Summary
Even though the oral microbiome is one of the most complex sites on the body it is an excellent model for narrow‐spectrum antimicrobial therapy. Current research indicates that disruption of the microbiome leads to a dysbiotic environment allowing for the overgrowth of pathogenic species and the onset of oral diseases. The gram‐negative colonizer, Porphyromonas gingivalis has long been considered a key player in the initiation of periodontitis and Streptococcus mutans has been linked to dental caries. With antibiotic research still on the decline, new strategies are greatly needed to combat infectious diseases. By targeting key pathogens, it may be possible to treat oral infections while allowing for the recolonization of the beneficial, healthy flora. In this review, we examine unique strategies to specifically target periodontal pathogens and address what is needed for the success of these approaches in the microbiome era.
Keywords: antibiotic resistance, antibiotics, microbiome, oral health, selective drug target
1. INTRODUCTION
Humans are colonized with over 100 trillion microbes, bringing new meaning to poet John Donne's line that “no man is an island, entire of itself.”1 Following the Human Genome Project, the Human Microbiome Project, sponsored by the US National Institute of Health, was undertaken to sequence and identify our resident microbiome in order to further characterize our collective genome. Currently, the Human Microbiome Project has characterized more than 70 million 16s ribosomal RNA sequences from 15 body sites.2 The focus on humans as supraorganisms has fueled further interest in the complex interactions between us and our microbiota. Recent studies suggest that our microbiome plays a significant role in our development and health.3 Some of these roles involve establishing our immune system after birth,4, 5 influencing how our brains process information,6 and aiding in digestion and nutrient acquisition.7 Disruption of this natural ecosystem has been linked to adverse health effects in various parts of the body. Antibiotic‐associated diarrhea caused by Clostridium difficile is a distinctive case of changes in the microbiome. The use of antimicrobials disrupts the normal gut microbiota, allowing for the overgrowth of C. difficile, resulting in fever, severe diarrhea, and colitis.8, 9
The importance of the oral microbiome to our health is no exception. It is a complex environment and one of the most diverse sites on the human body, containing up to 1000 phylotypes composed of viruses, protozoa, fungi, archaea and bacteria.2, 10 It is established through the colonization by bacteria of the hard and soft surfaces (eg teeth, surfaces of the tongue, and epithelium) of the oral cavity to form biofilms, more commonly known as dental plaque. However, not all of these microorganisms are considered pathogenic and many play a key role in maintaining both oral and systemic health. For one, commensal bacteria can prevent the colonization of pathogens, a phenomenon known as colonization resistance.11 These commensal organisms occupy the niche, limiting the available space and nutrients, and thereby preventing the establishment of foreign colonizers. As an indirect mechanism, commensal colonizers can produce antagonistic substances against pathogenic species. Many streptococcal species in the oral cavity can synthesize inhibitory substances that prevent colonization of other species. Streptococcus sanguinis produces hydrogen peroxide, which can inhibit the growth of methicillin‐resistant Staphylococcus aureus 12 and Streptococcus mutans, the major contributor to dental caries.13 Interestingly, studies also show that the oral microbiome plays a functional role in systemic health, possibly helping to regulate the levels of nitrite.14 This is beneficial, as metabolic studies show nitric oxide is important for maintaining cardiovascular health through improved mitochondrial function and reduced blood pressure.15 Decreased oral health has been linked to systemic co‐morbidities. Brushing and invasive dental procedures allow bacteria to enter the bloodstream and disseminate to other sites, such as the brain, lungs, and liver, linking oral diseases to a vast array of health issues from pregnancy complications to respiratory, cardiovascular, and cerebrovascular diseases.16
2. CHANGES IN THE MICROBIOME LEAD TO DISEASE
Typically, with proper oral hygiene, the oral microbiome exists in a beneficial or benign state. Changes to that natural state can lead to the onset of two of the most common bacterial infections, dental caries and periodontal disease.17 Host factors such as genetic susceptibility, smoking, diabetes mellitus, age and poor oral hygiene can lead to systemic changes (chronic inflammation and altered immune response) associated with the disruption of the oral ecosystem.18 Additionally, microbial interactions play a significant role. Based on the current keystone‐pathogen hypothesis, it has been shown that certain species can act as community activators by transforming the surrounding environment.19 These changes create a dysbiotic environment that decreases the healthy microbial diversity and favors the growth of oral pathogens that contribute to the development of oral diseases.
Periodontitis is associated with the keystone‐pathogen hypothesis. In clinically healthy hosts, the population is maintained in homeostasis, being predominately colonized by gram‐positive oral streptococci. Colonization by Porphyromonas gingivalis can selectively modify the host immune response to prevent cell clearance and allow for cytokines that are involved in bone resorption and tissue degradation to generate a nutrient‐rich, inflamed site. The now altered oral environment selects for asaccharolytic organisms, such as P. gingivalis, which can use available nutrients. In addition, the degeneration of alveolar bone provides new niches for pathogenic species to colonize.19 For dental caries, an increase in carbohydrate intake can disrupt the oral environment, selecting for cariogenic bacteria such as S. mutans, Streptococcus sobrinus, and Lactobacillus spp.20 These acidogenic bacteria metabolize dietary sugars, producing acid by‐products and creating a low‐pH environment that weakens the tooth enamel leading to tooth decay. This also contributes to dysbiosis, as the normal acid‐sensitive population is eliminated, further encouraging the colonization of cariogenic bacteria such as S. mutans.
