Abstract
Lentil is a major cool-season grain legume grown in South Asia, West Asia, and North Africa. Populations in developing countries of these regions have micronutrient deficiencies; therefore, breeding programs should focus more on improving the micronutrient content of food. In the present study, a set of 96 diverse germplasm lines were evaluated at three different locations in India to examine the variation in iron (Fe) and zinc (Zn) concentration and identify simple sequence repeat (SSR) markers that associate with the genetic variation. The genetic variation among genotypes of the association mapping (AM) panel was characterized using a genetic distance-based and a general model-based clustering method. The model-based analysis identified six subpopulations, which satisfactorily explained the genetic structure of the AM panel. AM analysis identified three SSRs (PBALC 13, PBALC 206, and GLLC 563) associated with grain Fe concentration explaining 9% to 11% of phenotypic variation and four SSRs (PBALC 353, SSR 317–1, PLC 62, and PBALC 217) were associated with grain Zn concentration explaining 14%, to 21% of phenotypic variation. These identified SSRs exhibited consistent performance across locations. These candidate SSRs can be used in marker-assisted genetic improvement for developing Fe and Zn fortified lentil varieties. Favorable alleles and promising genotypes identified in this study can be utilized for lentil biofortification.
Introduction
Lentil (Lens culinaris subsp. culinaris) is an annual, self-pollinating, herbaceous, cool-season grain legume originating from the Near East center of origin [1]. From the Mediterranean region, the crop spread to different parts of the world, which led to the evolution of six geographical groups based on morphology, physiology, and functional variation [2]. Pilosae-type lentil with low biomass, small seeds, short rudimentary tendrils, pubescent foliage, and precocity in flowering and maturity is grown in South Asia. Several research groups have recommended the introgression of the Mediterranean lines [3–6] and wild species for increasing the genetic diversity and broadening the genetic base of the pilosae-type lentil. This crop is used mainly for food and fodder. Lentil grains are rich source of protein, fiber, minerals and carbohydrates playing a crucial role in reducing micronutrient deficiency in developing countries [7]. The crop is usually grown in rotation with cereals to break disease cycles and to fix atmospheric nitrogen [8].
The global lentil production during 2014 was 4.95 million tons [9]. Major lentil-growing regions included South Asia, West Asia, and North Africa. Micronutrient deficiency is prevalent in these regions because of high population densities and poor resources. India is a major producer and consumer of lentil. In India, lentil is grown in 1.89 million hectare with a production of 1.13 million ton [9]. This crop is mainly grown in the rain-fed areas of Central India and parts of Eastern India on residual moisture from the rainy season. The productivity of lentil is low because of short growing period and moisture stress during flowering and fruiting. Earlier study [5] has reported narrow genetic base of Indian lentil.
Micronutrients represent the essential vitamins and minerals required for normal cellular and molecular functions. Micronutrient deficiency is widespread in developing countries because of the poor quality of diet, which consists mainly of staple crops and very small amounts of meat, pulses, and fruits owing to low income. Micronutrient deficiency in food crops is primarily because of the low natural levels of available micronutrients in the soil owing to the decreased use of animal manure, crop residue and compost [10], which further results in low availability of micronutrients to plants [11]. Fe has been reported as heme and non heme forms. Non vegetarian food is source of heme iron while non heme iron is found in plants. In human beings Fe is essential constituent of many proteins and enzymes and is involved in cell growth and differentiation and oxygen transport. The heme Fe bioavailability is 12%–25% and non-heme Fe bioavailability is less than 5% [12]. Fe deficiency results in fatigue due to decreased Fe delivery as well as poor immunity and performance [13]. Fe deficiency affects more than 30% people worldwide [14]. Fe deficiency results in the disruption of the optimal function of both endocrine and immune systems [15]. It can cause anemia, which increases the risk of hemorrhage and bacterial infections during childbirth, thereby resulting in maternal deaths [16]. Babies may be born prematurely and may be liable to infections, learning disabilities, and delayed development [17]. Almost 40% pregnant women and more than 40% children under the age of 5 years in developing countries are anemic [18]. Almost 50% of these anemia cases are estimated to be due to Fe deficiency. Globally, half of the cultivated soil is zinc (Zn) deficient [19]. Zn is an essential component of proteinases, dehydrogenases, and peptidases in plants and is found in soil in the form of Zn2+ and Zn (OH)+[20]. In humans, Zn is essential for the normal growth and development of fetuses and adolescent children [21]. Regular intake of Zn is required because the human body cannot store Zn [22]. Zn deficiency impairs immune functions and is associated with an increased risk of gastrointestinal infections [23,24]. It is also a contributing factor in child deaths due to diarrhea [25].
With the emergence of PCR technology [26, 27], a new era began in molecular biology. In lentil, PCR-based markers, such as RAPD, RFLP, AFLP, inter-simple sequence repeats (ISSRs), and sequence-tagged microsatellites, were initially used for phylogenetic studies, linkage map construction, and diversity analysis within cultivated and between different Lens species [28–33]. Later Simple sequence repeats (SSRs) were introduced. SSRs are short tandem repeats of 1–6 bp [34]. SSRs are multiallelic, hypervariable, and chromosome specific; exhibiting codominant inheritance [35,36]. SSRs are distributed throughout the genome in both coding and noncoding regions [37]. SSRs are also present in chloroplasts [38] and mitochondria [39]. However, the development of genomic microsatellite markers is expensive, time consuming, and labor intensive and requires prior knowledge of DNA sequences [40,41]. SSRs from expressed sequence tag (EST) are associated with the transcribed regions of the genome [42,43] and can be developed through mining EST databases. Compared with genomic SSRs, EST-SSRs have a high level of transferability across related species [44–50]. Only limited reports are available on the development of genomic SSRs [30–32] and EST-SSRs [51,33] and their use in diversity analysis.
Breeding for quantitative or complex traits is a tedious and time-consuming process. Both genotypic and environmental factors play a role in the expression of a phenotype. The identification of molecular markers in the late 1980s has led to the development of tools for the directed manipulation of quantitative traits in field crops. Collard et al. [52] proposed a scheme for quantitative trait loci (QTL) mapping in crop plants for their further use in marker-assisted selection. Linkage analysis (using biparental populations) and association mapping (using diverse germplasm lines) are tools for dissecting quantitative traits. For linkage analysis from biparental crosses, several types of mapping populations (F2, RILs, NILs, double haploid, and backcross) can be developed. The progenies of the developed populations are screened for a trait of interest. Bulk segregant analysis, suggested by Michelmore et al. [53], is used to identify markers linked with the trait of interest. Although QTL mapping is a crucial tool for tagging and mapping of genes in crop plants, the method has limitations such as few meiotic events, difficulty in building a segregating population, reduced diversity derived from two parents, high cost involved, low resolution, and simultaneous evaluation of few alleles [54, 55]. The association mapping (AM) approach, also known as linkage disequilibrium (LD) mapping [56, 57], has been initially used by geneticists to map and clone many genes governing complex traits in humans [58–60]. However, in recent years, plant geneticists have also used it for identifying QTL in different crops [61–67]. AM harnesses genetic diversity of natural population, for searching genotype-phenotype correlations among unrelated individuals. The main advantage of AM is likelihood for a higher resolution mapping because of the utilization of majority recombination events from a large number of meiosis throughout the germplasm development history [68]. The association mapping approach is based on the LD between the marker and the QTL, and it provides higher mapping resolution. The detection power of QTL depends on the LD between the QTL and the marker. A strong LD between the markers enhances the detection of QTL with a small effect [69]. The variance explained by QTL is underestimated if the LD between the marker and QTL is incomplete. In this study, we used association mapping for identifying QTL linked to grain Fe and Zn concentration in lentil. Inductively coupled plasma-mass spectrometry (ICP-MS) was used to estimate grain Fe and Zn concentration in a set of 96 Indian and Mediterranean lentil genotypes with 73 genomic and EST-SSRs. We then conducted an association mapping study by using a general linear model (GLM) to detect the significant loci responsible for natural variations of grain concentrations of Fe and Zn. We present results on significantly associated loci and favorable alleles. This approach can be used for identifying germplasm lines with desirable traits for accelerating lentil breeding through marker-assisted selection.
Material and methods
Plant material and field experiment
An AM panel of 96 diverse L. culinaris subspecies culinaris genotypes was used in this study (Table 1). The panel consisted of advanced breeding lines developed at different lentil breeding centers of India, released lentil varieties, and exotic germplasm lines obtained from the International Center for Agricultural Research in the Dry Areas (ICARDA). The genotypes were selected (based on origin and seed size) from global lentil germplasm maintained at Indian Agricultural Research Institute, New Delhi. The panel included both microsperma (small seeded) and macrosperma (large seeded) lentil types. The selected genotypes exhibited highly significant variation for both grain Fe and Zn concentration (S1 and S2 Tables) and almost normal distribution for these traits (S1 Fig). The genotypes of panel were grown at three geographical locations: (i) Delhi (North-West Plain Zone; 28°40′N 77°12′E, 218 meters above the sea level [masl]), during 2013–14 and 2014–15 (ii) Indore (Central Zone; 30.9°N 75.85°E, 244 masl, during 2013–14 and (iii) Dharwad (Peninsular Zone; 28°58′N 79°25′E 344 masl) during 2013–14. The soil characteristics (pH, EC, organic carbon content, available N, P and K and soil texture) of Delhi, Dharwad and Indore are presented in S3 Table. In previous season mungbean was planted across all the three locations in these plots. No micronutrient spray or basal application was given. Only fertilizer applied to mungbean crop was 100 kg Di ammonium Phosphate / ha. To ensure proper homogenization, the soil was pulverized and thoroughly mixed and the field was leveled at each location. The experiment was planted in two replication. From each replication in each location ten samples were drawn to estimate soil Fe and Zn reveal significant variation ensuring the plot homogeneity. The panel was planted in a randomized complete block design with two replicates per entry (3 rows per replication) with a plant distance of 5 × 30 cm and a row length of 4 m. Standard agronomic practices were followed for crop cultivation. Fe and Zn concentration in the soil were estimated using a procedure proposed by Singh et al. [70].
