Summary
Rabbit haemorrhagic disease virus (RHDV) is a lagovirus that can cause fatal hepatitis (rabbit haemorrhagic disease, RHD) with mortality of 80–90% in farmed and wild rabbits. Since 1986, RHDV has caused outbreaks in rabbits (Oryctolagus cuniculus) in Europe, but never in European brown hares (Lepus europaeus, EBH). In 2010, a new RHDV‐related virus, called RHDV2, emerged in Europe, causing extended epidemics because it largely overcame the immunity to RHDV present in most rabbit populations. RHDV2 also was identified in Cape hare (Lepus capensis subsp. mediterraneus) and in Italian hare (Lepus corsicanus). Here, we describe two distinct incidents of RHDV2 infection in EBH that occurred in Italy (2012) and Spain (2014). The two RHDV2 strains caused macroscopic and microscopic lesions similar to European brown hare syndrome (EBHS) in hares, and they were genetically related to other RHDV2 strains in Europe. EBHs are common in Europe, often sharing habitat with rabbits. They likely have been exposed to high levels of RHDV2 during outbreaks in rabbits in recent years, yet only two incidents of RHDV2 in EBHs have been found in Italy and Spain, suggesting that EBHs are not a primary host. Instead, they may act as spillover hosts in situations when infection pressure is high and barriers between rabbits and hares are limited, resulting in occasional infections causing EBHS‐like lesions. The serological survey of stocked hare sera taken from Italian and Spanish hare populations provided an understanding of naturally occurring RHDV2 infection in the field confirming its sporadic occurrence in EBH. Our findings increase the knowledge on distribution, host range and epidemiology of RHDV2.
Keywords: lagovirus, European brown hare, Lepus europaeus, rabbit haemorrhagic disease virus type 2, Spain, Italy
Introduction
Rabbit haemorrhagic disease (RHD) and European brown hare syndrome (EBHS) are caused by two highly related but phylogenetically distinct viruses, belonging to the genus Lagovirus of the family Caliciviridae. Historically, RHDV and EBHSV infections have been restricted to specific hosts both naturally and experimentally (Lenghaus et al., 1994; Lavazza et al., 1996; Bergin et al., 2009) and these viruses initially were considered genus‐specific.
RHDV was thought to infect almost exclusively wild and domestic European rabbits (Oryctolagus cuniculus), causing 80–90% mortality (OIE – Rabbit haemorrhagic Disease, [Link]), with just one report of infection in another lagomorph species, the Iberian hare (Lepus granatensis) (Lopes et al., 2014). RHDV strains can be divided into three phylogenetically distinct groups (Kerr et al., 2009) and at least six genogroups (G1–G6) based on temporal distribution (Le Gall‐Reculé et al., 2003). The genetic group G6 includes those isolates identified as the main antigenic variant called RHDVa (Capucci et al., 1998; Schirrmeier et al., 1999). In 2010, a new RHDV‐related pathogenic virus was first detected in France in wild and farmed rabbits (Le Gall‐Reculé et al., 2011), and rapidly spread throughout Europe, causing losses in farmed and wild rabbits in France (Le Gall‐Reculé et al., 2013), Italy (Puggioni et al., 2013), Portugal (Abrantes et al., 2013), Spain (Dalton et al., 2012, 2014), Germany (FLI information, 2013), the United Kingdom (Westcott et al., 2014), Malta (Lavazza, A. and Capucci, L., unpublished results, 2012), Norway (OIE, WAHID (World Animal Health Information Database), 2014a), Denmark (OIE, WAHID (World Animal Health Information Database), 2014b), Sweden (OIE, WAHID (World Animal Health Information Database), 2015), Switzerland (OIE, WAHID (World Animal Health Information Database), 2016a) and Finland (OIE, WAHID (World Animal Health Information Database), 2016b). This new virus, called RHDV2, has a sufficiently different antigenic profile from that of RHDV that it could be considered a distinct serotype. Moreover, among the specific and distinguishing characteristics of RHDV2 infection that include slightly longer disease duration, variable mortality rates (5–60%) and capacity to infect young rabbits (from 10 to 15 days of age), it also has the capacity to infect and cause an EBHS‐like disease in Cape hares (L. capensis subsp. mediterraneus) (Puggioni et al., 2013), which may be considered another primary host. In addition, a single sporadic case of infection and disease caused by RHDV2 was reported in the Italian hares (L. corsicanus) (Camarda et al., 2014).
EBHS was first reported in Sweden in the early 1980s (Gavier and Morner, 1991) and is restricted to Europe. EBHS has been endemic in Italy since the first description in the late 1980s (Poli et al., 1991). On the contrary, just one case has been reported and confirmed in Spain, in the Central Pyrenees in 1998 (Fernández de Luco et al., 2003). EBHSV was detected mainly in European brown hares (EBH, L. europaeus) but also in mountain hares (L. timidus) and Italian hares (L. corsicanus). EBHS has never been described in other European hare species such as L. granatensis and L. castroviejoi, both present on the Iberian Peninsula, or Cape hares in Sardinia (Italy) (Puggioni et al., 2013). However, field and experimental data recently have demonstrated that in Italy, the eastern cottontail (Sylvilagus floridanus) is susceptible to infection with EBHSV, occasionally resulting in EBHS‐like disease (Lavazza et al., 2015).
Infection of RHDV2 in some hare species and of EBHSV in cottontails suggests a broader susceptibility of different species within the order Lagomorpha to lagoviruses, particularly RHDV2, which also may pose a risk for EBH populations. Therefore, following the first RHDV2 detection in north‐eastern Italy in summer 2011 in industrial farmed rabbits, and after the first RHDV2 case occurred in wild rabbits in Trento Province on January 2012 (Paternolli, S. and Dellamaria, D., personal communication, 2012), targeted surveillance was implemented for wild lagomorphs. Any case of lagomorph mortality, including hares, was reported to veterinary authorities, and carcasses were submitted to the veterinary laboratories such as IZSLER, and the others of the national network, for necropsy, diagnosis and typing of lagoviruses (RHDV, RHDV2 and EBHS).
According to Dalton et al. (2014, 2012), RHDV2 (called RHDVb by these authors) was first detected in Spain in October 2011 and spread through a large number of Spanish provinces in a relatively short period of time, largely replacing the previously predominant G1 RHDV genotypes. Almost all of the studied outbreaks (94.5%) in Spanish farms in 2012 were caused by RHDV2, and only 3 of the 55 farms were affected by classic RHDV.
The objective of this study was to provide the first description of sporadic cases of RHDV2 infection in EBHs in Lombardia, northern Italy and Catalonia, north‐east Spain, using virological and pathological data from three European brown hares found dead with EBHS‐like disease. Serosurveys of EBHs in Italy and Spain are presented to provide an epidemiological context for these cases.
Materials and Methods
In November 2012, a wild adult hare found dead by a hunter near Cavernago (45°38′00″N 9°46′00″E) in the Province of Bergamo (Lombardy, northern Italy), was frozen and then transferred to IZSLER for laboratory investigations (307946/2012). In northern Italy, where EBHS is endemic since the mid‐1980s, EBH and mountain hare are the only hare species present. Identification of the individual as an adult EBH was achieved based on phenotypic and morphological characters as well as on the habitat type of the sampling area (plain landscape, 199 m.s.l.). Species identification was confirmed through the identification of specific hare IgG from liver extract using an in‐house MAb‐based anti‐Ig ELISA (Capucci, L., data not shown).
The frozen hare carcass was thawed at IZSLER for examination. Various tissue (kidney, lung, spleen, liver, gut) specimens were collected during necropsy for laboratory analyses. Bacteriological cultures from all tissues were performed in aerobic and anaerobic conditions on enrichment media (blood agar – OXOID) and selective media (Gassner Agar – OXOID), with incubation at 37° for 24 h to confirm or discard most common bacterial infections (Pasteurella sp., E. coli, Clostridia, Yersinia pseudotuberculosis, etc.). Parasitological investigations were carried out by preparing fresh smears of the gut content and by flotation. Portions of target organs (liver and spleen) were taken for lagovirus testing by sandwich ELISA. However, tissues were too autolysed for histopathological analysis.
