Abstract
Bioorthogonal catalysis broadens the functional possibilities of intracellular chemistry. Effective delivery and regulation of synthetic catalytic systems in cells is challenging due to the complex intracellular environment and catalyst instability. Here, we report the fabrication of protein-sized bioorthogonal nanozymes through the encapsulation of hydrophobic transition metal catalysts into the monolayer of water-soluble gold nanoparticles. The activity of these catalysts can be reversibly controlled by binding a supramolecular cucurbit[7]uril ‘gate-keeper’ onto the monolayer surface, providing a biomimetic control mechanism that mimics the allosteric regulation of enzymes. The potential of this gated nanozyme for use in imaging and therapeutic applications was demonstrated through triggered cleavage of allylcarbamates for pro-fluorophore activation and propargyl groups for prodrug activation inside living cells.
TOC image
Regulation of bioorthogonal catalysis in living systems is challenging because of the complex intracellular environment. Now, the activity of protein-sized bioorthogonal nanozymes has been regulated by binding a supramolecular cucurbit[7]uril ‘gate-keeper’ onto the monolayer surface, providing controlled activation of pro-fluorophores and prodrugs inside living cells for imaging and therapeutic applications.
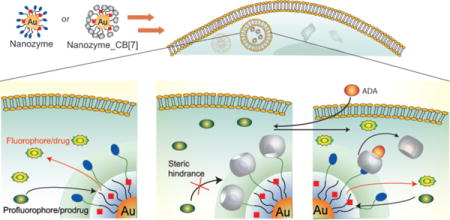
Bioorthogonal chemistry1–3 is a promising strategy for the intracellular generation of molecules for therapeutic4 and imaging applications5,6 unattainable through naturally occurring bioprocesses7,8. Transition metal catalysts (TMCs) are excellent candidates for use in bioorthogonal processes9–11, rapidly catalyzing transformations that cannot be performed via enzymatic processes12–15. However, the application of TMC-mediated reactions in living cells is challenging due to issues of biocompatibility, water solubility, catalyst stability, and rapid efflux of catalysts from living cells12,13.
Loading of TMCs into nanomaterial scaffolds can be used to provide water solubility and a protective environment for TMCs. Bradley et al.13 and Unciti-Broceta et al.16 used palladium-catalyst loaded polystyrene beads to catalyze reactions such as Suzuki-Miyaura coupling and alkylcarbamate/N-propargyl cleavage inside and outside cells, respectively. The particles used in these studies, however, were far larger than normal proteins, creating potential interference in cellular processes. Additionally, these particles did not provide the capability of mimicking allosteric regulation of enzymes, a key component in cellular homeostasis17. The integration of biomimetic size and controlled response into a bioorthogonal catalysis platform would provide new avenues for both therapeutics and integrated biological/abiotic cellular systems.
We have developed a family of gold nanoparticles (AuNPs) based on ~2 nm core size18–20 that feature biomimetic size, possess diverse functional properties21,22, and are efficiently transported into cells23,24. We report here the use of this AuNP structural motif to encapsulate25 hydrophobic TMCs, providing NP_Ru (Fig. 1) or NP_Pd. The resulting nanozymes26–30 feature surface moieties that can be reversibly functionalized using host-guest chemistry31,32 to provide NP_Ru_CB[7] or NP_Pd_CB[7]. Complexation of the monolayer terminal functionalities by cucurbit[7]uril (CB[7])33–35 in this system blocks access to the catalytic site, resulting in essentially complete inhibition of catalytic activity. The gatekeeper molecules36,37 can then be released from the AuNPs using competitive guests38, restoring catalytic activity (Fig. 1). The efficacy of this system was demonstrated in solution and in cells through two applications: 1) the gated generation of a fluorophore through deallylation of a non-fluorescent precursor and 2) the gated activation of a prodrug by cleaving the propargyl functionality that has been introduced to block the active side of original drug. Such gated control of catalysis has not been demonstrated in cells to date and is important because it allows the potential for multiple useful capabilities, such as switching on therapeutics at target tissues and regulation of activity to maintain homeostasis for long-term therapeutics. To date, supramolecular machines based on gating strategy were designed to entrap the guest molecules in the pore reservoir of silica nanoparticles and their release was studied mostly in test tubes39 or intracellularly37 in a few studies however, no gated control of substrate activation was demonstrated inside cells.
Figure 1. Bioorthogonal nanozyme design and supramolecular regulation of intracellular catalysis.
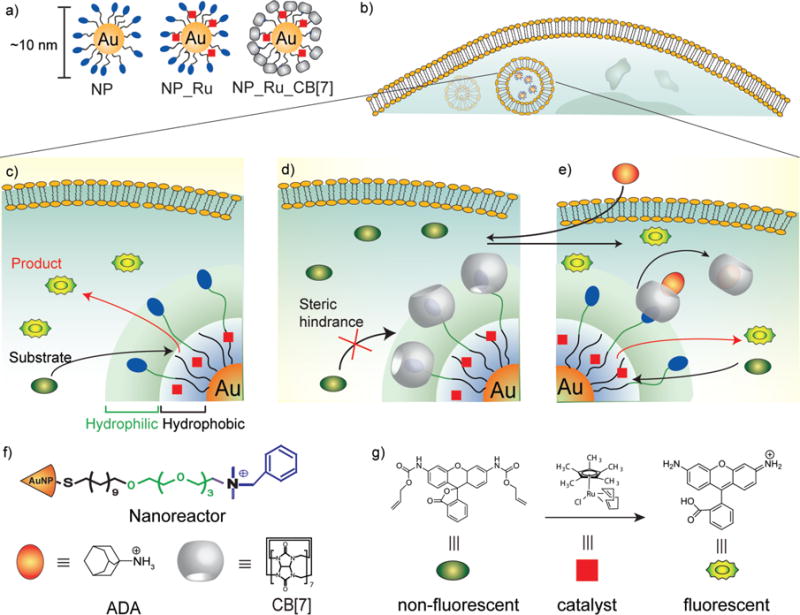
a, AuNP, catalyst-embedded AuNP, and CB[7]-capped catalyst-embedded AuNPs used in study. b, Endosomal uptake of nanozymes. c, Intracellular catalysis with NP_Ru converting substrate into product. d, CB[7] complexation with the ligand headgroup to provide NP_Ru_CB[7] inhibits catalyst activity. e, Nanozyme activity is restored through addition of the competitive guest 1-adamantylamine (ADA). f, Structures of the NP platform with the surface ligand bearing a dimethylbenzylammonium headgroup, CB[7] gatekeeper, and ADA, a competitive guest molecule for CB[7] binding. f, Structures of the pro-fluorescent substrate (rhodamine 110 derivative), fluorescent product (rhodamine 110) obtained after catalysis, and embedded catalyst for allylcarbamate cleavage.
