SUMMARY
The combinatorial expression of Hox genes along the body axes is a major determinant of cell fate and plays a pivotal role in generating the animal body plan. Loss of HOXA13 and HOXD13 transcription factors (HOX13) leads to digit agenesis in mice, but how HOX13 proteins regulate transcriptional outcomes and confer identity to the distal-most limb cells has remained elusive. Here, we report on the genome-wide profiling of HOXA13 and HOXD13 in vivo binding and changes of the transcriptome and chromatin state in the transition from the early to the late-distal limb developmental program, as well as in Hoxa13−/−; Hoxd13−/−limbs. Our results show that proper termination of the early limb transcriptional program and activation of the late-distal limb program are coordinated by the dual action of HOX13 on cis-regulatory modules.
In Brief
HOX13 homeobox transcription factors determine digit cell fate during limb bud development. Sheth et al. identify HOX13-dependent regulation of gene expression and chromatin state that suggests HOX13 activity at cis regulatory modules allows for a coordinated transition from the early to the late-distal program in the developing limb bud.
INTRODUCTION
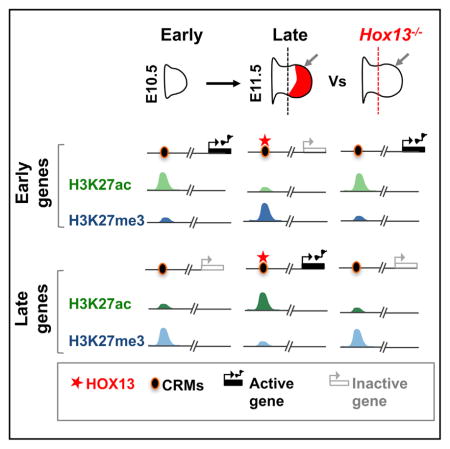
During organogenesis, appropriate spatial and temporal gene expression is necessary for the specification of individual cell identities and, ultimately, establishment of organ structure and function. As a paradigmatic system, the developing limb bud is used to understand the principles of pattern formation, the process through which cells are specified, determined, and subsequently differentiate to form a morphological structure (Tabin and Wolpert, 2007; Zeller et al., 2009; Tickle, 2015; Delgado and Torres, 2016). In mouse embryos, the forelimb bud at embryonic day 10.5 (E10.5) (hereafter referred to as “early”) consists primarily of undifferentiated mesenchymal cells enclosed in a layer of ectoderm, which undergo rapid proliferation and differentiation. By E11.5 (hereafter referred to as “late”), a basic skeletal pattern is laid down along the proximal-distal (P-D) axis (Zeller et al., 2009). The changes made to the early default program that, within 1 day, lead to the formation of the distal-most derived structures of the limb (the wrist and digits) offer a paradigm to study the dynamics of transcriptional regulatory networks in vivo.
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) of post-translationally modified lysine (K) 27 residues of histone H3 can reveal the genome-wide pattern of active- (H3K27ac) and polycomb-mediated repressed (H3K27me3) chromatin states (Johnson et al., 2007; Creyghton et al., 2010; Rada-Iglesias et al., 2011). Combining genome-wide analyses of histone modifications and transcription factor (TF) occupancy is useful to identify TF-mediated regulatory networks. Cell-culture-based studies provided evidence that specific TFs coordinate initiation and execution of cell-type-specific transcriptional programs (Xu et al., 2015; Takahashi and Yamanaka, 2016), known to be critical for organogenesis (Graf and Enver, 2009; Davidson, 2010). Studying TF-dependent regulatory networks in vivo can be challenging due to the activity of paralogous TFs, which are often co-expressed and functionally redundant. This difficulty applies in particular to the Hox cluster genes. The precise spatial and temporal expression of 5′HoxA and HoxD genes (HoxA/D9-13) is required to regulate growth and patterning along the P-D axis in the developing limb (Zakany and Duboule, 2007). HoxA and HoxD paralogous genes from group 9–10 are important for patterning the stylopod, while group 11 genes are crucial for the zeugopod, and the Hoxa13 and Hoxd13 genes (hereafter referred to as Hox13) are required for patterning the digits (Davis and Capecchi, 1996; Favier et al., 1996; Fromental-Ramain et al., 1996a, 1996b; Carpenter et al., 1997). While Hox13 are expressed in all presumptive digit cells, the remaining 5′HoxA/D genes, with the exception of Hoxa11, are expressed either partially or at low levels in digit cells (Woltering et al., 2014). This co-expression and functional redundancy have limited an in-depth understanding of cell- and tissue-type-specific programming as regulated by HOX proteins. Nonetheless, genetic analyses in mice have shown that loss of HOX13 leads to complete digit agenesis (Fromental-Ramain et al., 1996b), indicating that HOX13 plays a crucial role in triggering the digit-specific program. How HOX13 specifically acts to regulate the switch from the early to the late-distal limb programs, ultimately giving rise to the digits, remains unclear.
In this work, we show that Hox13 inactivation leads to the downregulation of genes preferentially expressed in late-distal wild-type (WT) limb buds, while a subset of genes transcribed at higher levels in early limb buds, but normally excluded from the WT digit territory, is upregulated. We show that genome-wide HOXA13 and HOXD13 binding is highly redundant and that these proteins preferentially target putative cis-regulatory modules (CRMs). Furthermore, our epigenetic profiling of early and late-distal WT limb cells shows that the early to late-distal transition is accompanied by rapid changes in regulatory activity. Importantly, we demonstrate that in the absence of HOX13, regulatory activities specific to late-distal limb cells is impaired while activities specific to the early regulatory program are maintained. These results, in conjunction with altered gene expression in Hox13−/− limbs, indicate that HOX13-dependent changes in cis-regulatory activities are essential for setting up the transcriptional program of presumptive digit cells.
RESULTS
Inactivation of Hox13 Alters the Transcriptional Program in Late-Distal Limb Cells
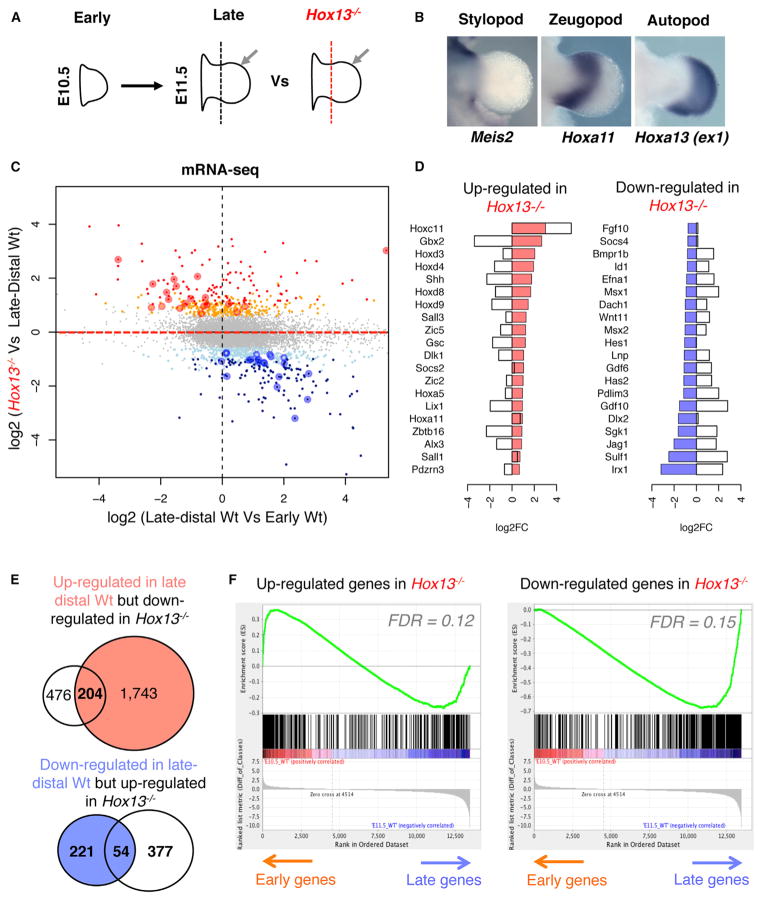
To examine the gene network underlying distal limb patterning and isolate the Hox13-dependent components of this network, we first performed expression profiling in early WT, late-distal WT, and late-distal Hox13−/− forelimb buds using RNA sequencing (RNA-seq) (Figure 1A). We performed the analysis at E11.5, when the morphology of the late-distal tissue in WT and double-mutant limb buds is indistinguishable. To ensure consistency in late-distal limb microdissection, we used a morphological landmark provided by the indentation at the proximal border of the handplate, such that the isolated tissue includes some zeugopod progenitors (Figure 1B). In order to increase the statistical robustness of our transcriptome analysis, we also considered recently published RNA-seq profiles obtained from early limb cells (Lewandowski et al., 2015) along with the profiles we generated in early, late-distal WT, and mutant limb buds (Table S1). Comparison of early and late-distal WT limb cells shows that 1,743 genes are upregulated in late-distal cells while 221 genes are downregulated (using lenient thresholds, i.e., 1.5 fold-change and p value ≤ 0.05; Table S2). Furthermore, comparing the late-distal WT and Hox13−/− cells shows 377 upregulated genes and 476 downregulated genes in Hox13−/− cells (Table S2). A cumulative plot of differentially expressed genes (DEGs) indicates that genes downregulated in Hox13−/− cells are preferentially expressed in late-distal WT cells compared to early WT cells (Figure 1C, lower right quadrant, and Figure 1E). Moreover, a subset of genes showing upregulation in Hox13−/− cells is transcribed at lower levels in the late-distal WT cells (Figure 1C upper left quadrant and Figure 1E). Indeed, many known limb-patterning genes that are expressed either in early limb and/or proximally restricted are upregulated in Hox13−/− limb buds (Figure 1D), including genes from the Hox clusters normally excluded from the presumptive digit cells (e.g., Hoxa11, Hoxc11, and Hoxd4-9; Figure 1D). The same trends are observed after performing gene set enrichment analysis (GSEA), i.e., upregulated genes in Hox13−/− are preferentially expressed in the early WT limbs, while downregulated genes are preferentially expressed in the late-distal WT cells (Figure 1F). Taken together, these results indicate that the transcriptional changes that occur during the transition from the early to late-distal programs in WT limbs are disrupted in the Hox13−/− limb.
