Summary
A number of long noncoding RNAs (lncRNAs) have been reported to regulate transcription via recruitment of chromatin-modifiers or bridging distal enhancer elements to gene promoters. However, the generality of these modes of regulation and the mechanisms of chromatin attachment for thousands of unstudied human lncRNAs remain unclear. To address these questions, we performed stringent nuclear fractionation coupled to RNA-seq. We provide genome-wide identification of human chromatin-associated lncRNAs, and demonstrate tethering of RNA to chromatin by RNAPII is a pervasive mechanism of attachment. We also uncovered thousands of chromatin-enriched RNAs (cheRNAs) that share molecular properties with known lncRNAs. Although distinct from noncoding RNAs derived from active enhancers, the production of cheRNAs is strongly correlated with the expression of neighboring protein-coding genes. This work provides an updated framework for nuclear RNA organization that includes a large chromatin-associated transcript population with enhancer-like properties, and may prove useful in de novo enhancer annotation.
Introduction
The advent of RNA-sequencing has revealed thousands of long noncoding RNA (lncRNA) species in the human genome (Cabili et al., 2011; Derrien et al., 2012; Ilott and Ponting, 2013), presenting the challenge of distinguishing functional lncRNAs from transcriptional noise. Several of these lncRNAs have been implicated in development and disease (Chalei et al., 2014; Dinger et al., 2008; Huarte et al., 2010; Sauvageau et al., 2013; Wang et al., 2011b), and the broader tissue-specific expression and conservation of the thousands of lncRNAs that have been discovered thus far suggest such functions may indeed be more general (Iyer et al., 2015; Mercer and Mattick, 2013; Necsulea et al., 2014; Ponting et al., 2009). From a small number of well-studied cases, it appears that lncRNAs may regulate local chromatin states, either by acting as intermediaries to recruit chromatin modulators (Chalei et al., 2014; Lai et al., 2013; Nagano et al., 2008; Rinn and Chang, 2012; Rinn et al., 2007) or by potentiating contacts between genes and distal enhancer-elements to promote transcriptional activation (Lai et al., 2013; Li et al., 2013; Yang et al., 2013). Yet it is unclear whether similar chromatin-based mechanisms of gene regulation are relevant for the vast majority of unstudied lncRNAs.
We sought to isolate the set of lncRNAs that are likely to function at the chromatin interface by using biochemical fractionation of the nuclear compartment coupled to RNA-seq. Similar nuclear extraction methods have been employed to great effect in studying transcription, mRNA processing and export (Bhatt et al., 2012; Dye et al., 2006; Pandya-Jones and Black, 2009; Weber et al., 2014; Wuarin and Schibler, 1994). We find that the bulk of annotated lncRNAs are chromatin-enriched, suggesting widespread roles in chromatin regulation. However, Gencode and Broad lncRNAs account for only a small portion of observed chromatin-enriched transcripts; the majority represent a distinct subclass of lncRNAs that we term “chromatin-enriched RNA” (cheRNA). Most cheRNAs are tethered to chromatin by RNA pol II (RNAPII) and their presence correlates with neighboring gene transcriptional activity at a level similar to, or better than the current state-of-the-art active enhancer annotations (ENCODE Project Consortium et al., 2012; Ernst et al., 2011; Rada-Iglesias et al., 2011; Zentner et al., 2011). Yet cheRNAs appear distinct from recently described bi-directional transcripts that emanate from canonical active enhancers (Andersson et al., 2014; Kim et al., 2010; Lam et al., 2014; Wang et al., 2011a).
Results
Nuclear fractionation quantitatively distinguishes cis and trans-acting lncRNAs from mRNAs
Our initial aim was to identify chromatin-associated lncRNAs. We first extracted HEK293 nuclei with a forcing buffer to separate soluble and loosely bound material from the chromatin pellet, which retains tightly-bound factors (Bhatt et al., 2012; Dye et al., 2006; Pandya-Jones and Black, 2009; Wuarin and Schibler, 1994) (Figure 1A). We then performed RNA-seq from three biological replicates of the resulting soluble-nuclear extract (SNE) and chromatin pellet extract (CPE), yielding greater than 49 million uniquely-mapped reads from each fraction replicate. De novo assembled transcripts (Kim et al., 2013; Trapnell et al., 2013) greater than 1,000 nucleotides from the CPE were added to the latest Gencode gene annotation (Harrow et al., 2012), and each transcript was scored for its abundance in the CPE relative to SNE fractions (Figure S1G).