Results and discussion
Design and synthesis of nanozymes
We used AuNPs with core diameters of ~2 nm as the scaffold for our catalysts, with the goal of creating protein-sized systems with functional monolayers. The monolayer coverage of the NP nanoreactor scaffold features three crucial components: 1) a hydrophobic alkane segment for catalyst encapsulation, 2) a tetra(ethylene glycol) unit to provide biocompatibility40, and 3) a dimethylbenzylammonium group to impart water solubility and bind the nontoxic CB[7] gatekeeper41,42 (Fig. 1f).
We chose ruthenium-catalyzed deallylation43,44 as a model bioorthogonal process to regenerate the fluorescence from an allylcarbamate caged fluorophore in solution and inside cells. To this end, we immobilized [Cp*Ru(cod)Cl] (Cp* = pentamethylcyclopentadienyl, cod = 1,5-cyclooctadiene) in the hydrophobic portion of the AuNP monolayer to provide NP_Ru. Transmission electron microscopy (TEM) images of the AuNPs before and after encapsulation of the catalyst indicated that no aggregation or decomposition of AuNP structure occurred after encapsulation (Supplementary Fig. S9), a result that was also confirmed by dynamic light scattering (Supplementary Fig. S10). The amount of ruthenium catalyst relative to AuNP was quantified using inductively coupled plasma mass spectrometry (ICP-MS, Supplementary Fig. S11)45 of 101Ru relative to 197Au, with 42 ± 3 catalyst molecules encapsulated per AuNP.
Catalytic efficacy of NP_Ru nanozymes in solution
The catalytic efficacy of the nanozymes in solution was assessed using the allylcarbamate cleavage of bis-N,N′-allyloxycarbonyl rhodamine 110 (Fig. 1g). Fluorescence started to increase immediately for NP_Ru after the addition of substrate, while no significant change in fluorescence was observed for NP_Ru_CB[7] (Supplementary Fig. S12). After 24 h bright fluorescence was observed using the NP_Ru nanozymes (Fig. 2d). However, only minimal fluorescence was observed for the NP_Ru_CB[7], which originated from the background of caged fluorophore as observed at 0 h (Supplementary Fig. S13). As expected no reaction occurred with the control particle NP that lacked embedded catalysts (Supplementary Fig. S14), as the [Cp*Ru(cod)Cl] was insoluble in water, preventing study of the catalyst alone. Additionally, fluorogenesis of NP_Ru and NP_Ru_CB[7] taken after 5 days confirmed the long term stability of the gated catalysis system in solution (Supplementary Fig. S15).
Figure 2. Catalytic activity of nanozymes in solution.
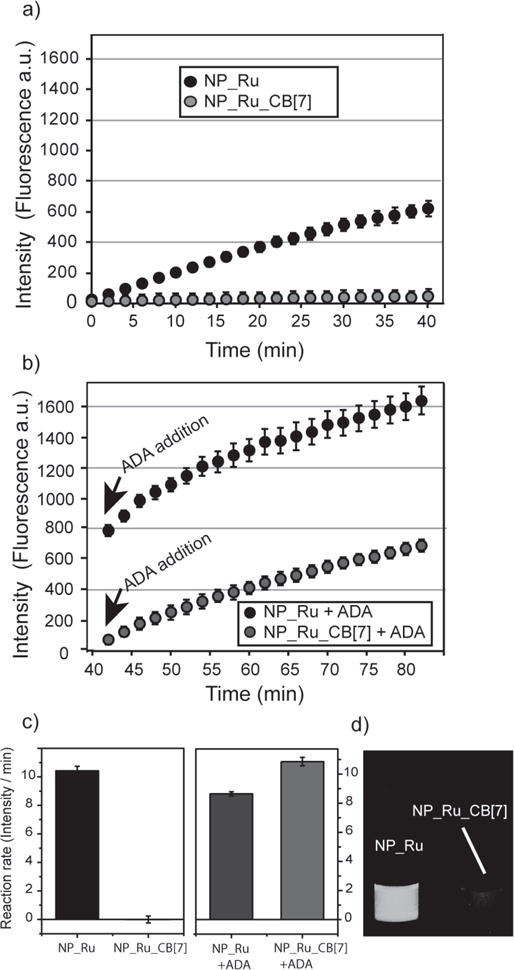
a, Fluorescence was generated by NP_Ru after the cleavage of profluorophore bis-Alloc-rhodamine 110, while NP_Ru_CB[7] showed no significant change. b, After adding ADA, catalytic activity of NP_Ru_CB[7] was restored and no significant effect was observed for the activity of NP_Ru. c, The reaction rates of NP_Ru and NP_Ru_CB[7] before and after adding ADA showing catalytic activity for NP_Ru_CB[7] was fully recovered after the addition of ADA. The reaction rate experiments were performed in triplicate. Error bars represent standard deviations of these measurements. d, Photo of the reaction mixtures in water with NP_Ru and NP_Ru_CB[7] under UV light.
