M, Calderwood S B. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosa J H. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullen J J. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 19.Green M H, Vermeulen C W. Res Microbiol. 1994;145:269–272. doi: 10.1016/0923-2508(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 20.Kluger M J, Rothenburg B A. Science. 1979;203:374–376. doi: 10.1126/science.760197. [DOI] [PubMed] [Google Scholar]
- 21.Escolar L, Perez-Martin J, de Lorenzo V. J Mol Biol. 1998;283:537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- 22.Tao X, Schiering N, Zeng H Y, Ringe D, Murphy J R. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 23.Rowland B M, Grossman T H, Osburne M S, Taber H W. Gene. 1996;178:119–123. doi: 10.1016/0378-1119(96)00349-6. [DOI] [PubMed] [Google Scholar]
- 24.Ochsner U A, Vasil M L. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan H M, Sun H C. Proc Natl Acad Sci USA. 1992;89:3217–3221. doi: 10.1073/pnas.89.8.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassett D J, Howell M L, Ochsner U A, Vasil M L, Johnson Z, Dean G E. J Bacteriol. 1997;179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratledge C, Dover L G. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 28.Lynch M C, Kuramitsu H K. Infect Immun. 1999;67:3367–3375. doi: 10.1128/iai.67.7.3367-3375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto Y, Higuchi M, Poole L B, Kamio Y. Biosci Biotechnol Biochem. 2000;64:1106–1109. doi: 10.1271/bbb.64.1106. [DOI] [PubMed] [Google Scholar]
- 30.Xiong A, Singh V K, Cabrera G, Jayaswal R K. Microbiology. 2000;146:659–668. doi: 10.1099/00221287-146-3-659. [DOI] [PubMed] [Google Scholar]
- 31.Quail M A, Jordan P, Grogan J M, Butt J N, Lutz M, Thomson A J, Andrews S C, Guest J R. Biochem Biophys Res Commun. 1996;229:635–642. doi: 10.1006/bbrc.1996.1856. [DOI] [PubMed] [Google Scholar]
- 32.Ohlsen K, Koller K P, Hacker J. Infect Immun. 1997;65:3606–3614. doi: 10.1128/iai.65.9.3606-3614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podbielski A, Woischnik M, Pohl B, Schmidt K H. Med Microbiol Immunol. 1996;185:171–181. doi: 10.1007/s004300050028. [DOI] [PubMed] [Google Scholar]
- 34.Federle M J, McIver K S, Scott J R. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaussee M S, Ajdic D, Ferretti J J. Infect Immun. 1999;67:1715–1722. doi: 10.1128/iai.67.4.1715-1722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konkel M E, Tilly K. Microbes Infect. 2000;2:157–166. doi: 10.1016/s1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- 37.Maurelli A T. Microb Pathog. 1989;7:1–10. doi: 10.1016/0882-4010(89)90106-x. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson B, Schwan T G, Rosa P A. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bono J L, Tilly K, Stevenson B, Hogan D, Rosa P. Microbiology. 1998;144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 40.Hurme R, Rhen M. Mol Microbiol. 1998;30:1–6. doi: 10.1046/j.1365-2958.1998.01049.x. [DOI] [PubMed] [Google Scholar]
- 41.Hoe N P, Goguen J D. J Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilar P S, Hernandez-Arriaga A M, Cybulski L E, Erazo A C, de Mendoza D. EMBO J. 2001;20:1681–1691. doi: 10.1093/emboj/20.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lafontaine E R, Sokol P A. J Bacteriol. 1998;180:2836–2841. doi: 10.1128/jb.180.11.2836-2841.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barton H A, Johnson Z, Cox C D, Vasil A I, Vasil M L. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu P V. J Infect Dis. 1973;128:506–513. doi: 10.1093/infdis/128.4.506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.