Abstract
Recent advances in molecular technology have revolutionized research on all aspects of the biology of organisms, including ciliates, and created unprecedented opportunities for pursuing a more integrative approach to investigations of biodiversity. However, this goal is complicated by large gaps and inconsistencies that still exist in the foundation of basic information about biodiversity of ciliates. The present paper reviews issues relating to the taxonomy of ciliates and presents specific recommendations for best practice in the observation and documentation of their biodiversity. This effort stems from a workshop that explored ways to implement six Grand Challenges proposed by the International Research Coordination Network for Biodiversity of Ciliates (IRCN‐BC). As part of its commitment to strengthening the knowledge base that supports research on biodiversity of ciliates, the IRCN‐BC proposes to populate The Ciliate Guide, an online database, with biodiversity‐related data and metadata to create a resource that will facilitate accurate taxonomic identifications and promote sharing of data.
Keywords: Cultivation, information resources, molecular phylogeny, morphology, nomenclature, phylogenetics, systematics, taxonomy, type specimens
THE nomenclature of ciliates and other heterotrophic protists (sensu Adl et al. 2012) is governed by the International Code of Zoological Nomenclature (“the Code”) that provides rules for naming taxa up to the level of family (ICZN 1999). The Code regulates nomenclatural issues, and it also establishes minimal standards for documenting newly described species. These are deliberately general and suited more to the study of animals than protists. In particular, the rule to deposit type specimens in permanently curated repositories (ICZN 1999, Art. 16.4.2 and Art. 75.3.7) has, over time, generated a database of material that not only has fixed names but also has formed the nucleus of large museum collections. These collections provide a wealth of information about the variability and biogeography of species and represent a potential source of DNA for investigations of molecular phylogenetics.
For ciliates and many other protists, the logistical difficulties imposed by their small size meant there was no convention for depositing type specimens until the latter part of the 20th century, when deposition of permanently stained preparations or other physical specimens (e.g. on SEM stubs) began to be required as type material for taxonomic descriptions or redescriptions. In addition, new methods of visualizing morphology and sequencing genes have been introduced over the last several decades, but there are no formal standards for application of these methods to taxonomic identifications or descriptions of ciliates, despite recent recommendations (Aescht 2001, 2008; Foissner 2002; ICZN 1999; Lynn and Simpson 2009; Santoferrara et al. 2016).
In 2012, the International Research Coordination Network for Biodiversity of Ciliates (IRCN‐BC), which is jointly funded by the NSF and NSFC, identified 20 Grand Challenges of Biodiversity of Ciliates (http://ircn-bc.org), of which six relate to the documentation of ciliate biodiversity:
Understand speciation in ciliates by investigating evolutionary rates and changes in morphology, development, life cycle strategies, and genetics underlying divergence of specific lineages
Determine the biogeographical distributions of species of ciliates, test hypotheses of endemicity, and identify mechanisms and rates of dispersal
Identify patterns of diversity and diversity “hot spots”, both geographic and ecological, for ciliates
Develop and implement a significant new paradigm for capturing and storing biological information about ciliates that can support integrative research on their biodiversity
Establish a repository (“protist museum”) to store DNA and fixed cells for use in molecular analyses
Establish a uniform protocol for describing or re‐describing species of ciliates, including minimal standards that must be met for publication and provide guidance on morphological and molecular characterization, sampling protocols, analytical methods, culturing, and long‐term storage of samples.
The first three challenges are theoretical and are supported by implementation of the last three. Furthermore, the latter three challenges all relate to α‐taxonomy (i.e. the description and redescription of species), the first step toward investigating biodiversity.
With this in mind, the present paper offers recommendations for using currently available methodologies and resources to generate a firm base of high‐quality primary data that will facilitate and encourage modern, integrative research into the biodiversity of ciliates. The intent is to: (i) supplement guidance contained in the most recent version of the Code (ICZN 1999) for describing and redescribing species, (ii) consolidate and update recommendations for taxonomic and observational procedures, and (iii) promote the creation of information resources that will support an integrative approach to research into biodiversity of ciliates.
Background
Historical overview
Since Antonie van Leeuwenhoek (1632–1723) made the first descriptions of ciliates (Corliss 2002; Dobell 1932), they have been discovered in almost every environment where there is sufficient water for their survival. They are common in almost all bodies of water below temperatures of about 45 °C and also live in habitats as diverse as desert sands (Foissner et al. 2002), deep‐sea hydrothermal vents (Coyne et al. 2013), polar sea ice (Petz et al. 1995), and East African soda lakes (Finlay et al. 1987). Approximately 8,000 free‐living and epibiotic species are recognized (Lynn 2008). Of these, roughly 4,800 are free‐living, but the total is debated (Agatha 2011; Finlay et al. 1996, 2004; Foissner et al. 2008). Other ciliates are symbionts, either commensals or parasites, mostly on the external surfaces of their hosts but sometimes internally. As with free‐living ciliates, the number of known symbiotic species is probably only a small fraction of their true diversity.
Toward the end of the last millennium, there was an estimated 4,300 described free‐living ciliate species (Finlay et al. 1996). In the new millennium, species of free‐living ciliates are being described at an average rate of ~50 per year (Foissner et al. 2008). In part, this is the result of recent explorations of poorly known or extreme environments (Agatha et al. 1990, 1993; Foissner et al. 2002, 2003; Petz et al. 1995), but a sharp increase in sampling of some understudied areas of the world, especially China and Korea, also has been a major factor. The body of morphological, environmental, and genetic information arising from such descriptions of ciliates is fragmentary relative to that of comparable‐sized groups of multicellular organisms (e.g. vertebrates). This constitutes a major impediment to developing integrative uses of data from investigations of ciliates to address large‐scale issues (e.g. impact of global climate change).
Over the last century, the stepwise introduction of new methods has played a major role in creating this heterogeneity of data by adding chronological “layers” of often disconnected information regarding taxa. In the 1920s–1950s, introduction of silver staining methods provided recognition of ciliary patterns and details of nuclear morphology (Chatton and Lwoff 1930; Cole and Day 1940; Fauré‐Fremiet 1950; Klein 1926); these observations allowed better α‐taxonomy and stimulated major revisions of ciliate taxonomy and classification (Corliss 1961; Jankowski 2007; Lynn 2008). Beginning in the 1950s, application of electron microscopy to investigate organelles and ultrastructures added another “layer” of data. The rapid development of PCR and gene sequencing during the past three decades fueled a leap in our ability to address taxonomic and phylogenetic questions about ciliates. However, they also contributed to the heterogeneity of data by promoting separate paths of training: molecular vs. morphological. Finally, information has been gathered on the ecophysiology, biogeography, and behavior of ciliates, which has been used to complement morphological, morphogenetic, and molecular characterization of taxa. Although all of the above methods continue to be applied to ciliate taxonomy, there have been few attempts to rationalize and combine their application.
The species concept applied to ciliates
As with other organisms, it has been difficult to settle on an acceptable definition of what constitutes a species of ciliate. Sonneborn (1957) suggested the “principle of minimal irreversible evolutionary divergence”. Although “irreversible” may no longer be appropriate as reversals over evolutionary distance occur (Seifert 2014), Sonneborn's concept can be amended to be consistent with α‐taxonomy by deleting “irreversible” (Schlegel and Meisterfeld 2003). Molecular, morphological, or other characters can then be used as evidence of evolutionary divergence, an approach that has been termed the “Pragmatic Species Concept” (Seifert 2014). Insofar as ciliate morphology is closely related to the function of the organism in nature, the morphospecies concept is as valid as any, and probably more pragmatic than any other. Finlay et al. (1996) defined the morphospecies as “a collection of forms that all fit into a defined range of morphological variation—forms that, so far as we can tell, occupy the same ecological niche”, and concluded that “at present, it is difficult to see any other method of differentiating and identifying free‐living ciliates”. Thus, the morphospecies concept is the one most widely adopted in the absence of any pragmatic, viable alternative.
Documenting and accessing data relating to biodiversity of ciliates
The goal of integrative biodiversity has been defined as delimiting the units of life's diversity from multiple and complementary perspectives such as phylogeography, comparative morphology, population genetics, ecology, development, and behavior (Dayrat 2005). There is, however, anecdotal evidence of a decline in taxonomic expertise in most countries of the world. This “taxonomic impediment” (de Carvalho et al. 2007) is a concern in respect to all eukaryotes and specifically threatens progress in ciliate biodiversity and evolution. At the same time, the exponential increase in metagenomics and environmental sequence data, combined with the development of methods for sharing information via the Internet and in open‐access journals, is providing opportunities for progress toward documenting ciliate biodiversity. One increasingly popular strategy for streamlining the formal description of larger numbers of new species combines concise morphological descriptions by expert taxonomists with DNA barcodes and high‐resolution digital imaging. The term “turbo‐taxonomy” has been coined for this approach (Butcher et al. 2012), which, however, might cause serious problems in the future. Consequently, there is a need to move toward closing the gaps in the taxonomic knowledge of ciliates. This will require a solid “systematics portal” to act as a foundation to make information (e.g. digitized images of type specimens, barcodes, and diagnoses) more accessible (Godfray 2007).
Progress with the Grand Challenges of Biodiversity of Ciliates hinges on both describing species remaining to be discovered and on redescribing the many species for which only scanty descriptions exist. Researchers require cross‐training in morphological and molecular methods and an understanding of ecological and behavioral aspects. Data must then be made available through systems for sharing, archiving, and searching taxonomic information (i.e. ones that are efficient, permanent, cost‐effective, and freely available world‐wide). In the following sections, we review key issues of taxonomic procedures as they relate to ciliates and recommend standards for generating the foundation of data needed for the integrative investigations of their biodiversity. These will be categorized as ones that MUST be done to satisfy basic requirements for taxonomic identification or description of ciliates, ones that SHOULD be done to ensure that the highest quality of taxonomic data are presented, and ones that COULD be done to maximize the value of data to integrative investigations.