Supramolecular control of catalysis through CB[7] complexation
With catalytic efficacy of the nanozymes established, we next explored CB[7] complexation of the ligand headgroups to act as the gatekeepers in NP_Ru_CB[7]. Isothermal titration calorimetry (ITC)46 indicated that 60 ± 5 CB[7] molecules bound per NP, with KD = 11.3 ± 2.4 μM (Supplementary Fig. S16). Complexation with CB[7] effectively shut down catalysis in NP_Ru_CB[7], as minimal fluorogenesis was observed with nanozymes after addition of CB[7] (Fig. 2d). Kinetic studies (Fig. 2a, c) verified this observation, indicating essentially complete inhibition of catalysis by CB[7] complexation. This inhibition was reversible: after the addition of 1-adamantylamine (ADA), a competitive guest molecule for CB[7], catalytic activity was completely restored (Fig. 2b, c). Control studies of the free catalyst in an acetone/water solution showed that catalyst efficiency was unaffected by either CB[7] or ADA (Supplementary Fig. S17), demonstrating that particle-CB[7] gating controls the catalytic process.
Kinetic analysis of the NP_Ru nanozymes using Lineweaver–Burk analysis
The analogy between controlled regulation of the nanozymes and their enzyme counterparts was explored through kinetic analysis of the nanozymes using Lineweaver–Burk analysis (LBA)47, 48. These studies indicate that CB[7] complexation results in competitive inhibition of reactor activity (Fig. 3 and Supplementary Fig. S18), presumably by blocking access to the [Cp*Ru(cod)Cl] “active site”. Little change in activity was observed for CB[7]:NPs ratios greater than ~80:1, consistent with the ITC results (Supplementary Fig. S19 and S20). Likewise, the value of Ki from the LBA (11.4 ± 4.3 μM) is essentially identical to the affinity obtained by ITC (11.3 ± 2.4 μM). Taken together, these studies demonstrate the direct correlation between CB[7]-AuNP equilibrium binding processes and kinetic behavior of the nanozyme.
Figure 3. Lineweaver-Burk plot showing competitive binding of CB[7] to nanozyme.
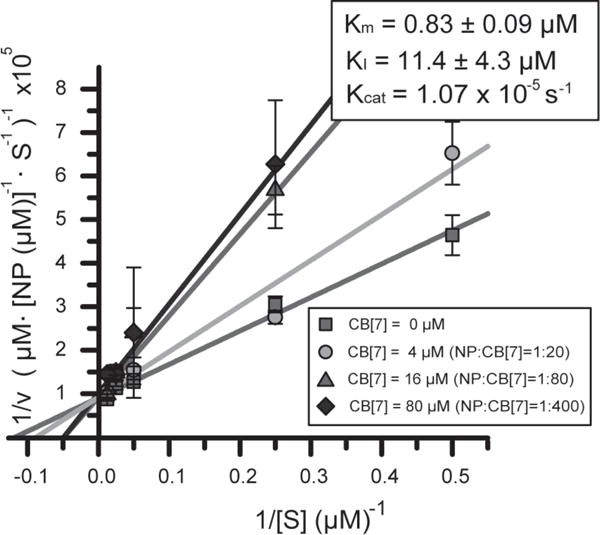
Kinetic studies of NP_Ru and NP_Ru_CB[7] in sodium phosphate buffer (5 mM, pH 7.4) indicate that CB[7] inhibits catalyst activity through a competitive inhibition mechanism, with CB[7] affinity and stoichiometry consistent with ITC binding studies. Numbers calculated on a per particle basis. Kinetic experiments with each nanozyme were repeated in triplicate. Error bars represent standard deviations of these measurements.
Catalytic efficacy of NP_Pd nanozymes in solution
To demonstrate the versatility of this catalysis platform, we chose a second catalyst: hydrophobic palladium catalyst: (1,1′- Bis(diphenylphosphino) ferrocene)palladium(II)dichloride. This palladium catalyst was encapsulated in the hydrophobic portion of the AuNP monolayer to provide NP_Pd. The amount of palladium catalyst relative to AuNP was quantified using ICP-MS of 106Pd relative to 197Au. It was calculated that 32 ± 1 catalyst molecules encapsulated per AuNP (Supplementary Fig. S21).
Similar to NP_Ru nanozyme, NP_Pd can also cleave the allylcarbamate of bis-N,N′-allyloxycarbonyl rhodamine 11013. After 6 h, fluorescence was generated for the NP_Pd (Supplementary Fig. S22), however; NP_Pd_CB[7] showed only a slight background fluorescence of caged fluorophore (Supplementary Fig. S22). Kinetic studies for NP_Pd and NP_Pd_CB[7] indicated the inhibition of catalysis by CB[7] complexation. Followed by the addition of ADA, the catalytic activity of nanozyme was restored (Supplementary Fig. S23). These findings supported that gated catalysis is efficiently working for different catalyst systems.
Supramolecular regulation of catalysis inside living cells
Having characterized the activity and gating of nanozyme catalysis in solution, we next studied the intracellular behavior of these nanozymes using HeLa cells. The cellular uptake of the nanozymes was quantified by tracking [197Au] using ICP-MS, with NP_Ru_CB[7] particles demonstrating a slightly more efficient (1.7-fold greater) uptake than uncomplexed NP-Ru (Supplementary Fig. S24). Significantly, no toxicity was observed at the concentrations used for our studies with either NP or NP_Ru (Supplementary Fig. S25).