Two adult EBHs (LE14020 and LE14021) were found dead in late February 2014 near Oristà (41°56′04.92″N 2°03′42.84″E) in Barcelona Province (Catalonia, north‐eastern Spain) and submitted to the Wildlife Diseases Research Group (SEFaS, Veterinary School, Universitat Autònoma de Barcelona) for pathological investigation. Both animals were free‐ranging wild hares from the same geographical area, where only EBH and not Iberian hares are present. Species identification to confirm they were EBH was made on the basis of phenotypic characteristics and morphometric measurements (Palacios, 1989) (Fig. 1a).
Figure 1.
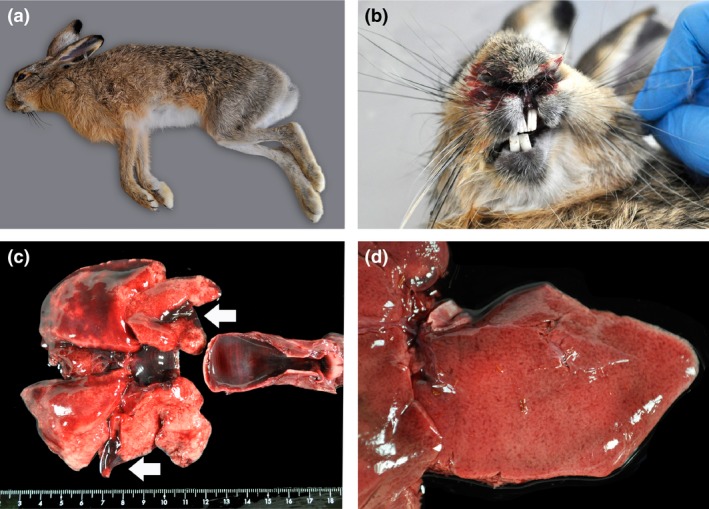
Specimen, epistaxis and macroscopic lesions in a brown hare found dead in Barcelona Province, Spain. (a) One of the European brown hares found dead in Barcelona Province, Spain; (b) epistaxis; (c) diffuse hyperaemia of the tracheal mucosa and multifocal haemorrhages in lungs (arrows); and (d) generalized reticular pattern in the liver suggestive of zonal vacuolar hepatocellular degeneration and necrosis (pictures taken after sampling the tissue).
Because necropsy findings were consistent with EBHS, additional laboratory investigations were focused on diagnosis of this disease. At necropsy, tissues samples (brain, trachea, lungs, heart, liver, spleen and kidneys) were collected, fixed in 4% neutral buffered formalin and prepared for routine microscopic examination at the Veterinary Faculty, Universitat Autònoma de Barcelona in Spain. Paraffin blocks of different organs were also sent to SVA (National Veterinary Institute in Uppsala, Sweden) for additional microscopic examination and polymerase chain reaction for EBHSV. Formalin‐fixed, paraffin embedded tissues were processed routinely for microscopic examination and stained with haematoxylin and eosin.
Four 6‐μm‐thick sections of paraffin blocks containing liver were processed for RNA extraction following the procedures described by Boom et al. (1990) and Gomez‐Laguna et al. (2010) with the following modifications. During paraffin removal, a single cycle of heating was followed by warming at 55°C, centrifugation and then placement on ice. Detection of EBHSV was carried out using nested RT‐PCR as described by Ros Bascuñana et al. (1997). Briefly, one step RT‐PCR was performed using the Qiagen kit following the manufacturer's instructions (PCR I) followed by a nested PCR (PCR II). Because PCR did not detect EBHSV but histological lesions were consistent with lagovirus infection, frozen liver samples were sent from Spain to the OIE Reference laboratory for RHD (IZSLER; Brescia, Italy) for further analysis.
To perform initial diagnosis of lagovirus and particularly to establish the presence of EBHSV in the livers of the three hares, two independent ELISA tests, both based on the use of specific monoclonal antibodies (MAbs) produced against RHDV, RHDVa, RHDV2 and EBHSV, were used; that is, (i) the ‘typing ELISA’ is a sandwich ELISA employing a panel of MAbs able to identify the lagovirus species present in the samples (RHDV or EBHSV) (Capucci et al., 1995; McIntosh et al., 2007); and (ii) the ‘subtyping ELISA’ based on the use of a group of RHDV2 specific MAbs to specifically identify RHDV2 (Le Gall‐Reculé et al., 2013; Puggioni et al., 2013).
Presence of EBHSV and RHDV2 RNA was tested by RT‐PCR. Total RNA was extracted from the liver of two hares from Spain and the one from Italy using Trizol Reagent (Qiagen, Milan, Italy), following the manufacturer's instructions. One step RT‐PCR was performed using the Qiagen kit (Qiagen, Milan, Italy). Primers EBHS‐148F (5′‐GTTGCAGCATCTGTTGCCACTGCGG‐3′) and EBHS‐578R (5′‐TGTACACACTCACGACGAGYGTTGGG‐3′) were used for EBHSV RNA detection. Primers based on the sequences of VP60 genes of RHDV2 available in GenBank: Fra109‐F (5′‐ACTACTAGCGTGGTCACCACC‐3′) and Fra567‐R (5′‐TTGTTATAAACGCTCAGGACCAAC‐3′) were used for RHDV2 RNA detection. The entire VP60 genes of each strain were amplified as previously described (Le Gall‐Reculé et al., 2013) and sequenced.
To perform phylogenetic analysis, the nucleotide sequences from the viruses in the Spanish and Italian hares were aligned by ClustalW with the VP60 gene sequences available in GenBank. The sequence panel included the following: (i) the RHDV2 strains identified, respectively, in France in 2010, in Italy from rabbits and other hare species in 2011–2012, and in Iberian Peninsula in 2011–2014; (ii) G1‐G6 RHDV genogroups; (iii) non‐pathogenic rabbit calicivirus (European RCVs and RCV‐A1); and (iv) the Italian prototype strain EBHSV BS89. The neighbour‐joining phylogenetic tree was generated using the Kimura 2‐parameter evolutionary model, implemented in MEGA 6 (Tamura et al., 2013). Bootstrap resampling was performed on 1000 replicates.
Serological surveys were performed on sera collected from hare population representative of those areas in Italy and Spain where the sporadic cases of RHDV2 in hares were detected. In particular, a total of 106 blood samples of EBHs were collected in Spain from north–east of Catalonia (Barcelona and Girona provinces) during the same period (hunting season 2014) when the two dead hares were collected. Specifically, hunters collected blood samples from shot animals and sent them to the laboratory. After centrifugation, sera were frozen (−20°C) and then sent to the OIE reference Laboratory for RHD where they were examined. A similar survey was conducted in Italy on a total of 407 EBH sera collected after the first occurrence of RHDV2 in Italy (June 2011) during hunting seasons 2012–2013 (n = 154) and 2013–2014 (n = 253). All of these samples were taken from live animals caught in areas managed as ‘breeding for restocking grounds’ (BfRG) in the provinces di Cremona, Brescia and Bergamo (Lombardy, northern Italy), close to the area where the RHDV2‐positive hare was found. In addition, we included 149 hare sera that were collected in the same geographical area in Lombardy (northern Italy), between 2007 and 2010 to represent control sera collected before the first detection of RHDV2 in Italy.