Taxonomic Procedures
Rules of nomenclature
Nomenclatural acts relating to taxa with ranks from subspecies to superfamily must follow the Code (ICZN 1999; http://iczn.org/iczn/index.jsp), which includes amendments (ICZN 2012) addressing issues associated with publication practices (inclusive of electronic‐only publications) and the use of a registry (ZooBank). Concerns over editorial competence in taxonomic matters and nomenclature for certain electronic‐only journals (Dubois et al. 2013) suggest that taxonomic work should be published in journals that print at least some multiple identical paper copies, unless they have a track record for handling taxonomic issues (i.e. major journals, published for many years). For taxonomic papers where longevity is required, taxa should be published in electronic‐only journals only if the journal has an affiliation with an archive that meets the requirements of Article 8.5.3.1 of the 2012 Code amendment (ICZN 2012). An example of such an archive is CLOCKSS (https://www.clockss.org/) which guarantees to make archived journal files available should the publisher be unable to continue supporting the journal. All nomenclatural acts should be added to ZooBank (Pyle and Michel 2008), the official registry of zoological nomenclature maintained by the ICZN (http://iczn.org/content/about-zoobank). For further discussion on the utility and application of the Code, see Dubois et al. (2016) and Dubois and Aescht (2016).
Key issues not regulated by the Code include what criteria define species‐ to family‐level taxa (see section on species concept above), taxa above family rank, and phylogenetic elements. The following should be consulted: Corliss (1972) provides practical guidelines for higher taxa of ciliates; Aescht (2001, 2008) provides a checklist of required and recommended procedures for complying with provisions of the Code when describing or redescribing species or genera of ciliates.
A type specimen is only the name‐bearer of a nominal species. Therefore, it should not be considered as source of all descriptive data needed in (re)identification, or documenting the range of variation of a species. While the type specimen is a nomenclatural concept, the (re)description refers to taxonomic functions. This is the main reason why the possibility of designating one or more hapantotypes should not be used unless there is a compelling reason for doing so, e.g. specimens at different stages in a life cycle. Although not a strict requirement of the Code, type specimens must be deposited in permanently curated repositories (e.g. museums), not in private or temporary collections. If name‐bearing types are deposited in a laboratory collection, there must be a formal agreement, cited in the original description along with the name of a secondary repository that specifies provisions for permanent curation by the institution in which the laboratory is housed and for passing types to the secondary institution if curation cannot be maintained. Paratype specimens should be placed in a different repository than the one that houses the holotype if it is deposited in any collection other than one in a major museum. Voucher specimens and associated data should be deposited when they are used for taxonomic works (e.g. revision of taxa); where possible this practice could be applied to nontaxonomic works where accurate identification of species may be important (e.g. some ecological investigations). A voucher specimen is defined as a nominated specimen representing the taxonomic unit mentioned in a work, which may or may not be any category of type, e.g. a specimen used in a physiological study or retained as evidence that it was found in a particular location (Hawksworth 2010). Two reprints or a pdf copy of the paper in which type or voucher specimens were designated should be sent to each repository, to preserve the link between specimen(s) and nomenclatural acts.
Composing descriptions and redescriptions
Winston (1999) is an invaluable guide to describing species and should be followed, as descriptions and redescriptions of ciliates should conform to standards applied to other eukaryotes. Recent descriptions and redescriptions of ciliates published in major journals of protistology should be consulted for guidance in formatting and inclusion of data. Descriptive terms must follow those currently accepted (for a glossary, see Lynn 2008), and their usage must be consistent within the description (i.e. avoid synonyms). An analysis of gene sequences should be included in a description of a new species or redescription of a known species, unless material that can be used for extraction of DNA is unavailable. If it is not possible to perform a molecular analysis, cells should be deposited in a publicly accessible repository. In fact, generally, for all type depositions, material should be deposited for later molecular analysis (Dayrat 2005).
Making accurate identifications
Many ciliates are known from only a short original description in the older literature, which is an impediment to accurate identifications. Comprehensive taxonomic sources exist only for few groups of ciliates (e.g. Berger 1999; Vďačný and Foissner 2012). Therefore, identifications of known ciliate species should be supported by detailed comparisons with the original descriptions that confirm its diagnostic features and add new information. If the specimens studied match only some of the published illustrations, these should be itemized. If a redescription is used instead of the original description, this should be justified. Establishment of new species should be accompanied by a justification for the generic affiliation and a comparison with morphologically similar species.
Depositing type and voucher specimens in collections
Stained permanent slides upon which descriptions of novelties are based and redescriptions where a neotype is designated must be deposited (Aescht 2008; Lynn and Simpson 2009). These slides should constitute a homogenous sample of the specimens used for the work and not specimens collected at different places or at different times whose conspecificity is uncertain although a heterogeneous sample might be designated as a syntype under the rules of the Code (Art. 73.2). One or more preparations of directly related individuals, e.g. representing different stages in the life cycle, may be designated as a hapantotype, which is the holotype of the nominal taxon (Art. 72.5.4 and Art. 73.3). The continued acceptance of the hapantotype concept, however, has recently been questioned (Dubois et al. 2016).
Permanent preparations must conform to the basic definition of a type specimen [for type definitions, see International Commission for Zoological Nomenclature (1999) and Aescht (2008)] by revealing sufficient diagnostic characters to allow unambiguous identification and, therefore, should be fixed and stained by a method that shows taxonomically critical features of the major taxon of ciliates to which the species belongs. At present, the two methods that fulfill these criteria most reliably are protargol and silver nitrate (Foissner 2014), although others may become appropriate. Some staining methods (e.g. ammoniacal silver carbonate) result in samples that fade and should not be used to prepare type specimens. Type slides cannot be a source of DNA for sequencing, so additional cells should be deposited for future analysis (e.g. in 95% alcohol at −20 °C or cryopreserved).
In general, voucher material representing populations examined for special purposes (e.g. DNA barcoding, functional ecology) should be submitted to a repository for long‐term preservation. This has not been a common practice for ciliates, with the consequence that much of the ecological literature contains taxonomic identifications that cannot be verified. At a minimum, vouchers could consist of photomicrographs or videos displaying the taxonomically relevant features plus measurements of the organisms. For documenting barcodes, preserved cells showing the taxonomically relevant structure should be made available. Several of the world's major museums, universities, and research institutes hold publicly accessible ciliate collections, of which the following are especially notable.
Biology Centre of the Upper Austrian Museum, Linz, Austria (http://www.landesmuseum.at/): approximately 3,500 slides (holotypes, syntypes, hapantotypes, neotypes, or paratypes) of 990 species representing approximately 420 genera and 150 families; about 20,000 voucher slides of protists
Laboratory of Protozoology, Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China (http://www.ouc.edu.cn/english/); about 2,500 slides of 830 nominal species, including 420 type slides
International Protozoan Type Slide Collection, Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, DC 20013‐7012, USA (http://invertebrates.si.edu/collections.htm); 518 items deposited as type material (470 microscope slides, six SEM stubs, 42 alcohol‐ or formalin‐preserved specimens) representing 119 holotypes, 135 paratypes, 34 lectotypes, 34 paralectotypes, 10 hapantotypes, 28 syntypes and 111 vouchers of more than 350 nominal species
Natural History Museum, London, U.K. (http://www.nhm.ac.uk/); approximately 500 slides (holotypes, hapantotypes, neotypes, paratypes, or vouchers) (http://www.nhm.ac.uk/research-curation/research/projects/protists/), about 22,000 slides of other protists and over 1,000 video sequences of protists (http://www.nhm.ac.uk/research-curation/research/projects/protistvideo/)
Muséum National d'Histoire Naturelle, Paris, France (http://www.mnhn.fr/fr); 114 type slides, most of which were collected in France
Zoological Survey of India, Kolkata, India (http://www.zsi.gov.in); approximately 1,130 slides, 49 holotypes and 104 paratypes
Australian Museum, Sydney, Australia (http://australianmuseum.net.au/); 59 lots of registered specimens, mainly epibiotic species from Australian freshwater crustaceans preserved in 70% ethanol but also including 13 type slides
National Marine Biodiversity Institute of Korea, Chungcheongnam‐do, Korea (http://www.mabik.re.kr/); slides of approximately 80 marine species, including 12 type slides (1 holotype and 11 paratypes) of ten species.
Deposition, Conservation, and Curation of Ancillary Material
General considerations
Beyond depositing material as outlined above, researchers can make supplementary material and data available to future investigators and the public. There are three major issues that must be considered when providing such ancillary materials. They should be:
Provided and maintained with forethought to the development of new technologies or methodologies
Offered by the curation facility in a form that will be available at no cost to stakeholders
Of sufficient quantity and quality to be of utility.
Photomicrographs, videos, or line drawings of cells
Type and voucher slides are useful to taxonomic specialists but are not readily accessible to other potential users, e.g. those not listed as “approved users” of certain museum collections. This issue could be addressed by making high‐resolution digital images of the specimen(s) accessible. Best practice would be to provide images of the specimen(s) taken at different focal planes to allow assembly into 3‐D composites. Such images may then be adaptable to more sophisticated future methodologies, in which physical type or voucher specimens will be accompanied by deposition of raw and 3‐D holographic images in databases (e.g. GBIF).
Here, we suggest that a single portal would facilitate a variety of integrative investigations, allowing newly collected material to be compared with type and voucher specimens, using bioinformatic and image‐resolution software or dedicated search engines. At present, this kind of ability is being created mainly for molecular data; however, morphological, morphogenetic, behavioral, or physical data have their own intrinsic value in terms of revealing ecological or physiological functionality and will become increasingly important as integrative approaches to the investigation of biodiversity continue to evolve.
Molecular specimens
Molecular data are now used in virtually all areas of biological research. Consequently, major natural history museums have established facilities to house collections of physical specimens that can be used in molecular investigations. Prominent among these are the American Museum of Natural History, New York (https://research.amnh.org/amcc/), the Smithsonian National Museum of Natural History, Washington, DC (http://www.mnh.si.edu/rc/biorepository/index.html), the Muséum National d'Histoire Naturelle, Paris (https://www.mnhn.fr/fr/collections/ensembles-collections/ressources-biologiques-cellules-vivantes-cryoconservees), the Natural History Museum, London (http://www.nhm.ac.uk/our-science.html), and the Museo Nacional de Ciencias Naturales, Madrid (http://www.mncn.csic.es/). These house permanently curated repositories for samples of frozen material and environmental samples that are maintained under controlled conditions to make them readily accessible to researchers. Molecular specimens are given an identifier, such as a registration or accession number, allowing further genetic analyses. In this respect, they are similar to traditional museum specimens.