Table 1. Source / Origin of 96 genotypes of lentil used in the study.
| S.No. | Genotypes | Source | S.No. | Genotypes | Source |
|---|---|---|---|---|---|
| 1 | L 404 | IARI, New Delhi | 49 | L 4590 | IARI, New Delhi |
| 2 | L 830 | IARI, New Delhi | 50 | PL 4 | GBPUAT, Pantnagar |
| 3 | L 4596 | IARI, New Delhi | 51 | L 4591 | IARI, New Delhi |
| 4 | L 4602 | IARI, New Delhi | 52 | LL 1203 | PAU, Ludhiana |
| 5 | L 4603 | IARI, New Delhi | 53 | RLG 147 | ARS, Durgapura |
| 6 | L 4618 | IARI, New Delhi | 54 | DL 11–4 | TCA, Dholi |
| 7 | L 4620 | IARI, New Delhi | 55 | KLS 113 | CSAU, Kanpur |
| 8 | L 4648 | IARI, New Delhi | 56 | NDL 11–1 | NDUAT, Faizabad |
| 9 | L 4649 | IARI, New Delhi | 57 | PL 122 | GBPUAT, Pantnagar |
| 10 | L 4650 | IARI, New Delhi | 58 | SKUAL 9 | Srinagar |
| 11 | L 4698 | IARI, New Delhi | 59 | L 4706 | IARI, New Delhi |
| 12 | L 5120 | IARI, New Delhi | 60 | DPL 15 | IIPR, Kanpur |
| 13 | L 5126 | IARI, New Delhi | 61 | LL 1210 | PAU, Ludhiana |
| 14 | L 5253 | IARI, New Delhi | 62 | KLB 345 | CSAU, Kanpur |
| 15 | ILL 7663 | ICARDA, Aleppo, Syria | 63 | PL 024 | GBPUAT, Pantnagar |
| 16 | L 7818 | IARI, New Delhi | 64 | PL 129 | GBPUAT, Pantnagar |
| 17 | L 7903 | IARI, New Delhi | 65 | IPL 324 | IIPR, Kanpur |
| 18 | DPL 15 | IIPR, Kanpur | 66 | IPL 406 | IIPR, Kanpur |
| 19 | DPL 21 | IIPR, Kanpur | 67 | L 4707 | IARI, New Delhi |
| 20 | DPL 58 | IIPR, Kanpur | 68 | LL 1204 | PAU, Ludhiana |
| 21 | PL 02 | GBPUAT, Pantnagar | 69 | LH 84–8 | HAU, Hisar |
| 22 | P 13129 | ICARDA, Aleppo, Syria | 70 | RVL 48 | Sehore |
| 23 | PL 101 | GBPUAT, Pantnagar | 71 | KLB 314 | CSAU, Kanpur |
| 24 | PL 639 | GBPUAT, Pantnagar | 72 | IPL 325 | IIPR, Kanpur |
| 25 | RL 1 | IARI, New Delhi | 73 | DPL 62 | IIPR, Kanpur |
| 26 | ILL 2581 | ICARDA, Aleppo, Syria | 74 | P 2102 | ICARDA, Aleppo, Syria |
| 27 | SKL 259 | IARI, New Delhi | 75 | P 2124 | ICARDA, Aleppo, Syria |
| 28 | EC 1 | IARI, New Delhi | 76 | P 2125 | ICARDA, Aleppo, Syria |
| 29 | 10-2-B-2 | IARI, New Delhi | 77 | P 2126 | ICARDA, Aleppo, Syria |
| 30 | 10-3-Y-26 | IARI, New Delhi | 78 | P 2127 | ICARDA, Aleppo, Syria |
| 31 | Globe mutant | IARI, New Delhi | 79 | P 2130 | ICARDA, Aleppo, Syria |
| 32 | Fasciated mutant | IARI, New Delhi | 80 | P 2205 | ICARDA, Aleppo, Syria |
| 33 | HM 1 | HAU, Hisar | 81 | P 2215 | ICARDA, Aleppo, Syria |
| 34 | MC 6 | IARI, New Delhi | 82 | P 2230 | ICARDA, Aleppo, Syria |
| 35 | K 75 | CSAU, Kanpur | 83 | P 2233 | ICARDA, Aleppo, Syria |
| 36 | VL 103 | VPKAS, Almora | 84 | P 2239 | ICARDA, Aleppo, Syria |
| 37 | FLIP 96–57 | ICARDA, Aleppo, Syria | 85 | P 3113 | ICARDA, Aleppo, Syria |
| 38 | LL 147 | PAU, Ludhiana | 86 | P 3204 | ICARDA, Aleppo, Syria |
| 39 | LL 931 | PAU, Ludhiana | 87 | P 3208 | ICARDA, Aleppo, Syria |
| 40 | LC 74-1-5-1 | IARI, New Delhi | 88 | P 3220 | ICARDA, Aleppo, Syria |
| 41 | LC 300–1 | IARI, New Delhi | 89 | P 13104 | ICARDA, Aleppo, Syria |
| 42 | PL 4 | GBPUAT, Pantnagar | 90 | P 13113 | ICARDA, Aleppo, Syria |
| 43 | LL 1231 | PAU, Ludhiana | 91 | P 13122 | ICARDA, Aleppo, Syria |
| 44 | IPL 221 | IIPR, Kanpur | 92 | P 13135 | ICARDA, Aleppo, Syria |
| 45 | VL 143 | VPKAS, Almora | 93 | P 13143 | ICARDA, Aleppo, Syria |
| 46 | PL 406 | GBPUAT, Pantnagar | 94 | P 14103 | ICARDA, Aleppo, Syria |
| 47 | PL 117 | GBPUAT, Pantnagar | 95 | P 14201 | ICARDA, Aleppo, Syria |
| 48 | IPL 220 | IIPR, Kanpur | 96 | P 15104 | ICARDA, Aleppo, Syria |
Analysis of grain sample for Fe and Zn concentration
The grains of each genotype were harvested separately at maturity. Care was taken to prevent contamination from dust and metallic equipment. Plants were hand-threshed to avoid any type of contamination. Then the grains were placed in a clean plastic tray manually (using contaminant-free gloves). The grains of each sample were washed with Milli-Q water and dried at 35°C for five days in a contamination-free uncorroded oven. From each sample, 10 g of grains was grounded manually into a fine powder using a mortar and pestle. The micronutrient analyses were performed at the Division of Soil Science and Agricultural Chemistry, IARI, New Delhi, India. The grain powder sample (0.5 g) was digested following the modified diacid protocol [70] by using a microwave digestion system (Multiwave ECO, Anton Paar, les Ulis, France). Fe and Zn concentration (in mg/kg seed) were estimated through ICP-MS (Perkin Elmer, model: NexION 300 ICP-MS, USA) by using an automatic sampling protocol. Two technical replications per biological replication were followed while estimating the concentration.
Genomic DNA extraction, purification and SSR amplification
Genomic DNA of the genotypes was extracted from 5 g of fresh leaf tissue by using the CTAB method proposed by Murray and Thompson [71]. The quality and quantity of DNA were determined using a spectrophotometer, and the samples were diluted to 10 ng/μL. The PCR mixture (20 μL) consisted of a 10X buffer (100 mM Tris-HCl, 15 mM MgCl2, and 500 mM KCl), 0.5 μM each of forward and reverse primers, 200 μM of each dNTP, 1 U of Taq DNA polymerase, PCR reagents, an EST-SSR or genomic SSR primer procured from Sigma-Aldrich (Spruce Street, St. Louis, USA) and approximately 40 ng of template genomic DNA. PCR was performed in a VeritiTM thermal cycler (Applied Biosystems, Life Technologies, Singapore) using the following temperature cycle: one denaturation cycle at 94°C for 4 min, followed by 30 cycles of 94°C for 1 min, annealing at 59°C–62°C (primer specific) for 30 sec, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The amplified products were electrophoresed for 3 h on 3% metaphor TM agarose gels (Lonza, Rockland, ME USA) at a constant voltage of 100 V in 1X TBE buffer. The gel was stained using ethidium bromide and visualized. The amplicons were photographed under UV light with a CCD camera attached to a gel documentation system (Alpha Imager) at 260 nm. A 50-bp DNA ladder (MBI, Fermentas, Vilnius, Lithuania) was used as a molecular size marker. Sixty genomic SSRs and 260 EST-SSRs were used to study polymorphism. Of the 73 polymorphic markers identified for genotyping, 20 were genomic SSRs [72,30, 31] and 53 were EST-SSRs [33, 51].
Diversity analysis and population structure
Polymorphism information content (PIC) was computed using the formula PIC = 1-ΣPi − ΣΣPi Pj, where “i” is the total number of alleles detected for the SSR marker, “Pi” is the frequency of the i allele in the set of 96 genotypes investigated, and j = I + 1 [73]. A binary matrix was then transformed to a genetic similarity matrix by using Jaccard’s coefficient. Unweighted neighbor joining (UNJ) method available in DARwin 5.0 (http://darwin.cirad.fr/) was used to visualize the dendrogram.
STRUCTURE 2.3.4 was used [74] to determine the number of subgroups in the population assuming prior values of k = 1 to 10. The data was analyzed at a run length of 2,50,000 as the burning period length followed by 2,50,000 Markov Chain Monte Carlo iterations by keeping α constant. Each k value was repeated 10 times with an admixture model and correlated the allele frequency. The optimum k value was determined by plotting the Ln P (D) value against the given k value. Structure harvester v 6.92 [75] was used for obtaining the optimum k value determined by plotting the Ln P (D) value against k. The highest plateau was observed at delta k = 6; hence, the number of inferred populations was assumed to be six for further analysis.
Association mapping and favorable allele mining
To identify QTL for grain Fe and Zn concentration, association analysis were performed using 73 SSRs. The LD was estimated between each pair of polymorphic loci by calculating the square of the correlation coefficient (r2) using TASSEL 3.01 [76] with 10,000 permutation. The General Linear Model (GLM with Q matrix to reduce false associations) was used to examine association between grain micronutrients and markers. An LD plot with p and r2 values was generated by TASSEL to depict the overall LD among the entire SSR set. The markers considered to be significantly associated with the trait are represented in Manhattan plot [76]. Quantile-quantile (QQ) plots of the expected and observed p values was plotted to evaluate the adequacy of controlling Type I error.
Favorable allele for a marker loci associated with grain Fe and Zn concentration were identified using formula suggested by Li et al. [77]. The phenotypic allele effect (ai) was calculated as ai = ∑xij/ni − ∑Nk/nk, where ai is the phenotypic effect of the ith allele, xij is the phenotypic value over the jth material with the ith allele, ni is the number of genotypes with the ith allele, Nk is the phenotypic value over all genotypes, and nk is the number of genotypes [78]. It represents a comparison of the average phenotypic value of genotypes with a specific allele with that of all genotypes. When, ai > 0, then this allele is supposed to have positive effect on the trait. When ai < 0, the allele gives a negative effect [79].
Results
Phenotypic variation in lentil germplasm
The variation in grain Fe and Zn concentration across locations and years are shown in the S1 Table. The average soil Fe and Zn concentration were 5.01 mg/kg and 1.68 mg/kg at Delhi (2013–14), 5.23 mg/kg and 1.62 mg/kg at Delhi (2014–15), 4.2 mg/kg and 0.62 mg/kg at Indore (2013–14) and 4.5 mg/kg and 0.985 mg/kg at Dharwad (2013–14), respectively. The mean grain Fe concentration ranged from 31.55 to 119.35 mg/kg and that of Zn ranged from 7.80 to 75.45 mg/kg.
In this study, efforts were made to identify lentil genotypes with high grain Fe and Zn concentration based on the multilocation data. The range for Fe concentration at Delhi was 38.4–119.35 mg/kg during 2013–2014 and 34.4–115.35 mg/kg during 2014–2015. Indore and Dharwad exhibited a range of 40.52–111.4 mg/kg and 31.55–106.05 mg/kg, respectively, during 2013–2014. Considering the performance of the studied genotypes in all the environments, L 4596 (122.46 mg/kg) was the most promising genotype, followed by L 5126 (114.96 mg/kg), whereas P 3226 had the lowest grain Fe concentration (27.52 mg/kg). The range for Zn concentration at Delhi was 12.3–74.15 mg/kg during 2013–2014 and 12.65–78.75 mg/kg during 2014–2015. Indore and Dharwad exhibited a range of 7.4–63.65 mg/kg and 22.25–62.95, respectively. Considering the data from the four environments, P 3220 (78.75 mg/kg) and P 13129 (70.45 mg/kg) were determined as the most promising genotype for the grain Zn concentration. The indigenous lines were found to be rich in grain Fe, whereas the exotic lines were found to be rich in grain Zn. A mean Fe grain concentration higher than 70 mg/kg seed was considered high, whereas that lower than 60 mg/kg seed was considered low. Similarly, a mean Zn grain concentration higher than 50 mg/kg seed was considered high and that lower than 40 mg/kg seed was considered low. Promising genotypes such as P 2130 (Fe: 85.75 mg/kg; Zn: 61.27 mg/kg), P 2126 (Fe: 102.26 mg/kg; Zn: 62.74 mg/kg), and L 4596 (Fe: 116.19 mg/kg; Zn: 47.38 mg/kg) were found for both grain Fe and Zn. The genotypic and phenotypic correlation matrix for grain Fe and Zn concentration for different geographical regions and years is presented in S2 Table. A significant genetic correlation was recorded for the grain Fe concentration among different geographical regions during 2013–2014. Similar results have been recorded for the grain Zn concentration. A low correlation was recorded between grain Fe and Zn concentration among different geographical locations and years. A histogram depicting the distribution of genotypes for grain Fe and Zn concentration is shown in S1 Fig.