All the sera were tested in two cELISAs specific for EBHSV and RHDV2, respectively. Methods were equivalent to those described and used to detect anti‐RHDV antibodies (OIE – Rabbit haemorrhagic Disease, [Link]) but employed specific immunological reagents towards EBHSV and RHDV2. Briefly, a hare anti‐EBHSV or a rabbit anti‐RHDV2 hyperimmune serum was adsorbed to the ELISA plates (Nunc Maxisorb) in standard carbonate buffer. Then, each serum, tested at four dilutions starting from 1/10 then 1/40, 1/160 and 1/640, was incubated with the specific virus used at a dilution giving 1.0–1.2 OD492. Finally, a specific HRP‐conjugated MAb semi‐quantified the virus bound to the solid phase. A serum sample was considered negative if its OD value at the 1/10 dilution was higher than 85% of the OD value at the same dilution of the negative control serum. Serum was considered doubtful (inconclusive result) if its OD value at the 1/10 dilution was equal to or higher than 75% of the OD value at the same dilution of the negative control serum. A serum sample was considered positive if its OD value at the 1/10 dilution was lower than 75% of the OD value at the same dilution of the negative control serum. The titre of a positive serum sample corresponded to the dilution causing a 40–60% reduction of the OD value of the negative control serum.
Results
In both incidents from Italy and Spain, EBHS was initially suspected. Macroscopic findings in the hare from Italy included tracheal congestion, disseminated visceral haemorrhages, moderate splenomegaly and a discoloured and friable liver. Hares from Spain displayed epistaxis, multifocal haemorrhages in the lungs, and slightly rounded margins, pallor and generalized reticular/zonal pattern in the liver (Fig. 1b, c, and d, respectively). In addition, no evidence of other infectious causes of death, including tularaemia, coccidiosis and toxoplasmosis, were seen macroscopically (both incidents) or microscopically (Spain) and routine laboratory examinations (Italy) ruled out the most common and typical bacterial and parasitic infections of hares. Two different diagnostic approaches were then taken to confirm a viral aetiology in the Italian and Spanish incidents, respectively. Organs (liver and spleen) from the Italian hare were immediately examined by the double independent immunological ELISAs, whereas the two Spanish hares were first analysed by histopathological examination, then by an EBHS‐specific PCR and finally by the lagovirus ELISAs. Tissues from the Spanish hares were moderately to severely autolysed, but microscopic examination could still detect extensive to massive hepatic necrosis consisting of both widespread single‐cell coagulative necrosis and multifocal areas of lytic necrosis (Fig. 2). Acidophilic bodies were also scattered throughout the sections. There was moderate to marked fatty degeneration of hepatocytes, and multinucleate hepatocytes occasionally were seen (8 per 10 high power fields). Epithelium of occasional proximal tubules in the kidney was degenerate, necrotic, and/or attenuated, and proteinaceous globules were seen in scant tubules, consistent with acute tubular nephrosis. Lungs were moderately to severely congested, and the tracheal submucosa was markedly expanded by congestion. Lesions in all described tissues were more severe in hare LE14020 than in hare LE14021. In addition, splenic pathology was only seen in hare LE14020 and consisted of moderate lymphocytolysis (necrosis) in the white pulp and moderate to severe fibrin deposition and necrosis in the red pulp. No significant findings were seen in the brain, heart, skeletal muscle or reproductive organs of either hare, but autolysis precluded detailed examination. RT‐PCR formalin‐fixed, paraffin embedded tissues did not detect EBHSV in the Spanish hares.
Figure 2.
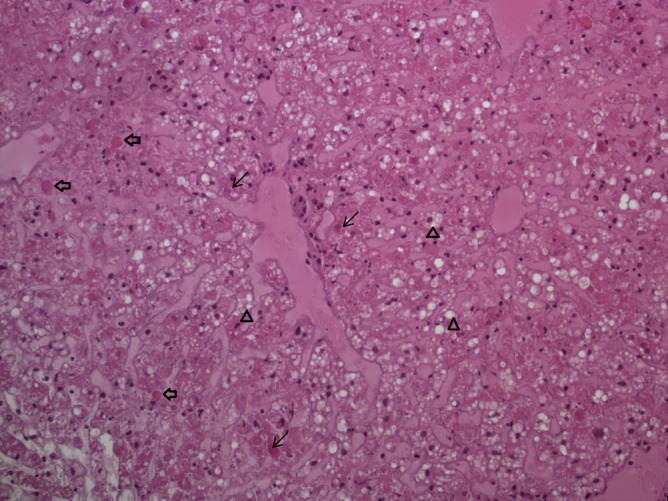
Microscopic lesions in the liver of a RHDV2 infected brown hare photomicrograph of the liver from one of the European brown hares (LE14021) found dead in Barcelona Province, Spain. Note the fatty degeneration of hepatocytes (▵ arrowheads), necrotic hepatocytes (← thin arrows) and acidophilic bodies (⇐ broad arrows). Haematoxylin–eosin staining, 200× magnification.
The results of the double independent immunological ELISAs testing indicated the presence of a high amount of lagovirus in all three liver samples. The lagovirus was further identified as RHDV2, not EBHSV, in all cases. In fact, the MAb reactivity pattern of the isolates showed the typical profile for RHDV2, being unreactive to MAbs specific for the classical RHDV, RHDVa and EBHSV, but highly reactive with the MAbs specific for RHDV2 (data not shown). This antigenic pattern was almost identical to those observed in the two previous cases of detection of RHDV2 in Italy, respectively, in Cape hares in Sardinia (Puggioni et al., 2013) and in Italian hares in Sicily (Camarda et al., 2014).
The RT‐PCR performed on the liver of the hare found dead in Italy with primers specific for EBHSV was negative, whereas the amplification with the specific primers for RHDV2 returned one 458‐bp amplicon.
The sequences of the entire VP60 gene from the two Spanish hares had 100% nt identity, and one was submitted to GenBank (Accession No. KT308116) together with the sequence of the Italian hare isolate (Accession No. KT308115). Phylogenetic analysis (Fig. 3) showed that the three RHDV2 isolates identified in EBHs clustered together with the previously identified RHDV2 strains, which formed a distinct clade, separated from the RHDV and RCV groups. The BLAST analysis of sequences (excluding primers) revealed that the Italian RHDV2 hare strain was 97.7% (98.8% at amino acid level) identical to the VP60 gene of the Spanish one. Moreover, the Italian RHDV2 hare strain was 97.2% (98.4% at amino acid level) identical to the VP60 gene of the isolate Ud11 (Accession No. JQ929052), which was the first RHDV2 identified in farmed rabbits in Italy in mid‐2011 (Le Gall‐Reculé et al., 2013). Regarding RHDV2 strains identified in other hare species in Italy, the Italian RHDV2_Bg12 strain of EBH (Accession No. KT308115) was more similar to the RHDV2_Sr12‐2 strain (Accession No. KC741409), identified in the Italian hare in Sicily in the same year (98.3% nucleotide identity and 99.6% at amino acid level) than to the RHDV2_Vs11‐2 strain (Accession No. KC345613) found in Cape hares in Sardinia the year before (97.2% nucleotide identity and 98.9% at amino acid level). The VP60 gene of the Spanish RHDV2_Barcelona14 hare isolates (Accession No. KT308116) was 98.6% (98.9% at amino acid level) identical to that of isolate RHDV‐N11 (Accession No KM87868), the first Spanish RHDV2/RHDVb strain isolated in rabbits in 2011 (Dalton et al., 2012), and belongs to the same cluster that also includes several RHDV2 strains identified in the Iberian peninsula in the same area and period (Abrantes et al., 2013; Dalton et al., 2014). Additionally, because of the occurrence of multiple recombinant events in RHDV2 strains on Iberian Peninsula during 2012–2014 (Lopes et al., 2015), phylogenetic and SimPlot analyses were performed on the 1000‐bp region upstream of the recombinant breakpoint (5305nt). The nt identity of approximately 99% with three strains (RHDV‐N11 KM87868, Zar11 KP129398 and Seg08‐12 KP129396) excluded the possibility that the Spanish hare isolate was a recombinant RHDV2 (data not shown).
Figure 3.