Figure 1. Comparative Analysis of the Transcriptome in Early, Late-Distal, and Hox13−/− Limb Buds.
(A) Schematics showing limb tissues used in this study: WT early limb (left), WT late-distal limb (middle; gray arrow), and Hox13−/− late-distal tissue (right; gray arrow). Dashed lines mark the position of the cut.
(B) In situ hybridization (ISH) showing (from left to right) Meis2, Hoxa11, and Hoxa13-exon1 expression marking the stylopod, zeugopod, and autopod territories, respectively.
(C) Scatterplot of differentially expressed genes between Hox13−/− and WT late-distal buds. Each dot represents a gene, shown in terms of log-fold-change in late-distal Hox13−/− limbs as compared to its log-fold-change in the late-distal versus early WT limbs. Genes showing a false discovery rate (FDR) ≤0.05 and at least a 2-fold up- or downregulation (termed stringent) are indicated in red or navy, respectively. Genes not significant to the stringent threshold but showing a p value ≤ 0.05 and at least a 1.5-fold change (termed lenient) are indicated in orange or light blue depending on the direction of the change.
(D) Examples of genes upregulated (left) and downregulated (right) in Hox13−/−. Bars show log2-fold-changes. Color-filled bars indicate the log fold-change in the mutant, while void bars indicate the difference between early and late-distal buds.
(E) Venn diagrams showing the overlap between late-distal genes and those downregulated in Hox13−/− (top) and between early genes and those upregulated in Hox13−/− (bottom).
(F) GSEA analysis using the upregulated (right) and downregulated genes (left) in Hox13−/− as gene set, over all the genes detectable by RNA-seq, sorted by fold-change in early versus late-distal samples (FDR based on permutations of the original data).
HOX13 Binds to CRMs in the Developing Limb Bud
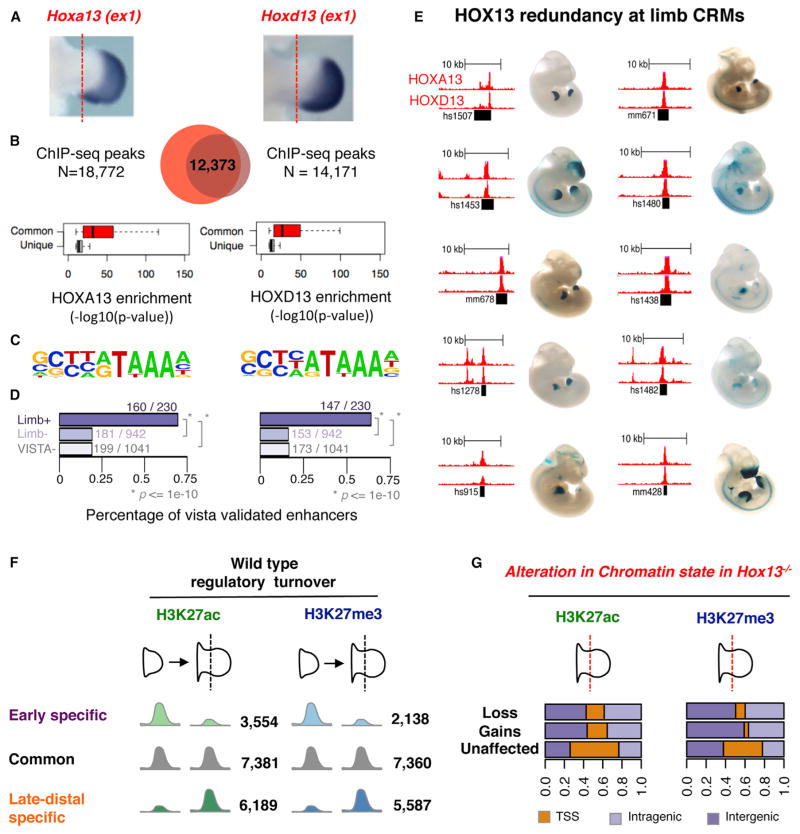
To investigate the possibility that the transcriptional changes in the Hox13−/− limb are the result of direct HOX13 activity, we first mapped genomic sites bound by HOXA13 and HOXD13 using ChIP-seq in late-distal WT cells at E11.5 (Figure 2A). To this aim, we used a previously validated HOXA13 antibody (Figure S1A; Knosp et al., 2004) and a newly generated HOXD13-specific antibody (Figure S1B). HOXA13 and HOXD13 ChIP-seq profiles reveal a comparable number of enriched regions (18,772 and 14,171, respectively; Figure 2B; Table S3) and show a very high degree of overlap (Figure 2B). Accordingly, HOXA13 and HOXD13 have a similar genome-wide distribution at promoters and intergenic and intragenic regions (Figure S2A; Table S3). The genomic regions co-occupied by both HOX13 TFs show a quantitatively higher enrichment for each TF compared to sites bound by only one of the two paralogous TFs (Figure 2B). Gene ontology analysis using the Genomic Regions Enrichment Annotations Tool (GREAT; McLean et al., 2010) shows that both HOXA13 and HOXD13 binding sites are closely associated with genes that are involved in limb and digit morphogenesis (Figure S2B). More importantly, de novo motif discovery (Heinz et al., 2010) returned an almost identical top-scoring motif for each dataset (Figure 2C). Furthermore, HOX13-occupied intergenic and intronic sites show a high degree of overlap with loci bound by p300 (Visel et al., 2009), a mark of active enhancers (Figures S2C and S2D). Accordingly, a number of experimentally validated, limb-specific enhancers are bound by HOX13 in vivo in WT cells (Figures 2D and 2E). HOX13 binding at the regions proximal to promoters (defined as regions ±2.5 kb from the known transcription start site [TSS]) is a mere 8%–10% of the total number of sites; the vast majority of HOX13-occupied genomic sites (90%–92%) are located outside these regions (Figure S2A). To focus on HOX13 action over putative regulatory regions, we excluded regions proximal to the promoter from further analyses. In light of the high degree of similarity between the two ChIP-seq profiles and the slightly higher signal-to-noise ratio of the HOXA13 dataset, we used only the set of HOXA13-occupied intergenic and intronic sites for further analyses (for consistency, it is referred to HOX13 hereafter). The high level of redundancy between HOXA13- and HOXD13-occupied sites, along with the marked functional redundancy observed via genetic analysis (Fromental-Ramain et al., 1996b), suggests that the presence of one paralog is sufficient to trigger and/or execute the digit patterning program.
Figure 2. Genome-wide Mapping of HOXA13 and HOXD13 Binding in Distal Limbs.
(A) ISH on E11.5 WT limbs using Hoxa13-exon1 and Hoxd13-exon1 probes. Red dashed lines indicate the proximal edge of the tissue used for ChIP-seq.
(B) Venn diagram showing the overlap between ChIP-seq peaks for HOXA13 and HOXD13. Boxplots showing enrichment (in terms of p value from MACS peak calling) for regions common to HOXA13 and HOXD13 (red) compared to regions unique for either TF (gray).
(C) The logo shows the most enriched motif from de novo motif discovery for HOXA13 (left) and HOXD13 (right).
(D) Fraction of VISTA elements (Visel et al., 2007) either positive in limb (limb+), negative in limb but positive in another tissue (limb−), or negative in all tissues (VISTA−) overlapping one or more HOXA13 or HOXD13 binding site. p values from Fisher’s exact test are indicated.
(E) Selection of ten VISTA elements (Visel et al., 2007) active in the late-distal limbs, showing binding by both HOXA13 and HOXD13 (ChIP-seq, red tracks).
(F) H3K27ac- and H3K27me3-enriched regions (intergenic and intronic peaks defined as farther than 2.5 kbp from the annotated TSS) in WT early and late-distal limbs.
(G) Genomic annotations of H3K27ac and H3K27me3 profiles, classified according to the gain or loss observed in Hox13−/−.