We then probed the catalytic activity of the nanozymes inside the living cells. HeLa cells were incubated with the nanozyme for 24 h in serum-containing media and then washed multiple times to remove adsorbed particles on the cell surface. Fresh media containing the substrate was added, followed by 24 h incubation and washing. As shown in Fig. 4a, flow cytometry indicated that there was a significant increase in fluorescence with NP_Ru relative to the control NP. Confocal microscopy (Fig. 4d) showed that the cells treated with NP_Ru had bright punctate fluorescence. This fluorescence co-localized with LysoTracker® (Supplementary Fig. S26), indicating that the deallylation reaction occurred in the endosomes and cleaved fluorophore stayed in the endosomes. This outcome is expected given the endosomal uptake pathway49,50 previously observed for similar nanoparticles, coupled with the limited membrane permeability of the cleaved dye51.
Figure 4. Triggered allylcarbamate cleavage in living cells using gated nanozymes.
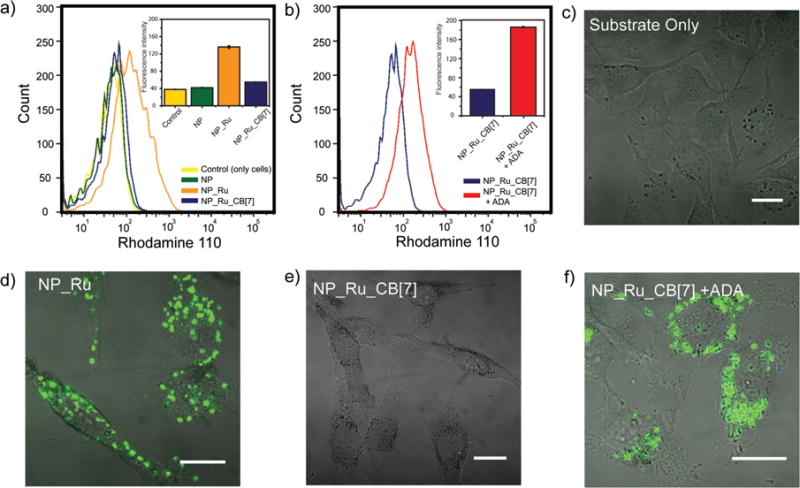
a, Flow cytometry of NP_Ru, NP_Ru_CB[7], and controls (only cell and NP) revealing NP_Ru showed significant increase in fluorescence while NP_Ru_CB[7] was completely inhibited. b, Addition of ADA to NP_Ru_CB[7] treated cells recovered the catalysis and resulted in increase in fluorescence. c–f, Confocal microscopy images of HeLa cells showing the supramolecularly regulated intracellular chemical reactions. A punctate fluorescence was observed for NP_Ru and NP_Ru_CB[7] + ADA treated cells as the indication of catalysis while no fluorescence was obtained for only substrate and NP_Ru_CB[7] (scale bars = 10 μm).
We next investigated the intracellular gating of the nanozyme by CB[7] complexation. Flow cytometry showed that the NP_Ru_CB[7] particles were completely inhibited, indicating intracellular stability of the complexes (Fig. 4a, e). Treatment with ADA (400 μM) restored nanozyme activity, with even higher activity observed in the cells after treatment with ADA than was observed with NP_Ru (Fig. 4b). This increased catalytic efficiency is echoed in the micrograph (Fig. 4f) and potentially arises from enhanced protection of the catalyst by the CB[7] coverage in serum and inside the cell.
We further tested the control of gated catalysis through the treatment of cells with free CB[7]. First, HeLa cells were incubated with the NP_Ru for 24 h and then after multiple washings, free CB[7] in serum-containing media was treated to cells for 24 h before the addition of the substrate. A significant decrease in the fluorescence was observed indicating the complexation of CB[7] with NP_Ru and blocking access to the catalytic site (Supplementary Fig. S27).
Palladium catalyst embedded nanozymes were likewise used for intracellular controlled activation of the caged fluorophore. NP_Pd effectively performed the intracellular catalysis, while as expected the gated NP_Pd_CB[7] did not. ADA addition into cells that were previously incubated with NP_Pd_CB[7] resulted in the recovery of intracellular catalysis (Supplementary Fig. S28).
Intracellular prodrug activation using NP_Pd nanozymes
Bioorthogonal activation of prodrugs using TMC-loaded nanomaterials was recently demonstrated by Unciti-Broceta and co-workers16 Although prodrug activation was successfully achieved, no gated control over this activation was shown. We have bioorthogonally demonstrated gated-activation of a prodrug inside cells using NP_Pd/NP_Pd_CB[7]. 5-fluorouracil (5FU) is a chemotherapeutic drug used in cancer treatment including breast, stomach, pancreatic, and skin cancers. Although it has an established history in cancer treatment, it shows toxic side effects due to its limited safety profile52. These side effects can be eliminated or at least minimized if a prodrug strategy coupled with gated-intracellular catalysis were to be available. By this gated catalysis, activation of prodrugs will be achieved on demand upon reaching to site of action thus eliminating the off target effects.
5FU can be converted into a prodrug via functionalization on its N1 position, as modification at this position will block the activity of 5FU and render it non-toxic16. We introduced a propargyl moiety to turn 5FU into an inactive prodrug ‘pro-5FU’ (Fig. 5a). First, through palladium mediated chemocatalysis the propargyl masking unit is intracellularly cleaved to yield 5FU followed by the enzymatic reactions to convert 5FU into the cytotoxic nucleotidic fluorouridine monophosphate via functionalization at the N1 position53. These active metabolites work through misincorporation into RNA and DNA molecules and irreversibly inhibit the nucleotide synthetic enzyme thymidylate synthase to disrupt cell functions and induce cytotoxicity. Overall, this process is an example of the integration of chemocatalysis and enzymatic biochemistry in a one-pot chemical sequence54,55.
Figure 5. Prodrug activation in living cells using supramolecularly controlled nanozymes.