3. THE CALL FOR TARGETED ANTIMICROBIAL THERAPY
Unfortunately, treatment for advanced oral diseases relies on the non‐specific removal of dental plaque.21 Although oral infections are associated with polymicrobial environments, the total eradication of the microbiome often leads to increased susceptibility to infection and disease re‐occurrence.22 The indiscriminate removal of the oral population allows for the recolonization of the pathogen and may lead to pathogenic species becoming dominant, decreasing the normal, healthy microbial diversity.23 It may be more prudent to only remove the dysbiotic species to allow for restoration of the normal population. In caries infections, clinical studies suggest that the absence of S. mutans provides a protective benefit. Children who are not colonized with S. mutans at an early age show a lower risk of caries development, which may contribute to the colonization resistance effect of a developed microbiome.24 Similarly, the presence of P. gingivalis is closely associated with clinical outcomes of periodontitis. Porphyromonas gingivalis has been found in 85% of patients with periodontitis and is isolated at higher levels from areas of disease progression compared with healthy sites.25, 26 In an animal periodontitis model, vaccinating against a major P. gingivalis virulence factor reduced alveolar bone loss, decreased pathogen colonization, and improved disease outcome.27 Clinical trials using human patients showed that repeated applications of monoclonal antibodies specific to a P. gingivalis protease complex prevented recolonization of the pathogen for approximately 9 months, leading to a significant improvement in oral health.28
By targeting key pathogens, such as P. gingivalis and S. mutans, it may be possible to treat periodontitis or dental caries while allowing for the protection of the beneficial, commensal flora and reducing the chance for antibiotic resistance. This brings in to play the rationale for alternative strategies such as targeted or pathogenic‐selective antimicrobials. Narrow‐spectrum production can be more cost effective long term and antibiotics designed against a limited population may also lead to faster discovery rates as targets will not need to be conserved or effective across a range of disparate species.29 With antibiotic resistance and the necessity of maintaining the healthy microbial population leaving few good treatment options available, novel approaches in antimicrobial development are imperative.
In this review, we examine targeted strategies explored in the oral cavity. It is impossible to say one species is completely responsible for periodontitis or dental caries, but studies have clearly linked certain pathogens to their progression, making the oral microbiome an excellent model for this strategy. Although not comprehensive and although the application of novel strategies within the oral microbiome is still in its infancy, we focus on a few studies aimed selectively against oral pathogens including targeted delivery systems and our own selective drug discovery project, exploiting pathogenic essential genes. We will also address what is needed for the successful development of novel antibiotics in the microbiome era.
4. STRATEGIES APPLIED AGAINST PERIODONTAL PATHOGENS
4.1. Targeting pathogenic essential genes
Essential genes are critical for the growth and survival of an organism and therefore present an ideal pool of potential drug targets. Due to essential genes being characteristically conserved,30 target‐based screens undertaken in the early years of the genomics era used a broad‐spectrum approach for drug discovery. A bioinformatics approach was applied to select genes conserved across a diverse population of species. It was assumed that if the target was essential in one species and conserved, it would be essential in other species. Unfortunately, this was not the case. In one prime example, GlaxoSmithKline initially selected more than 350 genes and screened between 260 000 and 530 000 compounds, which only yielded one lead compound against Staphylococcus aureus, targeting the fatty acid biosynthesis enzyme, FabI.31 The ability of some pathogens to use exogenous fatty acids or the presence of isozymes performing the same function limited the success rate. Yet the application of this approach could yield better results if applied in a narrow‐spectrum drug discovery campaign. Essential gene targets can be selected and separated by different categories aimed at a specific group, pathogen or environment; for instance, differences in essential genes required for cell‐wall biosynthesis in gram‐negative, gram‐positive, and mycobacteria. The cell wall components of gram‐negative bacteria contain lipopolysaccharides whereas gram‐positive bacteria express teichoic acids and Mycobacteria are composed of mycolyl‐arabinogalactan‐peptidoglycan. Another departure is the incorporation of meso‐diaminopimelate (m‐DAP) in the cell wall of gram‐negative bacteria whereas gram‐positive bacteria use lysine. Genes required for energy production in anaerobic vs aerobic respiration could also present selective targets. Some metabolic processes are crucial to cell survival only in vivo, resulting in a subset of conditionally essential genes. Nutrient‐deprived or nutrient‐replete environments may require a differential expression of vitamins, cofactors and amino acid biosynthetic pathways that are not assessed in normal laboratory conditions. Screening of Pseudomonas aeruginosa in sputum revealed essential genes not associated with minimal or rich media. These genes were connected to membrane synthesis including the outer membrane lipoprotein, OprI, and a hypothetical outer membrane chaperone.32 These variations in cell requirements can produce a different set of essential pathways and essential genes that may aid in selecting targets against a limited range of bacteria.
We undertook this approach by focusing on a pathogen‐selective target against the gram‐negative periodontal pathogen, P. gingivalis.33 We previously conducted a genome‐wide essential gene identification study in the gram‐positive, early oral colonizer Streptococcus sanguinis using systematic single and double gene deletions. Once experimentally identified, the essential genes were grouped by specific categories based on their KEGG functional annotation. When linked together, we were able to create a model of essential pathways and determined that essential genes were conserved within three major categories of biological function: maintenance of the cell envelope, energy production and processing of genetic information. The underlying benefit of this study was that we discovered general rules for essential genes, providing us with the basis for predicting essential genes in other organisms including Staphylococcus aureus, Bacillus subtilis, Streptococcus pneumoniae,34 and recently for P. gingivalis and Mycobacterium tuberculosis. Our essential gene predictions were confirmed and shown to be highly accurate in a comparison with genome‐wide gene knock‐out experimental data performed in other laboratories (>90% accuracy). Our prediction does not rely on sequence features among different species, but a gene's contribution to an essential end product. Even though the essential gene itself may differ, the overall essential function remains unchanged. We believe the reliance on genomic function and not sequence, addresses inconsistencies of essential gene data among different bacterial species as a result of genetic diversity while still allowing for an accurate prediction. This suggested that the general rule for essential genes identified through our experimental studies in S. sanguinis, could be universally applied for different bacterial species including gram‐negative bacteria.