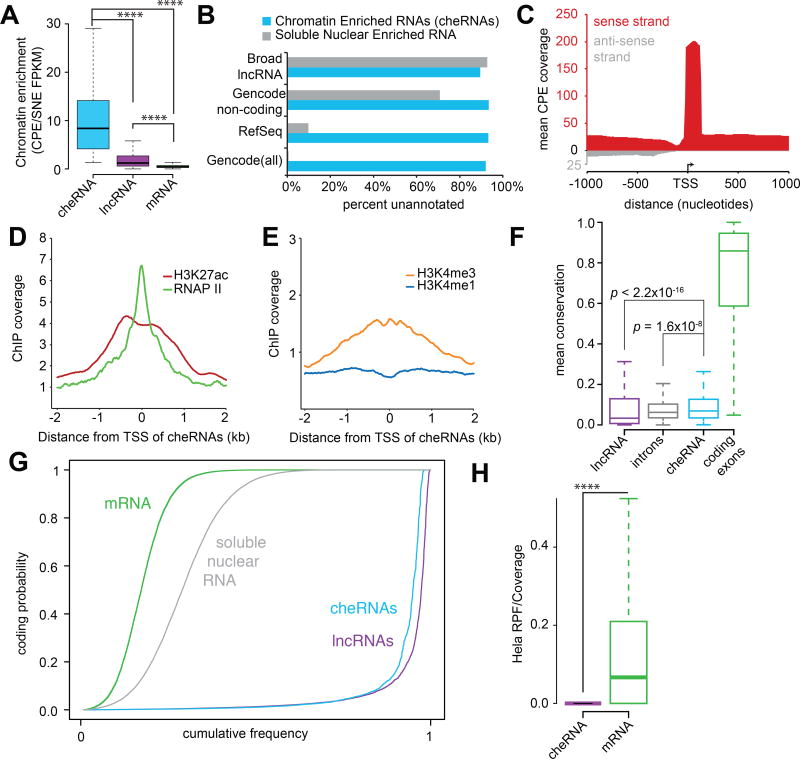
Figure 1. Nuclear fractionation isolates chromatin-associated RNA, which includes over half of annotated lncRNAs.
(A) Depiction of the nuclear fractionation procedure adapted from Wuarin and Schibler, 1994. Purified nuclei from Human Embryonic Kidney (HEK293) cells were extracted with a forcing urea/detergent buffer to yield a soluble-nuclear extract (SNE) and chromatin pellet extract (CPE). RNA from both fractions was isolated by Trizol in triplicate, ribosome-depleted and sequenced retaining strand information. (B) Scatter plot of relative RNA abundance in each of the two fractions (Y-axis CPE, X-axis SNE) presented on a log10 FPKM-scale with densities for each category plotted as curves along each axis, and a slope of 1 indicated by the dashed line. All assembled transcripts with FPKM>0 in both fractions (32,400) are represented in orange, with Gencode (v19) categories overlaid on top: 14,674 mRNAs (‘protein-coding’) in green, 4,829 pseudo genes in red, 2,409 antisense in blue, and 2,404 lncRNAs in purple.
We first validated our fractionation by confirming robust chromatin enrichment of two canonically chromatin associated lncRNAs, Xist and Kcnq1ot1 (Kalantry et al., 2009; Mohammad et al., 2010; Pandey et al., 2008; Penny et al., 1996; Plath et al., 2003), and soluble nuclear enrichment of the mRNAs beta-actin (Actb) and Gapdh (Figure S1A,B,D). In contrast, the trans-acting lncRNAs Hotair and Evf2/Dlx6-as displayed an intermediate level of solubility, consistent with proposed models that suggest both nuclear mobility and chromatin attachment (Berghoff et al., 2013; Bond et al., 2009; Rinn et al., 2007; Tsai et al., 2010)(Figure S1C,D). Collectively, these data indicate biochemical fractionation of nuclei coupled to RNA-seq can distinguish mRNAs, chromatin-enriched cis-acting lncRNAs, and trans-acting lncRNAs based on their sub-nuclear compartmentalization.
Applying this approach to Gencode noncoding RNAs (ncRNAs) revealed 57% of lncRNAs, 52% of antisense transcripts, and 28% of pseudo-genes are chromatin-enriched, in contrast to 16% of mRNAs. The relative abundance of each of these transcripts in the CPE versus SNE provides a valuable resource for further exploration of functional ncRNAs that are likely to operate at the chromatin-interface (Table S1). Surprisingly, we also observed that the size of the chromatin-associated population is substantially greater than previously appreciated (Figure 1B) (Bhatt et al., 2012), the overwhelming majority of which are not present in the Gencode annotation.