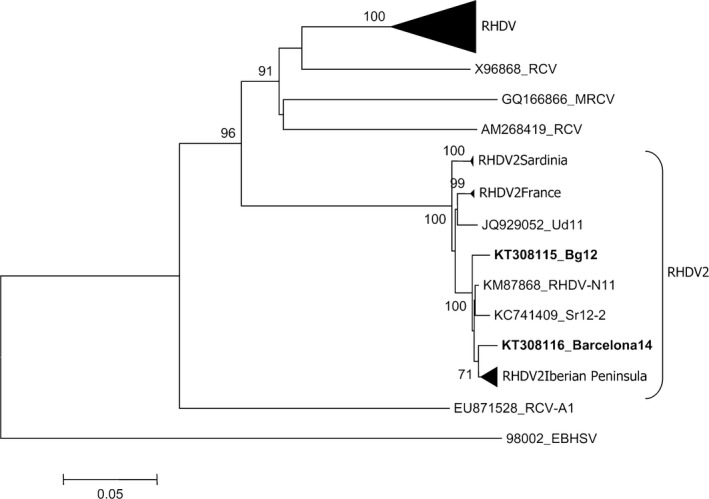
Phylogenetic analysis of VP60 sequences of RHDV2 strains identified in hares in Italy and Spain. For the phylogenetic analysis of the VP60 sequences, the neighbour‐joining method was applied based on the Kimura 2 parameters using the software package MEGA6 (Tamura et al., 2013). The European brown hare syndrome virus (EBHSV) strain BS89 (GenBank Accession No.: X98002) was used as an out‐group to root the tree. Bootstrap probability values above 70% for 1000 replicate runs are indicated at the nodes. Twenty‐one selected sequences representing previously described RHDV genogroups (G1–G6; GenBank a.n.: EF558575, Y15441, X87607, Y15426, AJ535094, AJ535092, Y15427, AJ495856, KC595270, KC345614, Z49271, Z29514, Y15440, L48547, AF402614, U49726, M67473, U54983, Y15425, EU250330, AJ302016), the sequences of the RHDV2 isolates identified in France (GenBank a.n.: HE819400, HE800530, HE819400), in Sardinia/Italy (GenBank Accession No.: KC345611, KC345613, JX106023, KC345612) (2010–2012) and in the Iberian Peninsula (GenBank a.n.: KM115712, KM115713, KM115714, KM115715, KM115716, KP129397, KM115697, KM115689) (2012–2014) have been collapsed. The accession numbers are indicated for the remaining RHDV2 sequences, including RHDV‐N11 (GenBank a.n.: KM87868), the first Spanish RHDV2 isolate; RHDV2_Sr12‐2 strain (GenBank a.n.: KC741409), the isolate from Italian hares in Sicily; and Ud11 (GenBank a.n.: JQ929052), the first RHDV2 isolated in Italy. The sequences of the Spanish (Barcelona14, GenBank a.n.: KT308116) and Italian (Bg12, GenBank a.n.: KT308115) RHDV2 hare strains are in bold. Four sequences of European (X96868_RCV, Italy 1995; GQ166866_MRCV, USA 2001; AM268419_RCV, France 2006) and Australian (EU871528_RCVA1) non‐pathogenic RCVs were also included.
The ELISA results of sera from the four different groups of hares collected in northern Italy (Lombardy) and north‐east Spain (Catalonia), expressed as a percentage of total sera, are summarized in Fig. 4. Negative and doubtful results were grouped together because, as often occurs with samples collected by hunters, the quantity and quality of the sera were poor, increasing the proportion of doubtful results. Panels A to C report the data from the Italian hares; sera prior to RHDV2 detection (archival sera from 2007 to 2010), and sera collected in 2012 and 2013, respectively. The overall distribution of titres is similar but some fluctuations are also evident.
Figure 4.
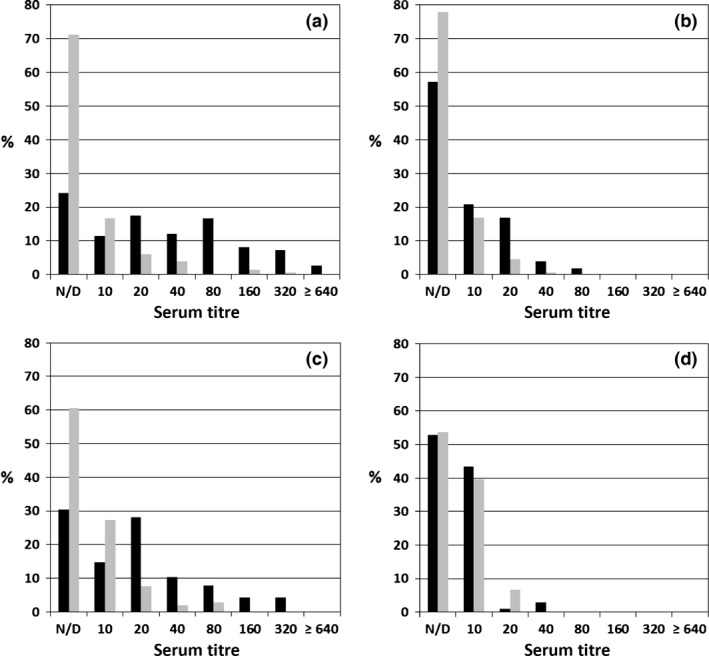
Results of serological surveys performed on Italian and Spanish European brown hare sera tested for EBHSV and RHDV2. Hare sera were tested in cELISAs for EBHSV (■ black bars) and RHDV2 ( grey bars). Titre values are reported as reciprocals (i.e. 20 means 1/20). The categories of negative and doubtful sera were grouped together. Panels summarize the titre distributions of: (a) 149 sera collected in northern Italy between 2007 and 2010 (pre‐RHDV2 period); (b) 154 sera collected in northern Italy during 2012; (c) 253 sera collected in northern Italy during 2013; (d). 106 sera collected in Spain during the same period (2014) and in the Barcelona Province where the two EBHs were found dead.
grey bars). Titre values are reported as reciprocals (i.e. 20 means 1/20). The categories of negative and doubtful sera were grouped together. Panels summarize the titre distributions of: (a) 149 sera collected in northern Italy between 2007 and 2010 (pre‐RHDV2 period); (b) 154 sera collected in northern Italy during 2012; (c) 253 sera collected in northern Italy during 2013; (d). 106 sera collected in Spain during the same period (2014) and in the Barcelona Province where the two EBHs were found dead.
Regarding EBHSV antibodies, the percentage of the negative/doubtful sera were doubled in 2012 compared to the previous years and this was also accompanied by a decrease of the average titre of positive sera, which were mainly distributed in the categories 1/10 and 1/20. This decreasing trend reversed in 2013, when the percentage of positive sera again approached that found during the period 2007–2010, with a corresponding increase of the category of titres between 1/40 and 1/320. Altogether the serological data, supported by the number of EBHSV cases reported in dead hares during the same time period from the field (data not shown), confirmed that during the end of 2012 and 2013, an increase of EBHSV infections occurred among hare populations in north Italy.
Regarding the presence of RHDV 2 antibodies in the three groups of Italian sera, we found a distribution similar to that detected for EBHSV, but with a higher percentage (approximately double) of negative/doubtful sera. Additionally, the average titres of RHDV2 positive sera were 4–8 times lower than those for EBHSV, especially during the periods of higher incidence of EBHS (2007–2010 and 2013). Of note, very few sera, and only within the pre‐RHDV2 group, showed an RHDV2 titre of 1/160. However, the corresponding titre in EBHSV of these sera were from 1/1280 to 1/2560 (RT > 4) suggesting that the competition for RHDV2 was due to cross‐reactive Ab. Using the same approach on sera from Spain (panel D), we obtained clearly different results: the prevalence of both EBHSV and RHDV2 antibodies was comparable, with about half of the sera included in the negative/doubtful category and the rest showing titres mainly of 1/10 and in few cases ≤ 1/40.