Reliable methods of preserving cells or extracted DNA and RNA that have been applied to ciliates (either as samples of whole cells or within an environmental sample) are FTA Cards (Whatman/GE Healthcare Life Sciences, Pittsburgh, PA; Hide et al. 2003) and RNAlater® (Ambion, Carlsbad, CA). Another method potentially for use with ciliates is DMSO/EDTA/saturated NaCl (DESS) (Gray et al. 2013). The range of materials that can be stored in natural history repositories and that could be used for ciliates includes: (i) specimens deep‐frozen in liquid nitrogen at −196 °C; (ii) whole cells stored at −80 °C; (iii) alcohol‐preserved specimens stored at −20 °C; (iv) DNA extracted from nuclei and mitochondria stored in water or buffers at −20 °C; (v) silica‐dried material stored at room temperature; (vi) extracted RNA stored at −80 °C; (vii) genomic phage/BAC libraries stored at −80 °C; (viii) expression (cDNA) libraries in bacterial culture; (ix) DNA/RNA samples and expression (cDNA) libraries on FTA cards stored at room temperature; and (x) lyophilized/freeze‐dried material stored at room temperature.
Molecular data and analyses
Although not regulated by the Code, additional deposition of genetic information in publicly available databases (e.g. NCBI GenBank) should be included in a description or redescription (e.g. Lynn and Simpson 2009). Most individual sequences that have been deposited are the gene coding for the small subunit ribosomal DNA (SSU rDNA), which is useful for ciliate identification and phylogenetic analysis. Currently, there are SSU rDNA sequences linked to approximately 850 named species of ciliates (Fig. 1). This represents only ~10% of the known ciliate species (Lynn 2008). Many of these are associated with deposited material, but most are not associated with physical specimens of any sort.
Figure 1.
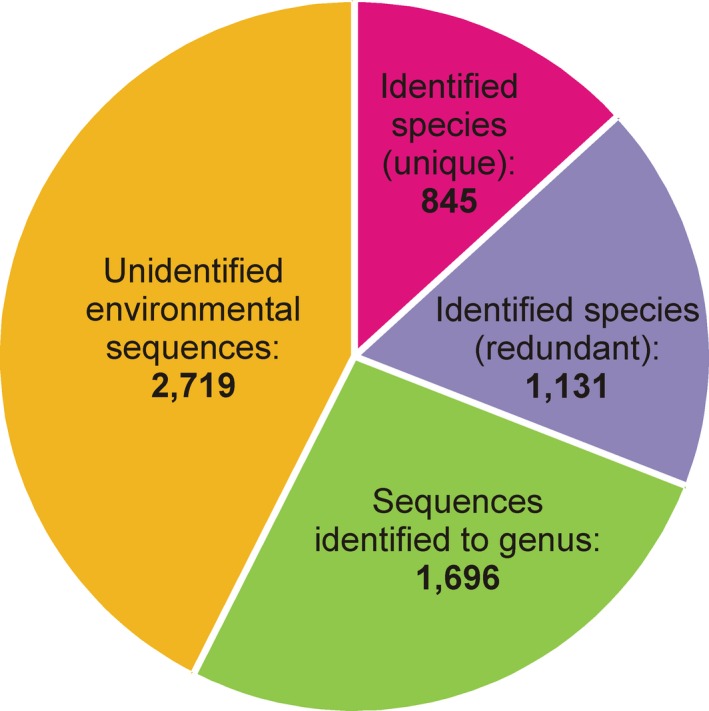
Ciliate sequences of the small subunit ribosomal RNA gene in the NCBI GenBank (http://www.ncbi.nlm.nih.gov; accessed on 6/4/2015).
To date, there is no easily searchable database for housing analyses of data such as alignments and phylogenetic trees. TreeBASE (http://treebase.org/treebase-web/about.html), intended to fill this role, is not searchable by taxonomic category or using names of species (Anwar and Hunt 2009). It is possible to link data to a publication as supplementary material, but this will scatter information among sources that are not easily searched. Hence, we suggest that the online Ciliate Guide (http://ciliateguide.myspecies.info/) should be used, where data can be linked to names of species or higher order taxa in a searchable database. Analytical material such as nexus files of alignments or Newick files of phylogenetic trees also should be deposited in TreeBASE, as it provides quality control (e.g. nexus files of alignments must be capable of being opened in Mesquite) and updating of information by curators.
Genetic data must be evaluated in context of morphological characters to address broad questions about complex processes that involve multiple factors such as evolutionary rates, convergent evolution, population structure, and functional ecology acting in concert. This is also true for relatively narrow avenues of inquiry such as α‐taxonomy. For example, the degree of divergence between sequences of a key gene (e.g. SSU rRNA, ITS, CO1), by itself, cannot substitute for actual characters because there is no generally accepted threshold value for the degree of divergence between congeneric species, including cryptic and pseudocryptic species or higher taxa. Analysis of DNA from environmental samples reveals a large and hidden diversity of ciliates that is rarely reported in morphological surveys of ecosystems studied so far.
Cultures and preserved viable cells
Research into some aspects of ciliate biodiversity is constrained by the difficulty of establishing and maintaining them in long‐term cultures or preserving cells in a viable state. Thus, unlike other microorganisms (e.g. bacteria, algae, fungi), there is no tradition for designating viable protozoan cells preserved in a metabolically inactive state or protozoan cultures as a name‐bearing type. Given the small number of ciliate strains in stable cultures and the technical difficulty of maintaining live cultures or preserved viable cells, it is premature to suggest a similar role for cultures of ciliates. In a few cases, however, long‐term maintenance of ciliate cells in an inactive but viable state has been achieved by cryopreservation (Daggett and Nerad 1992; Müller et al. 2008, 2010; Nerad and Daggett 1992; Simon and Hwang 1967). Also, few ciliate cultures (~700 strains belonging to 54 genera) are deposited in culture collections that accept protists (American Type Culture Collection/ATCC; www.lgcstandards-atcc.org/ and Culture Collection of Algae and Protozoa/CCAP; www.ccap.ac.uk), from which they are available as live cultures or cysts. Most cultured species are members of a few genera used extensively as model organisms (e.g. Tetrahymena, Paramecium, Euplotes). We recommend that researchers could isolate and deposit cultures of taxa they describe, and the following resources are available to this end: (i) formulations for media and protocols for starting and maintaining cultures (Day et al. 2007; Finlay et al. 2000; Lee and Soldo 1992; Nerad 1991; Tompkins et al. 1995); (ii) protocols for preserving dried cysts on filter paper (McGrath et al. 1977); and (iii) protocols for cryopreservation (Müller et al. 2008).
Biological Resource Centers (BRCs), such as the ATCC or the CCAP, play a key role in safeguarding and providing cultures, with the main objective being the preservation of the phenotypic and genotypic characteristics of the conserved taxa (Müller et al. 2008). In recent years, the role of BRCs has been extended, largely as a result of the rapid increase in metagenomics and environmental sequencing data. The CCAP has responded to this challenge by developing KnowledgeBase, for quality‐controlled biological material, hyperlinked to manually curated molecular, bibliographic, and taxonomic information (Gachon et al. 2013).
Morphological and Ontogenetic Characters
Documenting morphology and ontogeny
Virtually the entire body of knowledge about ciliates is linked to morphospecies. Therefore, morphological characters must be studied for species descriptions and redescriptions and should be included with other characteristics when investigating aspects of ciliate biodiversity. Best practice should be to measure, count, describe, and illustrate every observable detail of morphology and ontogeny, even though all data might not be published. Where known cryptic species complexes have been recognized by molecular characters (e.g. isozymes, cox1, ITS2; Nanney and McCoy (1976), Sonneborn (1957) and references in section below on DNA barcoding) a detailed morphological description should be carried out with the aim to search for new taxonomically relevant characters that can be analyzed by multivariate statistics (e.g. Gates and Berger 1974, 1976; Powelson et al. 1975).
If possible, clonal cultures should be used for observations to avoid reliance on data from field samples alone that create the risk of confusing morphologically similar species. Some species also have polymorphic life cycles that only can be revealed under culture conditions, including using different conditions (e.g. Tetrahymena aquasubterranea; Quintela‐Alonso et al. 2013). However, it should be noted that cells in culture may exhibit phenotypes not typically seen in nature, so culture conditions must be described.
A description or redescription must be based on deposited, fixed material, revealing the taxonomically relevant characters and also must be based on live observation (Foissner 2014), except when meeting either or both of these requirements is not reasonable and other material (physical or virtual) allowing a reinvestigation of the diagnostic features is deposited. Cells can shrink or swell during fixation, and the characters of some taxa can contract and/or disappear; therefore, live observations provide useful information (e.g. see Fig. 2). Information obtained from live observation should include:
Figure 2.