Stripping highly abundant mRNA from the chromatin pellet with urea was critical to identify CPE transcripts because it effectively magnified the coverage depth of low-abundance RNA species. Indeed, far fewer reads were consumed by exons from annotated genes in the CPE relative to the SNE and whole-cell RNA sequencing (Figure S2A, S3A). The resulting higher coverage of non-exonic portions of the transcriptome combined with the statistical power of our high-depth biological replicates enabled stringent assembly of chromatin-associated transcripts (see Figures S3A–C for examples). These findings are distinct from previous efforts to examine chromatin-associated noncoding RNA (Djebali et al., 2012; Mondal et al., 2010) in that the fractionation method we employed efficiently isolated tightly-chromatin associated RNAs from nucleoplasmic species (Bhatt et al., 2012; Dye et al., 2006; Wuarin and Schibler, 1994) (See Supplemental Discussion for detailed comparison, Figure S1A,S1I).
Chromatin-enriched RNA: a distinct subclass of lncRNAs
To further investigate the chromatin-associated RNA pool we focused on intergenic transcripts that were significantly enriched in the CPE (p<0.05; geometric mean normalization). We term the resulting set of 2,621 transcripts chromatin-enriched RNAs (cheRNAs), noting even more robust chromatin enrichment than Gencode lncRNAs (Figure 2A). Consistent with our analysis above, 81% of cheRNAs were absent in RefSeq (Pruitt et al., 2014), Broad lncRNA (Cabili et al., 2011), or Gencode (v19) (Harrow et al., 2012) annotations, whereas fewer than 1% of transcripts enriched in the soluble-nuclear extract were unique (Figure 2B, Table S2).
Figure 2. The properties of 2,621 chromatin enriched RNAs (cheRNAs).
(A) Although lncRNAs as a whole are chromatin-enriched relative to mRNAs (CPE/SNE mean=5.4, median=1.2), cheRNAs are defined by far more robust chromatin-enrichment (CPE/SNE mean=10.3, median=8.4). (B) Fraction of unique cheRNAs compared to latest Broad lncRNA, RefSeq and Gencode annotations. (C) Average coverage of stranded RNA-seq reads from combined CPE replicates that map to a 2,000 bp window centered on the putative TSSs of cheRNAs. Coverage in the same sense as the cheRNA annotation is depicted in red and antisense reads in grey. (D) Mean ChIP-seq coverage in HEK293 cells of RNA polymerase II (RNAPII) and H3K27 acetyl (H3K27ac), or (E) H3K4me3 and H3K4me1 profiles centered at the TSS of cheRNAs. (F) Boxplot of conservation measured by mean phastCons score (100 way vertebrate) of Gencode lncRNA exons, introns, cheRNAs, and mRNAs (each box spans the 25th to 75th percentile with the median indicated as a line). Comparisons of populations and p values measured by non-parametric one-sided Wilcoxon rank sum/Mann-Whitney U test. (G) Coding probability of cheRNAs (light blue), transcripts significantly enriched in the soluble-nuclear extract (grey, p<0.05), Gencode mRNAs (green) and lncRNAs (purple) assessed by CPAT. (H) Ribosome profiling coverage from HeLa cells (Guo et al., 2010) expressed as the density of ribosome-protected fragments (RPF) mapped to cheRNAs and mRNAs normalized to coverage in the total RNA-seq library.
To determine whether cheRNAs represent pervasive transcriptional ‘noise’ or directed transcription we analyzed their molecular properties and found them to be analogous to lncRNAs in a number of respects. CheRNAs exhibited a strong specific strand bias from their putative transcription start sites (TSSs) (Figure 2C, S2B), and fewer than 14% of cheRNAs are located within 500 bp of coding genes, arguing that the majority do not reflect the byproducts of divergent promoters, cryptic upstream promoters or read-through transcription from upstream genes (Core et al., 2008; He et al., 2008; Preker et al., 2008; Seila et al., 2008). Additionally, cheRNA TSSs displayed peaks of RNAPII, as well as histone modification patterns of histone 3 lysine 27 acetylation (H3K27ac), and a bias of histone 3 lysine 4 trimethylation (H3K4me3) over monomethylation (H3K4me1) (ENCODE Project Consortium et al., 2012; Grzybowski et al., 2015) similar to those observed for lncRNAs, (Guttman et al., 2009; Rinn and Chang, 2012)(Figure 2D,E). We also validated the TSS at these ChIP signatures for 9/10 cheRNAs by 5'RACE (Figure S2C, S3B,C), affording further evidence for their independent transcription.
