Abstract
Colorectal cancer (CRC) is the second most common cause of cancer death in the western world. An effective screening program leading to early detection of disease would severely reduce the mortality of CRC. Alterations in the gut microbiota have been linked to CRC, but the potential of microbial markers for use in CRC screening has been largely unstudied. We used a nested case–control study of 238 study subjects to explore the use of microbial markers for clbA+ bacteria harboring the pks pathogenicity island, afa‐C+ diffusely adherent Escherichia coli harboring the afa‐1 operon, and Fusobacterium nucleatum in stool as potential screening markers for CRC. We found that individual markers for clbA+ bacteria and F. nucleatum were more abundant in stool of patients with CRC, and could predict cancer with a relatively high specificity (81.5% and 76.9%, respectively) and with a sensitivity of 56.4% and 69.2%, respectively. In a combined test of clbA+ bacteria and F. nucleatum, CRC was detected with a specificity of 63.1% and a sensitivity of 84.6%. Our findings support a potential value of microbial factors in stool as putative noninvasive biomarkers for CRC detection. We propose that microbial markers may represent an important future screening strategy for CRC, selecting patients with a “high‐risk” microbial pattern to other further diagnostic procedures such as colonoscopy.
Keywords: gut microbiota, clbA, F. nucleatum, stool, screening, colorectal cancer
Short abstract
What's new?
Nobody looks forward to a colonoscopy, and now a pair of telltale bacteria could help people avoid them. Researchers know that microbial changes occur in colorectal cancer, and have hoped these microbial changes could provide less invasive screening tools to detect tumors. These authors conducted a nested case–control study investigating 3 bacterial markers in 238 patients. Two of the markers, clbA+ bacteria and Fusobacterium nucleatum, successfully predicted colorectal cancer with high sensitivity, particularly when tested together.
Abbreviations
- CRC
colorectal cancer
- E. coli
Escherichia coli
- DAEC
diffusely adherent E. coli
- F. nucleatum
Fusobacterium nucleatum
- FECSU
Faecal and Endoscopic Colorectal Study in Umeå
- CAMA
cancer‐associated microbial alterations
Colorectal cancer (CRC) is the second most diagnosed cancer in women, and the third in men worldwide.1, 2 The mortality of patients with metastatic disease is high, indicating the necessity of a good and reliable screening method to detect the tumor at an early operable stage. Colonoscopy is currently the most reliable method for detection of early staged CRC, but it is uncomfortable for the patients, time consuming and costly. Recent studies have shown that changes in the intestinal microbiota are associated with CRC.3, 4, 5, 6 A noninvasive screening method, analyzing cancer‐associated microbial alterations in stool, may have many benefits for both the health care system and the participating patients.
The gut microbiota plays many important roles in digestion, but has at the same time been implicated in diseases of the host. In the last years, accumulating evidence suggests that interactions between the mucosa and the microbiota are important, in both immunology and tumorigenesis (reviewed in Ref. 7). A number of studies have also provided mechanistic evidence that specific bacterial populations can change the milieu in the mucosa, promoting a proinflammatory response, and inducing double‐stranded DNA breakage and mutations that can lead to tumor initiation and progression.8, 9, 10, 11, 12, 13, 14, 15, 16, 17
Bacteria positive for clbA harbors the pks pathogenicity island and produces colibactin, a genotoxin capable of inducing double‐stranded DNA breaks and cellular senescence, leading to increased production of growth factors that can stimulate tumor growth.18, 19 Colibactin‐producing Escherichia coli (E. coli) and afa‐C+ diffusely adherent E. coli (DAEC) have both been linked to human CRC.20, 21, 22 Escherichia coli carrying pks have been found in 56–67% of human colorectal tumors compared to around 20% in controls.20, 22 DAEC carrying the afimbrial adhesin (afa‐1) operon were shown by Prorok‐Hamon et al. to be more common among E. coli strains isolated from CRC patients compared to controls.21 Compared to 17–36% of controls, 67–80% of colorectal tumors are found to be positive for afaC.21, 22 The afa‐1+ DAEC have the ability to adhere to, and invade epithelial cells and likely play a role in epithelial‐to‐mesenchymal transition.16
Fusobacterium nucleatum (F. nucleatum) is part of the commensal flora of the gut and oral cavity, but has been linked to a number of pathological conditions, including periodontitis, appendicitis, inflammatory bowel disease (IBD) and CRC.23, 24, 25, 26, 27, 28 F. nucleatum has been found in higher levels in CRC, and adenomas, compared to adjacent normal tissue.10, 23, 24, 25 It is a highly adhesive bacterial species and has the ability to invade colonic epithelial cells.13, 29 Additionally, recent studies showed that, in human CRC, high amounts of F. nucleatum in tumor tissue is correlated to low infiltration of T lymphocytes and poor patient prognosis.30, 31
In this study, we investigated the utility of microbial markers for clbA+ bacteria (clbA) and, afa‐1+ DAEC (afaC), and F. nucleatum, in CRC detection, using 238 human stool samples from the FECSU (the Faecal and Endoscopic Colorectal Study in Umeå) cohort.