Genetic diversity and population structure
A total of 220 alleles were generated through 73 polymorphic SSRs in the AM panel (Table 2). The number of alleles produced per locus ranged from 2 to 5, with an average of 2.97 alleles per locus. The highest number of alleles (5) was detected for SSRs PBALC 353 and SSR 33, whereas 17 SSRs exhibited only two alleles. The PIC value ranged from 0.08 to 0.68, with an average of 0.36, indicating that the SSRs used in the study were informative. Genomic SSRs produced a higher average number of alleles (Na), and PIC over EST SSRs (Table 2). SSR 33 produced the maximum number of alleles (5 with the highest PIC value of 0.68).
Table 2. List of the 73 EST and genomic SSRs primers used in present study.
| S.No | Primer | Primer sequence | Ta°C | Na | PIC |
|---|---|---|---|---|---|
| 1 | PLC5 |
F:CATTGCAGCTTATTCTCACAGC R:TGACCCATCCTCATCCTTAAAT |
60 | 4 | 0.63 |
| 2 | PLC10 |
F:TGCAACAAAGGACACTAGAGGTT R:ATTTCTTTCTCCCTAACCAGCC |
59 | 3 | 0.3 |
| 3 | PLC16 |
F:CGTTTGATCTTCTAAGCCCCTA R:AAGGGAAAGGATGTTTGACTTG |
59 | 3 | 0.41 |
| 4 | PLC17 |
F:AAGCTGAAGGAAATCAAAGTGG R:TCAACACACTCCATGTTTAGAGC |
59 | 3 | 0.44 |
| 5 | PLC21 |
F:AACTCGCATCCTCTTCACAACT R:GGACCTTTCCCTTGTAGTCACC |
58 | 3 | 0.24 |
| 6 | PLC30 |
F:TTGGTCAGGTTCTCAATCCTCT R:ACGGATGAACGCTTGTAAAGAA |
61 | 3 | 0.47 |
| 7 | PLC35 |
F:TTGCTTCCTCCTCTTCTCACTC R:AGCCTCAGTACCCTCCTCTTTT |
60 | 3 | 0.36 |
| 8 | PLC38 |
F:CCTGGAGAAGTCTGTGGAAGAT R:AGCTCTAGCATTTTGCATGTGA |
59 | 2 | 0.36 |
| 9 | PLC39 |
F:CAGAGAAATCCCCTGCTGAG R:CATGATTCCCATAGCCTTGC |
58 | 3 | 0.29 |
| 10 | PLC40 |
F:CAACTCGCATCCTCTTCACA R:CAAAGGGGTTGGAGTCGTAA |
60 | 4 | 0.43 |
| 11 | PLC42 |
F:AACCAATCATGGCTTCTGCT R:TTTCACCGTCTTTATGAACCA |
60 | 4 | 0.66 |
| 12 | PLC46 |
F:CAAACTGGAAGATGCTGCTG R:TGACCCATCCTCATCCTTAAA |
60 | 3 | 0.22 |
| 13 | PLC51 |
F:CCATGATGAGCCTTGAATGA R:TCTTCAATCTCCAGGAACACTTT |
62 | 4 | 0.52 |
| 14 | PLC60 |
F:TGCTTGGACCCTAAATTTGC R:AAGAAAAGGGCAACCACTGA |
60 | 3 | 0.29 |
| 15 | PLC62 |
F:AAGCCAACCATTTTTGCATC R:AGTAATCCTTTGGTGCTGCG |
58 | 4 | 0.53 |
| 16 | PLC63 |
F:TTGATGGCTATGGGAGTGGT R:TGGTCCCAACAAAATACCAA |
60 | 3 | 0.19 |
| 17 | PLC74 |
F:GATTTACCGATGGATCTTCA R:CTAAGGGAGAGAAAGAAAAGG |
61 | 2 | 0.08 |
| 18 | PLC77 |
F:GGAAAGAGCCAAGAAGTTG R:ACCCATCCTCATCCTTAAAT |
56 | 3 | 0.49 |
| 19 | PLC80 |
F:GCTAACAAACAACACCATGA R:GCATCTAAGTTCTTCAATCTCC |
58 | 3 | 0.25 |
| 20 | PLC81 |
F:GGGTAGAGTATTATTGAAGGTGG R:AGAATCGCTAGTTTAGAGCAAG |
60 | 3 | 0.44 |
| 21 | PLC83 |
F:GTTCGGTTTTGTTGGAAGTGAG R: TCCTTCTTTCAGCCATGAGATT |
60 | 3 | 0.35 |
| 22 | PLC88 |
F:CCAAAACAAGCACCAGTACAAG R:TAGAAGACGTTGGAGGAGAAGC |
59 | 3 | 0.42 |
| 23 | PLC95 |
F:TTCATTCTTGGGCTAGGGA R:TGCAGATGTGAAATACCTCAGT |
59 | 2 | 0.21 |
| 24 | PLC96 |
F:TTCATCGTCGTTAATCGGAAC R:GAGAGGAAGGACATTGGAAGAA |
59 | 2 | 0.22 |
| 25 | PLC98 |
F:GTGCGGTGTTGTTGTATTGATT R:TCTTTAGCTTCTTCCAAAACGG |
59 | 3 | 0.51 |
| 26 | PLC100 |
F:TGCTTTACTTCCTTCTCTCTTTGC R:TAAGCCATCCACTTGCATCC |
60 | 3 | 0.51 |
| 27 | PLC104 |
F:AGCTGTTGATTTTGGCGG R:CCGCAGATCCAGAAAAGAAG |
59 | 3 | 0.54 |
| 28 | PBALC2 |
F:GATGCGACGCAGAAGATTAAG R:TGACCATAACCATTCCTCTGAT |
59 | 2 | 0.24 |
| 29 | PBALC6 |
F:ATGATCCGAGTTTCCTGCA R:TACACCACCAACTTCCACCA |
60 | 3 | 0.24 |
| 30 | PBALC13 |
F:GCAGCAGCATGAGAAAATG R:ATTACTCGACGCCCCCTAGT |
60 | 3 | 0.33 |
| 31 | PBALC18 |
F:CGTTGGTGGTGCAGTATTTG R:CCATAAACAAGTGCAATCCAG |
60 | 2 | 0.2 |
| 32 | PBALC25 |
F:ACCCCTGCAAATGTCAAGAG R:AAATCCAAATGCATAACTTCATTG |
60 | 2 | 0.15 |
| 33 | PBALC29 |
F:TATGCCATTGGATGTGGTTG R:TATTCAGTTTCCGCCAAAGG |
60 | 3 | 0.22 |
| 34 | PBALC32 |
F:CTGGAGGGAAAAGATGACGA R:TTTCCCCAACTTTCCTAAGC |
60 | 4 | 0.45 |
| 35 | PBALC43 |
F:GCATGGTTAAGAAGAAGGGTGT R:TAACAACAAACAAGCGCATTA |
61 | 3 | 0.33 |
| 36 | PBALC203 |
F:CATAGTCAACACTTGGTCGTT R:GTCCACAATGAAACTCATCAC |
60 | 3 | 0.63 |
| 37 | PBALC205 |
F:TTGAGTTTGAGGATGAGGATA R:CATAAAACCCCAAACATTACA |
59 | 2 | 0.11 |
| 38 | PBALC206 |
F:GATCCTGTTTTATCCCATTGT R:ACAATCACTTAGCCAAAATCA |
59 | 2 | 0.29 |
| 39 | PBALC207 |
F:ATGGAACACAAACCAATACAC R:TGTGGTGTCCTTTGTAGAAGT |
59 | 2 | 0.44 |
| 40 | PBALC213 |
F:AAGTTTGGGATAAACCTTTTG R:CATCATGCTAAAATCAAAACC |
61 | 3 | 0.31 |
| 41 | PBALC216 |
F:AAATAGAAGTGGAGAGGCAAT R:TTCGTTCTTGAGTGATATCGT |
60 | 3 | 0.33 |
| 42 | PBALC217 |
F:TTACCAAGAAATTGAATACAGC R:AGTTTGAAAGGATCTCCAAAG |
60 | 3 | 0.17 |
| 43 | PBALC219 |
F:TAGCAAATGGACGTGTAGAGT R:GTGGTGCTCAATACACAATCT |
60 | 2 | 0.26 |
| 44 | PBALC224 |
F:CCACCCACTTACAAGTACAAA R:TAAATTGGTGGTGGTGAGTAA |
59 | 2 | 0.15 |
| 45 | PBALC238 |
F:CGCAATCCAAACCTAATCTAT R:TTCTAGGATGTGATTTTGGTG |
59 | 2 | 0.36 |
| 46 | PBALC250 |
F:TGCATTTACCATCATCTCTAAC R:TGATTGATTCGGTACTTTTTG |
60 | 3 | 0.48 |
| 47 | PBALC254 |
F:ATGTTAATAAGCAGCAGCAAC R:AAGTTGCATGTAACCACAAAC |
60 | 4 | 0.32 |
| 48 | PBALC260 |
F:GTGAACTACCTCTGTGAATGC R:AGGCGAAATTTCATCTTCTA |
60 | 4 | 0.4 |
| 49 | PBALC265 |
F:AACATAAAGGAGAGGGTCATC R:CATCTTGTCAACAATTCCTTC |
59 | 4 | 0.38 |
| 50 | PBALC347 |
F:CAAAAATGGCTACTTTGATTG R:GCTTCAGATCAACTGTCTCAG |
59 | 3 | 0.31 |
| 51 | PBALC353 |
F:CCATAACAGACAAAACCCTACT R:ATTCTCAAAGCCCATTTAGTT |
59 | 5 | 0.34 |
| 52 | PBALC742 |
F:AGCAAATTCTATTCCAACACA R:CCAATTCTACTTCCACCTTCT |
59 | 2 | 0.49 |
| 53 | PBALC762 |
F:TGATGGAACCAAACTTCTTTA R:TATCCTCCCTAAAATCAAAGG |
59 | 3 | 0.31 |
| Mean | 2.94 | 0.35 | |||
| 54 | GLLC106 |
F:ACGACAATCCTCCACCTGAC R:AACAAGGAAGGGGAGAGGAG |
56 | 3 | 0.18 |
| 55 | GLLC108 |
F:CGACAATCCTCCACCTGAC R:ACAAGGAAGGGGAGAGGAAG |
56 | 4 | 0.27 |
| 56 | GLLC527 |
F:GTGGGACGGTTTGAATTTGA R:GAACATAAAATGGGAGTGTCACAA |
56 | 3 | 0.44 |
| 57 | GLLC538 |
F:AAGGGAAGGAAAAGGGAAGT R:GCACGAAGAGGGTACGTAGG |
56 | 2 | 0.23 |
| 58 | GLLC541 |
F:TGGGCTCATTGAACCAAAAG R:CCCCCTTTTAAGTGATTTTCC |
56 | 3 | 0.47 |
| 59 | GLLC562 |