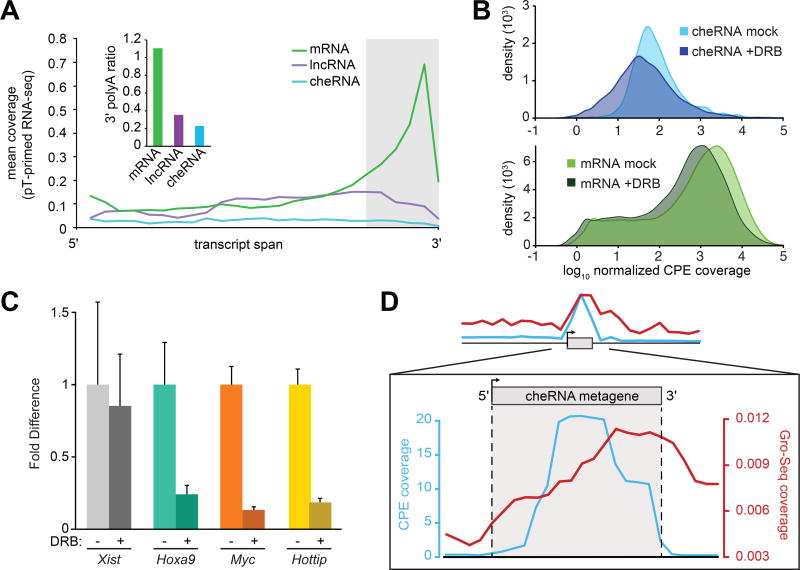
At the molecular level, cheRNAs display relatively modest conservation compared to coding exons, yet slightly greater mean conservation than both introns and lncRNA exons (Figure 2F). CheRNAs also exhibit negligible coding potential (Figure 2G) and ribosome profiling from HeLa cells (Guo et al., 2010) suggests that they are largely untranslated (Figure 2H). Relative to coding genes, cheRNAs are underspliced. Yet we do detect splice-junctions in approximately 1/4 of all cheRNAs comparable with lncRNA splicing in our data. To determine whether cheRNAs are 3' polyadenylated, a post-transcriptional processing step that is important for stability, export, and translation of mRNAs (Millevoi and Vagner, 2010), we performed RNA-seq with polyT-primed reverse transcription. As anticipated, reads that mapped to mRNAs were heavily biased towards their 3' ends. However this trend was not observed for either lncRNAs or cheRNAs (Figure 3A), in agreement with a previous observation that lncRNAs are over-represented in non-polyadenylated RNA sequencing libraries (Derrien et al., 2012).
Figure 3. CheRNAs are not polyadenylated and are tethered to chromatin by RNAP II.
(A) RNA-seq using polyT-primed reverse transcription, expressed as mean normalized tag counts across the length of each transcript. Inset: mean 3'-polyA ratio, computed from panel A as the normalized tag counts mapping to the last 20% divided by the first 80% of each transcript. (B) The distribution of cheRNA (blue) and mRNA (green) CPE RNA-seq reads treated with the RNA polymerase II elongation inhibitor DRB (darker shade, two hour incubation at 100 µM) and ‘mock’ treated (lighter shade) samples for two biological replicates. All reads in a given fraction were normalized to the amount of reads that mapped to XIST, which is not affected by DRB on a two hour time scale (see C). (C) Fold difference of each indicated RNA with DRB treatment compared to DMSO only (mock) normalized to 18S rRNA. Error bars indicate S.E.M. for three independent biological replicates. (D) Metagene analysis of read density contoured over all cheRNAs and flanking regions for the chromatin pellet in our HEK293 (CPE coverage, cyan) and global run-on (GRO-seq, red) from HEK293T (Liu et al., 2013).
Incomplete co-transcriptional processing, as evinced by occasional splicing and lack of polyadenylation, implicated RNAPII in cheRNA chromatin-attachment. To distinguish whether cheRNAs are maintained on chromatin by ongoing transcription or if the fully processed transcripts are tethered by independent mechanisms as may be the case for XIST and other lncRNAs (Engreitz et al., 2013; Grote et al., 2013; Huarte et al., 2010; Jeon and Lee, 2011; Martianov et al., 2007; Park et al., 2002; Quinn et al., 2014; Rinn et al., 2007), we re-examined the chromatin pellet after inhibiting RNAPII, which otherwise remains transcriptionally competent under similar preparation conditions (Core et al., 2008; Dye et al., 2006; Kimura et al., 1999). A two hour incubation with DRB, an inhibitor of RNAPII elongation (Yamaguchi et al., 1999; Zandomeni et al., 1982), caused a global reduction in the chromatin-pellet abundance of mRNAs and the majority of cheRNAs (Figure 3B). The cis-acting lncRNA HOTTIP (Wang et al., 2011b) was also depleted in the presence of DRB suggesting it’s chromatin association, like cheRNAs, is transcription dependent. In contrast, XIST, which is thought to be linked to chromatin via the YY1 transcription factor (Jeon and Lee, 2011), remained at equivalent levels (Figure 3C). While there has been much speculation as to how chromatin-associated lncRNAs are tethered to chromatin (Guttman and Rinn, 2012), we provide evidence that the majority of tightly chromatin-associated lncRNAs are connected by RNAPII transcription. Nevertheless, approximately 25% of cheRNAs maintained a +/−DRB ratio greater than one, suggesting that this subpopulation is adhered to chromatin by an RNAPII transcription-insensitive mechanism, or derives from another polymerase.