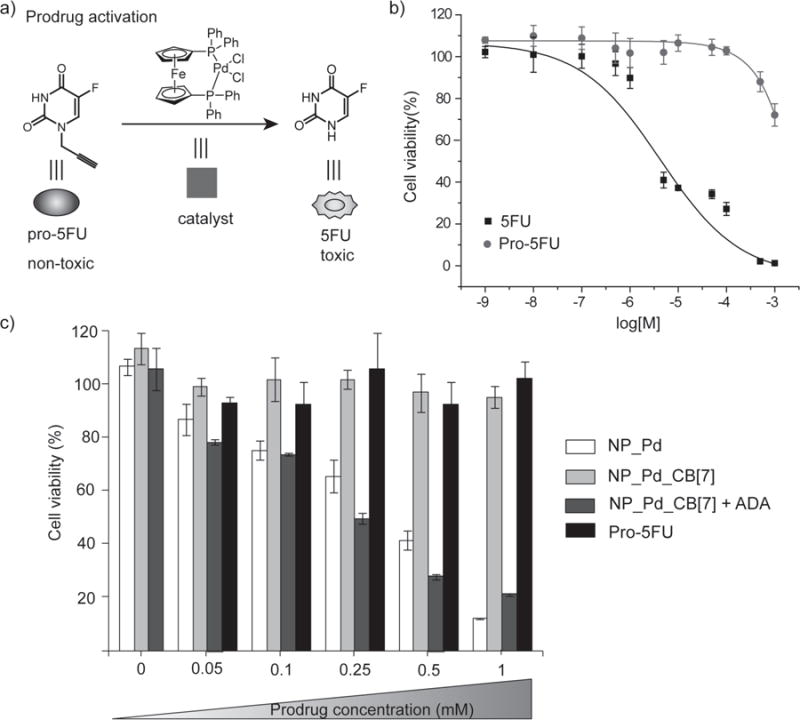
a, Structures of pro-5FU, 5FU and the palladium catalyst used for prodrug activation. b, Viability of cells treated with 5FU and pro-5FU at various concentrations, showing a nice therapeutic window was obtained between 5FU and pro-5FU. c, NP_Pd and ADA treated NP_Pd_CB[7] showed increasing intracellular toxicity as a result of more conversion of prodrug into 5FU drug at higher pro-5FU concentrations, while NP_Pd_CB[7] did not show any toxicity at any prodrug concentration used due to the blocking catalysis. Also, only prodrug, NP_Pd, NP_Pd_CB[7], and NP_Pd_CB[7] + ADA did not cause any toxicity into the system at zero prodrug concentration. Cell viability experiments were performed in triplicate. Error bars represent standard deviations of these measurements.
Cleavage of propargyl group on pro-5FU (1 mM) in the presence of NP_Pd (100 nM) was monitored using matrix-assisted laser desorption/ionization (MALDI)-MS. Most of the pro-5FU was converted into 5FU after 48 h (Supplementary Fig. S29). After confirming the cleavage, the toxicity profile of 5FU and pro-5FU were investigated by performing a cell viability assay. Cells were treated with various concentrations from 10 nM to 1 mM. While 5-FU showed toxicity as concentration increased, pro-5FU retained high cellular viability at all concentrations studied (Fig. 5b).
Cell viability studies for prodrug system in the presence of nanozymes were carried out using HeLa cells in a 96 well plate. Cells were first incubated with NP_Pd or NP_Pd_CB[7] at a concentration of 100 nM in serum-containing media for 24 h. Then, after multiple washings, cells were incubated with different concentrations of pro-5FU (0, 0.05, 0.1, 0.25, 0.5 and 1 mM) while some of the cells that were incubated with NP_Pd_CB[7], were treated with pro-5FU and ADA at the same time. Cells that were incubated with NP_Pd and NP_Pd_CB[7] + ADA showed elevated toxicity at higher concentration of pro-5FU while NP_Pd_CB[7] retained ~100 % cell viability even at higher concentration of pro-5FU (Fig. 5c). As expected, pro-5FU was not toxic at any concentrations used. Likewise, NP_Pd, NP_Pd_CB[7], and NP_Pd_CB[7] + ADA at zero pro-drug concentration were not toxic indicating toxicity was coming from the intracellular conversion of pro-5FU into 5FU using gated-catalysis but not the nanozyme itself.
In conclusion, we have described the fabrication of a AuNP based, bioorthogonal nanozyme that uses transition metal catalysis to effect transformations of imaging and therapeutic relevance without biological counterparts. These catalysts were built upon a platform featuring biomimetic size and surface functionality, making them attractive components for both in vitro and in vivo applications. The ability to control the activity of these nanozymes through host-guest interactions of CB[7] molecules with the benzyl headgroup of the AuNP ligands likewise provides an efficient and reversible means of regulating catalysis. This bioorthogonal catalysis can be employed not only in therapeutic applications such as the activation of prodrugs at the site of action but also in treating non-cancerous chronic diseases where the goal is not to kill the cell but to kill an infective agent or restore a malfunctioning pathway. This gated platform enables the introduction of an inactive reservoir of a bioorthogonal catalyst that would remain dormant between successive administrations of the substrate, thus reducing potential interference of the catalyst with cell constituents and protecting the metal from poisoning, release, systemic distribution and/or clearance. Furthermore, our protein-sized system demonstrates biomimetic behavior and yet performs totally abiotic chemistry that can be controlled intracellularly through a very simple host-guest feature. This system integrates biomimetic and bioorthogonal design elements to provide a new platform for imaging and therapeutic applications as well as pharmacological treatments integrating biological activity with man-made synthetic tools.
Methods
Synthesis of the ligands and the AuNPs (Fig. 1f) for the nanozyme are described in the Supplementary Information (Supplementary Fig. S1–S8). Details of the particle characterization methods, catalyst encapsulation process, ICP-MS sample preparation, kinetic studies and additional cell culture experiments can be found in the Supplementary Information.