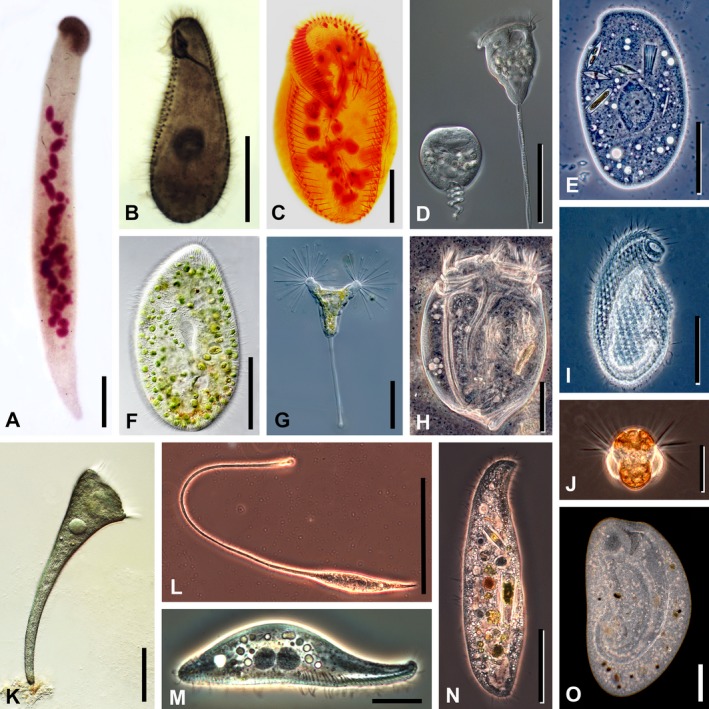
Photomicrographs of representative ciliate taxa (originals from D. J. Patterson). (A) Homalozoon vermiculare, a crawling, ribbon‐shaped haptorian predator. The cell has been fixed and stained with Feulgen reagent. The anterior mouth, at the top of the image, is underlain with a dense band of extrusomes that are used to capture food. The structures stained pink are the irregular ribbon of macronuclear nodules, the micronuclei, and a few developing extrusomes. Scale bar = 50 μm. (B) Tetrahymena pyriformis, a hymenostome ciliate. The cell has been fixed and stained with protargol that impregnates the bases of the cilia (basal bodies) and the nucleus. The somatic cilia are arranged in longitudinal rows; the oral cilia are densely packed and appear as short bands. A system of microtubules that forms the cytopharynx extends from the mouth into the cell. The macronucleus is the globular structure near the center of the cell. Scale bar = 20 μm. (C) Pattersoniella vitiphila, a hypotrich ciliate. Fixed cell stained with protargol. The C‐shaped collar around the front (top) of the cell is the adoral zone of membranelles (ciliary fans). The spots near the front of the cell, forming marginal bands and a curving row in the center of the cell, are cirri (ciliary bristles). The macronuclear nodules are the globular inclusions within the cell. Scale bar = 50 μm. (D) Vorticella convallaria, a peritrich ciliate. Two living cells under Nomarski differential interference contrast optics, the one to the right is extended and feeding, the one to the left is contracted. The feeding cilia (haplokinety and polykineties) extend around the edge of the anterior of the cell and curve into the mouth. The cells contain many food vacuoles. The stalk contains a spasmoneme that causes it to contract in a spiral fashion, while an elastic envelope acts as antagonist. Scale bar = 50 μm. (E) Trithigmostoma sp., a cyrtophorid ciliate. Living cell under phase‐contrast optics. The cell eats small algae, using the basket‐shaped mouth (upper right). Several ingested diatoms can be seen inside the cell. The large central structure is the macronucleus. There are multiple small contractile vacuoles in the cell that are used for osmoregulation. Scale bar = 50 μm. (F) Paramecium bursaria, a peniculid ciliate with endosymbiotic zoochlorellae. Living cell under Nomarski differential interference contrast optics. The cell surface is underlain with many rod‐shaped extrusomes (minute harpoon‐like structures). The pear‐shaped area in the center of the cell is the cytopharynx. Scale bar = 50 μm. (G) Acineta sp., a suctorian ciliate. Living cell under Nomarski differential interference contrast optics. This predator captures other ciliates and flagellates that swim into the extending sticky tentacles. Each tentacle is a mouth and has an expanded apex containing extrusomes that catch hold of any prey. The cell is attached to substrates by means of a stalk. Cilia are absent in the trophic cell, but present in the swarmers. Scale bar = 50 μm. (H) Eudiplodinium sp., an entodiniomorphid ciliate from the stomach of a bison. Living cell under phase‐contrast optics. The body is very stiff, sculpted with longitudinal grooves that form a rounded extension at the posterior end of the cell. There are two zones of cilia at the anterior end. The ciliate feeds on bacteria, many of which can be seen around the cell, and on small particles of wood. Two contractile vacuoles and associated vesicles lie over the macronucleus in the left cell half. Scale bar = 20 μm. (I) Cryptopharynx sp., a small and rarely encountered karyorelictean ciliate. Living cell under phase‐contrast optics. The mouth is the extension to the anterior right. The cilia form dimpled rows (kineties) that extend along the length of the body. Scale bar = 20 μm. (J) Mesodinium rubrum (=Myrionecta rubra), a small marine planktonic haptorian ciliate that contains endosymbiotic cryptomonad algae giving the cell its red color. Living cell under phase‐contrast optics. Stiff bundles of equatorial cilia extend from the cell and their action causes a jumping motion; further cilia lay adpressed to the posterior portion of the body. Scale bar = 20 μm. (K) Stentor coeruleus, a trumpet‐shaped heterotrich ciliate. Living cell under Nomarski differential interference contrast optics. The cell attaches to immersed surfaces via its posterior end and, when feeding, the cell has a characteristic conical form with the adoral zone of membranelles at the anterior of the cell. Cilia also extend from the remaining body surface. The blue color comes from minute inclusions (cortical granules) lying just under the cell surface. Scale bar = 100 μm. (L) Lacrymaria olor, a predatory haptorid ciliate. Living cell under phase‐contrast optics. The extensible and contractile anterior portion moves around, searching for prey, while the posterior end of the body often remains attached to the substrate. The mouth is at the tip of the extensible neck and is expanded because of the extrusomes that are used to kill prey—often small ciliates. Scale bar = 50 μm. (M) Litonotus sp., a pleurostomatid predatory ciliate. Living cell seen from the side under phase‐contrast optics. There are two large macronuclear nodules in the center of the cell. The mouth lies along the bottom edge of the cell, extending from near the middle to the anterior end, and is underlain with extrusomes that are used to kill prey. Somatic cilia extend from the ventral surface of the cell. Scale bar = 20 μm. (N) Loxodes sp., a karyorelictid ciliate from freshwater habitats. Living cell under phase‐contrast optics. The sickle‐shaped mouth region is located to the anterior right of the cell, which engulfs mostly algae, such as diatoms, and cyanobacteria. One of the macronuclear nodules is evident just above the mid‐line and in the left cell half; it has a dark core and a light peripheral region. Anteriorly and posteriorly to it are two vacuoles of a similar size but each with a bright refractile granule. These are Müller's bodies that inform the cell as to the direction of gravitational pull. Scale bar = 50 μm. (O) Bursaria sp., a huge colpodid ciliate. Living cell under dark field optics. There is a large, curved channel that leads form the anterior of the cell to the cytostome. The ciliate is mostly found in still‐freshwater habitats, where it feeds on planktonic algae. Scale bar = 50 μm.
Cell size and shape
Cilia lengths (oral, somatic, brush, etc.)
Cytoplasm color
Contractility and flexibility of cell
Presence, color, and arrangement of cortical granules
Presence of symbiotic algae, bacteria, or sequestered plastids
Presence, size, shape, and location of mature extrusomes
Presence, position, number, and shape of contractile vacuoles and the contractile vacuole pore
Composition, shape, and location of the nuclear apparatus.
Information obtained from live observation could also include:
Variations in cell size and shape associated with division, life cycles, or environmental conditions
Position and shape of the cytopyge
Presence of lipid droplets, crystals, or other inclusions
Contents of food vacuoles (indicating the role of the organism in the microbial food web—with the caveat that the types of food eaten may vary according to local conditions and may interact with abiotic factors to affect morphology or activity).
At present, staining with protargol or silver nitrate must be performed to provide fixed type material but where this is not reasonable, equivalent methods revealing the diagnostic features must be applied providing permanent type material (physical or virtual). Silver carbonate stains may be used to augment descriptions, but these fade and their results must be documented photographically (Fig. 2). In the future, it could be more appropriate to apply fluorescent staining of structures to augment descriptions (Arregui et al. 2002), enabling detailed examination by confocal microscopy. Information recorded from silver‐stained preparations should include:
Pattern of somatic ciliature to include general configuration, number, and orientation of kineties; number and spacing of kinetids in kineties, and structure of kinetid and brush (if present)
Pattern of oral ciliature (i.e. arrangement, number, and structure of ciliary organelles)
Structure of nuclear apparatus (i.e. number, shape, size, and location of nuclei and shape of nucleoli)
Miscellaneous features, such as positions and appearance of organelles (e.g. argyrophilic granules and fibrillar structures).
There is a range of other features that also may be examined. Ontogenesis (i.e. cell division including stomatogenesis, morphogenesis, and the regeneration of parental structures) should be included in descriptions when it provides taxonomically relevant features, e.g. the position of the oral primordium is diagnostic in oligotrichid ciliates (Agatha 2004) and the lack of a frontoventral cirrus in a certain position is informative in euplotids (Borror and Hill 1995); otherwise, the investigation of this process is recommended as even in well‐known ciliate groups, new patterns of cell division may be discovered (e.g. in halteriids by Song 1993). Resting cysts, if present, should be examined as morphological diversity occurs in some morphospecies, which may suggest the existence of pseudocryptic species (Foissner et al. 2002; Katz et al. 2005; Xu and Foissner 2005). Mating type interactions could provide insights into the breeding structure of a species (Nobili et al. 1978). Finally, some taxa (e.g. dileptids; Vďačný and Foissner 2012) display a regenerative capacity that should be documented.
Illustrating morphology
Observations that form the basis of descriptions or redescriptions must be documented by illustrations and should be accompanied by photomicrographs. A useful source regarding techniques to produce illustrations acceptable for publication is the Guild of Natural Science Illustrators (https://gnsi.org/). The handbook of the Guild (Hodges 2003) is the definitive source, but there are others (Ridgway 1978; Wood and McDonnell 1994; Zweifel 1988). Silver‐stained preparations should be used as the primary basis of illustrating specimens with line drawings. As photomicrographs of silver‐stained specimens show only one focal plane of the cell, image‐stacking software could be used to assemble several planes into a single composite image. Fluorescently labeled taxoids such as FLUTAX (Santa Cruz Biotechnology Inc., Dallas, TX) that bind specifically to microtubules could also be used, possibly in conjunction with confocal microscopy, to assess patterns of ciliature and infraciliature during the identification process (Arregui et al. 2002). The style for the arrangement of illustrations and plates, use of standard abbreviations, and other stylistic conventions should follow those in the recent literature.