Based on our prediction in P. gingivalis strain W83, 212 genes were essential. Of these, 46 genes fell into the maintenance of the cell envelope category. About 32% (7% of total predicted essential genes) of those genes were directly related to peptidoglycan biosynthesis whereas about 19% (4% of total) were related to fatty acid biosynthesis. Sixteen genes were involved in energy production with about 69% (5% of total) related to glycolysis. The majority of the predicted essential genes (148 genes) were grouped into the processing of genetic information category. About 9% (6% of total) were related to nucleotide biosynthesis including both purine and pyrimidine metabolism while 33% (24% of total) were related to ribosomal biosynthesis. Additionally, two genes were involved in the synthesis of cofactors for riboflavin. Although this prediction may not be comprehensive due to a lack of annotated biological data (ie hypothetical genes), the functional classification fell in line with recent experimental essential gene studies in P. gingivalis ATCC 33277.35, 36
As the healthy oral cavity comprises roughly 80% streptococcus species,37 it was theorized that selecting essential genes in P. gingivalis that were absent or not essential within S. sanguinis would present potential pathogen‐selective antimicrobial targets within the oral cavity. Based on the prediction, 68 essential genes were selective for P. gingivalis. Differences were mostly a result of alternative pathways and variations in nutritional requirements. A clear example is with regard to cell‐wall composition. Gram‐negative bacteria possess an outer membrane composed of lipopolysaccharide and lipoproteins with high lipid content. Streptococcus sanguinis expresses lipoteichoic acid on the cell membrane composed of high peptidoglycan content. Another key difference lay in terpenoid biosynthesis. The S. sanguinis uses the mevalonic acid pathway, producing terpenoids via the hydroxymethylglutaryl‐coenzyme A reductase pathway whereas P. gingivalis uses the alternate 2‐C‐methyl‐D‐erythritol 4‐phosphate/1‐deoxy‐D‐xylulose 5‐phosphate pathway or non‐mevalonate pathway. Several cell division and rod‐shape‐determining proteins were deemed essential for P. gingivalis. As it is believed that coccoid‐shape bacteria are the default morphology, these genes were unnecessary for streptococci. An interesting difference lay within lysine biosynthesis, which contained four pathway variants. The pathways differ by the substrate intermediates at the branch point of L‐2,3,4,5‐tetrahydrodipicolinate's (THDP) conversion to m‐DAP. The succinylase branch uses succinyl‐Coenzyme A to generate succinylated intermediates; similarly, the acetylase branch uses acetyl‐Coenzyme A to produce acetylated intermediates. These two variants are used by the majority of gram‐negative and gram‐positive bacteria. The aminotransferase branch, used by plants and methanococci, involves a single‐step amine transfer to produce the precursor of m‐DAP, LL‐DAP.38 For P. gingivalis, m‐DAP is directly produced by the enzyme meso‐diaminopimelate dehydrogenase (m‐Ddh, PG0806; GenBank ID: AAQ65966.1) in a single step.
It is important to note that although it is possible to identify certain essential pathways that diverge based on gram stain, nutritional requirements, or energy factors it may be difficult to select a single‐species essential gene target. Certain pathways or essential genes may be common to certain bacterial populations, limiting potential specificity. Our target, m‐Ddh, for example is not limited to P. gingivalis but is found in several other species of the Bacteroidetes phylum. However, this may be beneficial in certain environments or conditions, such as within the oral cavity, where it is clear that certain groups are evolutionarily related (Figure 1). Taking advantage of alternative pathways or isozymes and identifying a target like m‐Ddh that is evolutionarily conserved across certain populations would allow for narrow‐spectrum targeting, while sparing evolutionarily distant groups. A rational computer‐based methodology was undertaken for a high‐throughput virtual screening of small‐molecule libraries. The study identified three compounds with slight whole‐cell activity against P. gingivalis, but by taking advantage of an essential gene target limited to a subset of bacteria more potent inhibitors can be optimized and developed.
Figure 1.