Catalyst encapsulation into the monolayer of AuNPs
The catalyst ([Cp*Ru(cod)Cl] or 1,1′-Bis(diphenylphosphino) ferrocene)palladium(II)dichloride) and the AuNP were dissolved in an acetone/water mixture and then the acetone was slowly removed by evaporation. During the evaporation the catalyst was encapsulated in the particle monolayer, with excess catalyst precipitating and removed by filtration. The AuNPs were purified by multiple filtrations and dialyzed against water to remove free catalysts. The amount of encapsulated catalyst was measured by ICP-MS by tracking 101Ru relative to 197Au for NP_Ru and 106Pd relative to 197Au for NP_Pd.
Kinetic studies of nanozymes using Lineweaver–Burk analysis
In the titration of the reaction rate of the nanozyme using the CB[7], 400 nM of the AuNPs, 80 μM of the substrate, and set molar ratios of CB[7]s were used in sodium phosphate buffer (5 mM, pH 7.4). For the Lineweaver-Burk analysis, 400 nM of the AuNPs, 2, 4, 20, 40, and 80 μM of the substrate, and 0, 4, 16, and 80 μM of CB[7] were used. The concentration of the products was calculated based on the fluorescence standard curve of rhodamine 110 (Supplementary Fig. S30).
Catalytic activity of the nanozymes inside living cells
HeLa cells were grown in a cell culture flask using low-glucose Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2. For flow cytometry and confocal analysis, HeLa cells were seeded at 20,000 and 80,000 cells in 0.5 mL per well in 24-well plates and 1 ml in confocal dish 24 h prior to the experiment. During the experiment old media were replaced by 200 nM of the nanozymes and the nanozyme-bound CB[7] in serum-containing media and the cells were incubated for 24 h and washed with PBS buffer three times. 100 μM of the substrates were then added to the cells and incubated for 24 h. The cells were then washed with PBS three times. In the case of ADA treatment, cells were treated with 0.4 mM ADA solution (0.5 mL) for 24 h. For the cytometry, a Becton Dickinson LSR cytometer was used. A total of 10,000 events per sample were analyzed. A530/30 bandpass filter (FITC) was used for rhodamine 110. Confocal microscopy images were obtained on a Zeiss LSM 510 Meta microscope by using a 63× objective. The settings of the confocal microscope were as follows unless otherwise specified: green channel: λex=488 nm and λem=BP 505–530 nm; red channel: λex=543 nm and λem=LP 650 nm. Emission filters: BP=band pass, LP=high pass. LysoTracker® Red DND-99 was obtained from Invitrogen. 100 nM of LysoTracker® was incubated with cells for 30 min prior to the microscopy experiments.
Cytotoxicity measurements of 5FU versus pro-5FU
HeLa cells were seeded at 10,000 in 0.2 mL per well in 96-well plates 24 h prior to the experiment. Cells were washed with PBS buffer and then treated with 11 different concentrations ranging from 1 nM to 1 mM of 5FU or pro-5FU in triplicate. After 72 h of incubation, the cells were then completely washed off with PBS buffer three times and 10% Alamar Blue in serum containing media was added to each well (220 μL) and further incubated at 37 °C for 4 h. The cell viability was then determined by measuring the fluorescence intensity at 570 nm using a SpectraMax M5 microplate spectrophotometer.
Intracellular prodrug activation
HeLa cells were seeded at 10,000 in 0.2 mL per well in 96-well plates 24 h prior to the experiment. Cells were washed off and incubated with NP_Pd (100 nM) or NP_Pd_CB[7] (100 nM) in 10% serum containing media. After 24 h, cell were washed with PBS buffer three times and treated with pro-5FU at a concentration of 0, 0.05, 0.1, 0.25, 0.5 and 1 mM for 48 h. Some cells treated with NP_Pd_CB[7] were incubated with both pro-5FU and ADA (0.05 mM). After 48 h of incubation, the cells were then completely washed off and 10% Alamar Blue in serum containing media was added to each well (220 μL) and further incubated at 37 °C for 4 h. The cell viability was then determined by measuring the fluorescence intensity at 570 nm using a SpectraMax M5 microplate spectrophotometer.
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH (EB014277). T.M. is grateful to the Japan Society for the Promotion of Sciences for a Postdoctoral Fellowship for Research Abroad and for the Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation. We thank Professor Lyle Isaacs (University of Maryland) for providing cucurbit[7]uril.
Footnotes
Author contributions
G.Y.T., Y.J., and V.M.R. conceived and designed the experiments. G.Y.T., Y.J., B.D., T.M., R.M., R.D., S.T.K., Y,-C.Y., B.Y., and S.H. performed the experiments. All authors analyzed and discussed the data. G.Y.T., Y.J., B.D., and V.M.R. co-wrote the paper. V.M.R. revised the paper.
G.Y.T. and Y.J. contributed equally to this work.
Additional information
The authors declare no competing financial interests. Supplementary information that accompanies this paper is at www.nature.com/naturechemistry.