Morphometric data
Data obtained by morphometric studies must be derived from a sufficient number of appropriately oriented morphostatic specimens to provide information on the range of the characters and allow the species to be separated from other closely related taxa; this will vary depending on the taxon and the feature, but between 10 and 30 measurements should generally be appropriate. Data should be presented as standard descriptive statistics (e.g. mean, standard error, range). However, as measurements not provided in the description may emerge as diagnostic, the raw data should be submitted as supplementary material.
Molecular Characters
Using molecular data as taxonomic characters
Molecular phylogenies that disagree with classifications based on morphological characters create debate regarding the relative merits of these approaches, but in doing so they initiate the search for new characters. Certainly, the new molecular technologies cause similar large‐scale readjustments in the classification of ciliates like the previous introductions of new methods for studying morphological features. Molecular evidence is still relatively new, and the technology for obtaining it is evolving rapidly; therefore, pitfalls and limitations continue to emerge. Accordingly, use of molecular characters by themselves for defining taxa must be done with caution and only if every attempt to relate other features (described above) with molecular characters failed. There are already precedents for this, both in ciliates and in other groups of protists (Dunthorn et al. 2014; Hoef‐Emden and Melkonian 2003; Huss et al. 1999; Marin et al. 2003; Pröschold et al. 2001; Sun et al. 2012; Zhan et al. 2009).
In practice, molecular characters are not materially different from other categories of evolutionary evidence because they are constrained by the same analytical criteria. They must be evaluated critically like other characters (i.e. a priori vs. a posteriori weighting) and should be used to formulate phylogenetic hypotheses that can be tested by enlarging the dataset with further morphologic and molecular characters and taxa (Neff 1986). As with other sorts of characters, reliability of molecular characters must be evaluated by a thorough comparison within and between taxa and with cladistic analyses to discover elements such as homoplasies or polymorphisms, with the goal of detecting robust synapomorphies. Finally, molecular characters must be described, illustrated, and discussed.
DNA barcoding
Protists are so genetically diverse owing to their long independent evolutionary histories that it has been impossible to identify a single DNA barcode for all of them. Consequently, the Consortium for the Barcode of Life Protist Working Group (CBOL ProWG) was established in 2012 to identify standard barcode regions for protists and to assemble a reference library (Pawlowski et al. 2012). The CBOL ProWG suggests a two‐step barcoding approach of preliminary identification: a universal eukaryotic barcode (the pre‐barcode) followed by a species‐level assignment using a group‐specific barcode. Based on comparative studies and applying the CBOL (http://www.barcodeoflife.org/) selection criteria, the ProWG suggests the ~500 bp long variable V4 region of the 18S rDNA as the universal pre‐barcode, with group‐specific barcodes then being defined separately for each major protistan group (Pawlowski et al. 2012).
Ciliates exhibit a number of features that make it likely that a suitable DNA barcode will be identified. They are monophyletic and relatively well‐studied and have a complex, character‐rich morphology that allows most forms to be identified reliably to morphospecies (Corliss 2002). Despite these advantages, relatively few studies have focused on the DNA barcoding of ciliates (Barth et al. 2006; Chantangsi et al. 2007; Gentekaki and Lynn 2012; Guggiari and Peck 2008; Kher et al. 2011; Santoferrara et al. 2013; Stoeck et al. 2014; Strüder‐Kypke and Lynn 2010; Zhao et al. 2013). Owing to their genetic diversity, different coding regions may be needed as barcodes for different groups of ciliates. Potential candidates for barcoding markers include the coding regions for the small subunit ribosomal DNA (SSU rDNA, 18S rDNA), large subunit ribosomal DNA (LSU rDNA, 28S rDNA), internal transcribed spacers (ITS1 and ITS2), cytochrome b (cytb), and cytochrome c oxidase (cox1). Proofed genetic distances for members of individual major taxa should be established and adjusted taking into account the morphological data (Lee 2004; Lipscomb et al. 2003; Seberg et al. 2003; Will and Rubinoff 2004). Barcode data should be correlated with traditionally described taxa (see above) and the autecological information associated with them through names as part of the process of inferring ecological roles from occurrences documented by barcodes. Furthermore, as indicated above, voucher material showing the taxonomically relevant features should be deposited as a means of cross‐referencing a new barcode sequence, the extracted DNA from which the barcode sequence was derived also should be deposited, and fixed cells and/or live or cryopreserved cultures could be deposited.
Other Characters
Behavior
There there is a history dating back to Antonie van Leeuwenhoek, of complementing descriptions of protists with ancillary information on their behavior, but the Code does not explicitly support this approach. Some ciliates, however, exhibit characteristic behavior, such as jumping several “body lengths” in a fraction of a second, spiral swimming, or adhering to surfaces. Such characters could be included in descriptions. It is critical to note that such behaviors are dependent on the environment (food type and abundance, temperature, pH, substrate type, light, etc.), convergent behaviors occur across taxa, and behavior can be similar at least in congeners. Behavior may thus be a poor diagnostic feature. Anecdotal observations could include: patterns of crawling or swimming (e.g. helical trajectories), jumping and escape behavior, feeding, contractility, formation of stalks, temporary attachment, alternation of rests and movements. The speed of swimming could be roughly estimated, e.g. using a small strip of scaled paper placed beneath a Petri dish, or knowing the width of the field of view at lower microscope magnifications. Simple trajectories plus remarks on peculiarities of movement may be sufficient, but more sophisticated methods, such as image analysis of video sequences and computer modeling of cell motion, are available (Oliveira‐Pinto et al. 1993). In contrast, if a study fully characterizes the behavior of a taxon, then these data should be used to aid in the identification of live material and play a part in future research on functionality. We, therefore, encourage detailed studies of ciliate behavior and ecophysiology, which may then be tied to taxonomic descriptions. However, we caution against providing information as descriptive when it is not rigorously examined (with full documentation of methods and results).
Environmental data
Beyond describing the type environment, as outlined in the Code (Recommendation 76A), environmental factors can affect behavior and have a significant impact on growth/size. Therefore, physical data such as temperature, salinity, and pH of the water, sediment, or soil from which samples are taken should be recorded. Likewise, detailed culture conditions must be presented in the methods of described species (see above).
Information Resources
Overview
Both taxonomic and nontaxonomic data relating to biodiversity are often fragmentary and heterogeneous, making it difficult or impossible for researchers to identify and fill gaps in knowledge quickly and accurately. Information resources could reduce this issue by nourishing integrative biodiversity as an investigative approach, but data must be reliable and readily accessible to a broad, interdisciplinary research community. Furthermore, a good information resource should encourage an increasing array of links to other expert sources but not duplication of such resources. These are the aspirations we promote below.
Beyond type material, there are many objects that have value to taxonomy of ciliates (Table 1). However, at this time, no knowledge resource like GenBank or ZooBank exists for preserved cells, type slides, or other materials needed to ensure high‐quality taxonomic research on ciliates. Also it is difficult to find some published material, such as taxonomic treatments in older journals that include nomenclatural acts or descriptions/revisions plus illustrations that constitute the functional equivalent of type material.
Table 1.
Categories of information resources applicable to biodiversity of ciliates (modified from Baskauf 2010)
| Category of resource | Form of representation | Specific representations |
|---|---|---|
| Physical object | Material artifact | Film negative |
| Photographic print | ||
| Film movie | ||
| Living/viable culture | ||
| Preserved cell/DNA sample | ||
| Prepared slides | ||
| Fixed specimens | ||
| Hard‐copy field notes | ||
| Hard‐copy laboratory notes | ||
| Original illustrations | ||
| Information | Digital file | Image |
| Video | ||
| Specimen data | ||
| DNA/RNA sequence | ||
| Sequence alignment | ||
| Phylogenetic tree | ||
| Classification | ||
| Taxonomic synonymy | ||
| Geographic coordinates | ||
| Physical–chemical measurements |
There have been efforts to make some of these resources more available, through digitizing initiatives such as the Biodiversity Heritage Library (http://www.biodiversitylibrary.org/ and through taxon‐specific websites such as:
Ciliates in Activated Sludge (software package available for download from http://ciliateguide.myspecies.info/ciliates-activated-sludge)
Planktonic Ciliate Project (http://ciliate.zooplankton.cn)
The Microbial Digital Specimen Archives (Protist information Server: http://protist.i.hosei.ac.jp/pdb/images/menuE.html)
Free‐Living Ciliates in the Bohai and Yellow Seas, China (http://www2.ouc.edu.cn/akfs/ciliate/asp/)
Marine Benthic Ciliates (http://ciliate.myspecies.info/en).
However, the dispersed nature of the above resources reduces the potential to interrogate them. At the very least, as a community, we should supplement these efforts by digitizing items that are unavailable online and/or difficult to obtain in hard copy, following these guidelines:
Supplementary information and data linked with type or voucher specimens should be made available on a curated open‐access website
Sites providing access to metadata should be organized to allow remote interrogation (Gachon et al. 2013)
Documentation should be provided so that collections of specimens comply with the Convention on Biological Diversity (http://www.cbd.int/)
Standards for supplementary materials and data should be agreed upon by the research community
Reliable metadata should be made available via a dedicated web‐portal, e.g. The Global Genome Biodiversity Network [GGBN] Data Portal (http://data.ggbn.org) (Droege et al. 2014).
Development of resources for sharing information and identifying species
Here we suggest that integrative biodiversity of ciliates would benefit from development of a single online resource, following the criteria outlined above. There are four reasons for developing such a resource. It should:
Facilitate continual, accurate identification of ciliates collected from nature
Allow new taxonomic investigations to build on all prior work
Enable taxonomic data/metadata to be combined with reliable ecological and physiological data/metadata to facilitate large‐scale studies
Provide quality control for online materials from other web sites.