Material and Methods
Study cohort
The study is based on the Faecal and Endoscopic Colorectal Study in Umeå (FECSU) cohort of 1136 patients who went through colonoscopy at the University Hospital in Umeå, Sweden, between the years 2008–2013 (September 2008 to March 2013). Indications for colonoscopy were gastrointestinal symptoms of large bowel disease, visible bleeding and/or positive fecal hemoglobin (F‐Hb) test. A total of 2660 patients were scheduled for colonoscopy during the time period (see flow chart in Fig. 1). Independent of underlying indications, 1997 patients were invited to participate in the study. Exclusion criteria were planned colonoscopy within 1 week, dementia and low‐performance status, including mentally or physically disabled persons. Of the invited patients, 861 patients denied participation.
Figure 1.
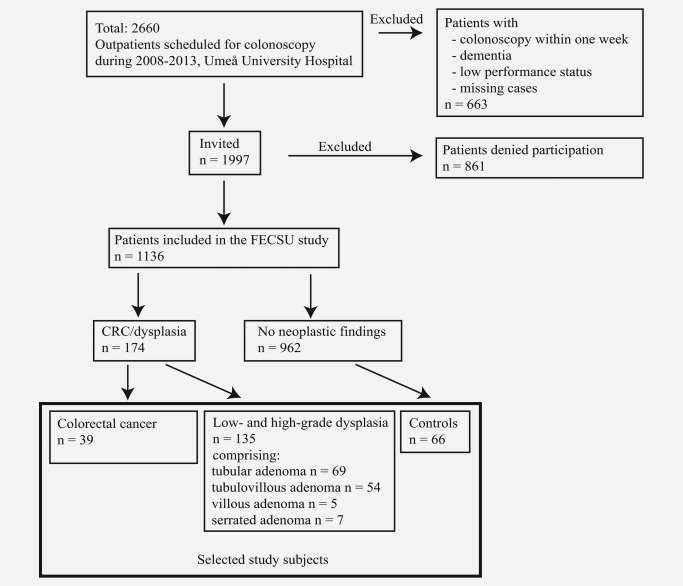
Flow chart describing the FECSU cohort and the selection of study subjects.
Patients were asked to leave stool samples before the precolonoscopy cleansing procedure started. Study information and tubes for stool sample collection were sent to the patients together with the invitation for the clinical colonoscopy examination. The colonoscopy was routinely performed at the endoscopy unit and the clinical findings were recorded. Biopsies were taken when clinically relevant. Lesions/findings were recorded by a pathologist, and the neoplastic lesions were further classified as low‐grade dysplasia, high‐grade dysplasia or adenocarcinoma according to the WHO classification of tumors of the digestive system.32 If several lesions were present, the most severe lesion was recorded for the patient.
The study protocol was approved by the Regional Ethical Review Board in Umeå, Sweden (Dnr 08–184 M; Dnr 07–045 M). All individuals in this study have signed a written consent form.
Selection of study subjects
Cases with CRC or dysplasia were identified by reading patient records including the pathology reports. Patients selected for the study included all the 39 identified cases of CRC, all the 135 cases of low‐ and high‐grade dysplasia and 66 controls. All study subjects included were adults with the youngest 34 years of age. As the number of cases with high‐grade dysplasia was low (only ten cases), both low‐ and high‐grade dysplasia were included in one group of dysplasia. Controls were selected from the group of FECSU patients where a biopsy was taken, but recorded with no neoplastic findings. Patients with IBD or findings of hyperplastic polyps were excluded from the controls. The controls were matched by age and gender to the CRC cases and a randomized subset of the dysplasia group.