Further support for polymerase-mediated adhesion derives from Global Run On Sequencing (GRO-seq) from the related HEK293-T cell line (Liu et al., 2013): we observe abundant nascent transcription at cheRNA loci to the exclusion of flanking regions (Figure 3D). Intriguingly, GRO-seq coverage was disproportionally enriched at the 3’ ends of cheRNAs following the peak of RNA-seq coverage, consistent with a model of paused polymerases tethering RNA to chromatin.
Collectively, these characteristics suggest that cheRNAs represent a subclass of regulated, conserved, and especially chromatin-enriched lncRNAs tethered to chromatin via ongoing or paused RNAPII transcription, thousands of which had previously escaped detection from conventional sequencing methods and annotations (Figure 2B).
CheRNA transcription correlates with proximal gene expression
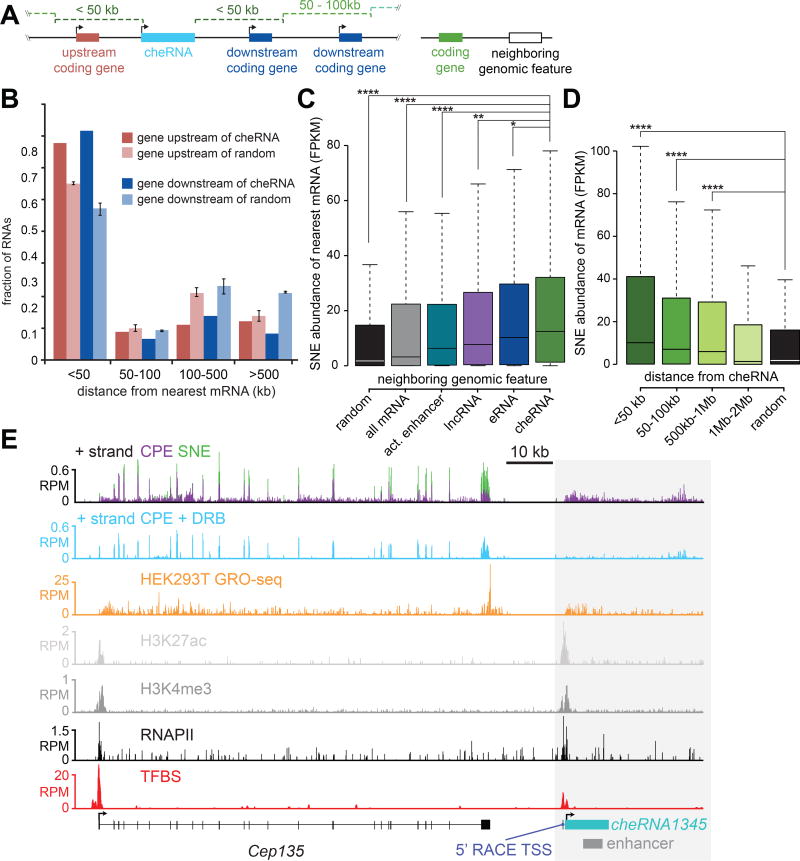
Inspired by several examples of lncRNAs altering the expression of neighboring genes in cis (Chalei et al., 2014; Li et al., 2013; Mohammad et al., 2010; Wang et al., 2011b; Ørom et al., 2010) we investigated a similar potential function for cheRNAs. Consistent with this possibility, the sites of cheRNA transcription are closer to coding genes than expected by chance (Figure 4A,B), and are strongly correlated with the expression of their nearest genes in the soluble-nuclear extract and total RNA (Figure 4C, S3D). Remarkably, proximity to cheRNAs was more highly correlated with the expression levels of nearby genes than the presence of nearby enhancers defined by the chromatin marks H3K4me1/H3K27Ac, expressed lncRNAs, or enhancer RNAs (eRNAs) (Figure 4C). Consistent with a possible local enhancer function affecting multiple genes, the expression of neighboring genes as a whole gradually decreased as a function of distance from cheRNAs (Figure 4D), but is more idiosyncratic on an individual basis (Figure S4B).