To this end, we propose development of The Ciliate Guide (http://ciliateguide.myspecies.info/), which is a Scratchpad/Biodiversity online resource, providing a nomenclatural template for keys, descriptions, and metadata to the level of species. We suggest that it should be used as the software platform for the following reasons:
Provides a taxonomic portal for entry of content
Provides an ability to create keys for identification
Provides the capacity to submit data and metadata
Provides guaranteed permanent hosting (EU‐sponsored Scratchpads project: http://scratchpads.eu/)
Is freely accessible
Can access material from independent sources
Is suited to a communal environment that allows a registered user to contribute materials that are vetted by a select group of authoritative users.
We also suggest that the IRCN‐BC should assume this task because of its broad, inclusive membership and focus on integrative biodiversity. Aside from providing a resource for identifying species and assembling data/metadata, The Ciliate Guide also can be used as a searchable database of physical specimens and type slides. Thus it can function as a “virtual museum”, making it much easier to locate specimens and link metadata to digitized images of type specimens. To develop The Ciliate Guide the IRCN‐BC will:
Establish a steering group to propose the initial requirements and set priorities
Configure The Ciliate Guide accordingly
Identify complementary initiatives and establish information‐flow
Establish a registration system allowing registered users to contribute, limiting editorial rights to a committee of experts
Promote participation to gather and review resources
Ensure an active update of the site
Identify and add features to accommodate developments in methodology and needs
Establish means to provide credit for contributions.
Conclusions
Technological advances over the last several decades have made it possible to achieve rapid progress in knowledge about the biodiversity of organisms. Research on ciliates and other protists has, of course, benefited from this trend, but not to the degree seen in multicellular taxa. In essence, research on ciliate biodiversity is constrained by a deficit in basic information about their biodiversity owing to historical difficulties in making systematic observations of microorganisms. The best remedy is an organized, sustained push to follow uniform standards of practice while assembling data that will form a future foundation. To this end, we have articulated needs in the form of Grand Challenges that are addressed by sets of specific recommendations for best practice and a proposal to populate a database on the biodiversity of ciliates. A regularly updated listing of recommendations accompanied by checklists for observational methodologies will be made available on the website of the IRCN‐BC (http://ircn-bc.org).
Acknowledgments
This review is the product of a workshop sponsored jointly by the IRCN‐BC and the British Society for Protist Biology that met at Royal Holloway, University of London, U.K. from 1–3 September 2014 (Dunthorn et al. 2015). The present paper reviews topics that were discussed and reflects the consensus of participants in the workshop. Funding for the workshop was provided by grant DEB 1136580 from the U.S. National Science Foundation to John Clamp and grant no. 31111120437 from the National Natural Science Foundation of China to Weibo Song. We thank Dr. K. Chandra, Director of ZSI, Kolkata, for providing details of the ZSI protozoa slide collection. We also thank Emily Perrier and the conference staff of Royal Holloway, University of London and Martin Carr, Treasurer of the British Society for Protist Biology (now called Protistology‐UK), for their efforts at making sure that all of the arrangements for the workshop went smoothly. We thank the editor of JEM and an anonymous reviewer for constructive comments.
Affiliations are listed at the end.
Literature Cited
- Adl, S. M. , Simpson, A. G. , Lane, C. E. , Lukeš, J. , Bass, D. , Bowser, S. S. , Brown, M. W. , Burki, F. , Dunthorn, M. , Hampl, V. , Heiss, A. , Hoppenrath, M. , Lara, E. , Le Gall, L. , Lynn, D. H. , McManus, H. , Mitchell, E. A. D. , Mozley‐Stanridge, S. E. , Parfrey, L. W. , Pawlowski, J. , Rueckert, S. , Shadwick, L. , Schoch, C. , Smirnov, A. & Spiegel, F. W. 2012. The revised classification of eukaryotes. J. Eukaryot. Microbiol., 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aescht, E. 2001. Catalogue of the generic names of ciliates (Protozoa, Ciliophora). Denisia, 1:1–350. [Google Scholar]
- Aescht, E. 2008. Annotated catalogue of “type material” of ciliates (Ciliophora) and some further protists at the Upper Austrian Museum in Linz (Austria) including a guideline for “typification” of species. Denisia, 23:125–234. [Google Scholar]
- Agatha, S. 2004. A cladistic approach for the classification of oligotrichid ciliates (Ciliophora: Spirotricha). Acta Protozool., 43:201–217. [PMC free article] [PubMed] [Google Scholar]
- Agatha, S. 2011. Global diversity of aloricate Oligotrichea (Protista, Ciliophora, Spirotricha) in marine and brackish sea water. PLoS One, 6:e22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agatha, S. , Spindler, M. & Wilbert, N. 1993. Ciliated protozoa (Ciliophora) from Arctic sea ice. Acta Protozool., 32:261–268. [Google Scholar]
- Agatha, S. , Wilbert, N. , Spindler, M. & Elbrächter, M. 1990. Euplotid ciliates in sea ice of the Weddell Sea (Antarctica). Acta Protozool., 29:221–228. [Google Scholar]
- Anwar, N. & Hunt, E. 2009. Improved data retrieval from TreeBASE via taxonomic and linguistic data enrichment. BMC Evol. Biol., 9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arregui, L. , Muñoz‐Fontela, C. , Serrano, S. , Barasoain, I. & Guinea, A. 2002. Direct visualization of the microtubular cytoskeleton of ciliated protozoa with a fluorescent taxoid. J. Eukaryot. Microbiol., 49:312–318. [DOI] [PubMed] [Google Scholar]
- Barth, D. , Krenek, S. , Fokin, S. I. & Berendonk, T. U. 2006. Intraspecific genetic variation in Paramecium revealed by mitochondrial cytochrome c oxidase I sequences. J. Eukaryot. Microbiol., 53:20–25. [DOI] [PubMed] [Google Scholar]
- Baskauf, S. J. 2010. Organization of occurrence‐related biodiversity resources based on the process of their creation and the role of individual organisms as resource relationship nodes. Biodivers. Inform., 7:17–44. [Google Scholar]
- Berger, H. 1999. Monograph of the Oxytrichidae (Ciliophora, Hypotrichia). Monogr. Biol., 78:1–1080. [Google Scholar]
- Borror, A. C. & Hill, B. F. 1995. The order Euplotida (Ciliophora): taxonomy, with division of Euplotes into several genera. J. Eukaryot. Microbiol., 42:457–466. [Google Scholar]
- Butcher, B. A. , Smith, M. A. , Sharkey, M. J. & Quicke, D. L. J. 2012. A turbo‐taxonomic study of Thai Aleiodes (Aleiodes) and Aleiodes (Arcaleiodes) (Hymenoptera: Braconidae: Rogadinae) based largely on COI barcoded specimens, with rapid descriptions of 179 new species. Zootaxa, 3457:1–232. [Google Scholar]
- de Carvalho, M. R. , Bockmann, F. A. , Amorim, D. S. , Branda, C. R. F. , de Vivo, M. , de Figueiredo, J. L. , Britski, H. A. , de Pinna, M. C. C. , Menezes, N. A. , Marques, F. P. L. , Papavero, N. , Cancello, E. M. , Crisci, J. V. , McEachran, J. D. , Schelly, R. C. , Lundberg, J. G. , Gill, A. C. , Britz, R. , Wheeler, Q. D. , Stiassny, M. L. J. , Parenti, L. R. , Page, L. M. , Wheeler, W. C. , Faivovich, J. , Vari, R. P. , Grande, L. , Humphries, C. J. , DeSalle, R. , Ebach, M. C. & Nelson, G. J. 2007. Taxonomic impediment or impediment to taxonomy? A commentary on systematics and the cybertaxonomic‐automation paradigm. Evol. Biol., 34:140–143. [Google Scholar]
- Chantangsi, C. , Lynn, D. H. , Brandl, M. T. , Cole, J. C. , Hetrick, N. & Ikonomi, P. 2007. Barcoding ciliates: a comprehensive study of 75 isolates of the genus Tetrahymena . Int. J. Syst. Evol. Microbiol., 57:2412–2425. [DOI] [PubMed] [Google Scholar]
- Chatton, E. & Lwoff, A. 1930. Imprégnation, par diffusion argentique, de l'infraciliature de ciliés marins et de d'eau douce, après fixation cytologique et sans desiccation. C. R. Séances Soc. Biol., 104:834–836. [Google Scholar]
- Cole, R. & Day, M. F. 1940. The use of silver albumose (Protargol) in protozoological technique. Abstract 47. J. Parasitol., 26(Suppl. 6):30–31. [Google Scholar]
- Corliss, J. O. 1961. The ciliated protozoa: Characterization, classification and guide to the literature. Pergamon Press, London and New York: p. 1–310. [Google Scholar]
- Corliss, J. O. 1972. Common sense and courtesy in nomenclatural taxonomy. Syst. Zool., 21:117–122. [Google Scholar]
- Corliss, J. O. 2002. A salute to Antony van Leeuwenhoek of Delft, most versatile 17th Century founding father of protistology. Protist, 153:177–190. [DOI] [PubMed] [Google Scholar]