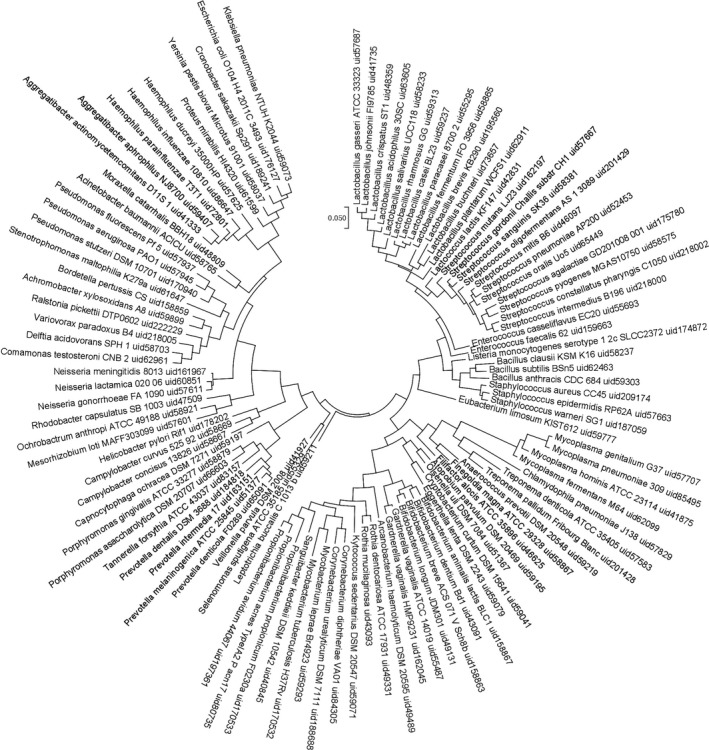
Evolutionary relationships of oral taxa classified by their essential genes. Oral microbial essential genes were identified by BLAST proteins of completed oral microbial genomes against the Database of Essential Genes (DEG). Candidate essential genes were then manually curated using our essential gene prediction model. Orthologous essential genes were identified among the oral microbial genomes and a phylogenetic tree was built based on the classification of the presence or absence of the essential genes
4.2. Selective delivery of antimicrobial peptides
Natural products have long proved to be an effective source of antimicrobials. As they are synthesized and produced by organic sources such as plants, algae, fungi and bacteria, they are naturally adapted for their environmental role. Around 1930, the modern “golden age” of antimicrobial research began, when Alexander Fleming made an accidental discovery from the fungus Penicillium notatum, resulting in the widespread use of penicillin Spanning a 40‐year period, researchers and pharmaceutical companies examined microbial and fungal metabolites for naturally occurring antibiotics leading to the discovery of the major antibiotic class scaffolds: cephalosporins, penicillins, quinolones, and macrolides, still used today. During this period, steady progress was made with improvements in the form of second‐ and third‐generation synthetic derivatives, resulting in various β‐lactams, sulfonamides, and aminoglycosides then tetracyclines, macrolides, and glycopeptides.39 By the 1980s, development began to slow. The ideal natural sources had been exhausted and any newly designed antimicrobials, by that point, were chemical modifications of common core scaffolds and targets, incrementally improving efficacy or the design of wholly synthetic compounds. However, as microbial resistance has reached a critical point, drug developers are starting to re‐examine natural compounds. Advances in screening along with tapping into previously unexplored or uncultivable sources has renewed interest in naturally derived antibiotic agents.40, 41, 42
Within the past 20 years, antimicrobial peptides (AMPs) have become of interest as a source of potential antimicrobial agents derived from natural sources. They comprise a diverse group of small‐molecular‐weight molecules that are a natural part of the host‐defense response in both eukaryotic and prokaryotic cells. These peptides are classified into subgroups based on their composition, size or structure and display broad‐spectrum activity against bacterial, parasitic, fungal, and viral infections. Around 40 different AMPs can be found in saliva, protecting the host against the overgrowth of oral pathogens and transient bacteraemia with various sensitivities.43 Their primary mechanism of action against bacteria involves the destabilization of the cell membrane orchestrated by electrostatic interactions between the negatively charged phospholipids of the bacterial cell membrane and the positively charged peptide. This interaction would result in the loss of membrane potential and integrity, leading to cell leakage and ending in cellular death. However, recent studies have shown that AMPs can kill by other mechanisms. Some AMPs can induce cellular damage by interferring with cell wall,44 DNA,45 and protein synthesis.46 Although antimicrobial peptides have proven to be effective, this process is non‐specific, targeting a broad range of gram‐negative and gram‐positive bacteria. To circumvent this, Eckert et al., designed a novel class of AMPs, which were named specifically targeted antimicrobial peptides (STAMPs), against the cariogenic pathogen, S. mutans.47 The STAMP, C16G2, consists of the non‐specific linear antimicrobial peptide that possesses broad‐spectrum killing activity. This is linked together with a targeted binding peptide containing a fragment of the S. mutans specific competence stimulation peptide (CSP). This targeting region increases binding onto the surface of the intended target, therefore increasing the antimicrobial activity and cell clearance.48 When tested, C16G2 showed strong bactericidal activity against several strains of S. mutans in both a planktonic and biofilm growth stage but did not affect the growth of S. sanguinis or Streptococcus gordonii even within a mixed‐species biofilm model.47 A short preliminary study in humans showed that treatment with the peptide decreased S. mutans colonization as well as reduced caries‐associated demineralization.49 Different antimicrobial peptides have been linked to the CSP targeting domain. Mai et al. tested the salt‐resistant AMP pleurocidin linked with the targeted CSP.50 As some AMPs have shown sensitivity at high salt concentrations,51 the use of a salt‐resistance AMP, derived from an oceanic source, may prove to be more effective in vivo where saline concentrations are typically higher. The targeted AMP, IMB‐2, killed >90% of S. mutans but only 20% of the other streptococci tested in a dual‐species culture. Another group used a similar approach against P. gingivalis. The sheep myeloid antimicrobial peptide, SMAP29, which has shown potent antimicrobial activity against Fusobacterium nucleatum, P. gingivalis, and several other periodontal organisms,52 was conjugated to a P. gingivalis surface‐specific IgG antibody.53 Treatment of P. gingivalis, Aggregatibacter actinomycetemcomitans, and Peptostreptococcus micros in a mixed‐species culture showed inconsistent but promising results. At higher concentrations, the IgG–SMAP29 conjugate killed all three species. However, it displayed slightly more specificity at lower doses with the viability of A. actinomycetemcomitans and Peptostreptococcus micros decreasing four to five times less than P. gingivalis. An attempt to improve the specificity of targeted SMAP28, a proline‐rich protein 1, which can bind to the fimbriae of P. gingivalis, was alternatively linked to the AMP.54 However, this did not appear to increase the antimicrobial activity against P. gingivalis or decrease activity against other species. Although successful with S. mutans and the species‐specific competence peptide, a conjugated, pathogen‐specific targeting domain may be difficult to design. However, due to the potency of AMPs, targeted AMPs present promising alternatives to conventional approaches.