Figure 4. Expression of genes positively correlates with proximity to cheRNAs.
(A) Schematic representation of a cheRNA locus with upstream and downstream coding genes; a more general representation of a cheRNA and a neighboring genomic feature. (B) Distribution of nearest gene distances to upstream and downstream cheRNAs (dark columns) compared to random distributions of coordinates with the same spans as cheRNAs (light columns, n = 3, error bars indicate S.E.M. for three trials). (C) Comparison of soluble-nuclear extract (SNE) expression (FPKM) of the nearest neighboring genes to the following genomic features: cheRNAs genes (green), randomly shuffled coordinates representing the same chromosomal spans as cheRNA genes (black), all mRNA (grey), active HEK293 enhancers (cyan) as defined by overlapping H3K27ac and H3K4me1 ChIP-seq peaks (Rada-Iglesias et al., 2011; Zentner et al., 2011), all expressed lncRNAs (2,404, purple), and eRNAs produced from active enhancers (316, blue). *p<0.05, ** p<1×10−10, ****p<2.2×10−16 computed with Mann–Whitney U test. (D) Similar to (C), expression of mRNAs that fall within the indicated distances from cheRNA genes. (E) RNA-seq (SNE, green; CPE, purple) and ChIP-seq contoured over the Cep135 - cheRNA1345 locus in HEK293. Experimentally validated transcription factor binding sites from multiple cell types (ENCODE Project Consortium et al., 2012) and a functional Vista Enhancer are also indicated.
As a representative example, we highlight cheRNA1345, which is produced from a locus ~15 kb downstream of Cep135 and overlaps with an experimentally validated tissue-specific enhancer element (Figure 4E) (Visel et al., 2007). Peaks of H3K4me3, H3K27ac, and RNAPII decorate the TSS, which we validated by 5' RACE, and also bears a number of validated transcription factor binding sites (Gerstein et al., 2012). Similar patterns are observed with other cheRNA loci (Figure S3A–C).
Approximately two-thirds of the 2,621 HEK293 cheRNAs also overlapped with putative enhancers culled from nine cell lines (Ernst et al., 2011), (empirical p<0.001). Despite cell type mismatch, the expression of genes near HEK293 cheRNA-enhancers in each of three ENCODE cell lines queried were greater than tissue-specific ‘weak’ enhancers, and comparable to ‘strong enhancers’ (Figure S3E). Notably, ‘strong enhancers’ are partially defined by high levels of H3K4me3 (Ernst et al., 2011) hinting that they might promote expression of cheRNAs, consistent with a recent unifying hypothesis of regulatory elements (Andersson et al., 2015).
Segmentation of cheRNAs based on orientation reveals important distinctions
Despite many shared properties and strong correlation to transcriptional status of neighboring genes, the set of cheRNAs is unlikely to be monolithic in function. Genes with overlapping antisense cheRNAs exhibited significantly lower expression as compared to all mRNAs (Figure S4A), consistent with models of transcriptional interference (Callen et al., 2004). Division of cheRNAs by strand sense and orientation relative to their nearest coding genes reveals an uneven distribution, with slight skew towards shared sense (60%), and a stronger bias towards being downstream (71%) (Figure S2D). CheRNAs downstream of their neighbors display even stronger expression correlation than the set as a whole, whereas the upstream cheRNAs are more weakly correlated on a whole (Figure S4A) despite notable counterexamples (Figure S4B). This composition raises the possibility that some cheRNAs in the same sense represent read-through from upstream genes (Iyer et al., 2015) or, to a lesser extent, cryptic initiation sites for downstream genes (Preker et al., 2008). Pervasive cheRNA biogenesis of this sort might account for the strong cis-expression correlation. Indeed, there are examples of cheRNAs spliced to proximal coding genes detectable in our data and in EST databases potentially accounting for ~5% of cheRNAs, comparable to the 9% of lncRNAs previously noted to be cryptic UTRs of proximal coding genes (Derrien et al., 2012). However, removing these cheRNAs from our analysis does not alter the observed correlation with nearby gene expression (Figure S4A), nor is the correlation exclusive to cheRNAs in the same sense as the nearest mRNA (Figure S4A). Further, within 1Mb there is little correlation between distance to the nearest neighboring gene and apparent cis-activation (Figure S4B). Several additional lines of evidence presented in Supplemental Discussion argue that the vast majority of cheRNAs are independently transcribed units, and that the apparent cis-enhancer effect is independent of cheRNAs that could potentially represent read-through, cryptic introns or extended UTRs from coding genes.