F:TGTGTAGGCACATCAACAAAA R:GGTGGGCATGAGAGGTGTTA |
55 | 4 | 0.47 |
| 60 | GLLC563 |
F:ATGGGCTCATTGAACAAAAG R:CCCCCTCTAAGAGATTTTCCTC |
56 | 3 | 0.59 |
| 61 | GLLC598 |
F:TGGGCTCATTGAACCAAAAG R:CCCCCTTCTAAGTGATTTTCC |
55 | 3 | 0.27 |
| 62 | GLLC609 |
F:GCGACATGGAATTGGATTTG R:GCACAAAGTCGAGGAGCCTA |
55 | 3 | 0.58 |
| 63 | GLLC614 |
F:AACCCCAGCCAGATCTTACA R:AAGGGTGGTTTTGGTCCTATG |
55 | 3 | 0.54 |
| 64 | SSR132RN |
F:CCAGAACAAACGTAAACC R:CTATCGCATATGAGTGAAC |
52 | 3 | 0.1 |
| 65 | SSR230 |
F:CCAACAACAATTCACCATAC R:AACATTGTACTGAGAGGTG |
53 | 3 | 0.51 |
| 66 | SSR317-1 |
F:GTGGGTGTAATTATTGCTAC R:GTATCAAACTTATGGTGAAATC |
53 | 3 | 0.57 |
| 67 | SSR66 |
F:GGTAGTGGTGAGGAATGAC R:GCATCACTGCAACAGACC |
55 | 3 | 0.4 |
| 68 | SSR72 |
F:CAAACAGTACAAGGAAAGGAG R:CTGACTGAGCTGCTTGAAC |
55 | 2 | 0.29 |
| 69 | SSR302 |
F:CAAGCCACCCATACACC R:GGGCATTAAGTGTGCTGG |
56 | 3 | 0.19 |
| 70 | SSR309-2 |
F:GTATGTCGTTAACTGTCGTG R:GAGGAAGGAAGTATTCGTC |
50 | 2 | 0.15 |
| 71 | SSR48 |
F:CATGGTGGAATAGTGATGGC R:CTCCATACACCACTCATTCAC |
57 | 3 | 0.55 |
| 72 | SSR33 |
F:CAAGCATGACGCCTATGAAG R:CTTTCACTCACTCAACTCTC |
56 | 5 | 0.68 |
| 73 | SSR233 |
F:CTTGGAGCTGTTGGTC R:GCCGCCTACATTATGG |
52 | 3 | 0.43 |
| Mean | 3.05 | 0.39 | |||
Ta = Annealing temperature, PIC = Polymorphism information content, Na = number of alleles
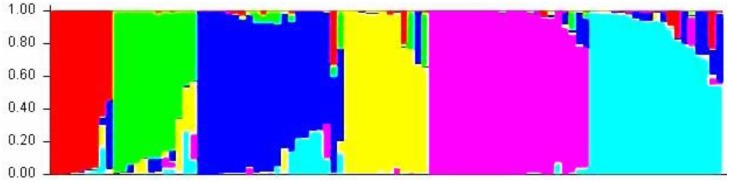
Admixture model-based simulations revealed six subgroups (SGs) in the panel. The six SGs represented as SG I, SG II, SG III, SG IV, SG V, and SG VI, which included 9, 12, 21, 12, 23, and 19 genotypes, respectively (Fig 1). SG I comprised of nine indigenous genotypes including released varieties and advanced breeding lines. SG II comprised of twelve genotypes including two exotic genotypes. SG III consisted of 21 genotypes including two exotic genotypes. SG IV comprised 12 indigenous genotypes developed at IARI, New Delhi. SG V included 23 exotic genotypes from ICARDA. SG VI comprised 19 indigenous genotypes. The grouping of genotypes in SG I, IV, V and VI was based on the origin of genotype. The SG II and III included mainly indigenous genotypes and few exotic genotypes.
Fig 1. STRUCTURE plot of the lentil association mapping population with K = 6 clusters based on all polymorphic SSRs.
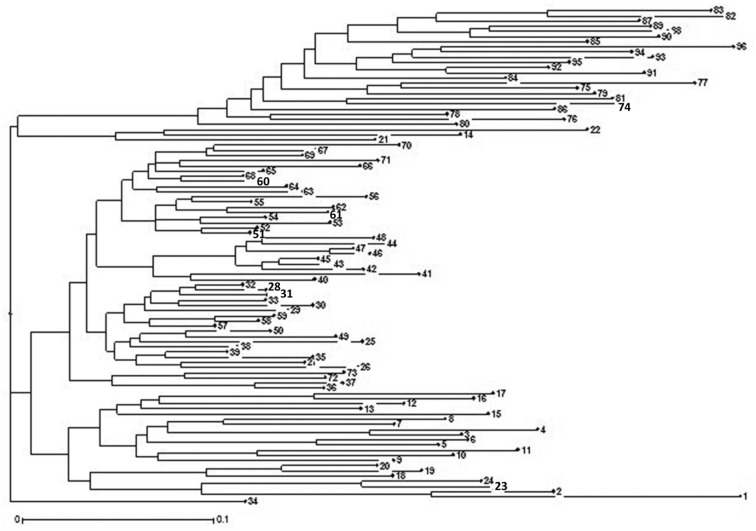
The UNJ separated the lentil genotypes into three clusters. Cluster I comprised 25 lentil genotypes including 23 exotic germplasm from ICARDA and two Indian lentil genotypes L 5253 and PL 02. Cluster II comprised 70 indigenous genotypes including advanced breeding lines and released varieties including breeding material from India. Cluster III comprised only one Indian genotype, namely MC 6. The genotypes within clusters I and II are further grouped into smaller subgroups on the basis of their origin and types. Most of the exotic genotypes from ICARDA were grouped in the upper branches of the dendrogram, whereas the advanced breeding lines developed at different lentil breeding centers of India, released lentil varieties, were grouped in lower branches (Fig 2).
Fig 2. Neighbour joining dendrogram of 96 genotype of lentil with 73 SSRs (Serial number of genotype in the figure corresponds with serial number and genotype in Table 1).
LD and marker trait association analysis
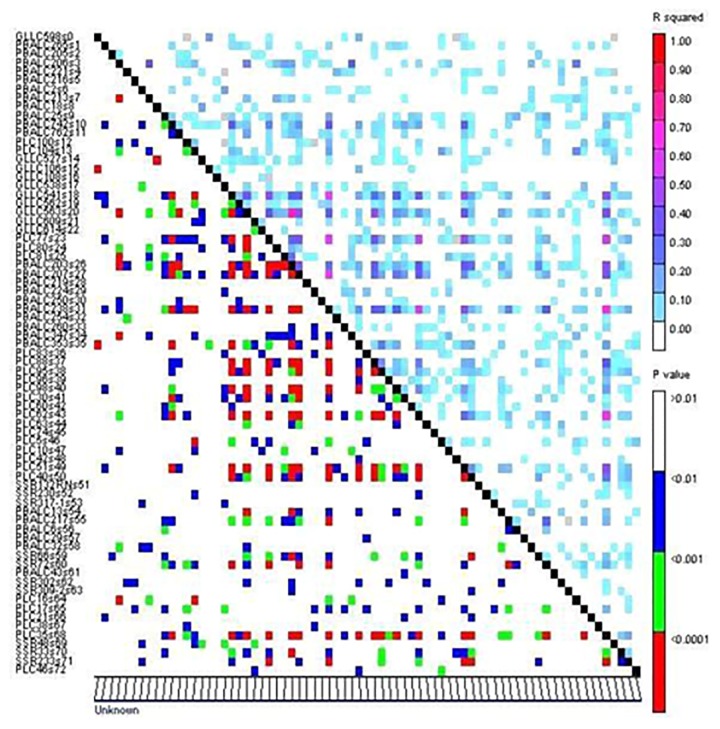
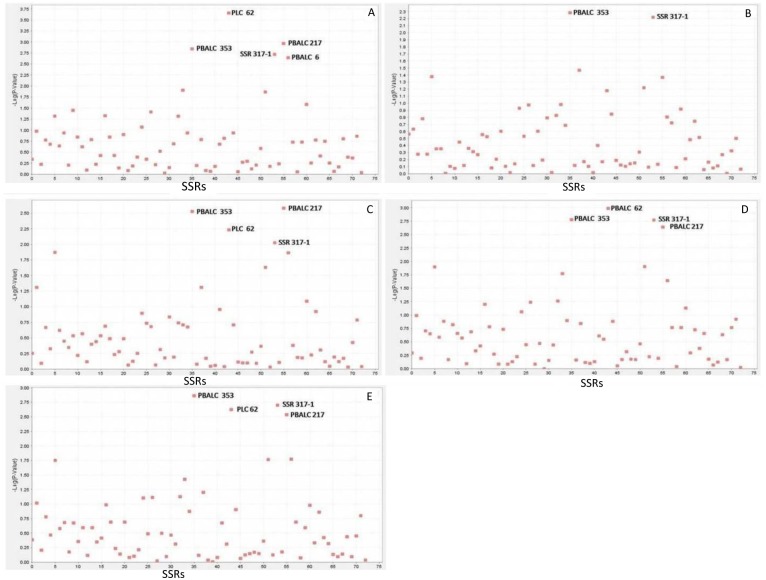
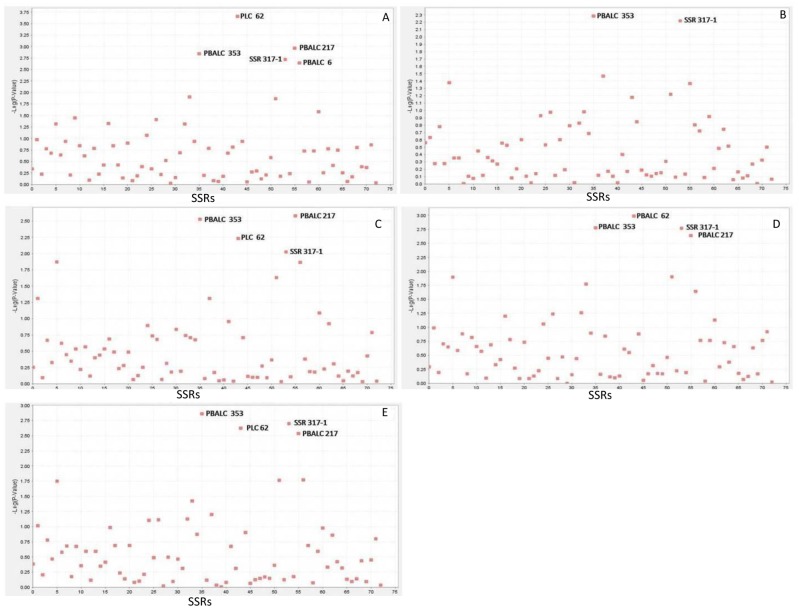
The LD patterns of all 2628 pairwise combinations of the 73 SSRs were assessed using TASSEL (Fig 3). The LD ranged from 0.0 to 0.70. The highest LD value was recorded between PLC 81 and GLLC 563, PLC 38, and PLC 60. In the present study, association mapping was used to identify linked markers for grain Fe and Zn concentration by using the GLM with the Q model. In total, eight SSRs (contributing to 8–22% of the phenotypic variation) were significantly associated with the grain Fe concentration, and five SSRs (contributing to 4–21% of the phenotypic variation) with the grain Zn concentration (Table 3). Environment-wise different SSRs associated with grain Fe and Zn concentration with -log10 p value >2 are presented in Figs 4 and 5. For the grain Fe concentration, the marker PBALC 13 was consistently identified in all four datasets; GLLC 563 in three datasets (Indore 2013–14, Delhi 2014–15 and Delhi 2013–14) and PBALC 206 (Dharwad 2013–14 and Delhi 2014–15) and PBALC 32 (Delhi 2013–14 and Delhi 2014–15) in two datasets each. For the grain Zn concentration, the marker PBALC 353 was consistently identified in all four datasets and SSR 317–1 (Delhi 2013–14, Delhi 2014–15, and Dharwad 2013–14), PLC 62 (Delhi 2013–14, Delhi 2014–15 and Indore 2013–14) and PBALC 217 (Delhi 2013–14, Delhi 2014–15 and Indore 2013–14) in three datasets each. Few SSRs exhibited a consistent association with grain Fe and Zn concentration across environments. Three SSRs (PBALC 13, PBALC 206, and GLLC 563) exhibiting an association with the grain Fe concentration revealed phenotypic variation of 11%, 9%, and 11%, respectively. SSRs PBALC 353, SSR 317–1, PLC 62, and PBALC 217 were found to be associated with the grain Zn concentration with the phenotypic variation of 21%, 18%, 14%, and 16%, respectively. The markers that were significantly associated with the trait were represented in Manhattan plots (Figs 4 and 5). A QQ plot is a graphical method of depicting the observed and expected probability distributions by plotting their quantiles next to each other. The results derived from the GLM analysis were also explained in these plots (S2 Fig). The hypothetical QQ plots of the marker-trait association study for grain Fe and Zn concentration are a good approximation of normality.