To better analyse the serological data, we used an additional approach, based on the following knowledge: (i) as the antigenic profile of RHDV2 and EBHSV is consistently different (Le Gall‐Reculé et al., 2013), the sets of antibodies induced in lagomorphs by infection with RHDV2 or EBHSV are largely distinct; (ii) the cELISAs for the two viruses are quite specific; in fact, when using these two methods for testing the convalescent sera of experimentally EBHSV‐infected hares which survived the infection, the ratio (RT) between the titres for EBHSV and RHDV2 antibodies range from 16 to 128; (iii) the specificity of the two cELISAs decreases when testing hare sera from the field, likely because of reinfections that favour the increase of a cross‐reactive subset of antibodies due to the existence of common epitopes, and also infection with antigenically related viruses; (iv) the specificity of EBHSV cELISA also decreases when titres for EBHSV antibodies range from 1/10 to 1/160, such as those mainly observed in Italy in 2012–2013. The percentage distribution of the RT values for the 4 different groups of hare sera is reported in Fig. 5, excluding the sera that were negative in both ELISAs, as this would falsely increase the RT 1 class, that is sera with the same ELISA titre for EBHSV and RHDV2. The sera of the pre‐RHDV2 group were equally divided into two main subsets: about half with RT in the range 8–64, containing the sera with high anti‐EBHSV titre, and half with RT ranging from 1 to 4, among which sera with a ratio of 1 were only 2.5%. The distributions of the 2012 and 2013 sera partially overlapped, with most sera having a ratio of 2 or 4. The main differences between these two groups were in the tails of the distribution: approximately 10% more sera of the 2013 group had a RT in the range of 8–64, compared to the 2012 group, which had an RT in the 0.5–1 range. This difference was simply due to an increase of EBHSV cases during 2013 (see above). Of note, in both these groups, some sera showed an RHDV2 titre that was double that for EBHSV (RT 0.5). The distribution of the Spanish sera was clearly different from that of the other three groups, being centred around RT 1, with nearly a symmetrical pattern for RT 0.5 and 2. Three per cent of the sera (1.8% if we consider all the sera tested) showed a ratio of 0.25, which was never found in sera from Italian hares. Overall, the data from the Spanish samples were in agreement with the epidemiological understanding that Spain is likely free from EBHSV.
Figure 5.
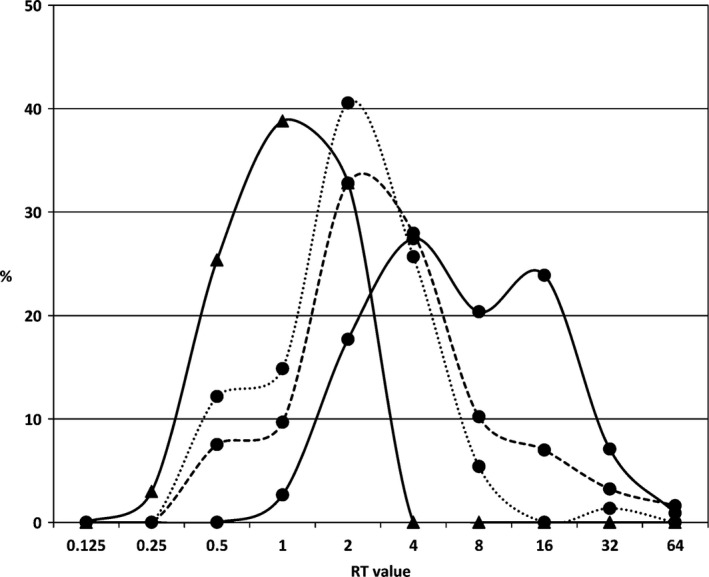
Distributions in percentage of ratio between EBHSV titre/RHDV2 titre (RT value) calculated for each serum belonging to the four epidemiological groups studied. The four curves represent the distributions in percentage of the RT values obtained by dividing the EBHSV cELISA titre with the RHDV2 cELISA titre for each serum belonging to a specific group. We did not include the sera that were negative to both ELISAs, which would have abnormally increased the category RT 1. Lines with circles (●) correspond to hare sera taken in Italy and triangles (▲) to hare sera from Spain. Black line (—): sera belonging to the pre‐RHDV2 group taken between 2007 and 2010; dot line (…): sera collected during 2012; dashed line (– –): sera collected during 2013. Category with ratio “1” does not include the sera that were negative to both ELISAs. In order to include in the analysis also the sera that were negative at one cELISA but positive to the other, we attributed to negative sera the formal value of 1/5. Therefore, for a serum negative on the RHDV2 cELISA but with a titre of 1/20 on the EBHSV cELISA we assigned a RT value of 4.
Discussion
The present study reports the occurrence of one and two cases of RHDV2 infection that affected European brown hares in Italy and Spain, respectively, causing a necrotizing hepatitis and other macroscopic and microscopic lesions similar to EBHS, the lagovirus disease that typically affects this species. However, using a specific immunoenzyme assay based on specific MAbs, and genome analysis, samples from three dead hares were found to contain the lagovirus RHDV2.
All the affected animals were wild and came from areas where EBH is the sole hare species present, living sympatrically with either large populations of wild rabbits (Catalonia, north‐east Spain) or in areas with scattered populations of wild rabbits and the presence of domestic and rural rabbits units as well as industrial rabbit farms (Po Valley, Lombardy, northern Italy). The main difference between the two situations is the differing presence of EBHS in hare populations of the respective countries. EBHS is endemic in Italy, where EBH represents the most widespread hare species. However, in Spain, the Iberian hare is the more widespread species (Palomo et al., 2007) and EBH populations are confined to the north‐east of the country. Therefore, because EBHSV has never been detected in Iberian hares (Gortàzar C., cited by Lopes et al., 2014), RHDV was found in just two Iberian hares in Portugal between 1996 and 1998 (Lopes et al., 2014), and EBHS was diagnosed in Spain only once (Fernández de Luco et al., 2003), the Spanish EBH populations, in contrast to those in northern Italy, must be considered most likely completely naïve and fully susceptible to virulent lagovirus infection, including EBHSV. The results of the sero‐epidemiological survey on Spanish hare sera taken in 2014 (see below for details) showed a lack of specific EBHSV immunity and thus confirmed this conclusion.
In the previous cases of RHDV2 in the two other hare species in Italy, a common origin of infection in hares and rabbits was concluded. For Cape hares in Sardinia, this was based on the verified direct epidemiological connection with RHDV2 outbreaks in rabbits (Puggioni et al., 2013); and in the case of Italian hares in Sicily, by direct contact with RHDV2 infected rabbits (Camarda et al., 2014), as well as on a 100% genomic identity between viral strains found in the two species. Conversely, in the present two incidents of RHDV2 in EBHs in Italy and Spain, the source of infection remains unknown and it was not possible to establish a common origin between rabbits and hares and/or to detect identical strains in the two species. However, these cases were coincident with several RHDV2 outbreaks detected during this period (2012–2014) in both countries among wild, domestic and farmed rabbits, which gave rise to documented epidemic waves of RHD (Le Gall‐Reculé et al., 2013; Dalton et al., 2014; Lopes et al., 2015). Why sporadic cases of RHDV2 occurred in an assumed resistant species remains elusive. Emergence and spread of the new virus was likely because of the full susceptibility of rabbits at the population level and by the limited cross‐protection induced by previously circulating RHDV strains. Consequently, it is feasible that the abundant sources of virus dissemination and high infection pressure in the environment during these epidemic waves favoured spillover events of infection of EBHs with RHDV2. The recent identification of RHDV in archival samples from two Iberian hares found dead in the 1990s in Portugal with signs of an EBHS‐like disease, further support this explanation (Lopes et al., 2014). These two cases were caused by RHDV G1 strains that were phylogenetically closely related to those circulating at that time and in the same areas in rabbit populations. Similar to our cases of RHDV2 in EBHs, these cases represented occasional findings within a sympatric lagomorph host species during an epidemic wave of an emerging virus in rabbits, with RHDV first identified in European rabbits in the late 1980s.