Discussion
Our study measured noncoding RNA adhesion to chromatin across the human genome. We find that the majority of known lncRNAs are chromatin-enriched, extending this property from the small set of well-studied lncRNAs (Chalei et al., 2014; Mohammad et al., 2010; Nagano et al., 2008; Rinn et al., 2007; Wang et al., 2011b; Ørom et al., 2010) to a more general principle, thereby providing a resource for future mechanistic studies (Table S1). We also observe that trans-acting lncRNAs exhibit intermediate levels of chromatin enrichment, suggesting either more labile chromatin attachment or reflecting two distinct pools of molecules: those bound to or searching for their target loci (Bond et al., 2009; Rinn et al., 2007). For a lncRNA of unknown function, the distinction between strong or intermediate enrichment may inform their mechanistic possibilities, suggesting whether cis or trans-acting lncRNA pathways are more likely.
A more holistic view of the nuclear transcriptome reveals two highly clustered populations corresponding to nuclear-soluble and chromatin-associated RNA (Figure 1B). While there have been hints of the existence and dimensions of this latter population (Bhatt et al., 2012; Derrien et al., 2012; Khalil et al., 2009), our findings establish this pool to be substantially larger than previously observed, consistent with hypotheses advocating a more widespread role of noncoding RNA in chromatin regulation (Bernstein and Allis, 2005; Mattick, 2004). Although many of these transcripts may be non-functional, the intergenic cheRNAs we focused on exhibited molecular properties similar to annotated lncRNAs (Derrien et al., 2012; Guttman et al., 2009; Rinn and Chang, 2012). Perhaps the most compelling case for function is our observation that proximity to a cheRNA locus was more strongly predictive of neighboring gene expression than any other class of noncoding RNA or enhancer annotation available in the HEK293 cell line (Figure 4C). Nevertheless, genetic perturbations and functional experiments of individual cheRNAs will be required to test their causality in proximal gene activation and examine the molecular mechanism(s) thereof. Even if these molecules themselves play no direct role in cis-enhancer function, their presence tethered to chromatin by transcription is sufficiently predictive of cis-gene activity, that determining chromatin-enriched transcripts in other cell types may aid in annotating active cis-enhancer elements in conjunction with existing methods (Andersson et al., 2014; ENCODE Project Consortium et al., 2012; Rada-Iglesias et al., 2011; Zentner et al., 2011).
We also identify that the predominant means of tethering of cheRNAs (including hundreds of annotated lncRNAs) is through active RNAPII. Although widely speculated (Bonasio et al., 2010; Guttman and Rinn, 2012; Quinodoz and Guttman, 2014; Rinn and Chang, 2012), we are not aware of tests of this proposed chromatin association beyond a few anecdotal cases (Mao et al., 2011; Simon et al., 2011). This result is consistent with the cis-activating correlation observed, leading to a model where ongoing or paused transcription of noncoding RNAs influences the expression of proximal genes (Bonasio et al., 2010; Guttman and Rinn, 2012). Conversely, the 25% of cheRNAs that remain on chromatin after pausing RNAPII transcription for two hours are compelling candidates for lncRNAs that may utilize additional methods of attachment (Jeon and Lee, 2011). Alternatively, this population could represent RNA projecting from RNAPIII or a more stably paused RNAPII, which may lose chromatin association on a longer timescale than sampled by our pharmacologic perturbation.
Given the annotated enhancer overlap and expression of nearby genes, we expected significant overlap of cheRNAs with the relatively new category of enhancer RNAs (eRNAs)(Andersson et al., 2014; Kim et al., 2010; Wang et al., 2011a). To our surprise, only 10.9% of cheRNAs overlapped a compendium of eRNAs derived from the majority of human tissues (Andersson et al., 2014), compared to 6.2% expected by chance. Although there appear to be functional similarities, there are several molecular characteristics that distinguish cheRNAs from the canonical definition of eRNAs. First, most eRNAs are bi-directionally transcribed from prototypical enhancers marked by the histone modifications H3K4me1 and H3K27ac (Kim et al., 2010; Lam et al., 2014; Wang et al., 2011a), while cheRNAs exhibited a specific strand bias (Figure 2C, S2B) and display H3K4 tri-methylation over monomethylation, more typical of mRNA and lncRNA TSSs (Figure 2D,E). To date, only one report has described a few hundred enhancers that produced largely unidirectional eRNA, although unlike cheRNAs, most were polyadenylated and appeared to have H3K4me1/HeK4me3 ratios greater than unity (Koch et al., 2011). Further, whereas eRNAs are generally considered short (median ~350 nt (Andersson et al., 2014)), CPE-specific transcripts which comprise the bulk of cheRNAs were bounded by a >1,000 nucleotide threshold to yield a median length of 2,110 nucleotides. Finally, although 58% of active HEK293 eRNAs displayed chromatin enrichment, this does not appear to be a defining characteristic of eRNAs (Figure S2E).