- Coyne, K. J. , Countway, P. D. , Pilditch, C. A. , Lee, C. K. , Caron, D. A. & Cary, S. C. 2013. Diversity and distributional patterns of ciliates in Guaymas Basin hydrothermal vent sediments. J. Eukaryot. Microbiol., 60:433–447. [DOI] [PubMed] [Google Scholar]
- Daggett, P.‐M. & Nerad, T. A. 1992. Long term maintenance of selected encysting ciliates by drying and cryopreservation In: Lee J. J. & Soldo A. T. (ed.), Protocols in Protozoology. Society of Protozoologists, Lawrence, KS: p. A93.1–A93.6. [Google Scholar]
- Day, J. G. , Achilles‐Day, U. , Brown, S. & Warren, A. 2007. Cultivation of algae and protozoa In: Hurst C. J., Crawford R. L., Garland J. L., Lipson D. A., Mills A. L. & Stetzenbach L. D. (ed.), Manual of Environmental Microbiology, 3rd ed ASM, Washington, DC: p. 79–92. [Google Scholar]
- Dayrat, B. 2005. Towards integrative taxonomy. Biol. J. Linn. Soc., 85:407–415. [Google Scholar]
- Dobell, C. 1932. Antony van Leeuwenhoek and his “little animals”. Dover Publications Inc., New York, NY: p. vii + 435. [Google Scholar]
- Droege, G. , Barker, K. , Astrin, J. J. , Bartels, P. , Butler, C. , Cantrill, D. , Coddington, J. , Forest, F. , Gemeinholzer, B. , Hobern, D. , Mackenzie‐Dodds, J. , O′Tuama, E. , Petersen, G. , Sanjur, O. , Schindel, D. & Seberg, O. 2014. The global genome biodiversity network (GGBN) data portal. Nucleic Acids Res., 42(D1):D607–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, A. & Aescht, E. 2016. Sessions 1–6 of the Linz Zoocode Committee (February–June 2016). Dumerilia, 6:38–70. [Google Scholar]
- Dubois, A. , Aescht, E. & Dickinson, E. C. 2016. Burning questions and problems of zoological nomenclature. The Linz International Workshop of Zoological Nomenclature (9–10 July 2014). Dumerilia, 6:24–34. [Google Scholar]
- Dubois, A. , Crochet, P.‐A. , Dickinson, E. C. , Nemésio, A. , Aescht, E. , Bauer, A. M. , Blagoderov, V. , Bour, R. , de Carvalho, M. R. , Laure, D. , Frétey, T. , Jaeger, P. , Koyamba, V. , Lavilla, E. O. , Löbl, I. , Malécot, V. , Schatz, H. & Ohler, A. 2013. Nomenclatural and taxonomic problems related to the electronic publication of new nomina and nomenclatural acts in zoology, with brief comments on optical discs and on the situation in botany. Zootaxa, 3735:1–94. [DOI] [PubMed] [Google Scholar]
- Dunthorn, M. , Otto, J. , Berger, S. A. , Stamatakis, A. , Mahé, F. , Romac, S. , de Vargas, C. , Audic, S. , Stock, A. , Kauff, F. & Stoeck, T. 2014. Placing environmental next‐generation sequencing amplicons from microbial eukaryotes into a phylogenetic context. Mol. Biol. Evol., 31:993–1009. [DOI] [PubMed] [Google Scholar]
- Dunthorn, M. , Zufall, R. A. & Lobban, C. 2015. Report of the 2014 joint workshop of the International Research Coordination Network for Biodiversity of Ciliates and the British Society for Protist Biology. Eur. J. Protistol., 51:118–119. [DOI] [PubMed] [Google Scholar]
- Fauré‐Fremiet, E. 1950. Morphologie comparée et systématique des ciliés. Bull. Soc. Zool. Fr., 75:109–122. [Google Scholar]
- Finlay, B. J. , Black, H. I. J. , Brown, S. , Clarke, K. J. , Esteban, G. F. , Hindle, R. M. , Olmo, J. L. , Rollett, A. & Vickerman, K. 2000. Estimating the growth of the soil protozoan community. Protist, 151:69–80. [DOI] [PubMed] [Google Scholar]
- Finlay, B. J. , Corliss, J. O. , Esteban, G. F. & Fenchel, T. 1996. Biodiversity at the microbial level: the number of free‐living ciliates in the biosphere. Q. Rev. Biol., 71:221–237. [Google Scholar]
- Finlay, B. J. , Curds, C. R. , Bamforth, S. S. & Bafort, J. M. 1987. Ciliated protozoa and other microorganisms from two African soda lakes (Lake Nakuru and Lake Simbi, Kenya). Arch. Protistenk., 133:81–91. [Google Scholar]
- Finlay, B. J. , Esteban, G. F. & Fenchel, T. 2004. Protist diversity is different? Protist, 155:15–22. [DOI] [PubMed] [Google Scholar]
- Foissner, W. 2002. Neotypification of protists, especially ciliates (Protozoa, Ciliophora). Bull. Zool. Nomencl., 59:165–169. [Google Scholar]
- Foissner, W. 2014. An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int. J. Syst. Evol. Microbiol., 64:271–292. [DOI] [PubMed] [Google Scholar]
- Foissner, W. , Agatha, S. & Berger, H. 2002. Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib Desert. Denisia, 5:1–1459. [Google Scholar]
- Foissner, W. , Chao, A. & Katz, L. A. 2008. Diversity and geographic distribution of ciliates (Protista: Ciliophora). Biodivers. Conserv., 17:345–363. [Google Scholar]
- Foissner, W. , Strüder‐Kypke, M. , Van der Staay, G. W. M. , Moon‐van der Staay, S.‐Y. & Hackstein, J. H. P. 2003. Endemic ciliates (Protozoa, Ciliophora) from tank bromeliads (Bromeliaceae): a combined morphological, molecular, and ecological study. Eur. J. Protistol., 39:365–372. [Google Scholar]
- Gachon, C. M. M. , Heesch, S. , Küpper, F. C. , Achilles‐Day, U. E. M. , Brennan, D. , Campbell, C. N. , Clarke, A. , Dorrell, R. G. , Field, J. , Gontarek, S. , Menendez, C. R. , Saxon, R. J. , Veszelovszki, A. , Guiry, M. D. , Gharbi, K. , Blaxter, M. & Day, J. G. 2013. The CCAP KnowledgeBase: linking protistan and cyanobacterial biological resources with taxonomic and molecular data. Syst. Biodivers., 11:407–413. [Google Scholar]
- Gates, M. A. & Berger, J. 1974. A biometric study of three strains of Tetrahymena pyriformis (Ciliatea: Hymenostomatida) . Can. J. Zool., 52:1167–1183. [DOI] [PubMed] [Google Scholar]
- Gates, M. A. & Berger, J. 1976. Morphometric inseparability of Paramecium primaurelia and P. pentaurelia . Trans. Am. Microsc. Soc., 95:507–514. [Google Scholar]
- Gentekaki, E. & Lynn, D. H. 2012. Spatial genetic variation, phylogeography and barcoding of the peritrichous ciliate Carchesium polypinum . Eur. J. Protistol., 48:305–313. [DOI] [PubMed] [Google Scholar]
- Godfray, H. C. J. 2007. Linnaeus in the information age. Nature, 446:259–260. [DOI] [PubMed] [Google Scholar]
- Gray, M. A. , Pratte, Z. A. & Kellogg, C. 2013. Comparison of DNA preservation methods for environmental bacterial community samples. FEMS Microbiol. Ecol., 83:468–477. [DOI] [PubMed] [Google Scholar]
- Guggiari, M. & Peck, R. 2008. The bacterivorous ciliate Cyclidium glaucoma, isolated from a sewage treatment plant: molecular and cytological descriptions for barcoding. Eur. J. Protistol., 44:168–180. [DOI] [PubMed] [Google Scholar]
- Hawksworth, D. L. 2010. Terms used in bionomenclature: The naming of organisms (and plant communities). Global Biodiversity Information Facility, Copenhagen: p. 1–215. [Google Scholar]
- Hide, G. , Hughes, J. M. & McNuff, R. 2003. A rapid and simple method of detection of Blepharisma japonicum using PCR and immobilisation on FTA paper. BMC Ecol., 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges, E. R. S. 2003. The guild handbook of scientific illustration. John Wiley & Sons, Hoboken, NJ: p. 1–656. [Google Scholar]
- Hoef‐Emden, K. & Melkonian, M. 2003. Revision of the genus Cryptomonas (Cryptophyceae): a combination of molecular phylogeny and morphology provides insights into a long‐hidden dimorphism. Protist, 154:371–409. [DOI] [PubMed] [Google Scholar]
- Huss, V. A. R. , Frank, C. , Harmann, E. C. , Hirmer, M. , Kloboucek, A. , Seidel, B. M. , Wenzeler, P. & Kessler, E. 1999. Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta). J. Phycol., 35:587–598. [Google Scholar]
- ICZN = International Commission on Zoological Nomenclature . 1999. International code of zoological nomenclature. Fourth edition adopted by the International Union of Biological Sciences. International Trust for Zoological Nomenclature, London: Tipografia La Garangola, Padova. p. xxix + 306. [Google Scholar]
- ICZN = International Commission on Zoological Nomenclature . 2012. Amendment of Articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bull. Zool. Nomencl., 69:161–169. [Google Scholar]
- Jankowski, A. V. 2007. [Phylum Ciliophora Doflein, 1901. Review of Taxa] In: Alimov A. F. (ed.), Protista: Handbook on Zoology. Part 2. Nauka, St. Petersburg: p. 415–976. (in Russian with English summary). [Google Scholar]
- Katz, L. A. , McManus, G. B. , Snoeyenbos‐West, O. L. O. , Griffin, A. , Pirog, K. , Costas, B. & Foissner, W. 2005. Reframing the ‘Everything is everywhere’ debate: evidence for high gene flow and diversity in ciliate morphospecies. Aquat. Microb. Ecol., 41:55–65. [Google Scholar]
- Kher, C. P. , Doerder, F. P. , Cooper, J. , Ikonomi, P. , Achilles‐Day, U. , Kupper, F. C. & Lynn, D. H. 2011. Barcoding Tetrahymena: discriminating species and identifying unknowns using the cytochrome c oxidase subunit I (cox‐1) barcode. Protist, 162:2–13. [DOI] [PubMed] [Google Scholar]
- Klein, B. M. 1926. Ergebnisse mit einer Silbermethode bei Ciliaten. Arch. Protistenk., 56:243–279. [Google Scholar]
- Lee, M. S. Y. 2004. The molecularisation of taxonomy. Invertebr. Syst., 18:1–6. [Google Scholar]