Stool/tissue collection and storage
With the envelope of invitation for colonoscopy examination that were sent to the patients, three tubes for stool sample collection were enclosed, along with information about the collection procedure; one tube containing 5 ml of preservative buffer, RNAlater ® (Ambion®), one tube for fecal hemoglobin (F‐Hb) analysis and a third tube for F‐calprotectin (data not presented). Stool samples were collected by the participant prior to the preparation for the colonoscopy procedure. The samples were stored at room temperature for a maximum of 7 days before being processed at the lab facility. RNAlater ® is bacteriostatic (the bacteria remain intact but do not grow). The adequacy of using RNAlater ® as a preservative was validated by comparison of DNA yield and quality of samples stored in RNAlater ® in room temperature for 5 days, to that of immediately frozen samples (Supporting Information, Table S1). The samples containing RNAlater ® were centrifuged for 20 min at 2000 rpm, and excess fluid was removed before storage at −80°C. F‐Hb was analyzed manually using a fecal immunochemical test (FIT), Analyz F.O.B Test (ANL products AB), according to manufacturer's instructions.
DNA extraction
Stool DNA (sDNA) was prepared from approximately 0.2 g stool using QIAamp® DNA Stool Mini Kit (Qiagen) according to the manufacturer's instructions. Double stranded DNA‐recovery was measured using Qubit® dsDNA BR Assay (Molecular probes) with the Qubit® 2.0 Fluorometer.
Detection of microbial markers in stool by real‐time qPCR
Quantitative PCR assays targeting the clbA gene and the afaC gene of the afa‐1 operon were used to detect pks+ bacteria and Afa‐1 adhesin‐expressing DAEC, respectively. Escherichia coli Nissle 1917 was used as a positive control for pks. Real‐time qPCR reactions were run in duplicates using the SYBR Green PCR kit on the 7900HT Fast Real‐Time PCR System (Applied Biosystems). The following cycling conditions were used: for clbA, 5 min at 95°C, 40 cycles of 15 sec at 95°C, 30 sec at 50°C and 1 min at 72°C; for afaC, 10 min at 95°C, and 40 cycles of 15 sec at 95°C, and 15 sec at 65°C; for the 16S rRNA gene, 5 min at 95°C, 40 cycles of 10 sec at 95°C, and 30 sec at 60°C. The quantification cycle (C q) was calculated using the SDS 2.4 software (Applied Biosystems). The performance of the PCR assays was checked by analyses of replicates, serial dilutions, melting curves and separation on agarose gels. Samples not amplified with the appropriate amplicon length and not within 38 cycles were considered negative for the microbial factors.
F. nucleatum was assessed by real‐time qPCR using the Microbial DNA qPCR Assay (Qiagen) containing a FAM‐labeled probe specific for F. nucleatum 16S rRNA gene (GeneBank Acc. FJ471654.1) according to manufacturer's instructions. F. nucleatum subsp. nucleatum Knorr (ATCC 25586 D‐5) was used as positive control. The reactions were run on the Applied Biosystems 7900HT Fast Real‐Time PCR System using the following cycling conditions; 95°C for 10 min, 40 cycles of 15 sec at 95°C, and 2 min at 60°C. Samples amplified within 38 cycles were considered to have template concentration positive for the specific sequence. The level of F. nucleatum in each sample was given as a relative quantification with the total microbial 16S rRNA gene DNA in each sample as reference. The relative levels of F. nucleatum were calculated using 2−Δ C q, giving ΔC q = − C q 16 S rRNA gene.
Two samples were excluded from qPCR analyses due to technical difficulties. The following primer sequences were used in this study: clbA , forward 5′‐ATGAGGATTGATATATTAATTGGACA‐3′ 33 and reverse 5′‐GGTTTGCCATATTTGCACGTAC‐3′ product size 233 bp, afaC ; forward 5′‐CGGCTTTTCTGCTGAACTGGCAGGC‐3′34 and reverse 5′‐CCGCTCAGCACGTATGTATGAACTC‐3′ product size 200 bp; 16S rRNA gene forward 5′‐CCATGAAGTCGGAATCGCTAG‐3′ and reverse 5′‐GCTTGACGGGCGGTGT‐3′ (16S rRNA Gene Universal Bacteria Control Primers from the NEBNext® Microbiome DNA Enrichment Kit (Biolabs)).