These data demonstrate that the emerging lagovirus RHDV2 that causes RHD in rabbits could also cause sporadic cases of disease in the EBH, especially in areas where the habitats of the two hosts overlap. Spillover is even more probable where the densities of the specific host, the rabbit, are high. This situation is more frequently observed in Spain than in Italy. The findings of sporadic cases of RHDV2‐induced disease in EBH further confirm that barriers against RHD viruses among species of the genus Lepus are not absolute, and moreover, they also confirm that EBHs are naturally susceptible to infection and development of disease. However, the ability of RHDV2 to infect members of the genus Lepus may vary considerably. In the outbreak involving Cape hares in Sardinia, a significant number of hares and rabbits were concurrently affected by the disease (Puggioni et al., 2013), indicating a quite high frequency of infection of hares. In the other cases, that is RHD in an Iberian hare (Lopes et al., 2014) and RHDV2 in both an Italian hare (Camarda et al., 2014) and EBH (this study), the virus spillovers to new species did not cause severe or easily detectable outbreaks, but rather only single, sporadic infections.
Considering the relatively high circulation of RHDV2 in rabbits throughout Europe, and the field surveillance conducted in Spain and Italy where different lagomorph live sympatrically, the few recorded cases of RHDV2 in EBH suggest that its susceptibility is likely low, and thus, only sporadic overt cases are expected. Similar to the two RHD cases in Iberian hares (1996–1998; Lopes et al., 2015) that have been the only documentation of RHDV in the genus Lepus for over 15 years, RHDV2 has apparently not caused epidemics in EBHs in the last 4 years. Following these two RHDV2 cases in EBH in Spain in 2014, and considering the reports of hunting associations reporting an important decrease of the hare population during the last hunting season, a specific surveillance was implemented during 2015 but only detected a few more cases of RHDV2 in EBH (Velarde, R., personal communication, 2016). Conversely, a similar survey conducted in 2015 in France detected many more cases of RHDV2 in EBH populations, showing a steady circulation of the virus in these populations (Le Gall‐Reculé, G., Guitton, J.S. and Marchandeau, S, personal communication, 2016).
The serological survey of stocked hare sera taken from Italian and Spanish hare populations provided an understanding of naturally occurring RHDV2 infection in the field. In particular, the availability of serological data from hare populations in Italy before the appearance of RHDV2 aided the understanding and interpretation of the final serological data. However, the serological results should be interpreted with caution due to the limitations of sera quality from wild animals sampled in the field. Interpretation of results is also hampered by the level of specificity of the RHDV2 and EBHSV cELISAs used, which is not absolute, even though they mainly detect antibodies binding the outer shell of the respective viruses. While sensitivity is quite good for sera from naïve animals infected for the first time with a specific virus, it decreases in cases of low cELISA titres and in field conditions where wild lagomorphs, and hares particularly, could be repeatedly re‐infected or infected with non‐pathogenic lagoviruses that could interfere with the serological results (Cooke et al., 2000; McPhee et al., 2009). Despite these limitations, the serological results suggest quite a clear epidemiological situation. In Italy, they confirm the endemic presence of EBHS, albeit with an incidence consistently lower than prior to 2000, when the anti‐EBHSV cELISA titres were mainly between 1/640 and 1/5120. Such values, typically found in hares recovered from EBHS, were quite frequent during epidemic waves of EBHS, especially in areas where severe outbreaks occurred (Scicluna et al., 1994). Our data also support the already observed periodic variation of the average EBHSV titres between different years, even if restricted to a low range of values, and confirm that, albeit still endemic, there was a decrease of EBHS incidence in the northern Italy in the last 10 years (Chiari et al., 2014). The results obtained by testing the same Italian hare sera with RHDV2 cELISA, indicated that RHDV2 was not widely distributed among hares, nor as a subclinical infection, but sporadic cases of infection not followed by disease and/or death could not be completely ruled out. In fact, approximately 10% of the sera from the 2012 and 2013 groups, all with titres <1/40, showed a RT between EBHSV and RHDV2 titres of 0.5. Thus, those hares with twice more antibodies for RHDV2 than to EBHSV should be suspected of being infected by RHDV2. Nevertheless, a 0.5 RT value could also be to the result of other technical and actual factors including: (i) the reproducibility of the cELISA is around 85–90%; that is, in case of repetition of the cELISA, 10–15% of tested sera may give a slightly different titre from the first analysis by a factor of 2. (ii) The specificity of the cELISAs decreases in old infections or reinfections when titres are generally low (<1/40), due to a quite complex and variable subset of serum IgG among hares. (iii) The previous infection with a non‐pathogenic (NP) lagovirus inducing a specific subset of cross‐reactive antibodies in hare sera that, in relation to the degree of genetic correlation of the NP lagovirus with EBHSV and RHDV2, interfere differently with the EBHSV and RHDV2 cELISAs.
Consequently, the serological survey on hares sampled in Italy after the appearance of RHDV2 indicated that RHDV2 was not distributed throughout the hare population, but did not completely eliminate the possibility of sporadic infections, which is in agreement with the occurrence of a few cases of the EBHS‐like diseases due to RHDV2 like those described here.
With regard to the Spanish situation, the symmetrical distribution around the value of RT 1 of sera collected from EBH in Spain on 2014 clearly indicated the absence of infection due to EBHSV, in agreement with the EBHS‐free status of the country. Regarding RHDV2 serology, the results were very similar to the Italian ones, suggesting that RHDV2 was not consistently present in the hare population but sporadic infection could not be definitively ruled out. Moreover, in the absence of EBHSV infection in Spain, we found that about 40% of sera were positive in the two cELISAs at very low titres. This situation is similar to that described in wild rabbits in Australia, which was attributed to the endemic presence of a NP calicivirus (RCVA1) (McPhee et al., 2009; Strive et al., 2009). This could be indicative of the existence of a NP lagovirus in the Spanish hare populations. Based on our serological results, this hypothetical virus should have a unique antigenic profile equally distant from those of EBHSV and RHDV2, and thus, it could even be related to the non‐pathogenic virus (HaCV) that we have recently found in an EBH farm in north Italy (Cavadini et al., 2015).
The results of this study demonstrate that the emerging lagovirus RHDV2 can infect and cause disease in the EBH, reinforcing the hypothesis that it is able to overcome species barriers. More difficult to explain are the reasons for this occurrence, how RHDV2 can overcome the species barriers, to which extent and frequency such spillover events occur and which factors could promote and eventually sustain the cross‐infection.
The phylogenetic analysis of the complete VP60 capsid gene of the RHDV2 Italian and Spanish strains from EBHs revealed a high homology among them, which clustered with all of the other RHDV2 isolates detected from RHD‐infected rabbits in the same areas and periods. Notwithstanding the absence of relevant genomic differences, it cannot be excluded that the capacity of this virus to infect new hosts could be due to virus‐related properties, for example a peculiar capsid structure that might facilitate the attachment and/or binding, the invasion, and/or the subsequent cellular stages of viral cycle. As RHDV2 is an almost ‘new’ rabbit lagovirus, and recent antigenic and genetic data indicate that the virus is still evolving (Capucci, L., unpublished results, 2016), it could be theoretically possible that future evolution of RHDV2 may result in new variants with the capacity of cross‐infecting other species of lagomorphs and with the acquisition of new ways of spreading via an expanded host range that could eventually allow the virus to persist in environments where wild rabbits are not present.
Other possible hypotheses, more host‐related and involving the increased susceptibility of certain individuals to the infection, also should be considered to explain the sporadic occurrence of clinical disease caused by RHDV2 in hares. Several studies indicate that, as for noroviruses (Tan et al., 2004), a specific binding between lagoviruses and glycans, particularly those of the histo‐blood group antigens (HBGAs), constituting the glycocalyx of the small intestinal epithelial cells, is the first step of the viral infection (Ruvoen‐Clouet et al., 2000; Guillon et al., 2009; Nyström et al., 2011). Because glycans can show variation within species as well as intraspecies polymorphism, they can surely act as the first element of a species barrier for lagoviruses. Therefore, another explanation for the sporadic occurrence of RHDV2‐induced disease in EBH could be linked to the presence of a minor subset of animals within the EBH population that have a HBGAs polymorphism to which RHDV2 can bind with a sufficient stability to allow the progress of infection.