References
- 1.Patterson DM, Nazarova LA, Prescher JA. Finding the Right (Bioorthogonal) Chemistry. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 2.Ramil CP, Lin Q. Bioorthogonal chemistry: strategies and recent developments. Chem Commun. 2013;49:11007–11022. doi: 10.1039/c3cc44272a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertozzi CR. A decade of bioorthogonal chemistry. Acc Chem Res. 2011;44:651–653. doi: 10.1021/ar200193f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koo H, et al. Bioorthogonal copper-free click chemistry in vivo for tumor-targeted delivery of nanoparticles. Angew Chem Int Ed. 2012;51:11836–11840. doi: 10.1002/anie.201206703. [DOI] [PubMed] [Google Scholar]
- 5.Devaraj NK, Thurber GM, Keliher EJ, Marinelli B, Weissleder R. Reactive polymer enables efficient in vivo bioorthogonal chemistry. Proc Natl Acad Sci USA. 2012;109:4762–4767. doi: 10.1073/pnas.1113466109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komatsu H, Shindo Y, Oka K, Hill JP, Ariga K. Ubiquinone-Rhodol (UQ-Rh) for Fluorescence Imaging of NAD(P)H through Intracellular Activation. Angew Chem Int Ed. 2014;53:3993–3995. doi: 10.1002/anie.201311192. [DOI] [PubMed] [Google Scholar]
- 7.Sletten EM, Bertozzi CR. From Mechanism to Mouse: A Tale of Two Bioorthogonal Reactions. Acc Chem Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxon E, Bertozzi CR. Cell Surface Engineering by a Modified Staudinger Reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Chen PR. Palladium-triggered deprotection chemistry for protein activation in living cells. Nature Chem. 2014;6:352–361. doi: 10.1038/nchem.1887. [DOI] [PubMed] [Google Scholar]
- 10.Sasmal PK, Streu CN, Meggers E. Metal complex catalysis in living biological systems. Chem Commun. 2013;49:1581–1587. doi: 10.1039/c2cc37832a. [DOI] [PubMed] [Google Scholar]
- 11.Unciti-Broceta A, Johansson EMV, Yusop RM, Sánchez-Martín RM, Bradley M. Synthesis of polystyrene microspheres and functionalization with Pd(0) nanoparticles to perform bioorthogonal organometallic chemistry in living cells. Nat Protoc. 2012;7:1207–1218. doi: 10.1038/nprot.2012.052. [DOI] [PubMed] [Google Scholar]
- 12.Streu C, Meggers E. Ruthenium-induced allylcarbamate cleavage in living cells. Angew Chem Int Ed. 2006;45:5645–5648. doi: 10.1002/anie.200601752. [DOI] [PubMed] [Google Scholar]
- 13.Yusop RM, Unciti-Broceta A, Johansson EMV, Sánchez-Martín RM, Bradley M. Palladium-mediated intracellular chemistry. Nature Chem. 2011;3:239–243. doi: 10.1038/nchem.981. [DOI] [PubMed] [Google Scholar]
- 14.Sasmal PK, et al. Catalytic Azide Reduction in Biological Environments. ChemBioChem. 2012;13:1116–1120. doi: 10.1002/cbic.201100719. [DOI] [PubMed] [Google Scholar]
- 15.Do JH, Kim HN, Yoon J, Kim JS, Kim HJ. A Rationally Designed Fluorescence Turn-On Probe for the Gold(III) Ion. Org Lett. 2010;12:932–934. doi: 10.1021/ol902860f. [DOI] [PubMed] [Google Scholar]
- 16.Weiss JT, et al. Extracellular palladium-catalysed dealkylation of 5-fluoro-1-propargyl-uracil as a bioorthogonally activated prodrug approach. Nat Commun. 2014;5:3277. doi: 10.1038/ncomms4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon HJ, Kuwabara J, Kim JH, Mirkin CA. Allosteric Supramolecular Triple-Layer Catalysts. Science. 2010;330:66–69. doi: 10.1126/science.1193928. [DOI] [PubMed] [Google Scholar]
- 18.De M, Ghosh PS, Rotello VM. Applications of Nanoparticles in Biology. Adv Mater. 2008;20:4225–4241. [Google Scholar]
- 19.Murphy CJ, et al. Gold Nanoparticles in Biology: Beyond Toxicity to Cellular Imaging. Acc Chem Res. 2008;41:1721–1730. doi: 10.1021/ar800035u. [DOI] [PubMed] [Google Scholar]
- 20.Boisselier E, Astruc D. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem Soc Rev. 2009;38:1759–1782. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- 21.Tonga GY, Saha K, Rotello VM. Interfacing Nanoparticles and Biology: New Strategies for Biomedicine. Adv Mater. 2014;26:359–370. doi: 10.1002/adma.201303001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mout R, Rotello VM. Bio and Nano Working Together: Engineering the Protein-Nanoparticle Interface. Isr J Chem. 2013;53:521–529. [Google Scholar]
- 23.Kim B, et al. Tuning payload delivery in tumour cylindroids using gold nanoparticles. Nat Nanotech. 2010;5:465–472. doi: 10.1038/nnano.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh P, et al. Intracellular Delivery of a Membrane-Impermeable Enzyme in Active Form Using Functionalized Gold Nanoparticles. J Am Chem Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim CK, et al. Entrapment of Hydrophobic Drugs in Nanoparticle Monolayers with Efficient Release into Cancer Cells. J Am Chem Soc. 2009;131:1360–1361. doi: 10.1021/ja808137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manea F, Houillon FB, Pasquato L, Scrimin P. Nanozymes: Gold-Nanoparticles-Based Transphosphorylation Catalysts. Angew Chem Int Ed. 2004;43:6165–6169. doi: 10.1002/anie.200460649. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, et al. Nanoparticle-based artificial RNA silencingmachinery for antiviral therapy. Proc Natl Acad Sci USA. 2012;109:12387–12392. doi: 10.1073/pnas.1207766109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pengo P, Baltzer L, Pasquato L. & Scrimin P. Substrate Modulation of the Activity of an Artificial Nanoesterase Made of Peptide-Functionalized Gold Nanoparticles. Angew Chem Int Ed. 2007;46:400–404. doi: 10.1002/anie.200602581. [DOI] [PubMed] [Google Scholar]