Statistical methods
Statistical analyses were performed using the IBM SPSS Statistics 23 (SPSS Inc.). χ 2 tests were used to compare categorical variables, unless expected frequencies were <5 when Fisher's exact test was used. The nonparametric Kruskal–Wallis H or Mann–Whitney U test was used to compare differences in continuous variables between groups. p values <0.05 were considered statistically significant. Area under the receiver operating characteristic (ROC) curve was calculated using the variable for F. nucleatum and cancer diagnosis, and the Youden's index was used to identify the cutoff for F. nucleatum levels that gave the most sensitive and specific assay to detect cancer. This cutoff was used in the F. nucleatum assay to identify stool samples as positive (with high levels of F. nucleatum) or negative (with low levels of F. nucleatum). In the CAMA test combining microbial markers or the test combining CAMA with F‐Hb, a negative test result was given to stool samples negative for both markers and a positive test result given to stool samples with one or both markers positive. Sensitivity was defined as the percent of CRC or dysplasia cases with a positive test result. Specificity was defined as the percent of nested controls with a negative test result.
Results
Patient characteristics
The overall study cohort included 1136 patients invited to colonoscopy after presenting with symptoms from the large bowel. A flow chart describing the collection of the study cohort with inclusion/exclusion criteria can be found in Fig. 1. Among these, 39 patients were diagnosed with CRC and 135 patients with different degrees of dysplasia, and were selected for further studies. Also included in the study were 66 matched controls, who underwent colonoscopy but with no pathological findings. The clinical characteristics of the included study subjects are presented in Table 1. Most of the cancers were found in stage II (53.8%) and in left colon (43.6%).
Table 1.
Clinical characteristics of study patientsa
| Total | Control | Dysplasia | Cancer | |
|---|---|---|---|---|
| n = 238 | n = 65 | n = 134 | n = 39 | |
| Age (%) | ||||
| 34–59 | 33 (13.9) | 6 (9.2) | 23 (17.2) | 4 (10.3) |
| 60–69 | 94 (39.5) | 22 (33.8) | 60 (44.8) | 12 (30.8) |
| 70–80 | 85 (35.7) | 26 (40.0) | 41 (30.6) | 18 (46.2) |
| >80 | 26 (10.9) | 11 (16.9) | 10 (7.5) | 5 (12.8) |
| Gender (%) | ||||
| Female | 103 (43.3) | 30 (46.2) | 54 (40.3) | 19 (48.7) |
| Male | 135 (56.7) | 35 (53.8) | 80 (59.7) | 20 (51.3) |
| Location (%) | n = 173 | |||
| Right colon | 49 (28.3) | n.a. | 37 (27.6) | 12 (30.8) |
| Left colon | 76 (43.9) | n.a. | 59 (44.0) | 17 (43.6) |
| Rectum | 48 (27.7) | n.a. | 38 (28.4) | 10 (25.6) |
| Stage (%) | ||||
| I | n.a. | n.a. | 2 (5.1) | |
| II | n.a. | n.a. | 21 (53.8) | |
| III | n.a. | n.a. | 8 (20.5) | |
| IV | n.a. | n.a. | 7 (17.9) |
Shown are patients with complete data sets for microbial markers.
Abbreviation: n.a., not applicable.
Bacteria positive for clbA are more abundant in stool of patients diagnosed with CRC
A qPCR‐assay targeting the clbA gene was used to detect colibactin‐producing bacteria in DNA from stool of study patients. clbA was more often detected in stool samples of CRC patients compared to patients with dysplasia (p = 0.004) or controls (p < 0.001) (Fig. 2 a). The difference in clbA detection frequency between patients with dysplasia and controls was of borderline significance (p = 0.055), but the stepwise increased frequency from controls, to dysplasia to cancers indicate that clbA may represent a useful marker for early changes leading to CRC. With a specificity of 81.5%, the clbA assay detected 56.4% of CRCs and 31.3% of dysplasias (Fig. 2 a and Table 2). The clinical characteristics of cancer patients in relation to clbA can be found in Supporting Information, Table S2.
Figure 2.
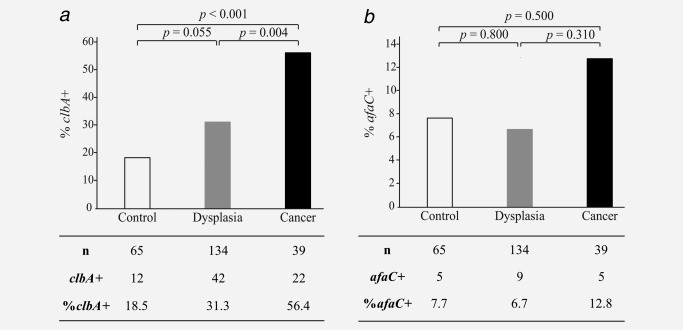
Bacteria carrying clbA are abundant in stool of CRC patients. Differences in absolute number (n) and percentage (%) of (a) clbA‐ and (b) afaC‐positive stool samples between controls, and patients diagnosed with dysplasia or cancer are illustrated.