Late-Distal-Specific Chromatin State at CRMs Is Dependent on HOX13-Binding
To investigate potential HOX13-dependent modulation of cis-regulatory activity in a systematic and unbiased way, we assessed H3K27ac and H3K27me3 by ChIP-seq in both early and late-distal WT limb buds (Table S1). Our analysis also included a previously published H3K27ac profile in early limbs (Cotney et al., 2013). To limit the analysis to putative CRMs, regions proximal to promoters were excluded from further analysis as in the HOX13 ChIP-seq (Figure S3). The distribution of putative CRMs, both active (H3K27ac-marked) and repressed (H3K27me3-marked), revealed a significant number of CRMs exclusive to either early or late-distal WT cells, while a third subset includes regions constitutively marked (hereafter termed common) in both early and late-distal cells (Figure 2F). The high rate of “regulatory turnover,” as defined by changes in H3K27ac or H3K27me3, indicates that de novo activation and repression of CRMs occurs during the transition from early to late-distal WT limb cells.
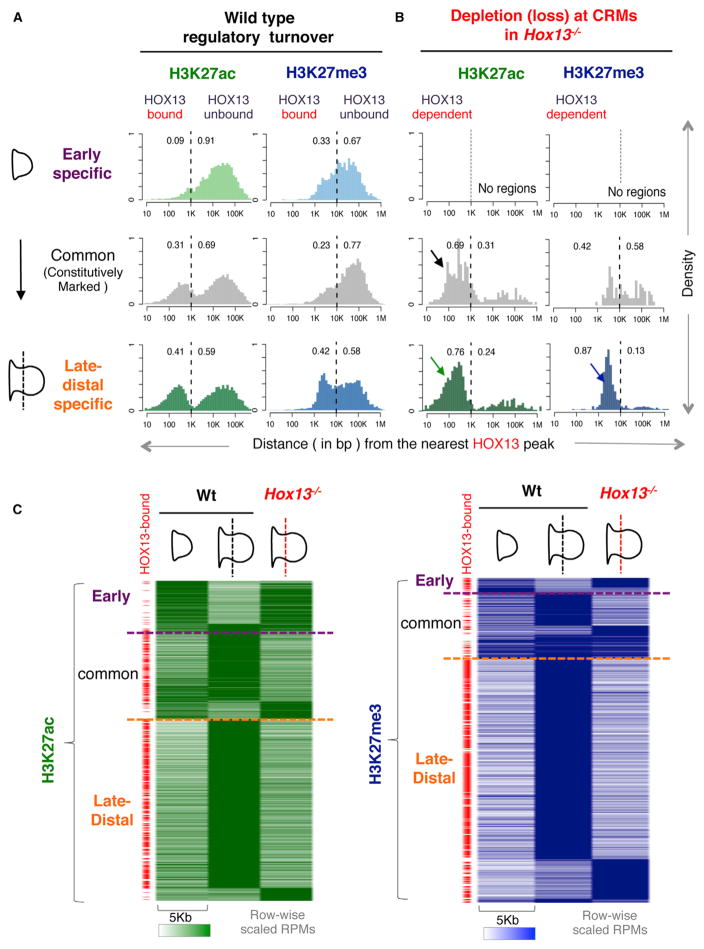
To identify CRMs whose functional state depends on HOX13, we also generated genome-wide maps of H3K27ac and H3K27me3 in the Hox13−/− late-distal cells. Most changes in chromatin state are found outside TSS (Figure 2G). Comparison of active and repressed CRMs using a highly conservative threshold (p value ≤ 1e-10) shows a substantial quantitative loss of H3K27ac and H3K27me3 marks in the absence of HOX13, mainly at CRMs that normally gain these marks specifically in late-distal cells (Figures 2G and S4A; Table S4). To contextualize the association between HOX13 binding and this differential regulatory activity, we plotted the distribution of CRMs relative to the nearest HOX13 peak (Figure 3A). The early-specific, common, and late-distal-specific CRMs are defined by the activity of CRMs in WT limb buds, in terms of H3K27ac and H3K27me3 patterns, as shown in Figure 2F. The distance distribution of common and late-distal-specific active (H3K27ac) CRMs show a bimodal shape, suggesting two well-defined enhancer populations in late-distal limbs: one directly bound by HOX13 (distance ≤1 kbp) and another one HOX13-unbound (distance >1 kbp) (Figure 3A). In contrast to the H3K27ac-marked regions, CRMs decorated by H3K27me3 are distributed farther away from HOX13-bound loci (distance ranging from 1 to 100 kbp from the nearest HOX13 peak in WT limb buds; Figure 3A). This is likely due to the large size of H3K27me3 regions, which are known to spread to surrounding regions, generating large domains. We next plotted the distribution of CRMs showing loss of, respectively, H3K27ac and H3K27me3 in Hox13−/− distal limbs (Figure 3B). Interestingly, the distance distribution shows that regions with H3K27ac loss in Hox13−/− limbs are predominantly CRMs bound by HOX13 (76% of the CRMs normally specifically active in late-distal cells and 69% of common CRMs; Figure 3B, black and green arrow, and Figure S4B). Loss of H3K27me3 in Hox13−/− limbs falls mainly into the group of CRMs normally repressed specifically in late-distal cells and located within 1–10 kb of HOX13-bound loci (Figure 3B, blue arrow, and Figure S4C). The changes in chromatin marks that occur in the absence of HOX13, at loci bound by HOX13 in WT limbs, reflect the importance of HOX13 binding for activation, maintenance, and repression of CRMs in late-distal limb cells.
Figure 3. Changes in Histone Marks from Early to Late-Distal Limbs Largely Occur at HOX13-Bound Loci.
(A) Histograms showing the distribution of the distances between the indicated subsets of H3K27ac and H3K27me3 regions and the nearest HOX13 peak summit.
(B) Same as (A) but considering H3K27ac or H3K27me3 loss in Hox13−/− distal limbs. In the absence of HOX13, H3K27ac loss occurs within 10 bp and 1 kb of the nearest HOX13 peak (green and black arrows), while depletions of H3K27me3 primarily occur within 1 kb and 10 kb of a HOX13 peak (blue arrow). The numbers on each panel show the fraction of HOX13-bound and unbound sites in each subset. For example, 41% of the late-distal specific H3K27ac are HOX13-bound CRMs, while 76% of late-distal-specific CRMs showing loss of H3K27ac in Hox13−/− limb buds are HOX13 bound.
(C) Heatmaps showing the cumulative chromatin profile across 5 kb from the center of the CRMs undergoing loss or gain of H3K27ac (left) or H3K27me3 (right) in Hox13−/−. HOX13-bound (defined according to A and B) regions are indicated by a red tag on the left side of both heatmaps.
Plotting the profile of CRMs showing chromatin state changes in the absence of HOX13 highlights two major subgroups (Figure 3C). First, a subset of early-specific CRMs shows sustained activity (H3K27ac) in late-distal mutant limb cells (Figure 3C). Consistent with HOX13 being primarily expressed at late stages, very few of these regions are bound by HOX13 (Figure 3C, red tags, and Figure S4B). Similarly, CRMs with subtle gains of activity in Hox13−/− cells at common and late-specific CRMs are infrequently bound by HOX13. Together, these data indicate that H3K27ac gains in Hox13−/− cells are most likely indirect. Second, the loss of HOX13 most significantly affects CRMs normally active in late-distal limbs, which show H3K27ac loss at constitutively marked and late-distal-specific CRMs (Figure S4A). These sites are almost invariably bound by HOX13 (Figure 3C, red tags, and Figure S4B). This loss reflects a failure to trigger and/or maintain the active state of these CRMs in the absence of HOX13. Loss of HOX13 also results in impaired repression at CRMs normally marked by H3K27me3 in late-distal limb cells (Figure S4A), and this impaired repression occurs at CRMs bound by HOX13 in WT cells (Figure 3C, red tags, and Figure S4B). Without HOX13, CRMs remain in a state that resembles their state in early limb buds (Figure 3C). Taken together, our data show that HOX13 binding at CRMs is required to control their activity, highlighting the direct role of HOX13 in effecting the regulatory program specific to late-distal limbs.