4.3. Inhibition of pathogenic biofilm
Biofilms are bacterial communities encased within an extracellular matrix composed of exopolysaccharides, proteins, lipids, DNA, and ions. These structures are ubiquitous in nature and represent a major health concern because they are a cause of persistent infections and are estimated to account for 80% of all bacteria‐related infections. They provide the bacteria with protection from external stresses and decrease their susceptibility to antimicrobial therapy and immune clearance, making treatment extremely difficult.55 The establishment of biofilms within the oral cavity is essential for the formation of dental plaque and the subsequent development of oral diseases. Oral streptococci, particularly S. mutans, can use sucrose obtained from the diet to synthesize extracellular and intracellular polysaccharides.56, 57 Bacterial exopolysaccharides are a primary component of the extracellular polymeric substances in biofilms and serve as a scaffold for the attachment of S. mutans to the tooth surface and allow colonization by other bacteria. Along with colonization and protection, the biofilm architecture facilitates an environment favorable to the survival of cariogenic pathogens. The development of three‐dimensional structures allows for the creation of water channels that connect to transport nutrients and signaling molecules and preserve the low‐pH conditions.58, 59
Molecules that could specifically inhibit the formation or disperse biofilms formed by S. mutans could be a unique strategy for controlling dental caries. There are several strategies to control biofilm development, such as inhibiting attachment, preventing cell–cell communication or promoting the dispersal of the exopolysaccharides.60 These inhibitors are typically derived from natural products such as garlic, ginseng, cranberry, and bacteriophages. This method has been shown to be effective against dental caries with several studies indicating anti‐biofilm activity within the oral cavity61, 62, 63; however, the compounds are non‐specific and run the risk of promoting dysbiosis. Recent studies by Garcia et al. identified a small‐molecule inhibitor that selectively dispersed biofilms formed by S. mutans but did not significantly affect the cell viability.64 The group screened 600 compounds and identified 3F1, based on 2‐aminoimidazole, a derivative of bromoageliferin from marine‐sponge products that may structurally mimic quorum‐sensing molecules, contributing to their ability to disperse biofilms.65, 66 These compounds have previously displayed anti‐biofilm activity against various microorganisms, including Pseudomonas aeruginosa, Acinetobacter baumannii, and Bordetella bronchispetica.67 However, the mechanism of action is unclear. The anti‐biofilm effect was still observed when tested against deletion mutants involved in the initial development of the biofilm such as the glucosyltransferase enzymes responsible for the synthesis of glucans and eDNA within the structure. It is speculated that the small molecule may interact with proteins related with the biofilm architecture. The compound did not affect in vitro biofilms formed by commensals S. sanguinis and S. gordonii nor the mutans family member S. sobrinus. More promising was the consistency of the oral microbiota when 3F1 was administered in a rat caries model. Sequencing following a 4‐week treatment showed fewer S. mutans compared with the no treatment control but no significant changes in the colonization at the phylum level. This work was based on their previous studies.65 A focused library of 506 small molecules based on the 2‐aminoimidazole backbone was used to identify eight compounds that inhibited S. mutans biofilm by at least 50%. These compounds altered the normal three‐dimensional structure, leading to a thinner structure, but did not affect the biofilm of S. sanguinis or S. gordonii. However, the most active compound did exhibit slight bactericidal activity against S. mutans, S. sanguinis and S. gordonii when cultured in a mixed‐species. Interestingly, these compounds appeared to affect the production of antigen I/II and the glucosyltransferases, indicating a potential mechanism of specificity. As the biofilm is a key virulence factor in the pathogenesis of S. mutans, identifying an agent that prevents biofilm establishment could be an effective strategy. However, it is not clear whether the dispersal of the biofilm without specifically inhibiting cell growth would lead to long‐term clinical benefits. Viable S. mutans within a dispersed biofilm may disseminate and colonize at other sites, leading to its re‐establishment within the oral cavity. Anti‐biofilm agents may be most effective in conjunction with targeted antimicrobials as the removal of the biofilm would allow for easier penetration of the drug.
5. FUTURE STRATEGIES FOR THE DEVELOPMENT OF TARGETED ANTIMICROBIAL THERAPY
As the microbiome is a complex system, it is necessary to thoroughly understand the microbial composition and its link to health and disease. Although the oral microbiome has been studied for years and there is a baseline for a healthy vs diseased population, other sites are not as clearly characterized. Unfortunately, many microorganisms cannot be cultured using standard laboratory techniques and this could leave out a significant contributor to the environmental ecology. Culture‐independent approaches offer a powerful tool to analyze diverse microbial communities.68 Next‐generation sequencing, metagenomics and metatranscriptomics can generate profiles based on a population or a specific species related to health vs disease state. Additionally, it can present transcriptional profiles based on gene expression and this could provide biomarkers.69 If an expression profile for a specific pathogen can be associated with an infection then a subset of gene targets can be identified. Although useful, there still may be issues with clinical practicality due to cost associations, limited reference databases and data analysis. The transcriptional profile could be beneficial for diagnostics. As time is essential when treating a patient and the identification of a positive culture can take days, most clinicians rely on broad‐spectrum antibiotics as a first line of defense. For targeted antimicrobial therapy to be effective, a system for the rapid identification of an etiological agent is necessary. By generating a model for expression in certain infectious diseases, sequencing and polymerase chain reaction‐based methods can facilitate an increase in personalized treatment.