Table 2.
Microbial alterations in stool of patients diagnosed with dysplasia or cancer
| Total | Control | Dysplasia | Cancer | p value | |
|---|---|---|---|---|---|
| clbA (%) | n = 238 | n = 65 | n = 134 | n = 39 | <0.001 |
| Negative | 162 (68.1) | 53 (81.5) | 92 (68.7) | 17 (43.6) | |
| Positive | 76 (31.9) | 12 (18.5) | 42 (31.3) | 22 (56.4) | |
| afaC (%) | 0.46a | ||||
| Negative | 219 (92.0) | 60 (92.3) | 125 (93.3) | 34 (87.2) | |
| Positive | 19 (8.0) | 5 (7.7) | 9 (6.7) | 5 (12.8) | |
| F. nucleatum (%) | <0.001 | ||||
| Low | 169 (71.0) | 50 (76.9) | 107 (79.8) | 12 (30.8) | |
| High | 69 (29.0) | 15 (24.3) | 27 (20.1) | 27 (69.2) | |
| F‐Hb (%) | n = 178 | n = 41 | n = 108 | n = 29 | <0.001 |
| Negative | 129 (72.5) | 37 (90.2) | 82 (75.9) | 10 (34.5) | |
| Positive | 49 (27.5) | 4 (9.8) | 26 (24.1) | 19 (65.5) |
Unless otherwise indicated, χ 2 test was used for categorical variables.
Fisher's exact test.
Abbreviation: F‐Hb, immunochemical fecal hemoglobin test.
DAEC carrying afa‐1 were detected in DNA from stool using a qPCR assay targeting the afaC gene in the afa‐1 operon. Very few of the stool samples were positive for afaC (Fig. 2 b). The stool samples originating from individuals diagnosed with CRC were slightly more frequently positive for afaC than samples from dysplasias or controls, but this was not statistically significant. With a specificity of 92.3%, the afaC assay detected only 12.8% of the cancers (Table 2). Notably, of the 5 afaC+ cancers, 4 were also clbA+, a pattern that was not found among the dysplasia or control groups (data not shown).
F. nucleatum is enriched in stool of patients with CRC
Quantitative PCR was applied to detect F. nucleatum in DNA from stool of study patients. F. nucleatum was found in stool of all patients, however, at varying levels (Fig. 3 a). Patients diagnosed with CRC displayed significantly higher levels of F. nucleatum in stool compared to patients with dysplasia (p < 0.001) and controls (p < 0.001). No difference was found in F. nucleatum DNA levels between patients with dysplasia and controls. The clinical characteristics of cancer patients in relation to F. nucleatum can be found in Supporting Information, Table S2.
Figure 3.
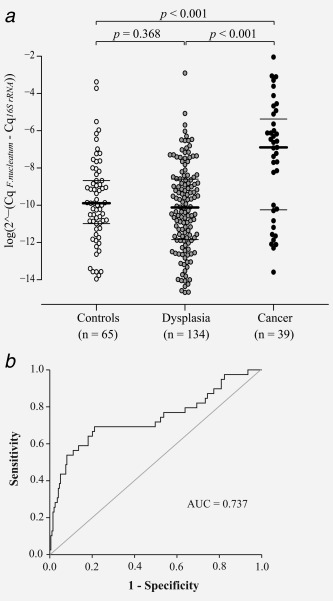
Increased levels of F. nucleatum are detected in stool of CRC patients. (a) A Beeswarm Boxplot is used to illustrate the relative levels of F. nucleatum in stool of control patients, and patients diagnosed with dysplasia or cancer. Horisontal lines indicate median (in bold) and quartiles. (b) An ROC curve displaying the specificity and the sensitivity for the F. nucleatum assay. The ROC curve was calculated using the variable for F. nucleatum and cancer/no cancer. The level of F. nucleatum in each sample is given as a relative quantification with the total microbial 16S rRNA gene DNA in each sample as reference 2∧ (−ΔCq), ΔCq = CqF. nucleatum − Cq16S rRNA gene).
The area under the ROC curve was 0.737 for detection of CRC (Fig. 3 b). A cutoff (0.00026) was selected that gave the most reliable analysis for detecting cancer in the study patients. With a specificity of 76.9%, the F. nucleatum assay detected 69.2% of CRCs and 20.1% of dysplasias (Table 2). At the selected cutoff, the F. nucleatum assay detected CRC with a higher sensitivity (69.2%) than clbA (56.4%) and the immunochemical F‐Hb test currently used in the clinic (65.5%). However, the F‐Hb test was more specific (90.2%) than F. nucleatum (76.9%), detecting <10% false positives (Table 2).