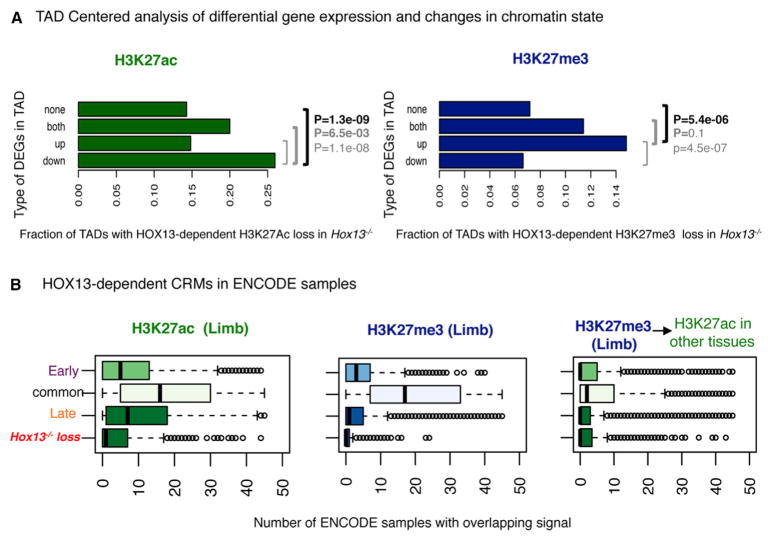
To confirm that HOX13-dependent changes in CRMs functional state occur in the vicinity of genes differentially expressed in Hox13−/− late-distal cells, we took advantage of the constraints imposed by topologically associated domains (TADs; Dixon et al., 2012; Nora et al., 2012). TAD boundaries have been shown to be mostly invariant across cell types and to favor enhancer-promoter interactions within defined chromatin territories. This framework allows us to examine the association between DEGs and the changes in chromatin state at CRMs within TADs. Based on the differential gene expression in the late-distal Hox13−/− and WT limbs, we divided TADs into four groups: (1) only upregulated genes, (2) only downregulated genes, (3) both up- and downregulated genes, and (4) no DEGs (Figure 4A). TADs with HOX13-dependent depletion of H3K27Ac at CRMs are primarily found in the group containing one or more downregulated genes (Figure 4A, left; compared to TADs with no DEGs, p = 1.3e-9, chi-square test). Similarly, TADs exhibiting significant HOX13-dependent depletion of H3K27me3 at CRMs are significantly enriched in the group containing one or more upregulated genes compared to TADs with no DEGs (Figure 4A, right; p = 5.4e-6, chi-square test). These associations suggest that a significant fraction of HOX13-dependent changes at CRMs occur within the regulatory landscape of genes with altered expression in Hox13−/− limbs and thus likely represent bona fide effectors of the HOX13-dependent transcriptional program in presumptive digit cells.
Figure 4. TAD-Centered Distribution of HOX13-Dependent CRMs and DEGs.
(A) TADs (Dixon et al., 2012) classified based on the differential expression of genes in Hox13−/− limbs. As compared to TADs showing no de-regulation in gene expression, those with downregulated gene(s) in Hox13−/− late-distal buds show significantly more HOX13-dependent loss of acetylation (p = 1.3e-9, chi-square test). Similarly, TADs with upregulated gene(s) show HOX13-dependent loss of H3K27me3 in the mutant (p = 5.4e-6).
(B) Boxplots showing the total number of non-limb ENCODE samples that show an enrichment for the indicated mark for different subsets of regions as defined throughout this study. The subset of regions with H3K27Ac or H3K27me3 loss in Hox13−/− shows the highest tissue-specificity. The rightmost panel shows the subset of genes with HOX13-dependent H3K27me3 loss in the late-distal limbs but enriched for H3K27Ac in other tissues.
To gain further insights into the specificity of the identified HOX13-dependent CRMs, we studied the extent to which these regions are enriched for H3K27ac and H3K27me3 in other tissues. We compared our data to publicly available datasets from the ENCODE Consortium (Consortium, 2012) across multiple tissues and developmental stages (Figure 4, 45 samples; see Supplemental Experimental Procedures). Regardless of the number of Hox-positive tissues in these datasets, HOX13-dependent CRMs are more specific to the limb than any other limb CRM (Figure 4). Interestingly, those regions showing H3K27me3 loss in the absence of HOX13 show putative enhancer activity (H3K27ac) in tissues other than limbs (Figure 4B, right), further suggesting that these CRMs are specifically repressed in late-distal limbs.
Collectively, our comparative genomic analyses in WT and Hox13−/− limbs show that, in the absence of HOX13, the late-distal-specific changes at CRMs are impaired while early-specific CRMs remain active. These complementary changes corroborate the idea that HOX13 binding at CRMs is essential to mediate the transition from the early- to the late-distal-specific transcriptional program that underlies digit formation.
HOX13-Dependent Regulatory Modulation and Spatial Alteration of Gene Expression
We then performed in situ hybridization to assess whether HOX13-dependent changes in chromatin state also lead to altered spatial distribution of the putative target expression. For example, Goose-coid (Gsc), Odz4 (Tenm4), and Sall1 are transcriptionally upregulated in Hox13−/− limbs (Table S2). In WT limbs, their expression at late stages is mainly restricted to the presumptive zeugopod (Figure 5A; Table S2). Accordingly, H3K27ac profiles highlight active CRMs in the vicinity of these genes in both early and late-distal WT cells (as our dissected tissue contains also zeugopod cells) (Figure 5A). In contrast, H3K27me3 profiles show repressive activity restricted to late-distal WT cells (Figure 5A) and a significant loss at these CRMs in the absence of HOX13. These H3K27me3 losses are accompanied by discrete H3K27ac gains in the surrounding regions (Figure 5A). Consistent with these changes, expression of Gsc, Odz4, and Sall1 expression expands to the distal-most limb cells in Hox13−/− limb buds. The loss of spatial restriction in Hox13−/− limb buds (Figure 5A) indicates that HOX13 binding at CRMs is required to prevent the expression of these genes in the autopod cells and provides further evidence that, in the absence of HOX13, the early-proximal regulatory program fails to terminate in late-distal cells. On the other hand, Hes1, Jag1, and Sgk1 are all downregulated in Hox13−/− buds (Table S2). Experimentally validated enhancers (Visel et al., 2007) in their vicinity recapitulate the expression of these genes in the presumptive digit territory (Figure 5B). Some of the surrounding CRMs show a HOX13-dependent loss of H3K27ac (Figure 5B) and directly overlapped the aforementioned in vivo validated enhancers (Figure 5B). Consistent with the loss of activity observed at these CRMs, expression of the corresponding genes is downregulated distally in the absence of HOX13 (Figure 5B). The near-perfect correlation between HOX13-dependent changes at CRMs in the late-distal limb cells and the spatial changes in gene expression observed at these loci strongly suggests that HOX13 regulate transcriptional output by modulating cis-regulatory activity in presumptive digit cells.
Figure 5. Hox13-Dependent Gene Regulation in Presumptive Digit Cells.
(A) UCSC genome browser views of Gsc, Odz4 and Sall1 genomic regions. (Top) ChIP-seq tracks for HOXA13 and HOXD13. (Left) Schematics of the tissue used for ChIP-seq. Below the H3K27ac and H3K27me3 profiles in Hox13−/− limb buds, green and red boxes indicate gains and losses of histone marks between WT and Hox13−/− late-distal limbs. The darkened regions spanning all tracks indicate genomic regions with no detectable change in H3K27ac or H3K27me3. Loss of spatial restriction in Hox13−/− forelimb buds (bottom). Hox13−/− fore- and hindlimbs are shown for Sall1. WT buds at slightly different developmental stages (E11.25–E11.5) are shown in order to match the morphology of mutant buds.
(B) UCSC genome browser views of Hes1, Jag1, and Sgk1 genomic regions. Track annotations are the same as in (A). Validated enhancer elements are highlighted as colored circles. VISTA identifiers (Visel et al., 2007) are indicated along with the lacZ staining of the validated enhancer and ISH on WT and Hox13−/−− limbs.
HOX13-Dependent Self-Regulatory Mechanisms at the Hox Loci
Previous expression studies in 5′ Hox mutants (Hoxa13−/−; HoxDdel(11–13)/del(11–13)) pointed to the existence of a HOX13-dependent self-regulatory process in the control of Hox expression in developing limbs (Sheth et al., 2014). Expression of HoxD genes (Hoxd3 to Hoxd11) in early limb buds and the presumptive zeugopod domain is controlled from the 3′ side of the cluster (Tarchini and Duboule, 2006; Andrey et al., 2013). In digit cells, the 3′ regulatory region is silenced, while the newly activated 5′ regulatory region controls the expression of Hoxd9 to Hoxd13 (Kmita et al., 2002; Spitz et al., 2003; Montavon et al., 2011; Andrey et al., 2013). Our ChIP-seq data show that the 3′ (early/zeugopod) and 5′ (digit) regulatory landscapes are highly occupied by HOX13 in late-distal limb cells (Figure 6A). On the 3′ side, regions specifically repressed in the late-distal WT cells show loss of H3K27me3 in the absence of HOX13 (Figure 6A, red bars). This includes an experimentally validated element (Visel et al., 2007), the activity of which is normally restricted to the proximal limb bud (Figure 6A). Moreover, in the absence of HOX13, gain of H3K27Ac is observed at CRMs that are active in early limb buds (Figure 6A). In contrast, in the absence of HOX13, H3K27ac is significantly depleted over the entire 5′ regulatory landscape, without any gain in H3K27me3 (Figure 6A). Consistent with these changes, Hoxd4, which in WT limb buds is exclusively under the control of 3′ regulation (Andrey et al., 2013), is ectopically expressed in the presumptive digit domain of Hox13−/− limbs (Figure 6A). HOX13 inactivation also results in altered spatial expression of Hoxd9-Hoxd11, with a single expression domain in the mutant instead of two distinct domains (presumptive forearm and digits) in the WT (Figure 6A). Although not part of the HoxD cluster per se, Evx2 is under the control of the 5′ regulatory region (Spitz et al., 2001) and, much like Hoxd13-exon1, is robustly expressed in WT digit cells (Figure 6A). In Hox13−/− limb buds, expression of Evx2 and Hoxd13-exon1 is significantly reduced in late-distal cells (Figure 6A; in situ hybridization [ISH]). Together, these results show that HOX13 is essential for the silencing of the 3′ regulation (early and zeugopod program) and for promoting the activity of the 5′ regulation (digit program) underlying HoxD expression. Consistent with our data, similar results regarding the role of HOX13 on the regulation of HoxD genes were recently reported (Beccari et al., 2016).