Fig 3. Linkage disequilibrium patterns among 96 genotypes genotyped with 73 SSRs.
The squared correlation coefficients (r2) for each pair of markers are presented in the upper triangle and their corresponding p values in the lower triangle.
Table 3. List of significantly associated marker with grain Fe and Zn concentration.
| Trait | SSR marker | Location | Year | P value | r2 value |
|---|---|---|---|---|---|
| Iron | PBALC13 | Dharwad | 2013–14 | 0.0030 | 0.0897 |
| Delhi | 2014–15 | 0.0021 | 0.0963 | ||
| Indore | 2013–14 | 0.0006 | 0.116 | ||
| Delhi | 2013–14 | 0.0021 | 0.0963 | ||
| Combined all location years | 0.0006 | 0.1168 | |||
| PBALC 221 | Dharwad | 2013–14 | 0.0053 | 0.1065 | |
| PBALC 206 | Dharwad | 2013–14 | 0.0063 | 0.0767 | |
| Delhi | 2014–15 | 0.0011 | 0.1073 | ||
| Combined all location years | 0.0019 | 0.09719 | |||
| PBALC 32 | Delhi | 2014–15 | 0.0047 | 0.1505 | |
| Delhi | 2013–14 | 0.0047 | 0.1505 | ||
| GLLC 563 | Delhi | 2013–14 | 0.0048 | 0.1084 | |
| Indore | 2013–14 | 0.0012 | 0.1354 | ||
| Delhi | 2014–15 | 0.0048 | 0.1084 | ||
| Combined all location years | 0.0043 | 0.1104 | |||
| PBALC 265 | Indore | 2013–14 | 0.0031 | 0.2276 | |
| PBALC 207 | Indore | 2013–14 | 0.0039 | 0.0852 | |
| PBALC 203 | Indore | 2013–14 | 0.0054 | 0.1061 | |
| PBALC 265 | Indore | 2013–14 | 0.0031 | 0.2276 | |
| Zinc | PBALC 353 | Dharwad | 2013–14 | 0.0052 | 0.1835 |
| Indore | 2013–14 | 0.0030 | 0.1960 | ||
| Delhi | 2014–15 | 0.0014 | 0.2115 | ||
| Delhi | 2013–14 | 0.0017 | 0.2081 | ||
| Combined all location years | 0.0013 | 0.2121 | |||
| SSR 317–1 | Dharwad | 2013–14 | 0.0061 | 0.1631 | |
| Delhi | 2014–15 | 0.0019 | 0.1877 | ||
| Delhi | 2013–14 | 0.0017 | 0.1901 | ||
| Combined all location years | 0.0020 | 0.1868 | |||
| PLC 62 | Delhi | 2014–15 | 0.0002 | 0.190 | |
| Indore | 2013–14 | 0.0059 | 0.1263 | ||
| Delhi | 2013–14 | 0.0010 | 0.1606 | ||
| Combined all location years | 0.0024 | 0.1443 | |||
| PBALC 217 | Delhi | 2014–15 | 0.0011 | 0.1803 | |
| Indore | 2013–14 | 0.0026 | 0.1624 | ||
| Delhi | 2013–14 | 0.0023 | 0.1652 | ||
| Combined all location years | 0.0029 | 0.1605 | |||
| PBALC 6 | Delhi | 2014–15 | 0.0022 | 0.123 |
Fig 4. Manhattan plot depicting association of 73 SSRs markers with grain iron concentration for (A) Delhi (2013–14) (B) Dharwad (2013–14) (C) Indore (2013–14) (D) Delhi (2014–15) (E) Combined all location year.
Fig 5. Manhattan plot depicting association of 73 SSRs markers with grain zinc concentration for (A) Delhi (2013–14) (B) Dharwad (2013–14) (C) Indore (2013–14) (D) Delhi (2014–15) (E) Combined all location years.
Favorable allele mining
Phenotypic allele effects (ai) were calculated for SSRs associated with grain Fe and Zn concentration. The ai value greater than zero indicates that the allele has a positive effect, whereas the value of ai less than zero indicates that the allele has a negative effect. The favorable alleles identified for the grain Fe concentration include PBALC 13–1, PBALC 206–1, GLLC 563–1, GLLC 563–2, and PBALC 32–1, of which PBALC 13–1 exerted the maximum positive effect on a phenotype. Similarly, the favorable alleles identified for the grain Zn concentration include PBALC 353–3, PLC 62–3, and PLC 62–4. PLC 62–4 had the maximum positive effect on the grain Zn concentration. The representative genotypes of favorable alleles for grain Fe and Zn concentration are listed in Table 4.
Table 4. Favorable alleles for grain Fe and Zn concentration.
| Trait | Favorable allele | ai * | No. of genotype | Representative genotypes |
|---|---|---|---|---|
| Fe | PBALC13-1 | +10.66 | 17 | L404, L 830, L 4596, L4602, L4603, L4698, DPL21, PL02,P2124, P2125,P2126, P2127, P2130,P3113,P3204,P3208,P15104 |
| PBALC206-1 | +9.73 | 17 | L404, L 830, L 4596, L4602, L4603, L4620, L4648, L4649, L4650,L4698, L5120, L5126, L5253, ILL7663,L7818,L7903 | |
| GLLC563-1 | +2.62 | 20 | L404, L 830, L 4596, L4602, L4603, L4620, L4648, L4649, L4650,L4698, L5120, L5126, L5253, ILL7663, DPL15, DPL21, DPL58 | |
| GLLC563-2 | +7.72 | 23 | P2102,P2124,P2125,P2126,P2127,P2130,P2205,P2215,P2230,P2233,P2239,P3113,P3204,P3208,P3220,P13104,P13113, P13122,P13135,P13143, P14103,P14201,P15104 | |
| PBALC32-1 | +10.58 | 18 | L404, L 830, L 4596, L4602, L4603, L4618, L4620, L4648, L4649, L4650,L4698, LL1231, P2102, P2124,P2125,P2126,P2127,P2130 | |
| Zn | PBALC353-3 | +8.54 | 18 | P13129,PL129,P2102,P2124,P2126,P2130,P2215,P2230,P2233, P3113,P3208,P13104, P13113, P13143, PL02,L4698,L5253 |
| PLC62-2 | +3.53 | 10 | DPL15,DPL21,DPL58,PL101,PL639,RL1,ILL2581, SKL259,L4590,PL4 | |
| PLC62-4 | +9.84 | 7 | P2124,P2126,P2130,P3113,P3204,P3208,P3220 |
*ai = the phenotypic effect of allelic variation
Discussion
Genetic variation in grain Fe and Zn concentration
Breeding micronutrient-rich crops is required to combat micronutrient deficiencies in humans [80]. The characterization of genotypes for micronutrient concentrations in different environments is essential for identifying genotypes rich in Fe and Zn. In the present study, high genetic variation for grain Fe and Zn concentration was recorded. Approximately 29.1% of genotypes had a high mean grain Fe concentration (>70 mg/kg) and 31.2% of genotypes had a low mean grain Fe concentration (<60 mg/kg). Similarly, 22.9% of genotypes had a high mean grain Zn concentration (>50 mg/kg) and 20.8% of the genotypes had a low mean grain Zn concentration (<40 mg/kg). The results indicated that the grain Fe concentration was higher in the indigenous genotypes than in the exotic genotypes. By contrast, the exotic genotypes were relatively richer in grain Zn concentrations than were the indigenous genotypes. The germplasm has been characterized for grain Fe and Zn concentration in wheat [81, 82], rice [83] and chickpea [84, 85]. A wide range of variability in grain Fe and Zn concentration in lentil [86, 87] indicated the role of genotypes and environmental interactions in the expression of these traits. The variation in grain micronutrient concentration of the lentil genotypes at different locations and years in the present study was due to the sensitivity of genotypes toward variations in weather and soil conditions. Soil parameters affect the availability of Zn uptake in plants and control the amount of organic matter and the grain Zn concentration in soil solutions [88]. By hybridizing indigenous Fe-rich grain varieties with exotic Zn-rich grain varieties, the concentration of both micronutrients can be improved simultaneously and the genetic base of indigenous genotypes can be broadened. Biofortified lentil varieties can be sustainable as well as cost-effective for alleviating the micronutrient deficiency, thereby complementing the process of food fortification. Thavarajah et al. [89] reported that 100 g of dry lentil can provide the recommended daily allowance of micronutrients (Fe and Zn) in adults. Therefore, lentil can be consumed as a whole grain to meet the daily requirement of grain micronutrient concentration in humans.
Correlation is a crucial aspect of the crop improvement program and is used to improve correlated traits simultaneously or reduce undesirable effects of some traits on another. We observed a positive correlation (r = 0.11) for grain Fe and Zn concentration. Similar results have been reported in other studies [90, 91]. Fe and Zn are found in similar types of foods, and their absorption mechanisms are affected by food compounds. Therefore, the biochemical status of Fe and Zn may be interconnected [92]. A significant correlation between grain Fe and Zn concentration was reported in fieldpea [93], barley [94] wheat [95] and maize [96, 97]. The positive association between these traits suggests some common genetic mechanisms for the uptake, accumulation, and concentration of Fe and Zn. Correlation is very useful in the crop improvement program as a breeder can improve the micronutrient concentration of both the traits (Fe and Zn) simultaneously. Although the levels of Zn and Fe in grains are positively related, fertilization of one element did not affect the grain concentration of the other [98, 99].