A test combining markers of microbial alterations in stool predicts CRC
To improve the CRC detection assay, the two microbial markers detecting clbA and F. nucleatum were combined in a single test here termed the cancer‐associated microbial alterations (CAMA) test, where one or more positive markers predicts CRC. The afaC assay was excluded from the test since afaC was detected in very few cases and did not significantly differ between controls and cancer. At a specificity of 63.1%, the CAMA test detected CRC with a sensitivity of 84.6% (Table 3). Combining the CAMA test with the immunological F‐Hb test, slightly increased sensitivity (89.7%) for detection of CRC, but at the same time specificity (61.0%) was slightly decreased.
Table 3.
A test combining microbial markers and F‐Hb in CRC screening
| Total | Control | Dysplasia | Cancer | p value | |
|---|---|---|---|---|---|
| CAMA (%) | n = 238 | n = 65 | n = 134 | n = 39 | <0.001 |
| Negative | 118 (49.6) | 41 (63.1) | 71 (53.0) | 6 (15.4) | |
| Positive | 120 (50.4) | 24 (36.9) | 63 (47.0) | 33 (84.6) | |
| CAMA/F‐Hb (%) | n = 178 | n = 41 | n = 108 | n = 29 | <0.001a |
| Negative | 70 (39.3) | 25 (61.0) | 42 (38.9) | 3 (10.3) | |
| Positive | 108 (60.7) | 16 (39.0) | 66 (61.1) | 26 (89.7) |
A positive score was given to stool samples positive for one or both of clbA and F. nucleatum. Unless otherwise indicated, χ 2 test was used for categorical variables.
2Fisher's exact test.
Abbreviation: CAMA, cancer‐associated microbial alterations.
DISCUSSION
In this nested case–control study, we explored the utility of using fecal microbial markers of microbial alterations in CRC detection. We found that individual markers for clbA+ bacteria and F. nucleatum were more highly abundant in stool of patients with CRC compared to controls, and could predict cancer with a sensitivity of 56.4% and 69.2%, respectively. The specificity of both assays was close to 80%. Combining the two markers into the CAMA test increased sensitivity to 84.6%, but with the drawback of reduced specificity. When combining CAMA and the immunochemical F‐Hb test, the sensitivity was even higher 89.7%, but the specificity was slightly reduced. Our findings support the potential role of microbial factors in stool as putative noninvasive biomarkers for CRC detection.
We chose a nested case–control model, as it is generally more efficient than a case–control design with the same number of selected controls. However, one limitation of this study is that it is not randomized, as all patients selected for colonoscopy presented with symptoms from the large bowel. The controls, even though recorded without disease, may therefore not have represented a true healthy population. Another limitation is the lack of a validation cohort. The CAMA test was able to detect cancer with a high sensitivity, suggesting it to be a good test to detect cancer. In this study it was found to be more sensitive than the F‐Hb test currently used in clinic. The relatively low specificity of the test would however result in around 35–40% of tested patients being diagnosed with a “high‐risk” flora but without showing signs of dysplasia or neoplasia. This may cause psychological burden for the screening participants and will require continuous follow up by further tests and colonoscopy. It will be with great interest that we follow up on the currently healthy patients diagnosed with a “high risk” microbial pattern, to find out whether or not these patients later on will develop disease. Further studies and verifications in additional cohorts are required to understand the full potential of microbial markers in CRC progression and screening.
Advantages of the microbial marker test is that it represents a cost‐efficient, straight forward, and noninvasive procedure. It is likely that a higher number of patients would agree to a screening procedure leaving stool samples compared to screening by colonoscopy. The stool samples were collected prior to the precolonoscopy cleansing procedure and stored in a chemical stabilizer with bacteriostatic activity, which according to our evaluation preserves microbial DNA quality. The test might, however, be sensitive to antibiotic treatment and stool sampling, as it is a single test, randomly sampled by the patient from a small amount of stool. Data on ongoing antibiotic treatment has been collected for all patients in the FECSU cohort, but no patients included in this study were registered with ongoing antibiotic treatment. However, we cannot exclude long‐term changes in the composition of the microbiota associated with previous antibiotic treatment.