Figure 6. Impact of HOX13 Inactivation on the Regulation of the HoxA and HoxD Gene Clusters.
(A) UCSC genome browser view of the regulatory landscape surrounding the HoxD cluster (purple shadow). Black lines indicate the TAD boundaries in ES cells as defined by Dixon et al. (Dixon et al., 2012). ChIP-seq tracks for HOXA13 and HOXD13 are shown on top. Schematics for the dissected tissues used for H3K27ac and H3K27me3 profiling are shown on the left. Dark green and red boxes indicating gains and losses of histone marks between WT and Hox13−/− late-distal limb buds are shown below the Hox13−/− H3K27ac and H3K27me3 profiles. Previously validated 3′ (early/zeugopod) and 5′ (digit) enhancers (Gonzalez et al., 2007; Montavon et al., 2011; Andrey et al., 2013) are highlighted (black circles) at the top of the panel. (Bottom left) lacZ expression in the proximal limb triggered by a validated VISTA element. (Bottom right) Hoxd ISH in Hox13−/− limbs. (Note: the HoxD locus is shown with Hoxd1 on the left and Hoxd13 on the right.)
(B) UCSC genome browser view of the regulatory landscape surrounding the HoxA cluster (Berlivet et al., 2013; Woltering et al., 2014). The HoxA cluster is highlighted in purple. All tracks and annotations are the same as in (A). Previously validated enhancers on the 5′ side (Berlivet et al., 2013) are shown above the HOXA13 track as black circles. (Bottom left) LacZ expression driven by the indicated VISTA element (Visel et al., 2007). (Bottom right) ISH for Hoxa5 and Hoxa11 in Hox13−/− limbs.
Reminiscent of HoxD regulation, in digit cells, multiple enhancers on the 5′ side of the HoxA cluster control the expression of Hoxa13 (Berlivet et al., 2013; Woltering et al., 2014). Our results show that in contrast to the HoxD cluster, the regulatory landscapes on both sides of the HoxA cluster are decorated with H3K27ac in early limb buds (Figure 6B). Moreover, the digit regulatory landscape located 5′ of the HoxA cluster is largely unchanged in Hox13−/− limbs (Figure 6B). However, compared to late-distal WT cells, a significant gain in H3K27ac and corresponding decrease in H3K27me3 is observed on the 3′ side of the cluster in the absence of HOX13. This 3′ domain contains multiple previously validated enhancers (Visel et al., 2007) showing activity in the proximal limb buds (Figure 6B). However, none of the HoxA genes are transcriptionally downregulated in Hox13−/− limb buds (Table S2). Rather, expression of both Hoxa5 and Hoxa11 is gained in presumptive digit cells (Figure 6B). These results suggest that the region 3′ of the HoxA cluster could act as a regulatory domain involved in fine-tuning HoxA expression in developing limb buds. Our data also indicate that this region is largely de-commissioned in presumptive digit cells through a HOX13-dependent mechanism. In addition, we found that, in distal cells, HOX13 transcription factors are bound at all Hox clusters, with the most significant binding in the HoxA cluster and the lowest occupancy levels in the HoxB cluster (Figures S5 and S6). However, the only cluster within which changes at CRMs is observed in the absence of HOX13 is HoxC; our results show a de novo gain of H3K27ac in the area surrounding Hoxc11 (Figure S6A). We validated a putative CRM bound by HOX13 in the vicinity of Hoxc11, which triggers consistent expression in the fore- and hindlimb buds (Figure S6A). We observed a gain of Hoxc11 expression in the presumptive digit cells in Hox13−/− limb, both by ISH (Figure S6A) and RNA-seq (Figure 1D), consistent with the gain of H3K27ac observed in the region. Moreover, Hoxc11 expression in the hindlimb bud, which is restricted to the presumptive zeugopod domain in WT limb buds, shows a complete loss of spatial restriction in Hox13 mutants (Figure 6A). Altogether, these results show that at Hox loci and beyond, HOX13 mediates the transition from the early to the late-distal limb program, controlling the spatial expression patterns of target genes by regulating the activity of cis-regulatory modules.
DISCUSSION
During embryonic development, distinct TF combinations are produced in different tissues and cell types, resulting in a unique code of TFs that ultimately determines whether a given gene is turned on or off in a given tissue or cell type or at a specific time during development. In this work, we used the developing limb bud as a model system to understand how HOX13 regulate the transcriptional program in the distal-most limb cells, a process that is essential for digit formation. Indeed, genetic analyses have shown that Hoxa13 and Hoxd13 inactivation in mice causes complete digit agenesis (Fromental-Ramain et al., 1996b). However, little is known about the in vivo genomic targets of these transcription factors.
To gain insights into how HOX13 triggers the digit-specific program, we generated and analyzed genome-wide maps of (1) genomic sites bound by HOXA13 and HOXD13 in vivo (2) changes in chromatin marks and gene expression during the transition from the early to the late-distal limb program and (3) changes in chromatin marks and gene expression in late-distal limb cells lacking HOX13 function. First, our results show that the dynamic changes in gene expression occurring during the transition from the early to late-distal WT limb programs are accompanied by substantial changes in regulatory activity, as indicated by de novo activation and repression of CRMs. These results are consistent with the idea that transient regulatory activities control tissue- and cell-type-specific programming during development (Nord et al., 2013). The temporal mapping of regulatory activity itself constitutes a useful resource for the analysis of genetic regulations involved in limb development. Second, we observed that HOX13 primarily bind to putative CRMs that are associated with either activation or repression in late-distal WT limb buds. We show that loss of HOX13 results in alterations of the functional chromatin state at these CRMs in distal cells: late-distal-specific activation and repression of CRMs are impaired, while early-specific CRM activity is retained. This result indicates that in the absence of HOX13, the early limb program fails to terminate, while the implementation of the late-distal regulatory program is compromised. The strong correlation between altered chromatin state and gene expression changes in Hox13−/− limb buds reveals that HOX13-dependent establishment and/or maintenance of the functional chromatin state determines transcriptional outcomes in presumptive digit cells. Importantly, despite the presence of several other HOX proteins in distal limb cells, the activity of a large number of CRMs is impaired in Hox13−/− limbs, thereby revealing the primary role of HOX13 in mediating the regulatory switch underlying the digit patterning program.
It has previously been shown that precocious, ectopic expression of HOX13 in developing limbs has deleterious effects on the proximal limb (Goff and Tabin, 1997; Peichel et al., 1997; Williams et al., 2006). This is in agreement with the posterior prevalence model (Duboule and Morata, 1994), which proposes a functional dominance of posterior (5′) Hox over anterior (3′) ones. In this study, we used well-established histone modifications, H3K27ac deposited by p300 at active enhancers (Jin et al., 2011) and H3K27me3 deposited by the PRC2 complex (Cao et al., 2002), to analyze the active and repressed chromatin states of CRMs bound by HOX13. We observed CRMs with both active and repressed chromatin marks in late-distal WT limb buds that lose these marks in the absence of HOX13, suggesting that HOX13 may be directly involved in the recruitment of activator and repressor complexes. In this view, the suppressive effect of precocious and ectopic Hox13 expression could be the result of HOX13 ability to alter the functional states of cis-regulatory modules. HOX13 may have a pioneering-like effect and act on previously inaccessible regulatory sequences (Zaret and Carroll, 2011; Drouin, 2014). Additional studies of enhancer-chromatin state, including an assessment of DNA accessibility, will be required in order to assess a “pioneering” role for HOX13.
Mutations in HOX proteins are known to cause the transformation of one homologous structure into a different one, a phenomenon known as homeotic transformation. For example, during Drosophila development, Antennapedia (antp) controls the formation of legs, while its loss of function transforms legs into ectopic antennae (Struhl, 1981). Similarly, in mice, the Hox10 paralogs are expressed in the developing lumbar region and are important to turn off the rib program, as their inactivation leads to ectopic ribs (Wellik and Capecchi, 2003). In contrast, while loss of HOX13 result in disruption of the distal limb program, there is no transformation of digits into a more proximal limb structure. The rudimentary cartilage condensation that forms in the distal part of Hox13−/− limb buds does not have any structural identity (Fromental-Ramain et al., 1996b). Accordingly, our RNA-seq data show a mixed transcriptional identity in Hox13−/− late-distal cells that is neither completely proximal nor distal. These observations further support the idea that HOX13 plays a critical role in setting the cis-regulatory networks that underlie the digit-specific developmental program.