SSR allelic diversity and population structure
SSR markers due to their codominant nature have been widely utilized for genetic diversity, gene tagging, and linkage mapping in numerous crop plants including lentil [31,32,72,100]. In this study, the genetic variability and population structure were analyzed among 96 germplasm with 73 polymorphic SSRs. The mean PIC value was 0.36. Twenty genomic SSRs exhibited higher mean PIC of 0.39 whereas fifty three EST SSRs exhibited mean PIC of 0.35. EST-SSRs located in the proximity of the coding region are associated with the expressed gene/QTL; hence, they are superior markers [101]. The PIC is a critical factor for selecting SSRs for the characterization of germplasm and tagging of genes [102]. PIC offers a more accurate assessment of diversity than does the raw number of alleles because PIC takes into account the relative frequencies of each allele [103]. It also indicates the ability to discriminate among genotypes. Kushwaha et al. [104] reported that markers with a PIC value ranging between 0.4 and 0.8 can be considered as informative exhibiting high polymorphism. The PIC observed in the current study were comparable with those reported in previous study by Andeden et al. [105], where PIC ranged from 0.13 to 0.79. In our study, the genetic diversity of genomic SSRs was higher than that of EST-SSRs and these findings are consistent with those of previous studies on lentil [100] and barley [106]. A lower level of polymorphism in EST-SSRs may be due to selection against variation in the conserved regions. PIC values of the SSRs PBALC 13, PBALC 206, and GLLC 563 associated with the grain Fe concentrations were 0.33, 0.29, and 0.59, respectively. Similarly, the PIC values of the SSRs PBALC 353, SSR 317–1, PLC 62, and PBALC 217 associated with the grain Zn concentration were 0.34, 0.57, 0.53, and 0.17, respectively.
The population structure explained the presence of six subgroups in the AM panel. (Fig 2). The subgroup SG 5 consisted of 23 genotypes from the Mediterranean region. This group separated the Mediterranean genotypes from the remaining genotypes from India. The initial report [5] indicated that lentil genotypes from India have a narrow genetic base and are genetically more isolated than that of the the ICARDA genotypes. However in the last two decades Mediterranean material has been used for broadening the genetic base. Previous studies on lentil diversity by using molecular markers have revealed mostly two distinct groups, namely South Asian and all other origins [86,107,108] results from PCoA and cluster analyses also demonstrated a narrower genetic variability among the Indian material. Similarly Mekonnen et al. [109] and Koul et al. [110] reported five subgroups in the lentil germplasm. The genotyping of available genetic diversity has demonstrated the need for incorporation of the exotic germplasm into breeding programs for broadening the genetic base.
Association analysis and favorable allele identification
In the present study, we identified markers that are consistent across environments. Three SSRs were associated with the grain Fe concentration and four SSRs with the grain Zn concentration. After the validation of these trait-associated SSRs, the markers can be used for identifying genes/QTLs regulating grain Fe and Zn concentration. Furthermore, they can be useful in the marker-assisted genetic enhancement programs. The markers exhibiting an association in more than two environments were considered more reliable than the markers present in a particular environment. The SSRs PBALC 13 and GLLC 563 accounted for phenotypic variation of 11%, whereas PBALC 206 explained 9% of phenotypic variation for the grain Fe concentration. For the grain Zn concentration, PBALC 353 exhibited the highest phenotypic variation (21%), followed by SSR 317–1 (18%), PBALC 217 (16%), and PLC62 (14%). Similar level of trait variation for grain Fe and Zn concentration was recorded in fieldpea [93] and chickpea [84]. The size of our lentil germplasm panel was adequate. Even with small population size the high quality extensive phenotyping offers a reasonable basis for the association of mapping studies [111]. Association mapping studies in bean [112], peanut [113], barley [114], tomato [115] and sugarcane [111] also used approximately 90 genotypes. Atwell et al. [63] reported that for some of the traits, significant results can be achieved in a population size of less than 100. The putative functions of PBALC 217 reported by Kaur et al. [51] were assigned through comparison with the non redundant sequence database at the National Center for Biotechnology Information by using the basic local alignment search tool (BLAST) BLASTX program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). This SSR exhibited homology with dormancy or auxin-associated protein in Medicago. truncatula (E value = 3e-42). Similarly, Upadhayay et al. [85] reported the auxin/IAA as a known/putative function of SNP (CakSNP1628/SNP55) associated with the seed Fe concentration.
By using genomic SSRs, association mapping has been successfully demonstrated in rice [116], wheat [117] and chickpea [118]. Our study in lentil using SSRs is the first attempt to identify QTLs or genes for grain Fe and Zn concentration. Gupta et al. [119] used 50 SSRs for the mapping QTLs for agronomic traits in foxtail millet. Gorafi et al. [82] used 70 SSRs for the identification of linked markers for grain Fe and Zn concentration in wheat. Lou et al. [120] used 90 SSRs for the mapping QTLs for agronomic traits in Fescue. Gyawali et al. [121] used 84 SSRs for association mapping in Brassica napus.
The association mapping approach was useful in identification of marker loci linked with the grain Fe and Zn concentration. This approach also aided in mining alleles and further utilizing these favorable alleles with the maximum positive effect in marker-assisted selection, as suggested by Wan [122]. The identified favorable alleles can be used in the lentil breeding program for improving grain Fe and Zn concentration. For the grain Fe concentration, the maximum positive effect for the favorable allele PBALC 13–1 was recorded in seventeen genotypes (L 404, L 830, L 4596, L 4602, L 4603, L 4698, DPL 21, PL 02, P 2124, P 2125, P 2126, P 2127, P 2130, P 3113, P 3204, P 3208, and P 15104). Similarly, for the grain Zn concentration, the maximum positive effect for the favorable allele PLC 62–4 was recorded in seven genotypes (P 2124, P 2126, P 2130, P 3113, P 3204, P 3208, and P 3220) originating from ICARDA. These genotypes can be utilized in a hybridization program for simultaneous improvement of grain Fe- and Zn-rich varieties. Similar studies have also been performed in other crops such, wheat [106] and tomato [115].
Biofortification (breeding micronutrient-rich crops) of lentil can be achieved through plant breeding without affecting the yield or quality. The process like transfer of other traits is tedious and involves screening of germplasm, hybridization, study of mode of inheritance, molecular marker-assisted selection, and regular phenotyping of segregating populations. The approach is a sustainable and cost-effective solution for delivering micronutrients [123] and has emerged as an agriculture-based strategy to fulfill the nutritional requirement of malnourished people throughout the world. Considerable knowledge has been obtained on the molecular mechanisms affecting the accumulation of Fe [124] and Zn [125] in plants. In the future, these studies can be applied to develop crops with enhanced mineral concentration with the help of biotechnological tools in conventional breeding. In this study, markers were identified as being linked to grain Fe and Zn concentration. These identified SSRs can be further validated and deployed in marker-assisted selection for developing grain Fe and Zn rich lentil varieties. Superior positive alleles can be pyramided by hybridization for enhancement of grain Fe and Zn concentration.
Supporting information
(TIF)
(TIF)
(XLS)
(XLSX)
(XLSX)
Acknowledgments
Authors are thankful to Director, Joint Director Research and Head, Division of Genetics, for providing the necessary facilities for research. The authors also acknowledge the partial funding support received from Harvest Plus through ICARDA for carrying out the research work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research work was funded through ICAR-IARI in house project CRSIARISIL2014007239 and partial funding for this work was received from Harvest Plus through ICARDA. There was no additional funding received for this study.
References
- 1.Zohary D. Monophyletic vs. polyphyletic origin of the crops on which agriculture was founded in the Near East. Genet. Resour. Crop Evol. 1999; 46:133–142. [Google Scholar]
- 2.Cubero J I. Origin, domestication and evolution In: Webb C, Hawtin GC, editors. Lentils. CAB: London, UK; 1981. pp. 15–38. [Google Scholar]
- 3.Ladizinski D, Braun D, Muehlbauer FJ. The biological species of the genus Lens. Bot. Gaz. 1984; 145:235–261. [Google Scholar]
- 4.Erskine W. Lessons for breeders from land races of lentil. Euphytica. 1997; 93: 107–112. [Google Scholar]
- 5.Erskine W, Chandra S, Chaudhry M, Malik IA, Sarker A, Sharma B, et al. A bottleneck in lentil: widening its genetic base in South Asia. Euphytica. 1998; 101: 207–211. [Google Scholar]
- 6.Rahman MM, Sarker A, Kumar S, Ali A, Yadav NK, Rahman ML. Breeding for short season environments In: Erskine W, Muehlbauer F, Sarker A, Sharma B, editors. The lentil: botany, production and uses. Wallingford: CAB International; 2009. pp.121–136. [Google Scholar]
- 7.Kumar S, Rajendran K, Kumar J, Hamwieh A, Baum M. Current knowledge in lentil genomics and its application for crop improvement. Front. Plant Sci. 2015; 6: 78 10.3389/fpls.2015.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanyolac B, Ozatay S, Kahraman A, Muehlbauer F. Linkage mapping of lentil (Lens culinaris L.) genome using recombinant inbred lines revealed by AFLP, ISSR, RAPD and some morpho logical markers. J. Agric. Biotech. Sustainable Dev. 2010; 2: 1–6. [Google Scholar]
- 9.FAO 2014. http://www.fao.org/faostat/en/#data/QC
- 10.Fageria NK, Baligar VC, Claerk RB. Micronutrients in crop production. Adv. Agron. 2002; 77: 85–268. [Google Scholar]
- 11.Fageria NK, Baligar VC. Ameliorating soil acidity of tropical Oxisols by liming for sustainable crop production. Adv. Agron. 2008; 99: 345–399. [Google Scholar]
- 12.West KP, Stewart CP, Caballero B, Black RE. Nutrition in Global Health: Diseases, Programs, Systems, and Policies. Merson MH, Black RE, Mills AJ, editors. Burlington: Jones and Bartlett Learning; 2012. pp. 271–304. [Google Scholar]
- 13.Haas JD, Brownlie T. Iron deficiency and reduced work capacity: a critical review of the research to determine a causal relationship. J. Nutr. 2001; 131: 676–690. [DOI] [PubMed] [Google Scholar]
- 14.De Benoist B, McLean E, Egli I, Cogswell M. Worldwide prevalence of anaemia 1993–2005: WHO global database on anaemia. Geneva: WHO; 2008. [Google Scholar]
- 15.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011; 474: 327–336. 10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rush D. Nutrition and maternal mortality in the developing world. Am. J. Clinical Nutr. 2000; 72:212s–240s. [DOI] [PubMed] [Google Scholar]