In addition, other factors in some circumstances could impair the capacity of single hare individuals to naturally resist the infection with viruses such as RHDV2. These factors could have an immune‐suppressive effect and may be the consequence of other concurrent subclinical infections, overt diseases and parasitic infestations, or the result of malnutrition, social competition, habitat detriment, etc.
Irrespective of whether host factors or the aetiological agent is mainly responsible for the ability of RHDV2 to overcome species boundaries, particular conditions, such as direct and repeated contacts with infected rabbits and/or high infectious doses, are likely necessary for spillover of RHDV2 from rabbits to hares. The fact that RHDV2 has been present in Spain and Italy since at least 2011 and it is now the main RHDV type identified with numerous outbreaks in different regions, including Catalonia and Lombardia where the three RHDV2 positive hares were found, that has resulted in situations of high infection pressure.
In conclusion, because we have demonstrated that RHDV2 has the capacity also to infect EBH, even if sporadically, the implementation of suitable surveillance programmes in Europe for domestic and wild lagomorph populations is warranted. The scope should be to detect and diagnose cases of lagovirus infection in all lagomorph species, that is European rabbit, the different hare species and also cottontail, and to achieve a precise genetic and antigenic characterization of the causative strains. Such surveillance is also necessary to define the current circulation of EBHSV, as recent studies, supported by our serological data, indicate a progressive decrease in the last years of the number of cases of EBHS, as well as the detection of lower titres in field collected samples and slightly lower EBHS seroprevalences in wild hares in most European countries (Chiari et al., 2014). Therefore, hare populations are less protected by specific immunity and thus more exposed to the emergence of new EBHS epidemics in the case of viral dissemination. The field evolution and circulation of lagoviruses should be strictly monitored to promptly assess if, in particular, RHDV2 is capable of more easily and frequently infecting other lagomorphs, such as the EBH. This in turn might lead to a wider and more rapid spread of RHDV2 and, at the same time, seriously threaten vulnerable or endangered Leporidae species.
Acknowledgements
We thank (i) the gamekeepers of the Cos d'Agents Rurals from the Catalonia government, for their invaluable collaboration in the wildlife passive surveillance programme in Catalonia and for reporting and bringing cases to our facilities at the veterinary faculty (SEFaS). This passive surveillance programme is supported by the Subdirecció General d'Activitats Cinegètiques i Pesca Continental from the Ministry of Agriculture, Livestock, Fisheries and Food of the Catalonian government; (ii) Åsa Hagström, NVI, Sweden, for conducting EBHSV PCR. Funding for analyses at NVI, Sweden, was partially provided by the Swedish Environmental Protection Agency; (iii) the technicians of the Proteomic Unit and of the Electron Microscopy Laboratory of IZSLER for their precious technical assistance.
References
- Abrantes, J. , Lopes A. M., Dalton K. P., Melo P., Correia J. J., Ramada M., Alves P. C., Parra F., and Esteves P. J., 2013: New variant of rabbit hemorrhagic disease virus, Portugal, 2012–2013. Emerg. Infect. Dis. 19, 1900–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergin, I. L. , Wise A. G., Bolin S. R., Mullaney T. P., Kiupel M., and Maes R. K., 2009: Novel calicivirus identified in rabbits, Michigan, USA. Emerg. Infect. Dis. 15, 1955–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom, R. , Sol C. J., Salimans M. M., Jansen C. L., Wertheim‐van Dillen P. M., and van der Noordaa J., 1990: Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarda, A. , Pugliese N., Cavadini P., Circella E., Capucci L., Caroli A., Legretto M., Mallia E., and Lavazza A., 2014: Detection of the new emerging rabbit hemorrhagic disease type 2 virus (RHDV2) in Sicily from rabbit (Oryctolagus cuniculus) and Italian hare (Lepus corsicanus). Res. Vet. Sci. 97, 642–645. [DOI] [PubMed] [Google Scholar]
- Capucci, L. , Frigoli G., Ronsholt L., Lavazza A., Brocchi E., and Rossi C., 1995: Antigenicity of the Rabbit Hemorrhagic Disease Virus studied by its reactivity with monoclonal antibodies. Virus Res. 37, 221–238. [DOI] [PubMed] [Google Scholar]
- Capucci, L. , Fallacara F., Grazioli S., Lavazza A., Pacciarini M. L., and Brocchi E., 1998: A further step in the evolution of rabbit hemorrhagic disease virus: the appearance of the first consistent antigenic variant. Virus Res. 58, 115–126. [DOI] [PubMed] [Google Scholar]
- Cavadini, P. , Molinari S., Pezzoni G., Chiari M., Brocchi E., Lavazza A., and Capucci L., 2015: Identification of a new non‐pathogenic lagovirus in Lepus europaeus. In: Proceedings of the 10th International Congress for Veterinary Virology and 9th Annual Epizone Meeting “Changing Viruses in a Changing World” Montpellier, France, August 31st – September 3rd 2015, p 76–77.
- Chiari, M. , Ferrari N., Giardiello D., Avisani D., Zanoni M.‐G., Alborali G. L., Guberti V., and Lavazza A., 2014: Temporal dynamics of European brown hare syndrome infection in Northern Italian brown hares (Lepus europaeus). Eur. J. Wildl. Res. 60, 891–896. [Google Scholar]
- Cooke, B. D. , Robinson A. J., Merchant J. C., Nardin A., and Capucci L., 2000: Use of ELISAs in field studies of rabbit haemorrhagic disease (RHD) in Australia. Epidemiol. Infect. 124, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton, K. P. , Nicieza I., Balseiro A., Muguerza M. A., Rosell J. M., Casais R., Álvarez Á. L., and Parra F., 2012: Variant rabbit hemorrhagic disease virus in young rabbits, Spain. Emerg. Infect. Dis. 18, 2009–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton, K. P. , Nicieza I., Abrantes J., Esteves P. J., and Parra F., 2014: Spread of new variant RHDV in domestic rabbits on the Iberian Peninsula. Vet. Microbiol. 169, 67–73. [DOI] [PubMed] [Google Scholar]
- Fernández de Luco, D. , Arnal M. C., Gortázar C., and Gavier‐Widen D., 2003: Síndrome de la liebre parda europea (EBHS) en una liebre norteña (Lepus europaeus) del Pirineo Aragonés. Proceedings of the XV Reunión de la Sociedad Española de Anatomía Patológica Veterinaria, Córdoba, Spain, June 2003, pp 33.
- FLI information , 2013. Hämorrhagische Kaninchenkrankheit, Neue Variante des RHD‐Virus nun auch in Deutschland entdeckt. Available at http://www.ljv-nrw.de/media/1399925596_fli_information_rhd_virus20131021.pdf (accessed October 21, 2013).