F‐Hb tests are currently the most commonly used noninvasive screening method in CRC and are generally found to be more specific than our CAMA test.35 A major advantage of microbial markers in relation to F‐Hb tests is that they could detect also nonbleeding lesions. Microbial markers such as for F. nucleatum have also been suggested to be useful for detecting serrated polyps, not fully efficiently detected by F‐Hb tests.36, 37 F. nucleatum levels in stool have previously been assessed as a noninvasive biomarker in CRC. In a study by Kostic et al., a significant stepwise increase in F. nucleatum levels was found from controls to adenomas to carcinomas, suggesting the potential of F. nucleatum as a marker for detection of early changes.10 In another study by Flanagan et al., F. nucleatum was found, like in our study, to be more abundant in stool of carcinomas but with no significant difference between adenomas (or dysplasias) and controls.38 In our study, clbA turned out to be the most promising marker for early detection, as the prevalence increased already with dysplastic lesions. Further studies of microbial factors as early detection markers are required to elucidate these differences.
As recently described in the bacterial driver‐passenger model, some bacteria are likely procarcinogenic and involved in CRC development while others defined as passengers may be involved in later stages of tumor progression.39 Colibactin‐producing E. coli can increase the mutation rate of infected cells,9 and could therefore be an example of a bacterial driver. On the other hand, a change in the microbiota could instead be a consequence of epithelial changes following CRC progression. F. nucleatum has been shown to bind to epithelial cells through a Fap2/Gal‐GalNAc interaction, where Gal‐GalNAc is overexpressed in CRC cells compared to non‐neoplastic epithelial cells.40 These findings suggest that F. nucleatum may be a bacterial passenger. Further studies, including the evaluation of bacterial markers in tumor tissue, are needed to elucidate the roles of microbial shifts in CRC development and progression.
Models of microbial alterations combining different “high‐risk” bacteria may improve the specificity of diagnostic tests for CRC. A few metagenomics studies have addressed variations of the microbiome in stool of patients with colorectal adenomas or carcinomas for potential use as noninvasive screening markers for CRC.41, 42, 43, 44 In a study by Yu et al., compositional differences for several bacterial species were identified in patients with CRC compared to controls. Using qPCR, the specific markers buturyl‐CoA dehydrogenase (F. nucleatum) and rpoB (Parvimonas micra) were further found to be highly enriched in stool of patients with early stages of CRC.42 In a study by Zackular et al., they identified a panel of microbial markers that was differentially expressed in controls, adenomas and carcinomas. These changes in the gut microbiome could significantly complement the ability of clinical characteristics, and the gFOBT test to identify the different patient diagnoses.44 Furthermore, Wong et al. very recently showed that quantitation of fecal F. nucleatum improved and had a complementary value added to the FIT test.45 Therefore, combining different microbial markers of “high‐risk” flora with F‐Hb tests, clinical characteristics and tumor‐specific DNA, RNA or protein biomarkers in stool may be a putative screening strategy in the future to more accurately identify patients in early stages of disease progression.
In conclusion, we suggest that analyses of markers of microbial alterations in stool may be putative noninvasive diagnostic markers for CRC. We suggest that detection of a “high‐risk” microbial pattern in stool may identify patients with increased risk of developing CRC. Future studies combining different microbial markers and as well as other biomarkers in a true population‐based setup may lead to important advances in CRC screening. Further studies are also needed to address the role of the microbiota in cause or consequence of tumor progression. These studies may lead to important understandings of the role of the microflora in progression of CRC and the identification of important microbial markers for detection of early disease.
CONTRIBUTORS
Study concept and design: VE, MLW, JR, OA, PK, RP; acquisition of data: ALB, VE, CZ; data analyses: ALB, VE, SE, PL; drafting of the manuscript: ALB, VE, CZ, MLW, SE, RP. Critical revision of the manuscript for important intellectual content: PL, PK, OA, JR. All authors approved the final version of the manuscript.
Supporting information
Supporting Information Table 1.
Supporting Information Table 2.
Acknowledgements
The authors are grateful to all the patients who participated in the study. They are very thankful to the staff of the Endoscopy unit, Umea University Hospital, Umea, Sweden, for invaluable assistance. They thank Kerstin Näslund for technical assistance and Robin Myte for help with illustrations. The study sponsors had no role in study design, data collection, analysis and interpretation of the data.