- 17.Failla ML. Trace elements and host defense: Recent advances and continuing challenges. J Nutr. 2003; 133:1443S–1447S. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization. Iron deficiency anaemia: assessment, prevention and control: a guide for programme managers. Geneva: WHO; 2001. http://www.who.int/iris/handle/10665/66914 [Google Scholar]
- 19.Graham RD. Micronutrient deficiencies in crops and their global significance In Alloway BJ editors. Micronutrient deficiencies in global crop production.Netherlands: Springer; 2008. pp. 41–61. [Google Scholar]
- 20.Fageria NK, Moraes MF, Ferreira EPVB, Knupp AM. Biofortification of trace elements in food crops for human health. Commun. Soil Sci. Plan. Ana. 2012; 43: 556–570. [Google Scholar]
- 21.Gernand AD, Schulze KJ, Stewart CP, West KP, Christian P. Micronutrient deficiencies in pregnancy worldwide: health effects and prevention. Nat. Rev. Endocrinol. 2016; 12: 274–289. 10.1038/nrendo.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rink L, Gabriel P. Zinc and the immune system. Proc. Nutr. Soc. 2000; 59: 541–552. [DOI] [PubMed] [Google Scholar]
- 23.Black RE, Victora CG, Walker SP, Bhutta ZA, Christian P, De Onis M, et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet. 2013; 382: 427–451. 10.1016/S0140-6736(13)60937-X [DOI] [PubMed] [Google Scholar]
- 24.Wessells K R, Brown K. Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS One. 2012; 7, e50568 10.1371/journal.pone.0050568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajait C, Thawani V. Role of zinc in pediatric diarrhea. Indian J Pharmacol. 2011; 43:232–235. 10.4103/0253-7613.81495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullis KB, Faloona FA. Specific Synthesis of DNA in vitro via a Polymerase-Catalyzed Chain Reaction. Method Enzymol. 1987; 155: 335–350. [DOI] [PubMed] [Google Scholar]
- 27.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, et al. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985; 230: 1350–1354. [DOI] [PubMed] [Google Scholar]
- 28.Ford R, Pang ECK, Taylor PWJ. Diversity analysis and species identification in Lens using PCR generated markers. Euphytica. 1997; 96: 247–255. [Google Scholar]
- 29.Sharma SK, Knox MR, Ellis THN. AFLP analysis of diversity and phylogeny of Lens and its comparison with RAPD analysis. Theor. Appl. Genet. 1996; 93: 751–758. 10.1007/BF00224072 [DOI] [PubMed] [Google Scholar]
- 30.Hamwieh A, Udupa SM, Choumane W, Sarker A, Dreyer F, Jung C, et al. A genetic linkage map of Lens sp. based on microsatellite and AFLP markers and the localization of Fusarium vascular wilt resistance. Theor. Appl. Genet. 2005; 110: 669–677. 10.1007/s00122-004-1892-5 [DOI] [PubMed] [Google Scholar]
- 31.Hamwieh A, Udupa M, Sarker A, Jung C, Baum M. Development of new microsatellite markers and their application in the analysis of genetic diversity in lentils. Breed. Sci. 2009; 59: 77–86. [Google Scholar]
- 32.Verma P, Sharma TR, Srivastava PS, Abdin MZ, Bhatia S. Exploring genetic variability within lentil (Lens culinaris Medik.) and across related legumes using a newly developed set of microsatellite markers. Mol. Biol. Rep. 2014;41:5607–5625. 10.1007/s11033-014-3431-z [DOI] [PubMed] [Google Scholar]
- 33.Jain N, Dikshit HK, Singh D, Singh A, Kumar H. Discovery of EST-Derived Microsatellite Primers in the Legume Lens culinaris (Fabaceae). Appl. Pl. Sci. 2013; 1:1200539 10.3732/apps.1200539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldstein DB, Roemer GW, Smith DA, Reich DE, Bergman A, Wayne RK. The use of microsatellite variation to infer population structure and demographic history in a natural model system. Genetics. 1999; 151: 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006; 9: 615–629. 10.1111/j.1461-0248.2006.00889.x [DOI] [PubMed] [Google Scholar]
- 36.Ellis JR, Burke JM. EST-SSRs as a resource for population genetic analyses. Heredity. 2007; 99: 125–132. 10.1038/sj.hdy.6801001 [DOI] [PubMed] [Google Scholar]
- 37.Jarne P, Lagoda PJ. Microsatellites, from molecules to populations and back. Trends Ecol. Evol. 1996; 11: 424–429. [DOI] [PubMed] [Google Scholar]
- 38.Powell W, Morgante M, Mcdevitt R, Vendramin GG, Rafalski JA. Polymorphic simple sequence repeat regions in chloroplast genomes–applications to the population-genetics of pines. Proc. Natl. Acad. Sci. 1995; 92:7759–7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soranzo N, Provan J, Powell W. An example of microsatellite length variation in the mitochondrial genome of conifers. Genome. 1999; 42:158–161. [PubMed] [Google Scholar]
- 40.Paniego N, Echaide M, Munoz M, Fernandez L, Torales S, Faccio P, et al. Microsatellite isolation and characterization in sunflower (Helianthus annuus L.). Genome. 2002; 45: 34–43. [DOI] [PubMed] [Google Scholar]
- 41.Varshney RK, Thiel T, Stein N, Langridge P, Graner A. In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol. Bio. Lett. 2002; 7: 537–546. [PubMed] [Google Scholar]
- 42.Nagaraj SH, Gasser RB, Ranganathan S. A hitchhiker’s guide to expressed sequence tag (EST) analysis. Briefings Bioinformatics. 2007; 8: 6–21. [DOI] [PubMed] [Google Scholar]
- 43.Liang X, Chen X, Hong Y, Liu H, Zhou G, Li S, et al. Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol. 2009; 9:35 10.1186/1471-2229-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, et al. SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet. 2001; 102:865–870. [Google Scholar]
- 45.Decroocq V, Fave MG, Hagen L, Bordenave L, Decroocq S. Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor. Appl. Genet. 2003; 106: 912–922. 10.1007/s00122-002-1158-z [DOI] [PubMed] [Google Scholar]
- 46.Liewlaksaneeyanawin C, Ritland CE, El-Kassaby YA, Ritland K. Single-copy, species-transferable microsatellite markers developed from loblolly pine ESTs. Theor. Appl. Genet. 2004; 109: 361–369. 10.1007/s00122-004-1635-7 [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez MV, VazPatto MC, Huguet T, Cubero JI, Moreno MT, Torres AM. Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theor. Appl. Genet. 2005; 110: 1210–1217. 10.1007/s00122-005-1951-6 [DOI] [PubMed] [Google Scholar]
- 48.Pashley CH, Ellis JR, McCauley DE, Burke MJ. EST databases as a source for molecular markers: lessons from Helianthus. J. Hered. 2006; 97: 381–388. 10.1093/jhered/esl013 [DOI] [PubMed] [Google Scholar]
- 49.Choudhary S, Sethy NK, Shokeen B, Bhatia S. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor. Appl. Genet. 2009;118: 591–608. 10.1007/s00122-008-0923-z [DOI] [PubMed] [Google Scholar]
- 50.Mnejja M, Garcia-Mas JJ, Audergon M, Arus P. Prunus microsatellite markers transferability across rosaceous crops. Theor. Appl. Genet. 2010; 6: 689–700. [Google Scholar]
- 51.Kaur S, Cogan NO, Pembleton LW, Shinozuka M, Savin KW, Materne M, et al. Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genomics. 2011; 12:265 10.1186/1471-2164-12-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005; 142: 169–196. [Google Scholar]
- 53.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. 1991; 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stich B, Maurer HP, Melchinger AE, Frisch M, Heckenberger M, van der Voort JR, et al. Comparison of linkage disequilibrium in elite European maize inbred lines using AFLP and SSR markers. Mol. Breed. 2006;17: 217–226. [Google Scholar]
- 55.Jannink JL, Walsh B. Association mapping in plant populations In: Kang MS, editors. Quantitative Genetics, Genomics and Plant Breeding. UK: CAB International; 2002. pp. 59–68. [Google Scholar]
- 56.Chakraborty R, Weiss KM. Admixture as a tool for finding genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. 1988; 85: 9119–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Natl. Genet. 1999; 22: 139–144. [DOI] [PubMed] [Google Scholar]
- 58.Weiss KM, Clark AG. Linkage disequilibrium and mapping of human traits. Trends Genet. 2002; 18:19–24. [DOI] [PubMed] [Google Scholar]
- 59.Taniguchi H, Lowe CE, Cooper JD, Smyth DJ, Bailey R, Nutland S, et al. Discovery, linkage disequilibrium and association analyses of polymorphisms of the immune complement inhibitor, decay-accelerating factor gene (DAF/CD55) in type 1 diabetes. BMC Genet. 2006; 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to Linkage Disequilibrium using haplotype tags: A class of tests and the determinants of statistical power. Hum. Hered. 2003;56: 18–31. 10.1159/000073729 [DOI] [PubMed] [Google Scholar]
- 61.Gupta PK, Rustgi S, Kulwal PL. Linkage disequilibrium and association studies in higher plants: Present status and future prospects. Plant Mol. Biol. 2005; 57: 461–485. 10.1007/s11103-005-0257-z [DOI] [PubMed] [Google Scholar]
- 62.Cross AJ, Leitzmann MF, Gail MH, Hollenbeck AR, Schatzkin A, Sinha R. A prospective study of red and processed meat intake in relation to cancer risk. PLoS Med. 2007; 4 (12):e325 10.1371/journal.pmed.0040325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010; 465: 627–631. 10.1038/nature08800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rafalski JA. Association genetics in crop improvement. Curr. Opin. Plant Biol. 2010; 13: 174–180. 10.1016/j.pbi.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 65.Neumann K, Kobiljski B, Dencic S, Varshney RK, Borner A. Genome wide association mapping: a case study in bread wheat (Triticum aestivum L.). Mol. Breed. 2010; 27: 37–58. [Google Scholar]
- 66.Maccaferri M, Sanguineti MC, Demontis A, El-Ahmed A, Garcia del Moral L LG, Maalouf F, et al. Association mapping in durum wheat grown across a broad range of water regimes. J. Exp. Bot. 2011; 62: 409–438. 10.1093/jxb/erq287 [DOI] [PubMed] [Google Scholar]
- 67.Le Gouis J, Bordes J, Ravel C, Heumez E, Faure S, Praud S, et al. Genome-wide association analysis to identify chromosomal regions determining components of earliness in wheat. Theor. Appl. Genet. 2012; 124: 597–611. 10.1007/s00122-011-1732-3 [DOI] [PubMed] [Google Scholar]
- 68.Abdurakhmonov IY, Abdukarimov A. Application of association mapping to understanding the genetic diversity of plant germplasm resources. Int. J Plant Genom. 2008; 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Inghelandt D, Melchinger AE, Martinant JP, Stich B. Genome-wide association mapping of flowering time and northern corn leaf blight (Setosphaeria turcica) resistance in a vast commercial maize germplasm set. BMC Plant Bio. 2011; 12: 56 10.1186/1471-2229-12-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh D, Chonkar PK, Dwivedi BS. Manual on soil, plant and water analysis. New Delhi: Westville Publishers; 2005. [Google Scholar]
- 71.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980; 8:4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saha GC, Sarker A, Chen W, Vandemark G J, Muehlbauer FJ. Identification of markers associated with genes for rust resistance in Lens culinaris Medik. Euphytica. 2010;175: 261–265. [Google Scholar]
- 73.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980; 32: 314–331. [PMC free article] [PubMed] [Google Scholar]
- 74.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multi locus genotype data. Genetics. 2000; 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Earl DA, VonHoldt BM. Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour.2012; 4: 359–361. [Google Scholar]