- Gavier, D. , and Morner T., 1991: Epidemiology and diagnosis of the European brown hare syndrome in Scandinavian countries: a review. Rev. Sci. Tech. Off. Int. Epiz. 10, 453–458. [DOI] [PubMed] [Google Scholar]
- Gomez‐Laguna, J. , Carrasco L., Ramis G., Quereda J. J., Gomez S., and Pallares F. J., 2010: Use of real‐time and classic polymerase chain reaction assays for the diagnosis of porcine tuberculosis in formalin‐fixed, paraffin‐embedded tissues. J. Vet. Diagn. Invest. 22, 123–127. [DOI] [PubMed] [Google Scholar]
- Guillon, P. , Ruvoen‐Clouet N., Le Moullac‐Vaidye B., Marchandeau S., and Le Pendu J., 2009: Association between expression of the H histo‐blood group antigen, alpha1,2fucosyltransferases polymorphism of wild rabbits, and sensitivity to rabbit hemorrhagic disease virus. Glycobiology 19, 21–28. [DOI] [PubMed] [Google Scholar]
- Kerr, P. J. , Kitchen A., and Holmes E. C., 2009: Origin and phylodynamics of rabbit hemorrhagic disease virus. J. Virol. 83, 12129–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazza, A. , Scicluna M. T., and Capucci L., 1996: Susceptibility of hares and rabbits to the European brown hare syndrome virus (EBHSV) and rabbit haemorrhagic disease virus (RHDV) under experimental conditions. J. Vet. Med. B 43, 401–410. [DOI] [PubMed] [Google Scholar]
- Lavazza, A. , Cavadini P., Barbieri I., Tizzani P., Pinheiro A., Abrantes J., Esteves P. J., Grilli G., Gioia E., Zanoni M.‐G., Meneguz P. G., Guitton J.‐S., Marchandeau S., Chiari M., and Capucci L., 2015: Field and experimental data indicate that the eastern cottontail (Sylvilagus floridanus) is susceptible to infection with European brown hare syndrome (EBHS) virus and not with rabbit haemorrhagic disease (RHD) virus. Vet. Res. 46, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall‐Reculé, G. , Zwingelstein F., Laurent S., de Boisséson C., Portejoie Y., and Rasschaert D., 2003: Phylogenetic analysis of rabbit haemorrhagic disease virus in France between 1993 and 2000, and the characterisation of RHDV antigenic variants. Arch. Virol. 148, 65–81. [DOI] [PubMed] [Google Scholar]
- Le Gall‐Reculé, G. , Zwingelstein F., Boucher S., Le Normand B., Plassiart G., Portejoie Y., Decors A., Bertagnoli S., Guérin J. L., and Marchandeau S., 2011: Detection of a new variant of rabbit haemorrhagic disease virus in France. Vet. Rec. 168, 137–138. [DOI] [PubMed] [Google Scholar]
- Le Gall‐Reculé, G. , Lavazza A., Marchandeau S., Bertagnoli S., Zwingelstein F., Cavadini P., Martinelli N., Lombardi G., Guérin J. L., Lemaitre E., Decors A., Boucher S., Le Normand B., and Capucci L., 2013: Emergence of a new lagovirus related to Rabbit Haemorrhagic Disease Virus. Vet. Res. 44, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenghaus, C. , Westbury H., Collins B., Ratnamoban N., and Morrissy C., 1994: Overview of the RHD Project in Australia. In: Williams R., and Munro R. (eds), Rabbit Haemorrhagic Disease: Issues in Assessment for Biological Control. pp. 104–129. Bureau of Resource Sciences, Australian Government Printing Service, Canberra. [Google Scholar]
- Lopes, A. M. , Marques S., Silva E., Magalhães M. J., Pinheiro A., Alves P. C., Le Pendu J., Esteves P. J., Thompson G., and Abrantes J., 2014: Detection of RHDV strains in the Iberian hare (Lepus granatensis): earliest evidence of rabbit lagovirus cross‐species infection. Vet. Res. 45, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, A. M. , Dalton K. P., Magalhães M. J., Parra F., Esteves P. J., Holmes E. C., and Abrantes J., 2015: Full genomic analysis of new variant rabbit hemorrhagic disease virus revealed multiple recombination events. J. Gen. Virol. 96, 1309–1319. [DOI] [PubMed] [Google Scholar]
- McIntosh, M. T. , Behan S. C., Mohamed F. M., Lu Z., Moran K. E., Burrage T. G., Neilan J. G., Ward G. B., Botti G., Capucci L., and Metwally S. A., 2007: A pandemic strain of calicivirus threatens rabbit industries in the Americas. Virol. J. 4, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee, S. R. , Butler K. L., Kovaliski J., Mutze G., Capucci L., and Cooke B. D., 2009: Antibody status and survival of Australian wild rabbits challenged with rabbit haemorrhagic disease virus. Wildl. Res. 36, 447–456. [Google Scholar]
- Nyström, K. , Le Gall‐Reculé G., Grassi P., Abrantes J., Ruvoën‐Clouet N., Le Moullac‐Vaidye B., Lopes A. M., Esteves P. J., Strive T., Marchandeau S., Dell A., Haslam S. M., and Le Pendu J., 2011: Histo‐blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain‐dependent manner. PLoS Pathog. 7, e1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE, 2012: Rabbit haemorrhagic disease , Chapter 2.6.2 In: Biological Standards Commission ; the World Organisation For Animal Health (OIE) , OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, pp 941–955. OIE, Paris, France: (NB: Version adopted in May 2016). [Google Scholar]
- OIE, WAHID (World Animal Health Information Database) , 2014a: Rabbit haemorrhagic Disease, Norway. Available at http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=15783 (accessed June 4, 2014).
- OIE, WAHID (World Animal Health Information Database) , 2014b: Rabbit haemorrhagic Disease, Denmark. Available at http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=16687 (accessed December 5, 2014).
- OIE, WAHID (World Animal Health Information Database) , 2015: Rabbit haemorrhagic Disease, Sweden. Available at http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=18693 (accessed September 18, 2015).
- OIE, WAHID (World Animal Health Information Database) , 2016a: Rabbit haemorrhagic Disease, Finland. Available at http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=20200&newlang=en (accessed May 17, 2016).
- OIE, WAHID (World Animal Health Information Database) , 2016b: Rabbit haemorrhagic Disease, Switzerland. Available at http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=20217&newlang=en (accessed May 24, 2016).
- Palacios, F. , 1989: Biometric and morphologic features of the genus Lepus in Spain. Mammalia 53, 227–264. [Google Scholar]
- Palomo, L. J. , Gisbert J., and Blanco J. C., 2007: Atlas y Libro Rojo de los mamíferos terrestres de España. Ministerio de Medio Ambiente, Madrid, Spain. [Google Scholar]
- Poli, A. , Nigro M., Gallazzi D., Sironi G., Lavazza A., and Gelmetti D., 1991: Acute hepatosis in the European Brown Hare (Lepus europaeus) in Italy. J. Wildl. Dis. 27, 621–629. [DOI] [PubMed] [Google Scholar]
- Puggioni, G. , Cavadini P., Maestrale C., Scivoli R., Botti G., Ligios C., Le Gall‐Reculé G., Lavazza A., and Capucci L., 2013: The new French 2010 variant of the rabbit hemorrhagic disease virus causes an RHD‐like disease in the Sardinian Cape hare (Lepus capensis mediterraneus). Vet. Res. 44, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros Bascuñana, C. , Nowotny N., and Belak S., 1997: Detection and differentiation of Rabbit Hemorrhagic Disease and European Brown Hare Syndrome Viruses by amplification of VP60 genomic sequences from fresh and fixed tissue specimens. J. Clin. Microbiol. 35, 2492–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvoen‐Clouet, N. , Ganiere J. P., Andre‐Fontaine G., Blanchard D., and Le Pendu J., 2000: Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo‐blood group family. J. Virol. 74, 11950–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirrmeier, H. , Reimann I., Kollner B., and Granzow H., 1999: Pathogenic, antigenic and molecular properties of rabbit haemorrhagic disease virus (RHDV) isolated from vaccinated rabbits: detection and characterisation of antigenic variants. Arch. Virol. 144, 719–735. [DOI] [PubMed] [Google Scholar]
- Scicluna, M. T. , Capucci L., and Lavazza A., 1994: European brown hare syndrome in northern Italy: results of a virological and serological survey. Rev. Sci. Tech. Off. Int. Epiz. 13, 893–904. [DOI] [PubMed] [Google Scholar]
- Strive, T. , Wright J. D., and Robinson A. J., 2009: Identification and partial characterization of a new lagovirus in Australian wild rabbits. Virology 384, 97–105. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher G., Peterson D., Filipski A., and Kumar S., 2013: MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M. , Hegde R. S., and Jiang X., 2004: The P domain of norovirus capsid protein forms dimer and binds to histo‐blood group antigen receptors. J. Virol. 78, 6233–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott, D. , Frossard J. P., Everest D., Dastjerdi A., Duff J. P., Steinbach F., and Choudhury B., 2014: Incursion of RHDV2‐like variant in Great Britain. Vet. Rec. 174, 333. [DOI] [PubMed] [Google Scholar]