Alternative strategies to classical approaches should be thoroughly considered. As previously stated, although essential gene targets may have limitations against broad‐spectrum screening, it could be used as a successful application when targeting a limited range of species. The targeting of in vivo or conditionally essential genes as well as pathogen‐specific virulence factors offers another subset of targets. Due to environmental variations, colonization, immune evasion or nutrient acquisition, genes crucial for the survival within the host can be an important resource for selective targeting. Combining this method with novel delivery systems would allow for increased specificity. A small‐molecule inhibitor, AMP or other antimicrobial agents could be conjugated to a pathogen‐specific siderophore, packaged within a phage delivery system or linked to a targeting moiety similar to STAMP technology. CRISPR‐Cas (Clustered Regularly Interspaced Short Palindromic Repeat Spacers) systems offer another unique approach to target specific bacteria. CRISPR can be designed to target a specific essential gene. Recent studies have shown that CRISPR RNA systems have the ability to distinguish between different strains of the same species within a mixed population.70
A strong emphasis needs to be placed on research and development. Scientific challenges, compounded by the regulatory and financial burden of antibiotic research, have promoted many pharmaceutical companies to exit this area. As antibiotics are short‐term prescriptions, the investment put in is not returned. Recently, the US Department of Health and Human Resources has proposed a model of dissociating profit by units sold allowing the pharmaceutical company to be rewarded with a known financial return. Although the amount would have to be substantial, if successful, the promise of profit would help to drive development. Companies also need to develop a large multi‐disciplinary team. As technology continues to advance, new approaches can be used. Experts in a vast network of fields such as bioinformatics, sequencing, biochemistry, microbiology, and medicinal chemistry are needed from target selection, assay development, and screening to lead identification and hit optimization. Regardless of the challenges, we must embrace new strategies if we are to move forward in this new era.
6. CONCLUSIONS
The oral community is a complex environment that has evolved into a highly regulated state of harmony between hundreds of microbial species. A disturbance in that natural balance alters the surrounding environment, favoring the colonization of pathogenic species. This change in bacterial composition is correlated with the onset of disease. We now know dental caries and periodontal disease are two diseases related to these modulations in the microbiome homeostasis with S. mutans and P. gingivalis playing critical roles. Conventional methods for controlling or reducing plaque biofilm result in the total eradication of the microbiome. Unfortunately, this only fuels the imbalance by removing the beneficial colonizers and allowing for pathogenic microorganisms to re‐colonize. Therefore, clinical treatment should aim to re‐establish the equilibrium by targeting key pathogens (Figure 2).
Figure 2.
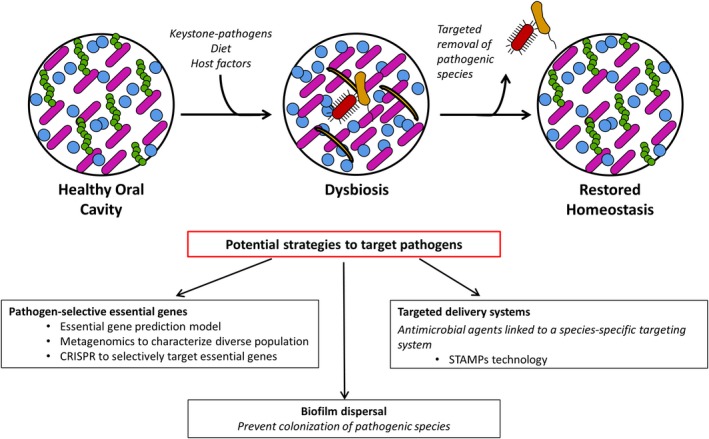
Targeted antimicrobial treatment to restore the microbial homeostasis. Various factors including manipulation by keystone‐pathogens and changes in diet and other host factors can alter the oral environment leading to changes in the microbial population. The altered environment selects for the growth of certain species, reducing the healthy, microbial diversity. Potential strategies such as the identification of pathogen‐selective essential gene targets, the dispersal of the pathogenic biofilm or the selective delivery of antimicrobial agents could be used to only eliminate the dysbiotic or pathogenic species. This would allow for the re‐establishment of the healthy microbial population and restore homeostasis
Although we highlighted studies for specifically targeting S. mutans and P. gingivalis, it is important to note that the aim of targeted therapeutics would be to re‐establish homeostasis. These strategies could be applied to other acidogenic or periodontitis‐associated bacteria. The use of CSP for STAMP technology may not be effective for all strains of S. sobrinus, another contributor to dental caries, but a bactericidal agent could be linked with other species‐specific delivery systems. Targeted therapy has become fundamental in other areas such as cancer therapy. Similarly, treatment for disease and other infections should adopt an individualized treatment geared towards the specific infection and patient. The development of targeted strategies antibiotics may prove more effective as previously discounted drugs with limited activity can be revisited, screening and testing may be more feasible in a smaller subset and different approaches can be used based on the pathogen of interest. Therefore, it is imperative that we look at where we are and ask how we can further progress species‐selective antibiotic drug development.
ACKNOWLEDGEMENTS
The authors thank Dr. Xiangzhen Kong for preparation of Figure 1. This work was supported by the National Institutes of Health, USA, grants R01DE018138 and R01DE023078 (P.X.) and F31DE024038 (V.S.).