- Lee, J. J. & Soldo, A. T. 1992. Protocols in protozoology. Society of Protozoologists, Lawrence, KS. [Google Scholar]
- Lipscomb, D. , Platnick, N. & Wheeler, Q. 2003. The intellectual content of taxonomy: a comment on DNA taxonomy. Trends Ecol. Evol., 18:65–66. [Google Scholar]
- Lynn, D. H. 2008. The ciliated protozoa. Characterization, classification and guide to the literature, 3rd ed Springer, Dordrecht: p. xxxiii + 605. [Google Scholar]
- Lynn, D. H. & Simpson, A. G. B. 2009. Describing new taxa of unicellular protists. J. Eukaryot. Microbiol., 56:403–405. [DOI] [PubMed] [Google Scholar]
- Marin, B. , Palm, A. , Klingberg, M. & Melkonian, M. 2003. Phylogeny and taxonomic revision of plastid‐containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist, 154:99–145. [DOI] [PubMed] [Google Scholar]
- McGrath, M. S. , Daggett, P.‐M. & Nerad, T. A. 1977. Studies on the preservation of the ciliate Didinium nasutum . Trans. Am. Microsc. Soc., 96:519–525. [Google Scholar]
- Müller, H. , Achilles‐Day, U. E. M. & Day, J. G. 2008. Cryopreservation of resting cysts of the freshwater ciliate Meseres corlissi by conventional two‐step methods and one‐step vitrification protocols. CryoLetters, 29:329–338. [PubMed] [Google Scholar]
- Müller, H. , Achilles‐Day, U. E. M. & Day, J. G. 2010. Tolerance of the resting cysts of Colpoda inflata (Ciliophora, Colpodea) and Meseres corlissi (Ciliophora, Spirotrichea) to desiccation and freezing. Eur. J. Protistol., 46:133–142. [DOI] [PubMed] [Google Scholar]
- Nanney, D. L. & McCoy, J. W. 1976. Characterization of the species of the Tetrahymena pyriformis complex. Trans. Am. Microsc. Soc., 95:664–682. [PubMed] [Google Scholar]
- Neff, N. A. 1986. A rational basis for a priori character weighting. Syst. Biol., 35:110–123. [Google Scholar]
- Nerad, T. A. 1991. ATCC catalogue of protists. ATCC, Rockville, MD. [Google Scholar]
- Nerad, T. A. & Daggett, P.‐M. 1992. Cryopreservation of non‐encysting ciliates. In: Lee J. J. & Soldo A. T. (ed.), Protocols in Protozoology. Society of Protozoologists, Lawrence, KS: p. A92.1–A92.4. [Google Scholar]
- Nobili, R. , Luporini, P. & Dini, F. 1978. Breeding system, species relationships and evolutionary trends in some marine species of Euplotidae (Ciliata Hypotrichida) In: Battaglia B. & Beardmore J. (ed.), Marine Organisms: Genetics, Ecology and Evolution. Plenum Press, New York, NY: p. 591–616. [Google Scholar]
- Oliveira‐Pinto, F. , Ricci, N. & Nieri, L. 1993. Simulation of creeping movements of ciliated protozoa. J. Theor. Biol., 163:365–372. [Google Scholar]
- Pawlowski, J. , Audic, S. , Adl, S. , Bass, D. , Belbahri, L. , Berney, C. , Bowser, S. S. , Cepicka, I. , Decelle, J. , Dunthorn, M. , Fiore‐Donno, A. M. , Gile, G. H. , Holzmann, M. , Jahn, R. , Jirku, M. , Keeling, P. J. , Kostka, M. , Kudryavtsev, A. , Lara, E. , Lukeš, J. , Mann, D. G. , Mitchell, E. A. D. , Nitsche, F. , Romeralo, M. , Saunders, G. W. , Simpson, A. G. B. , Smirnov, A. V. , Spouge, J. L. , Stern, R. F. , Stoeck, T. , Zimmermann, J. , Schnidel, D. & de Vargas, C. 2012. CBOL Protist Working Group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol., 10:e1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petz, W. , Song, W. & Wilbert, N. 1995. Taxonomy and ecology of the ciliate fauna (Protozoa, Ciliophora) in the endopagial and pelagial of the Weddell Sea, Antarctica. Stapfia, 40:1–23. [Google Scholar]
- Powelson, E. E. , Gates, M. A. & Berger, J. 1975. A biometrical analysis of 22 stocks of four syngens of Paramecium aurelia . Can. J. Zool., 53:19–32. [DOI] [PubMed] [Google Scholar]
- Pröschold, T. , Marin, B. , Schlösser, U. G. & Melkonian, M. 2001. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist, 152:265–300. [DOI] [PubMed] [Google Scholar]
- Pyle, R. L. & Michel, E. 2008. ZooBank: developing a nomenclatural tool for unifying 250 years of biological information. Zootaxa, 1950:39–50. [Google Scholar]
- Quintela‐Alonso, P. , Nitsche, F. , Wylezich, C. , Arndt, H. & Foissner, W. 2013. A new Tetrahymena (Ciliophora, Oligohymenophorea) from groundwater of Cape Town, South Africa. J. Eukaryot. Microbiol., 60:235–246. [DOI] [PubMed] [Google Scholar]
- Ridgway, J. L. 1978. Scientific illustration. Stanford University Press, Redwood City, CA: p. 1–187. [Google Scholar]
- Santoferrara, L. F. , Bachy, C. , Alder, V. A. , Gong, J. , Kim, Y.‐O. , Saccà, A. , da Silva Neto, I. D. , Strüder‐Kypke, M. C. , Warren, A. , Xu, D. , Yi, Z. & Agatha, S. 2016. Updating biodiversity studies in loricate protists: the case of the tintinnids (Alveolata, Ciliophora, Spirotrichea). J. Eukaryot. Microbiol., 63:651–656. [DOI] [PubMed] [Google Scholar]
- Santoferrara, L. F. , McManus, G. B. & Alder, V. A. 2013. Utility of genetic markers and morphology for species discrimination within the order Tintinnida (Ciliophora, Spirotrichea). Protist, 164:24–36. [DOI] [PubMed] [Google Scholar]
- Schlegel, M. & Meisterfeld, R. 2003. The species problem in protozoa revisited. Eur. J. Protistol., 39:349–355. [Google Scholar]
- Seberg, O. , Humphries, C. J. , Knapp, S. , Stevenson, D. W. , Petersen, G. , Scharff, N. & Andersen, N. M. 2003. Shortcuts in systematics? A commentary on DNA‐based taxonomy. Trends Ecol. Evol., 18:63–65. [Google Scholar]
- Seifert, B. 2014. A pragmatic species concept applicable to all eukaryotic organisms independent from their mode of reproduction or evolutionary history. Soil Org., 86:85–93. [Google Scholar]
- Simon, E. M. & Hwang, S. W. 1967. Tetrahymena – effect of freezing and subsequent thawing on breeding performance. Science, 155:694–696. [DOI] [PubMed] [Google Scholar]
- Song, W. 1993. Studies on the cortical morphogenesis during cell division in Halteria grandinella (Müller, 1773) (Ciliophora, Oligotrichida). Chin. J. Oceanol. Limnol., 11:122–129. [Google Scholar]
- Sonneborn, T. M. 1957. Breeding systems, reproductive methods, and species problems in protozoa In: Mayr E. (ed.), The species problem. A symposium presented at the Atlanta meeting of the American Association for the Advancement of Science December 28–29, 1955. American Association for the Advancement of Science, Washington, DC, 50:155–324. [Google Scholar]
- Stoeck, T. , Przyboś, E. & Dunthorn, M. 2014. The D1‐D2 region of the large subunit ribosomal DNA as barcode for ciliates. Mol. Ecol. Resour., 14:458–468. [DOI] [PubMed] [Google Scholar]
- Strüder‐Kypke, M. C. & Lynn, D. H. 2010. Comparative analysis of the mitochondrial cytochrome c oxidase subunit I (COI) gene in ciliates (Alveolata, Ciliophora) and evaluation of its suitability as a biodiversity marker. Syst. Biodivers., 8:131–148. [Google Scholar]
- Sun, P. , Clamp, J. , Xu, D. , Kusuoka, Y. & Miao, W. 2012. Vorticella Linnaeus, 1767 (Ciliophora, Oligohymenophora, Peritrichia) is a grade not a clade: redefinition of Vorticella and the families Vorticellidae and Astylozoidae using molecular characters derived from the gene coding for small subunit ribosomal RNA. Protist, 163:129–142. [DOI] [PubMed] [Google Scholar]
- Tompkins, J. , De Ville, M. M. , Day, J. G. & Turner, M. F. 1995. Culture Collection of Algae and Protozoa catalogue of strains. CCAP, Ambleside. [Google Scholar]
- Vďačný, P. & Foissner, W. 2012. Monograph of the Dileptids (Protista, Ciliophora, Rhynchostomatia). Denisia, 31:1–529. [Google Scholar]
- Will, K. W. & Rubinoff, D. 2004. Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics, 20:47–55. [DOI] [PubMed] [Google Scholar]
- Winston, J. E. 1999. Describing species. Practical taxonomic procedure for biologists. Columbia University Press, New York, NY: p. xx + 518. [Google Scholar]
- Wood, P. & McDonnell, P. 1994. Scientific illustration: A guide to biological, zoological, and medical rendering techniques, design, printing, and display. John Wiley & Sons, Hoboken, NJ: p. 1–176. [Google Scholar]
- Xu, K. & Foissner, W. 2005. Descriptions of Protospathidium serpens (Kahl, 1930) and P. fraterculum n. sp. (Ciliophora, Haptoria), two species based on different resting cyst morphology. J. Eukaryot. Microbiol., 52:298–309. [DOI] [PubMed] [Google Scholar]
- Zhan, Z. , Xu, K. , Warren, A. & Gong, Y. 2009. Reconsideration of phylogenetic relationships of the subclass Peritrichia (Ciliophora, Oligohymenophorea) based on small subunit ribosomal RNA gene sequences, with the establishment of a new subclass Mobilia Kahl, 1933. J. Eukaryot. Microbiol., 56:552–558. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Gentekaki, E. , Yi, Z. & Lin, X. 2013. Genetic differentiation of the mitochondrial cytochrome oxidase c subunit I gene in genus Paramecium (Protista, Ciliophora). PLoS One, 8:e77044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel, F. 1988. A handbook of biological illustration, 2nd ed University of Chicago Press, Chicago, IL: p. 1–152. [Google Scholar]