Variations in Hox regulation likely account for species-specific morphological differences and the evolution of structural novelties such as the tetrapod digits (Woltering and Duboule, 2010; Freitas et al., 2012; Gehrke et al., 2015; Kherdjemil et al., 2016). Our results show that in developing limbs, HOX13 binding at Hox-regulatory regions determines the spatial parameters of Hox cluster gene expression. Consistent with our results, Beccari et al. recently showed that the switch from the proximal to the distal regulation of HoxD genes requires HOX13 function (Beccari et al., 2016). At Hox loci and beyond, the unique alteration of chromatin state at CRMs and the corresponding changes in gene expression observed in this study provide a foundation for understanding the tissue-specific regulatory mechanisms driven by HOX13 proteins. Digit number variations are observed between different tetrapod species and between the fore- and hindlimbs of individual species (Cooper et al., 2014; Lopez-Rios et al., 2014; Pieretti et al., 2015). It is possible that HOX13-dependent modulation of cis-regulatory activities contributes to the variation in digit phenotypes. In this view, the core function of HOX13, i.e., the regulation of specific CRMs to terminate the early limb program and implement the digit-specific program, would be conserved, while species-specific differences in HOX13 binding would contribute to refining digit number.
EXPERIMENTAL PROCEDURES
Mice
Hoxa13+/− and Hoxd13+/− mice (Fromental-Ramain et al., 1996b; Kmita et al., 2000) were maintained in a mixed background. Noon of the day of the vaginal plug was considered as E0.5. Mice and embryos were genotyped by PCR using genomic DNA extracted from tail biopsy specimens and yolk sacs, respectively. Mice work at Institut de Recherches Cliniques de Montréal (IRCM) was reviewed and approved by the IRCM animal care committee (protocols 2012-28 and 2014-14). Transgenic animal work at Lawrence Berkeley National Laboratory (LBNL) was reviewed and approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee.
RNA Preparation, Sequencing, and Data Analysis
The indentation at the proximal border of the forming handplate at E11.5 (dotted lines in Figures 1A and 2A) was used as morphological landmark to ensure reproducible micro-dissections of distal forelimbs (dls). Micro-dissected E11.5 WT and mutant dls were stored at −80°C in QIAGEN RNAlater until genotyped. For each genotype, RNA was extracted from pools of micro-dissected tissues (n = 4) with the QIAGEN RNeasy micro kit following the manufacturer’s protocol. For each genotype, the quality of total RNA from two independent pools of limbs was analyzed on RNA 6000 Pico kit (Agilent 2100 bioanalyzer), followed by ribosomal RNA depletion using Ribo-Zero Magnetic Gold Kit (Epicenter). Sequencing was done on a HiSeq 2000 instrument with a paired-end 50 cycles protocol. Paired-end reads were aligned to the mm9 reference genome and the Mus musculus transcriptome (iGenome refGene GTF) using TopHat v2.0.13 (Kim et al., 2013). EdgeR (Robinson et al., 2010) was used to assess differential expression in E10.5 and E11.5 dl buds, as well as in E11.5 WT and Hox13−/− samples. After normalization and filtering, a stringent set and a lenient set of differentially expressed genes (DEGs) were defined (see Supplemental Experimental Procedures).
Gene Set Enrichment Analysis
GSEA (Subramanian et al., 2005) was run using genes ranked based on differential expression in E10.5 and E11.5 dl buds. DEGs between WT and Hox13−/− E11.5 late-dl buds were used as gene sets to compute the enrichments. Statistical significance was estimated empirically using 1,000 randomization of the original data.
HOXA13 and HOXD13 Antibodies
Murine HOXD13 antibody was produced following the protocol previously used to generate the HOXA13 antibody (Knosp et al., 2004). The HOXD13 immunizing peptide, consisted of a 16-amino-acid region with two additional lysine residues added to the C terminus (VGLQQNALKSSPHASL+KK) to facilitate full-length coupling to the KLH carrier protein (Knosp et al., 2004). GFP immunoprecipitation was made using lysates from E11.5 Hoxa13GFP/GFP limbs with a protein immunoprecipitation kit # 26149 (Thermo Fisher/Pierce) and a GFP antibody (AB3080, EMD/Millipore).
Chromatin Immunoprecipitation and Sequencing
Chromatin was cross-linked using formaldehyde (Lee et al., 2006) except for the HOXA13 and HOXD13 ChIPs, for which cross-linking was performed as described previously (Iraci et al., 2011). Chromatin was sonicated to obtain fragments with an average size ranging between 100 and 600 bp. Protein A and G Dynabeads (Invitrogen) were incubated for 6 hr at 4°C with 5 μg H3K27ac (ab-4729, Abcam), H3K27me3 (07-449, Millipore), or HOXA13 (Knosp et al., 2004) and our HOXD13 antibodies. Details of the immunoprecipitation can be found in the Supplemental Experimental Procedures. Sequencing was done on an Illumina HiSeq 2000 sequencer in a 50-cycle paired-end configuration.
Chip-Seq Data Analyses
Short reads were aligned to the mm9 genome using Bowtie v1.1.0 (Langmead et al., 2009)(see Supplemental Experimental Procedures and Table S1). Peak calling was performed using Model-Based Analysis for ChIP-Seq (MACS) v1.4 (Zhang et al., 2008) with matched input DNA as control. Hypergeometric Optimization of Motif EnRichment (HOMER; Heinz et al., 2010) was used to annotate enriched regions to the nearest RefSeq gene. Differential peak calling between chromatin profiles (H3K27ac and H3K27me3) of E10.5 and E11.5 late-dl buds was not performed directly due to major technical differences between the samples. We thus devised an alternative strategy to classify each peak as specific to E10.5, specific to E11.5 distal, or common to both (see Supplemental Experimental Procedures). For chromatin profiles from WT and Hox13−/− E11.5 distal buds, differential peak calling was performed using MACS v1.4, as these profiles had very comparable signal-to-noise ratios. HOMER was used to perform enrichment analysis for known transcription-factor-binding sites as well as de novo motif discovery.
Analyses of the Chromatin State of Regulatory Elements across Multiple Tissues and Stages during Mouse Development
Aligned reads from H3K27ac and H3K27me3 ChIP-seq experiments were downloaded from the ENCODE Data Coordination Center (DCC) website (https://www.encodeproject.org). Peak calling was performed using MACS v1.4 (Zhang et al., 2008). The complete list of datasets and parameters used for this analysis can be found in Supplemental Experimental Procedures.
Pre-filtering of Genomic Datasets
Every element mapping to chromosome X or Y, as well as those mapping to the mitochondrial genome, were filtered a priori. This filter was applied to both ChIP-seq peaks and genes.
Statistics
R was used to compute statistics and generate plots. Relevant p values are indicated in each panel. While a p value threshold of 1e-10 was applied to ChIPs performed on E11.5 late-distal limb buds, regions were considered enriched if showing a p value ≤ 1e-5 when looking at ChIPs performed on E10.5 limb buds. This choice was motivated by the very different signal-to-noise ratio shown by the samples from the two developmental stages considered. Several biological and technical factors could account for this difference. Most importantly, E10.5 samples were processed in different labs (see Table S1 for details) and sequencing facilities.
Supplementary Material
Supplemental Figure S1. Specificity assay for HOXA13 and HOXD13 antibody, Related to Figure 2. (A) Western blot analysis of homozygous Hoxa13GFP limb bud lysate using the HOXA13 antibody (αHOXA13) revealed a predominant 64 kDa band consistent with the predicted size of the HOXA13-GFP fusion protein as reported (Knosp et al., 2004). (B) áGFP immunoprecipitation of homozygous Hoxa13GFP limb bud lysate followed by immuno-blotting using the HOXA13 antibody (αHOXA13) confirms the predominant protein detected by áHOXA13 is HOXA13. (C) Western blot analysis of homozygous Hoxa13GFP limb bud lysate using the HOXD13 antibody (αHOXD13) revealed a predominant ~36 kDA band which is consistent with the predicted mass of the murine HOXD13 protein. (D) áGFP immunoprecipitation of homozygous Hoxa13GFP limb bud lysate followed by immuno-blotting using αHOXD13 revealed no detection of the 64 kDA HOXA13-GFP fusion protein, confirming no cross-reactivity between αHOXD13 and the HOXA13 protein.
Supplemental Figure S2. Genome-wide analysis of HOXA13 and HOXD13 binding in distal limb cells, Related to Figure 2. (A) Pie charts showing the genomic distribution of HOXA13 (left) and HOXD13 (right) ChIP-seq peaks. (B) Barplots showing the ten most enriched terms for Gene ontology Biological Process (yellow) and Mouse Phenotypes according to MGI annotation (purple) as assessed by GREAT on the complete sets of HOXA13 (left) or HOXD13 (right) peaks. (C–D) Venn diagrams showing the overlap between intergenic and intronic peaks of HOXA13, HOXD13 and p300 peaks from Visel et al,(Visel et al., 2009).
Supplemental Figure S3. Pie charts showing the genomic distribution of H3K27ac and H3K27me3 in Early and late-distal wild-type cells. Related to Figure 3.