- 29.Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42:6060–6093. doi: 10.1039/c3cs35486e. [DOI] [PubMed] [Google Scholar]
- 30.Natalio F, et al. Vanadium pentoxide nanoparticles mimic vanadium haloperoxidases and thwart biofilm formation. Nat Nanotechnol. 2012;7:530–535. doi: 10.1038/nnano.2012.91. [DOI] [PubMed] [Google Scholar]
- 31.Hastings CJ, Backlund MP, Bergman RG, Raymond KN. Enzyme-like Control of Carbocation Deprotonation Regioselectivity in Supramolecular Catalysis of the Nazarov Cyclization. Angew Chem Int Ed. 2011;50:10570–10573. doi: 10.1002/anie.201105325. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh S, Isaacs L. Biological Catalysis Regulated by Cucurbit[7]uril Molecular Containers. J Am Chem Soc. 2010;132:4445–4454. doi: 10.1021/ja910915k. [DOI] [PubMed] [Google Scholar]
- 33.Masson E, Ling X, Joseph R, Kyeremeh-Mensah L, Lu X. Cucurbituril chemistry: a tale of supramolecular success. RSC Adv. 2012;2:1213–1247. [Google Scholar]
- 34.Lee JW, Samal S, Selvapalam N, Kim HJ, Kim K. Cucurbituril Homologues and Derivatives: New Opportunities in Supramolecular Chemistry. Acc Chem Res. 2003;36:621–630. doi: 10.1021/ar020254k. [DOI] [PubMed] [Google Scholar]
- 35.Kim K, et al. Functionalized cucurbiturils and their applications. Chem Soc Rev. 2007;36:267–279. doi: 10.1039/b603088m. [DOI] [PubMed] [Google Scholar]
- 36.Ambrogio MW, Thomas CR, Zhao YL, Zink JI, Stoddart JF. Mechanized Silica Nanoparticles: A New Frontier in Theranostic Nanomedicine. Acc Chem Res. 2011;44:903–913. doi: 10.1021/ar200018x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H, et al. Glutathione-Induced Intracellular Release of Guests from Mesoporous Silica Nanocontainers with Cyclodextrin Gatekeepers. Adv Mater. 2010;22:4280–4283. doi: 10.1002/adma.201001417. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, et al. The Cucurbit[n]uril Family: Prime Components for Self-Sorting Systems. J Am Chem Soc. 2005;127:15959–15967. doi: 10.1021/ja055013x. [DOI] [PubMed] [Google Scholar]
- 39.Khashab NM, et al. pH-Responsive mechanised nanoparticles gated by semirotaxanes. Chem Commun. 2009:5371–5373. doi: 10.1039/b910431c. [DOI] [PubMed] [Google Scholar]
- 40.Hong R, et al. Control of Protein Structure and Function through Surface Recognition by Tailored Nanoparticle Scaffolds. J Am Chem Soc. 2004;126:739–743. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]
- 41.Kim C, Agasti SS, Zhu Z, Isaacs L, Rotello VM. Recognition-mediated activation of therapeutic gold nanoparticles inside living cells. Nature Chem. 2010;2:962–966. doi: 10.1038/nchem.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angelos S, et al. pH Clock-Operated Mechanized Nanoparticles. J Am Chem Soc. 2009;131:12912–12914. doi: 10.1021/ja9010157. [DOI] [PubMed] [Google Scholar]
- 43.Alcaide B, Almendros P, Alonso JM. A Practical Ruthenium-Catalyzed Cleavage of the Allyl Protecting Group in Amides, Lactams, Imides, and Congeners. Chem Eur J. 2006;12:2874–2879. doi: 10.1002/chem.200501227. [DOI] [PubMed] [Google Scholar]
- 44.Sasmal PK, Carregal-Romero S, Parak WJ, Meggers E. Light-Triggered Ruthenium-Catalyzed Allylcarbamate Cleavage in Biological Environments. Organometallics. 2012;31:5968–5970. [Google Scholar]
- 45.Zhu ZJ, et al. Determination of the Intracellular Stability of Gold Nanoparticle Monolayers Using Mass Spectrometry. Anal Chem. 2012;84:4321–4326. doi: 10.1021/ac203408v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon WS, et al. Complexation of Ferrocene Derivatives by the Cucurbit[7]uril Host: A Comparative Study of the Cucurbituril and Cyclodextrin Host Families. J Am Chem Soc. 2005;127:12984–12989. doi: 10.1021/ja052912c. [DOI] [PubMed] [Google Scholar]
- 47.Lineweaver H, Burk D. The Determination of Enzyme Dissociation Constants. J Am Chem Soc. 1934;56:658–666. [Google Scholar]
- 48.Miller DJ, Surfraz MBU, Akhtar M, Gani D, Allemann RK. Removal of the phosphate group in mechanism-based inhibitors of inositol monophosphatase leads to unusual inhibitory activity. Org Biomol Chem. 2004;2:671–688. doi: 10.1039/b312808c. [DOI] [PubMed] [Google Scholar]
- 49.Verma A, et al. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nature Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu Z, Biswas A, Zhao M, Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chem Soc Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 51.Cai SX, et al. Design and Synthesis of Rhodamine 110 Derivative and Caspase-3 Substrate for Enzyme and Cell-Based Fluorescent Assay. Bioorg Med Chem Lett. 2001;11:39–42. doi: 10.1016/s0960-894x(00)00590-4. [DOI] [PubMed] [Google Scholar]
- 52.Saif MW, Choma A, Salamone SJ, Chu E. Pharmacokinetically guided dose adjustment of 5-fluorouracil: a rational approach to improving therapeutic outcomes. J Natl Cancer Inst. 2009;101:1543–1552. doi: 10.1093/jnci/djp328. [DOI] [PubMed] [Google Scholar]
- 53.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nature Rev. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 54.Wang ZJ, Clary KN, Bergman RG, Raymond KN, Toste FD. A supramolecular approach to combining enzymatic and transition metal catalysis. Nature Chem. 2013;5:100–103. doi: 10.1038/nchem.1531. [DOI] [PubMed] [Google Scholar]
- 55.Marr AC, Liu S. Combining bio- and chemo-catalysis: from enzymes to cells, from petroleum to biomass. Trends Biotechnol. 2011;29:199–204. doi: 10.1016/j.tibtech.2011.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