Stone VN, Xu P. Targeted antimicrobial therapy in the microbiome era. Mol Oral Microbiol. 2017;32:446–454. https://doi.org/10.1111/omi.12190
REFERENCES
- 1. Donne J. Meditation XVII In: Alford H, ed. The Works of John Donne, Vol. III. London: John W. Parker; 1839:574‐5. [Google Scholar]
- 2. Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mayer EA, Knight R, Mazmanian SK, Cryan JF, Tillisch K. Gut microbes and the brain: paradigm shift in neuroscience. J Neurosci. 2014;34:15490‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krajmalnik‐Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. 2012;27:201‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile . Gastroenterology. 2014;146:1547‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khanna S, Pardi DS, Aronson SL, et al. The epidemiology of community‐acquired Clostridium difficile infection: a population‐based study. Am J Gastroenterol. 2012;107:89‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192:5002‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uehara Y, Kikuchi K, Nakamura T, et al. H2O2 produced by viridans group streptococci may contribute to inhibition of methicillin‐resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin Infect Dis. 2001;32:1408‐13. [DOI] [PubMed] [Google Scholar]
- 13. Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187:7193‐203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kapil V, Haydar SM, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. Physiological role for nitrate‐reducing oral bacteria in blood pressure control. Free Radical Biol Med. 2013;55:93‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lundberg JO, Carlstrom M, Larsen FJ, Weitzberg E. Roles of dietary inorganic nitrate in cardiovascular health and disease. Cardiovasc Res. 2011;89:525‐532. [DOI] [PubMed] [Google Scholar]
- 16. Babu NC, Gomes AJ. Systemic manifestations of oral diseases. J Oral Maxillofacial Pathol. 2011;15:144‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Albandar JM. Epidemiology and risk factors of periodontal diseases. Dent Clin North Am. 2005;49:517‐532, v‐vi. [DOI] [PubMed] [Google Scholar]
- 18. AlJehani YA. Risk factors of periodontal disease: review of the literature. Int J Dentistry. 2014;2014:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Hajishengallis G, Darveau RP, Curtis MA. The keystone‐pathogen hypothesis. Nat Rev Microbiol. 2012;10:717‐725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Selwitz RH, Ismail AI, Pitts NB. Dental caries. Lancet. 2007;369:51‐59. [DOI] [PubMed] [Google Scholar]
- 21. Dar‐Odeh NS, Abu‐Hammad OA, Al‐Omiri MK, Khraisat AS, Shehabi AA. Antibiotic prescribing practices by dentists: a review. Ther Clin Risk Manag. 2010;6:301‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol. 2015;6:1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perez F, Pultz MJ, Endimiani A, Bonomo RA, Donskey CJ. Effect of antibiotic treatment on establishment and elimination of intestinal colonization by KPC‐producing Klebsiella pneumoniae in mice. Antimicrob Agents Chemother. 2011;55:2585‐2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Law V, Seow WK, Townsend G. Factors influencing oral colonization of mutans streptococci in young children. Aust Dent J 2007;52:93‐100, quiz 159. [DOI] [PubMed] [Google Scholar]
- 25. Haffajee AD, Socransky SS, Patel MR, Song X. Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 2008;23:196‐205. [DOI] [PubMed] [Google Scholar]
- 26. How KY, Song KP, Chan KG. Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line. Front Microbiol 2016;7:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Page RC, Lantz MS, Darveau R, et al. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis . Oral Microbiol Immunol. 2007;22:162‐168. [DOI] [PubMed] [Google Scholar]
- 28. Booth V, Ashley FP, Lehner T. Passive immunization with monoclonal antibodies against Porphyromonas gingivalis in patients with periodontitis. Infect Immun. 1996;64:422‐427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bax R, Green S. Antibiotics: the changing regulatory and pharmaceutical industry paradigm. J Antimicrob Chemother. 2015;70:1281‐1284. [DOI] [PubMed] [Google Scholar]
- 30. Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962‐968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29‐40. [DOI] [PubMed] [Google Scholar]
- 32. Lee SA, Gallagher LA, Thongdee M, et al. General and condition‐specific essential functions of Pseudomonas aeruginosa . Proc Natl Acad Sci USA. 2015;112:5189‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stone VN, Parikh HI, El‐rami F, et al. Identification of small‐molecule inhibitors against meso‐2, 6‐diaminopimelate dehydrogenase from Porphyromonas gingivalis . PLoS ONE. 2015;10:e0141126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu P, Ge X, Chen L, et al. Genome‐wide essential gene identification in Streptococcus sanguinis . Sci Rep. 2011;1:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hutcherson JA, Gogeneni H, Yoder‐Himes D, et al. Comparison of inherently essential genes of Porphyromonas gingivalis identified in two transposon‐sequencing libraries. Mol Oral Microbiol. 2016;31:354‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis . BMC Genomics. 2012;13:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599‐1607. [DOI] [PubMed] [Google Scholar]
- 38. Pavelka MS Jr, Jacobs WR Jr. Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis . J Bacteriol. 1996;178:6496‐6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiong ZQ, Wang JF, Hao YY, Wang Y. Recent advances in the discovery and development of marine microbial natural products. Mar Drugs. 2013;11:700‐717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong WR, Oliver AG, Linington RG. Development of antibiotic activity profile screening for the classification and discovery of natural product antibiotics. Chem Biol. 2012;19:1483‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gorr SU, Abdolhosseini M. Antimicrobial peptides and periodontal disease. J Clin Periodontol. 2011;38(Suppl 11):126‐141. [DOI] [PubMed] [Google Scholar]