Supplemental Figure S4. Changes in CRM activity in Hox13−/− limbs occur primarily at HOX13-bound loc Related to Figure 5. (A) Total number of H3K27ac and H3K27me3 early, common and late-distal CRMs showing significant changes in Hox13−/− limb buds. (B) Stacked barplots showing the fraction of H3K27ac HOX13-bound regions in the three specific subsets of CRMs considered. All indicates the distribution of the entire subset of regions and is considered as the random expectation for any subset drawn from this set. Compared to this group, a significant, strong enrichment for late-distal specific acetylations was identified among the regions undergoing a loss of the mark in the Hox13−/− limb buds (p ≪ 1e-10, Chi-squared test). The actual numbers of regions are indicated in the plot. (C) Same as (B) but for the fraction of H3K27me3 HOX13-bound regions. A strong, significant enrichment for late-distal specific methylations was observed for those regions undergoing a loss of the mark in Hox13−/− limb buds, as compared to the entire set of HOX13-bound regions (p ≪ 1e-10, Chi-squared test).
Supplemental Figure S5. Mapping of the HOX13-bound loci and analysis of CRMs activity at the HoxA and HoxB clusters. Related to Figure 6. UCSC genome browser snapshots of the HoxA (A) and HoxB (B) gene clusters. HOXA13 and HOXD13 profiles are shown on top. In both cases, the Hox cluster itself does not show a significant gain or loss of H3K27ac and/or H3K27me3 at intergenic and intronic regions in Hox13−/− limb buds (note that regions overlapping TSS +/− 2.5 kbp are excluded from the analysis; a highly conservative threshold, p-value < 1e-10, was used for differential gain and loss of chromatin marks).
Supplemental Figure S6. Mapping of the HOX13-bound loci and analysis of CRMs activity at the HoxC and HoxD clusters. Related to Figure 2. UCSC genome browser snapshots of the HoxC (A) and HoxD (B) gene clusters. HOXA13 and HOXD13 profiles are shown on top. The HoxD cluster itself does not show a significant gain or loss of H3K27ac and/or H3K27me3 at intergenic and intronic regions within the cluster (note that regions overlapping TSS +/− 2.5 kbp are excluded from the analysis; a highly conservative threshold, p-value < 1e-10, was used for differential gain and loss of chromatin marks). On the other hand, an ectopic gain of activity (H3K27ac) is observed at regions within the HoxC cluster in Hox13−/− limb buds (black arrow). A validated enhancer located near Hoxc11 (bottom, left) resembles the expression of Hoxc11 in the Wt limb buds by ISH (bottom, right). The loss of spatial restriction of Hoxc11 is observed in fore- and hindlimb buds of Hox13−/− embryos as shown by ISH.
Table S1. Related to Figures 1 and 2. The high-throughput datasets referred to in this study are comprehensively listed. Previously published datasets are highlighted in grey.
Table S2. Related to Figure 1. For each gene, the table is showing the results for the analysis of differential expression between E11.5 late-distal and e10.5 proximal limb buds (blue columns), and between E11.5 late-distal Hox13−/− and wild-type limb buds (orange-red columns). For each comparison, this information is provided (edgeR): 1) log-fold-change; 2) p-value; 3) Benjamini-Hochberg q-value. For each comparison, four TRUE/FALSE flags are then indicating if the gene is significantly up- or down- regulated, using either a set of lenient or a set of stringent thresholds (see Materials and Methods).
Genome-wide HOXA13 and HOXD13 bound regions (as identified by ChIP-seq) are listed. For each region, mm9 coordinates are provided along with the MACS score (−10*log10(p-value)) and information about the nearest transcriptional start site.
Genome-wide maps for early-, late-distal and common H3K27ac and H3K27me3 regions. TSS-proximal (within +/− 2.5 kbps of RefSeq gene TSSs) and TSS-distal regions are provided separately. For each region, the score from MACS (−10*log10(p-value)) is indicated for: 1) early (E10.5) sample; 2) late-distal (E11.5) sample; 3) significant increases in the enrichment for the mark in Hox13−/− limb buds; 4) significant decreases in the enrichment for the mark in Hox13−/− limb buds. Distance to the nearest HOX13 bound site (as identified by ChIP-seq) and corresponding MACS score are indicated.
Highlights.
The early transcription program persists in late-distal limb cells lacking HOX13
Expression of late-distal-specific genes largely relies on HOX13 function
HOX13 regulates H3K27 modification at regulatory elements
HOX13 coordinates the early to the late-distal program transition in the limb bud
Acknowledgments
We thank P. Chambon and D. Duboule for providing the mutant mice lines. This work was supported by the Canada Research Chair Program (grant RCHS0192) and the Canadian Institutes for Health Research (CIHR; grants CIHR MOP-115127 and 126110 to M.K., CIHR MOP-123213 and CEERC EPI-120608 to J.D.) and Shriners Hospital (research grant 85400 to H.S.S.). A.V. was supported by National Institutes of Health grants R24HL123879, U01DE024427, R01HG003988, U54HG006997, and UM1HL098166. R.S. was supported by a CIHR postdoctoral fellowship. I.B. is supported by NIH grants to A.V. D.L. was supported by a fellowship from the Fonds de recherche du Québec en Santé. M.O. was supported by a Swiss National Science Foundation (SNSF) fellowship. S.N. is supported by CIHR grants to J.D. Research conducted at the E.O. Lawrence Berkeley National Laboratory was performed under Department of Energy contract DE-AC02-05CH11231 (University of California).
Footnotes
ACCESSION NUMBERS
The accession number for the raw datasets reported in this paper is GEO: GSE81358.
Supplemental Information includes Supplemental Experimental Procedures, six figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.11.039.
AUTHOR CONTRIBUTIONS
R.S., J.D., and M.K. designed the Project. R.S. performed the experiments. I.B. performed the bioinformatics analysis. D.L. performed the initial bioinformatics analysis. S.N. generated the e10.5 datasets. M.O. generated the transient transgenic embryos. H.L.C. and H.S.S. generated the Hoxd13 antibody. R.S. wrote the paper with I.B., S.N., and M.K. with inputs from all authors, especially A.V. and J.D.
References
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- Beccari L, Yakushiji-Kaminatsui N, Woltering JM, Necsulea A, Lonfat N, Rodríguez-Carballo E, Mascrez B, Yamamoto S, Kuroiwa A, Duboule D. A role for HOX13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev. 2016;30:1172–1186. doi: 10.1101/gad.281055.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 2013;9:e1004018. doi: 10.1371/journal.pgen.1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carpenter EM, Goddard JM, Davis AP, Nguyen TP, Capecchi MR. Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development. 1997;124:4505–4514. doi: 10.1242/dev.124.22.4505. [DOI] [PubMed] [Google Scholar]
- Consortium T.E.P.; ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Sears KE, Uygur A, Maier J, Baczkowski KS, Brosnahan M, Antczak D, Skidmore JA, Tabin CJ. Patterning and post-patterning modes of evolutionary digit loss in mammals. Nature. 2014;511:41–45. doi: 10.1038/nature13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Capecchi MR. A mutational analysis of the 5′ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development. 1996;122:1175–1185. doi: 10.1242/dev.122.4.1175. [DOI] [PubMed] [Google Scholar]
- Delgado I, Torres M. Gradients, waves and timers, an overview of limb patterning models. Semin Cell Dev Biol. 2016;49:109–115. doi: 10.1016/j.semcdb.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin J. Minireview: pioneer transcription factors in cell fate specification. Mol Endocrinol. 2014;28:989–998. doi: 10.1210/me.2014-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Favier B, Rijli FM, Fromental-Ramain C, Fraulob V, Chambon P, Dollé P. Functional cooperation between the non-paralogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development. 1996;122:449–460. doi: 10.1242/dev.122.2.449. [DOI] [PubMed] [Google Scholar]
- Freitas R, Gómez-Marín C, Wilson JM, Casares F, Gómez-Skarmeta JL. Hoxd13 contribution to the evolution of vertebrate appendages. Dev Cell. 2012;23:1219–1229. doi: 10.1016/j.devcel.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Dolle P, Chambon P. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development. 1996a;122:461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dollé P, Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996b;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- Gehrke AR, Schneider I, de la Calle-Mustienes E, Tena JJ, Gomez-Marin C, Chandran M, Nakamura T, Braasch I, Postlethwait JH, Gómez-Skarmeta JL, Shubin NH. Deep conservation of wrist and digit enhancers in fish. Proc Natl Acad Sci USA. 2015;112:803–808. doi: 10.1073/pnas.1420208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff DJ, Tabin CJ. Analysis of Hoxd-13 and Hoxd-11 misexpression in chick limb buds reveals that Hox genes affect both bone condensation and growth. Development. 1997;124:627–636. doi: 10.1242/dev.124.3.627. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Duboule D, Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev Biol. 2007;306:847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraci N, Diolaiti D, Papa A, Porro A, Valli E, Gherardi S, Herold S, Eilers M, Bernardoni R, Della Valle G, Perini G. A SP1/MIZ1/MYCN repression complex recruits HDAC1 at the TRKA and p75NTR promoters and affects neuroblastoma malignancy by inhibiting the cell response to NGF. Cancer Res. 2011;71:404–412. doi: 10.1158/0008-5472.CAN-10-2627. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kherdjemil Y, Lalonde RL, Sheth R, Dumouchel A, de Martino G, Pineault KM, Wellik DM, Stadler HS, Akimenko MA, Kmita M. Evolution of Hoxa11 regulation in vertebrates is linked to the pentadactyl state. Nature. 2016;539:89–92. doi: 10.1038/nature19813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M, Kondo T, Duboule D. Targeted inversion of a polar silencer within the HoxD complex reallocates domains of enhancer sharing. Nat Genet. 2000;26:451–454. doi: 10.1038/82593. [DOI] [PubMed] [Google Scholar]