CheRNAs are more similar in molecular properties to the transcriptionally activating ncRNA-a subset of lncRNAs (Lai et al., 2013; Ørom et al., 2010), however, only a few (n=20) of these overlap cheRNAs, whereas more overlap transcripts significantly enriched in the SNE (n=338). Despite these distinctions, their apparent functional similarity is a compelling reason for evaluating potential mechanistic commonalities, and perhaps redefining the eRNA or ncRNA-a categories to include chromatin-enriched transcripts. Going forward, the challenge will be to determine how these and myriad other noncoding RNA species contribute to the complexity of transcriptional control in humans and other multicellular organisms.
Experimental Procedures
Fractionation of human nuclei was performed following Bhatt et al., 2012, however this was performed in three biological replicates of 10–20 × 106 HEK293 cells using TruSeq Stranded Total RNA preparation (Illumina). Soluble-Nuclear Extract (SNE) replicate reads were assembled and merged relative to the Gencode (v19) annotation, and Chromatin Pellet Extract (CPE) reads were assembled into de novo transcripts using Cufflinks2 (Trapnell et al., 2013). Poly-dT primed RNAseq was performed on HEK293 Total RNA with strand-specific library preparation adapted from (Cloonan et al., 2008; Zhao et al., 2010), but with a polyT primer for first strand cDNA synthesis. DRB treatment was accomplished as previously described for two hours (Riising et al., 2014), followed by nuclear fractionation as above.
Supplementary Material
Acknowledgments
We would like to thank Peter Faber and Erika Hanson of the University of Chicago Functional Genomics Core for Illumina sequencing. This work was supported by awards to A.J.R. (Junior Investigator) and M.S.W. (Scholar) from the Chicago Biotechnology Consortium with support from The Searle Funds at The Chicago Community Trust and the Ellison Foundation (New Scholar in Aging). We would also like to acknowledge the Farnham and Weissman labs, and ENCODE for contributing ChIP-seq data sets in HEK293 cells. We thank the reviewers for prompting us to subdivide the cheRNA set by relative orientation to proximal genes as well as examine the extent of direct connection to neighbouring transcriptional units in the same sense.
Footnotes
Accesion numbers
The Gene Expression Ominbus for RNA-seq data sets is GSE66478. ChIP-seq for HEK293 H3K4me3:GSE60378, and HEK293-T Gro-seq:GSE51633. Encode accessions for data sets H3K4me1: ENCSR000FCG, H3K27ac: ENCSR000FCH, and RNAPII: ENCSR000EZA.
Supplemental Information includes Extended Experimental Procedures, four figures and four tables and can be found with this article online at
References
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Sandelin A, Danko CG. A unified architecture of transcriptional regulatory elements. Trends in Genetics. 2015:1–8. doi: 10.1016/j.tig.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140:4407–4416. doi: 10.1242/dev.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes & Development. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong A-J, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, Ponting CP. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowski AT, Chen Z, Ruthenburg AJ. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell. 2015;58:886–899. doi: 10.1016/j.molcel.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilott NE, Ponting CP. Predicting long non-coding RNAs using RNA sequencing. Methods. 2013;63:50–59. doi: 10.1016/j.ymeth.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Tao Y, Roeder RG, Cook PR. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol Cell Biol. 1999;19:5383–5392. doi: 10.1128/mcb.19.8.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, la Chapelle, de AL, Heidemann M, Hintermair C, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MTY, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3' end processing regulation. Nucleic Acids Research. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. Rna. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, La Cruz, de CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Research. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent Transcription from Active Promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.a. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Research. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011a;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011b;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandomeni R, Mittleman B, Bunick D, Ackerman S, Weinmann R. Mechanism of action of dichloro-beta-D-ribofuranosylbenzimidazole: effect on in vitro transcription. Proc Natl Acad Sci USA. 1982;79:3167–3170. doi: 10.1073/pnas.79.10.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song J-J, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.