- 44. Chen X, Hirt H, Li Y, Gorr SU, Aparicio C. Antimicrobial GL13K peptide coatings killed and ruptured the wall of Streptococcus gordonii and prevented formation and growth of biofilms. PLoS ONE. 2014;9:e111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Limoli DH, Rockel AB, Host KM, et al. Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. PLoS Pathog. 2014;10:e1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mardirossian M, Grzela R, Giglione C, et al. The host antimicrobial peptide Bac71‐35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem Biol. 2014;21:1639‐1647. [DOI] [PubMed] [Google Scholar]
- 47. Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. Targeted killing of Streptococcus mutans by a pheromone‐guided “smart” antimicrobial peptide. Antimicrob Agents Chemother. 2006;50:3651‐3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He J, Yarbrough DK, Kreth J, Anderson MH, Shi W, Eckert R. Systematic approach to optimizing specifically targeted antimicrobial peptides against Streptococcus mutans . Antimicrob Agents Chemother. 2010;54:2143‐2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eckert R, Sullivan R, Shi W. Targeted antimicrobial treatment to re‐establish a healthy microbial flora for long‐term protection. Adv Dent Res. 2012;24:94‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mai J, Tian XL, Gallant JW, et al. A novel target‐specific, salt‐resistant antimicrobial peptide against the cariogenic pathogen Streptococcus mutans . Antimicrob Agents Chemother. 2011;55:5205‐5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta‐defensin‐1 is a salt‐sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553‐560. [DOI] [PubMed] [Google Scholar]
- 52. Weistroffer PL, Joly S, Srikantha R, Tack BF, Brogden KA, Guthmiller JM. SMAP29 congeners demonstrate activity against oral bacteria and reduced toxicity against oral keratinocytes. Oral Microbiol Immunol. 2008;23:89‐95. [DOI] [PubMed] [Google Scholar]
- 53. Franzman MR, Burnell KK, Dehkordi‐Vakil FH, Guthmiller JM, Dawson DV, Brogden KA. Targeted antimicrobial activity of a specific IgG‐SMAP28 conjugate against Porphyromonas gingivalis in a mixed culture. Int J Antimicrob Agents. 2009;33:14‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bratt CL, Kohlgraf KG, Yohnke K, Kummet C, Dawson DV, Brogden KA. Communication: Antimicrobial Activity of SMAP28 with a targeting domain for Porphyromonas gingivalis . Probiotics Antimicrob Proteins. 2010;2:21‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114‐122. [DOI] [PubMed] [Google Scholar]
- 56. Bowen WH. Do we need to be concerned about dental caries in the coming millennium? Crit Rev Oral Biol Med. 2002;13:126‐131. [DOI] [PubMed] [Google Scholar]
- 57. Koo H, Xiao J, Klein MI. Extracellular polysaccharides matrix – an often forgotten virulence factor in oral biofilm research. Int J Oral Sci. 2009;1:229‐234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Flemming H‐C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623‐633. [DOI] [PubMed] [Google Scholar]
- 59. Xiao J, Koo H. Structural organization and dynamics of exopolysaccharide matrix and microcolonies formation by Streptococcus mutans in biofilms. J Appl Microbiol. 2010;108:2103‐2113. [DOI] [PubMed] [Google Scholar]
- 60. Rabin N, Zheng Y, Opoku‐Temeng C, Du Y, Bonsu E, Sintim HO. Agents that inhibit bacterial biofilm formation. Future Med Chem. 2015;7:647‐671. [DOI] [PubMed] [Google Scholar]
- 61. Yang H, Bi Y, Shang X, et al. Antibiofilm activities of a novel chimeolysin against Streptococcus mutans under physiological and cariogenic conditions. Antimicrob Agents Chemother. 2016;60:7436‐7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murugan K, Sekar K, Sangeetha S, Ranjitha S, Sohaibani SA. Antibiofilm and quorum sensing inhibitory activity of Achyranthes aspera on cariogenic Streptococcus mutans: an in vitro and in silico study. Pharm Biol. 2013;51:728‐736. [DOI] [PubMed] [Google Scholar]
- 63. Otsuka R, Imai S, Murata T, et al. Application of chimeric glucanase comprising mutanase and dextranase for prevention of dental biofilm formation. Microbiol Immunol. 2015;59:28‐36. [DOI] [PubMed] [Google Scholar]
- 64. Garcia SS, Blackledge MS, Michalek S, et al. Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. J Dent Res. 2017;96(7):807‐814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu C, Worthington RJ, Melander C, Wu H. A new small molecule specifically inhibits the cariogenic bacterium Streptococcus mutans in multispecies biofilms. Antimicrob Agents Chemother. 2011;55:2679‐2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kaufmann GF, Sartorio R, Lee SH, et al. Revisiting quorum sensing: discovery of additional chemical and biological functions for 3‐oxo‐N‐acylhomoserine lactones. Proc Natl Acad Sci USA. 2005;102:309‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rogers SA, Melander C. Construction and screening of a 2‐aminoimidazole library identifies a small molecule capable of inhibiting and dispersing bacterial biofilms across order, class, and phylum. Angew Chem. 2008;47:5229‐5231. [DOI] [PubMed] [Google Scholar]
- 68. Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525‐552. [DOI] [PubMed] [Google Scholar]
- 69. Dix A, Hünniger K, Weber M, Guthke R, Kurzai O, Linde J. Biomarker‐based classification of bacterial and fungal whole‐blood infections in a genome‐wide expression study. Front Microbiol. 2015;6:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome‐targeting CRISPR‐Cas systems. MBio. 2014;5:e00928‐00913. [DOI] [PMC free article] [PubMed] [Google Scholar]