- Kmita M, Fraudeau N, Hérault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420:145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- Knosp WM, Scott V, Bächinger HP, Stadler HS. HOXA13 regulates the expression of bone morphogenetic proteins 2 and 7 to control distal limb morphogenesis. Development. 2004;131:4581–4592. doi: 10.1242/dev.01327. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski JP, Du F, Zhang S, Powell MB, Falkenstein KN, Ji H, Vokes SA. Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev Biol. 2015;406:92–103. doi: 10.1016/j.ydbio.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J, Duchesne A, Speziale D, Andrey G, Peterson KA, Germann P, Unal E, Liu J, Floriot S, Barbey S, et al. Attenuated sensing of SHH by Ptch1 underlies evolution of bovine limbs. Nature. 2014;511:46–51. doi: 10.1038/nature13289. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL, Prabhakaran B, Vogt TF. The mouse Ulnaless mutation deregulates posterior HoxD gene expression and alters appendicular patterning. Development. 1997;124:3481–3492. doi: 10.1242/dev.124.18.3481. [DOI] [PubMed] [Google Scholar]
- Pieretti J, Gehrke AR, Schneider I, Adachi N, Nakamura T, Shubin NH. Organogenesis in deep time: A problem in genomics, development, and paleontology. Proc Natl Acad Sci USA. 2015;112:4871–4876. doi: 10.1073/pnas.1403665112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth R, Bastida MF, Kmita M, Ros M. “Self-regulation,” a new facet of Hox genes’ function. Dev Dyn. 2014;243:182–191. doi: 10.1002/dvdy.24019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Peichel C, Vogt TF, Duboule D, Zákány J. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 2001;15:2209–2214. doi: 10.1101/gad.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Struhl G. A homoeotic mutation transforming leg to antenna in Drosophila. Nature. 1981;292:635–638. doi: 10.1038/292635a0. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin C, Wolpert L. Rethinking the proximodistal axis of the vertebrate limb in the molecular era. Genes Dev. 2007;21:1433–1442. doi: 10.1101/gad.1547407. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Tickle C. How the embryo makes a limb: determination, polarity and identity. J Anat. 2015;227:418–430. doi: 10.1111/joa.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Williams ME, Lehoczky JA, Innis JW. A group 13 homeodomain is neither necessary nor sufficient for posterior prevalence in the mouse limb. Dev Biol. 2006;297:493–507. doi: 10.1016/j.ydbio.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Duboule D. The origin of digits: expression patterns versus regulatory mechanisms. Dev Cell. 2010;18:526–532. doi: 10.1016/j.devcel.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Noordermeer D, Leleu M, Duboule D. Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol. 2014;12:e1001773. doi: 10.1371/journal.pbio.1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Du Y, Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Specificity assay for HOXA13 and HOXD13 antibody, Related to Figure 2. (A) Western blot analysis of homozygous Hoxa13GFP limb bud lysate using the HOXA13 antibody (αHOXA13) revealed a predominant 64 kDa band consistent with the predicted size of the HOXA13-GFP fusion protein as reported (Knosp et al., 2004). (B) áGFP immunoprecipitation of homozygous Hoxa13GFP limb bud lysate followed by immuno-blotting using the HOXA13 antibody (αHOXA13) confirms the predominant protein detected by áHOXA13 is HOXA13. (C) Western blot analysis of homozygous Hoxa13GFP limb bud lysate using the HOXD13 antibody (αHOXD13) revealed a predominant ~36 kDA band which is consistent with the predicted mass of the murine HOXD13 protein. (D) áGFP immunoprecipitation of homozygous Hoxa13GFP limb bud lysate followed by immuno-blotting using αHOXD13 revealed no detection of the 64 kDA HOXA13-GFP fusion protein, confirming no cross-reactivity between αHOXD13 and the HOXA13 protein.
Supplemental Figure S2. Genome-wide analysis of HOXA13 and HOXD13 binding in distal limb cells, Related to Figure 2. (A) Pie charts showing the genomic distribution of HOXA13 (left) and HOXD13 (right) ChIP-seq peaks. (B) Barplots showing the ten most enriched terms for Gene ontology Biological Process (yellow) and Mouse Phenotypes according to MGI annotation (purple) as assessed by GREAT on the complete sets of HOXA13 (left) or HOXD13 (right) peaks. (C–D) Venn diagrams showing the overlap between intergenic and intronic peaks of HOXA13, HOXD13 and p300 peaks from Visel et al,(Visel et al., 2009).
Supplemental Figure S3. Pie charts showing the genomic distribution of H3K27ac and H3K27me3 in Early and late-distal wild-type cells. Related to Figure 3.
Supplemental Figure S4. Changes in CRM activity in Hox13−/− limbs occur primarily at HOX13-bound loc Related to Figure 5. (A) Total number of H3K27ac and H3K27me3 early, common and late-distal CRMs showing significant changes in Hox13−/− limb buds. (B) Stacked barplots showing the fraction of H3K27ac HOX13-bound regions in the three specific subsets of CRMs considered. All indicates the distribution of the entire subset of regions and is considered as the random expectation for any subset drawn from this set. Compared to this group, a significant, strong enrichment for late-distal specific acetylations was identified among the regions undergoing a loss of the mark in the Hox13−/− limb buds (p ≪ 1e-10, Chi-squared test). The actual numbers of regions are indicated in the plot. (C) Same as (B) but for the fraction of H3K27me3 HOX13-bound regions. A strong, significant enrichment for late-distal specific methylations was observed for those regions undergoing a loss of the mark in Hox13−/− limb buds, as compared to the entire set of HOX13-bound regions (p ≪ 1e-10, Chi-squared test).
Supplemental Figure S5. Mapping of the HOX13-bound loci and analysis of CRMs activity at the HoxA and HoxB clusters. Related to Figure 6. UCSC genome browser snapshots of the HoxA (A) and HoxB (B) gene clusters. HOXA13 and HOXD13 profiles are shown on top. In both cases, the Hox cluster itself does not show a significant gain or loss of H3K27ac and/or H3K27me3 at intergenic and intronic regions in Hox13−/− limb buds (note that regions overlapping TSS +/− 2.5 kbp are excluded from the analysis; a highly conservative threshold, p-value < 1e-10, was used for differential gain and loss of chromatin marks).
Supplemental Figure S6. Mapping of the HOX13-bound loci and analysis of CRMs activity at the HoxC and HoxD clusters. Related to Figure 2. UCSC genome browser snapshots of the HoxC (A) and HoxD (B) gene clusters. HOXA13 and HOXD13 profiles are shown on top. The HoxD cluster itself does not show a significant gain or loss of H3K27ac and/or H3K27me3 at intergenic and intronic regions within the cluster (note that regions overlapping TSS +/− 2.5 kbp are excluded from the analysis; a highly conservative threshold, p-value < 1e-10, was used for differential gain and loss of chromatin marks). On the other hand, an ectopic gain of activity (H3K27ac) is observed at regions within the HoxC cluster in Hox13−/− limb buds (black arrow). A validated enhancer located near Hoxc11 (bottom, left) resembles the expression of Hoxc11 in the Wt limb buds by ISH (bottom, right). The loss of spatial restriction of Hoxc11 is observed in fore- and hindlimb buds of Hox13−/− embryos as shown by ISH.
Table S1. Related to Figures 1 and 2. The high-throughput datasets referred to in this study are comprehensively listed. Previously published datasets are highlighted in grey.
Table S2. Related to Figure 1. For each gene, the table is showing the results for the analysis of differential expression between E11.5 late-distal and e10.5 proximal limb buds (blue columns), and between E11.5 late-distal Hox13−/− and wild-type limb buds (orange-red columns). For each comparison, this information is provided (edgeR): 1) log-fold-change; 2) p-value; 3) Benjamini-Hochberg q-value. For each comparison, four TRUE/FALSE flags are then indicating if the gene is significantly up- or down- regulated, using either a set of lenient or a set of stringent thresholds (see Materials and Methods).
Genome-wide HOXA13 and HOXD13 bound regions (as identified by ChIP-seq) are listed. For each region, mm9 coordinates are provided along with the MACS score (−10*log10(p-value)) and information about the nearest transcriptional start site.
Genome-wide maps for early-, late-distal and common H3K27ac and H3K27me3 regions. TSS-proximal (within +/− 2.5 kbps of RefSeq gene TSSs) and TSS-distal regions are provided separately. For each region, the score from MACS (−10*log10(p-value)) is indicated for: 1) early (E10.5) sample; 2) late-distal (E11.5) sample; 3) significant increases in the enrichment for the mark in Hox13−/− limb buds; 4) significant decreases in the enrichment for the mark in Hox13−/− limb buds. Distance to the nearest HOX13 bound site (as identified by ChIP-seq) and corresponding MACS score are indicated.