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 3. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sobhani I, Tap J, Roudot‐Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 2011;6:e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 2012;6:320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One 2011;6:e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gagniere J, Raisch J, Veziant J, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol 2016;22:501–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation‐driven potential of carcinogenesis via IL‐1, COX‐2, and IL‐8. Mol Cancer 2010;9:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cuevas‐Ramos G, Petit CR, Marcq I, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA 2010;107:11537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe 2013;14:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nesic D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature 2004;429:429–33. [DOI] [PubMed] [Google Scholar]
- 12. Raisch J, Rolhion N, Dubois A, et al. Intracellular colon cancer‐associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX‐2 expression. Lab Invest 2015;95:296–307. [DOI] [PubMed] [Google Scholar]
- 13. Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E‐cadherin/beta‐catenin signaling via its FadA adhesin. Cell Host Microbe 2013;14:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shenker BJ, Ojcius DM, Walker LP, et al. Aggregatibacter actinomycetemcomitans cytolethal distending toxin activates the NLRP3 inflammasome in human macrophages, leading to the release of proinflammatory cytokines. Infect Immun 2015;83:1487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 2009;15:1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cane G, Ginouves A, Marchetti S, et al. HIF‐1alpha mediates theGalNAc induction of IL‐8 and VEGF expression on infection with Afa/Dr diffusely adhering E. coli and promotes EMT‐like behaviour. Cell Microbiol 2010;12:640–53. [DOI] [PubMed] [Google Scholar]
- 17. Graillot V, Dormoy I, Dupuy J, et al. Genotoxicity of cytolethal distending toxin (CDT) on isogenic human colorectal cell lines: potential promoting effects for colorectal carcinogenesis. Front Cell Infect Microbiol 2016;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nougayrede JP, Homburg S, Taieb F, et al. Escherichia coli induces DNA double‐strand breaks in eukaryotic cells. Science 2006;313:848–51. [DOI] [PubMed] [Google Scholar]
- 19. Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence‐associated secretory phenotype. Gut 2014;63:1932–42. [DOI] [PubMed] [Google Scholar]
- 20. Arthur JC, Perez‐Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer‐inducing activity of the microbiota. Science 2012;338:120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prorok‐Hamon M, Friswell MK, Alswied A, et al. Colonic mucosa‐associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Viljoen KS, Dakshinamurthy A, Goldberg P, et al. Quantitative profiling of colorectal cancer‐associated bacteria reveals associations between Fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One 2015;10:e0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012;22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCoy AN, Araujo‐Perez F, Azcarate‐Peril A, et al. Fusobacterium is associated with colorectal adenomas. PLoS One 2013;8:e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Swidsinski A, Dorffel Y, Loening‐Baucke V, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 2011;60:34–40. [DOI] [PubMed] [Google Scholar]
- 27. Signat B, Roques C, Poulet P, et al. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 2011;13:25–36. [PubMed] [Google Scholar]
- 28. Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa‐derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 2011;17:1971–8. [DOI] [PubMed] [Google Scholar]
- 29. Manson McGuire A, Cochrane K, Griggs AD, et al. Evolution of invasion in a diverse set of Fusobacterium species. mBio 2014;5:e01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol 2015;1:653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. IARC . WHO classification of tumours of the digestive system, 4th edn. Lyon, France: International Agency for Research on Cancer (IARC), 2010. [Google Scholar]
- 33. Putze J, Hennequin C, Nougayrede JP, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun 2009;77:4696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bilge SS, Clausen CR, Lau W, et al. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea‐associated Escherichia coli to HEp‐2 cells. J Bacteriol 1989;171:4281–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JK, Liles EG, Bent S, et al. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta‐analysis. Ann Intern Med 2014;160:171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer 2015;137:1258–68. [DOI] [PubMed] [Google Scholar]
- 37. van Doorn SC, Stegeman I, Stroobants AK, et al. Fecal immunochemical testing results and characteristics of colonic lesions. Endoscopy 2015;47:1011–7. [DOI] [PubMed] [Google Scholar]
- 38. Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014;33:1381–90. [DOI] [PubMed] [Google Scholar]
- 39. Tjalsma H, Boleij A, Marchesi JR, et al. A bacterial driver‐passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Micro 2012;10:575–82. [DOI] [PubMed] [Google Scholar]
- 40. Abed J, Emgard JE, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor‐expressed Gal‐GalNAc. Cell Host Microbe 2016;20:215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goedert JJ, Gong Y, Hua X, et al. Fecal microbiota characteristics of patients with colorectal adenoma detected by screening: a population‐based study. EBioMedicine 2015;2:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non‐invasive biomarkers for colorectal cancer. Gut 2015. [DOI] [PubMed] [Google Scholar]
- 43. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early‐stage detection of colorectal cancer. Mol Syst Biol 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zackular JP, Rogers MA, Ruffin MTt, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014;7:1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wong SH, Kwong TN, Chow TC, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table 1.
Supporting Information Table 2.