- 76.Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007; 23:2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 77.Li X, Zhou Z, Ding J, Wu Y, Zhou B, Wang R, et al. Combined linkage and association mapping reveals QTL and candidate genes for plant and ear height in maize. Front. Plant. Sci. 2016; 7: 833 10.3389/fpls.2016.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mei H, Zhu X, Zhang T. Favorable QTL alleles for yield and its components identified by association mapping in Chinese Upland cotton cultivars. PLoS One. 2013; 26;8(12):e82193 10.1371/journal.pone.0082193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang T, Qian N, Zhu X, Chen H, Wang S, Mei H, Zhang Y. Variations and transmission of QTL alleles for yield and fiber qualities in upland cotton cultivars developed in China. PLoS One. 2013; 8(2), p.e57220 10.1371/journal.pone.0057220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bashir K, Takahashi R, Nakanishi H, Nishizawa NK. The road to micronutrient biofortification of rice: progress and prospects. Front. Plant Sci. 2013; 4:15 10.3389/fpls.2013.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rawat N, Tiwari VK, Singh N, Randhawa GS, Singh K, Chhuneja P, et al. Evaluation and utilization of Aegilops and wild Triticum species for enhancing iron and zinc content in wheat. Genet. Resour. Crop Evol. 2009; 56: 53–64. [Google Scholar]
- 82.Gorafi YS, Ishii T, Kim JS, Elbashir AAE, Tsujimoto H. Genetic variation and association mapping of grain iron and zinc contents in synthetic hexaploid wheat germplasm. Plant Genet. Res. 2016; 1–9. 10.1017/S1479262116000265 [DOI] [Google Scholar]
- 83.Norton GJ, Douglas A, Lahner B, Yakubova E, Guerinot ML, Pinson SRM, et al. (2014) Genome Wide Association Mapping of Grain Arsenic, Copper, Molybdenum and Zinc in Rice (Oryza sativa L.) Grown at Four International Field Sites. PLoS One. 9(2): e89685 10.1371/journal.pone.0089685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diapari M, Sindhu A, Bett K, Deokar A, Warkentin TD, Tar’an B. Genetic diversity and association mapping of iron and zinc concentrations in chickpea (Cicer arietinum L.). Genome. 2014; 57: 1–10. [DOI] [PubMed] [Google Scholar]
- 85.Upadhyaya HD, Bajaj D, Das S, Kumar V, Gowda CLL, Sharma S, et al. Genetic dissection of seed-iron and zinc concentrations in chickpea. Sci. Rep. 2016; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumar S, Hamweih A, Manickavelu A, Kumar J, Sharma TR, Baum M. Advances in lentil genomics In: Gupta S, Nadarajan N, Gupta DS, editors. Legumes in Omics Era. New York: Springer Science; 2014. pp. 111–130. [Google Scholar]
- 87.Thavarajah P, Wejesuriya A, Rutzke M, Glahn R P, Combs GF Jr, Vandenberg A. The potential of lentil (Lens culinaris L.) as a whole food for increased selenium, iron, and zinc intake: preliminary results from a 3 year study. Euphytica. 2011; 180: 123–128. [Google Scholar]
- 88.Alloway BJ. Micronutrient and crop production: An introduction In: Alloway BJ, editors. Micronutrient deficiencies in global crop production. Netherlands: Springer; 2008. pp. 1–39. [Google Scholar]
- 89.Thavarajah D, Thavarajah P, Sarker A, Vandenberg A. Lentils (Lens culinaris Medikus Subspecies culinaris): a whole food for increased iron and zinc intake. J Agri. Food Chem. 2009; 57: 5413–5419. [DOI] [PubMed] [Google Scholar]
- 90.Çakmak İ, Torun A, Millet E, Feldman M, Fahima T, Korol A, et al. Triticum dicoccoides: an important genetic resource for increasing zinc and iron concentration in modern cultivated wheat. Soil Sci. Plant Nutri.2004; 50: 1047–1054. [Google Scholar]
- 91.Peterson CJ, Johnson VA, Mattern PJ. Influence of cultivar and environment on mineral and protein concentrations of wheat flour, bran and grain. Cereal Chem. 1986;63: 183–186. [Google Scholar]
- 92.Lim K, Booth A, Szymlek-Gay EA, Gibson RS, Bailey KB, Irving D, et al. Associations between Dietary Iron and Zinc Intakes, and between Biochemical Iron and Zinc Status in Women. Nutrients. 2015; 7: 2983–2999. 10.3390/nu7042983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diapari M, Sindhu A, Warkentin TD, Bett K, Taran B. Population structure and marker-trait association studies of iron, zinc and selenium concentrations in seed of field pea (Pisum sativum L.). Mol. Breed. 2015; 35:1–14. [Google Scholar]
- 94.El-Haramein FJ, Grando S. Determination of iron and zinc content in food barley. In: ed. Ceccarelli S, Grando S, editors. Proceedings of the 10th International Barley Genetics Symposium, Alexandria: Egypt; 2010. pp. 603–606.
- 95.Genc Y, Verbyla AP, Torun AA, Cakmak I, Willsmore K, Wallwork H, et al. Quantitative trait loci analysis of zinc efficiency and grain zinc concentration in wheat using whole genome average interval mapping. Plant Soil. 2009; 314: 49–66. [Google Scholar]
- 96.Jin T, Jinfeng Z, Jingtang C, Liying Z, Yongfeng Z, Yaqun H. The genetic architecture of zinc and iron content in maize grains as revealed by QTL mapping and meta-analysis. Breeding Sci. 2013; 63: 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mallikarjuna MG, Thirunavukkarasu N, Hossain F, Bhat JS, Jha SK, Rathore A, Agrawal PK, Pattanayak A, Reddy SS, Gularia SK, Singh AM. Stability Performance of Inductively Coupled Plasma Mass Spectrometry-Phenotyped Kernel Minerals Concentration and Grain Yield in Maize in Different Agro-Climatic Zones. PloS one. 2015;10(9):e0139067 10.1371/journal.pone.0139067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cakmak I, Pfeiffer WH, McClafferty B. Biofortification of durum wheat with zinc and iron. Cereal Chem. 2010;87:10–20. [Google Scholar]
- 99.Wei Y, Shohag MJI, Yang X, Yibin Z. Effects of foliar iron application on iron concentration in polished rice grain and its bioavailability. J. Agric. Food Chem. 2012a; 60: 11433–11439. [DOI] [PubMed] [Google Scholar]
- 100.Dikshit HK, Singh A, Singh D, Aski MS, Prakash P, Jain N, et al. Genetic Diversity in Lens Species Revealed by EST and Genomic Simple Sequence Repeat Analysis. PloS one. 2015; 10, p.e0138101 10.1371/journal.pone.0138101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laborda PR, Oliveira KM, Garcia AAF, Paterniani MEAGZ, Dde Souza AP. Tropical maize germplasm: what can we say about its genetic diversity in the light of molecular markers? Theor. Appl. Genet. 2005; 111: 1288–1299. 10.1007/s00122-005-0055-7 [DOI] [PubMed] [Google Scholar]
- 102.Andersen JR, Lübberstedt T. Functional markers in plants. Trends Plant Sci. 2003; 8:554–560. 10.1016/j.tplants.2003.09.010 [DOI] [PubMed] [Google Scholar]
- 103.Peng JH, Lapitan NLV. Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct. Integr. Genom. 2005; 5: 80–96. [DOI] [PubMed] [Google Scholar]
- 104.Kushwaha UKS, Ghimire SK, Yadav NK, Ojha BR. Genetic Relatedness of Lentil (Lens Culinaris L.) Germplasm by using SSR Markers. Int. J. Appl. Sci. Biotech. 2013; 1: 16–26. [Google Scholar]
- 105.Andeden EE, Derya M, Baloch FS, Kilian B, Ozkan H. Development of SSR markers in lentil. In: Proceedings of Plant and Animal Genome Conference XXI P0351. San Diego: CA; 2013.
- 106.Zhang M, Mao W, Zhang G, Wu F. Development and characterization of polymorphic EST-SSR and genomic SSR markers for Tibetan annual wild barley. PloS One. 2014; 9, p.e94881 10.1371/journal.pone.0094881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ferguson ME, Robertson LD, Ford-Lloyd BV, Newbury HJ, Maxted N. Contrasting genetic variation amongst lentil landraces from different geographical origins. Euphytica. 1998; 102: 265–273. [Google Scholar]
- 108.Alo F, Furman BJ, Akhunov E, Dvorak J, Gepts P. Leveraging genomic resources of model species for the assessment of diversity and phylogeny in wild and domesticated lentil. J. Hered. 2011; 102: 315–329. 10.1093/jhered/esr015 [DOI] [PubMed] [Google Scholar]
- 109.Mekonnen F, Mekbib F, Kumar S, Ahmed S, Sharma TR. Phenotypic variability and characteristics of lentil (Lens culinaris Medik.) germplasm of Ethiopia by multivariate analysis. J Agri. Crop Res. 2014; 2:104–116. [Google Scholar]
- 110.Koul PM, Sharma V, Rana M, Chahota RK, Kumar S, Sharma TR. Analysis of genetic structure and interrelationships in lentil species using morphological and SSR markers. 3 Biotech. 2017. 1;7(1):83 10.1007/s13205-017-0683-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Racedo J, Gutiérrez L, Perera MF, Ostengo S, Pardo EM, Cuenya MI, et al. Genome-wide association mapping of quantitative traits in a breeding population of sugarcane. BMC Plant Biol. 2016; 16:142 10.1186/s12870-016-0829-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Galeano CH, Cortés AJ, Fernández AC, Soler Á, Franco-Herrera N, Makunde G, et al. Gene-based single nucleotide polymorphism markers for genetic and association mapping in common bean. BMC Genet. 2012;13: 48 10.1186/1471-2156-13-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang ML, Sukumaran S, Barkley NA, Chen Z, Chen CY, Guo B, et al. Population structure and marker–trait association analysis of the US peanut (Arachis hypogaea L.) mini-core collection. Theor Appl.Genet. 2011; 123: 1307–1317. 10.1007/s00122-011-1668-7 [DOI] [PubMed] [Google Scholar]
- 114.Gutiérrez L, Cuesta-Marcos A, Castro AJ, von Zitzewitz J, Schmitt M, Hayes PM. Association mapping of malting quality quantitative trait loci in winter barley: positive signals from small germplasm arrays. Plant Genome. 2011; 4: 256–272. [Google Scholar]
- 115.Ruggieri V, Francese G, Sacco A, D’Alessandro A, Rigano MM, Parisi M,et al. An association mapping approach to identify favourable alleles for tomato fruit quality breeding. BMC Plant Bio. 2014; 14:1 10.1186/s12870-014-0337-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu YY, He JB, Li AH, Fang NY, He WW, Dang LL, et al. Population structure analysis and association mapping of blast resistance in indica rice (Oryza sativa L.) landraces. Genet. Mole. Res. 2016; 15 10.4238/gmr.15038254 [DOI] [PubMed] [Google Scholar]
- 117.Zorić M, Dodig D, Kobiljski B, Quarrie S, Barnes J. Population structure in a wheat core collection and genomic loci associated with yield under contrasting environments. Genetica. 2012; 140:259–275. 10.1007/s10709-012-9677-2 [DOI] [PubMed] [Google Scholar]
- 118.Kujur A, Bajaj D, Upadhyaya HD, Das S, Ranjan R, Shree T, Saxena MS, Badoni S, Kumar V, Tripathi S, Gowda CL. A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea. Scient rep. 2015; 5:11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gupta S, Kumari K, Muthamilarasan M, Parida SK, Prasad M. Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant cell reports. 2014; 1; 33(6):881–93. 10.1007/s00299-014-1564-0 [DOI] [PubMed] [Google Scholar]
- 120.Lou Y, Hu L, Chen L, Sun X, Yang Y, Liu H, et al. Association Analysis of Simple Sequence Repeat (SSR) Markers with Agronomic Traits in Tall Fescue (Festuca arundinacea Schreb.). PLoS One. 2015; 10, p.e0133054 10.1371/journal.pone.0133054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gyawali S, Harrington M, Durkin J, Horner K, Parkin IA, Hegedus DD, et al. Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol. Breed. 2016; 36: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wan JM. Perspectives of molecular design breeding in crops. Acta Agron. Sin. 2006; 32: 455–462. [Google Scholar]
- 123.White PJ, Broadley MR. Biofortifying crops with essential mineral elements. Trends Plant Sci. 2005; 10: 586–593. 10.1016/j.tplants.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 124.Bauer P, Thiel T, Klatte M, Bereczky Z, Brumbarova T, Hell R, et al. Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiol. 2004; 136: 4169–4183. 10.1104/pp.104.047233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hacisalihoglu G, Kochian LV. How do some plants tolerate low levels of soil zinc? Mechanisms of zinc efficiency in crop plants. New Phytol. 2003; 159: 341–350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(XLS)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.