Abstract
The coronary vasculature is an essential vessel network providing the blood supply to the heart. Disruptions in coronary blood flow contribute to cardiac disease, a major cause of premature death worldwide. The generation of treatments for cardiovascular disease will be aided by a deeper understanding of the developmental processes that underpin coronary vessel formation. From an ENU mutagenesis screen, we have isolated a mouse mutant displaying embryonic hydrocephalus and cardiac defects (EHC). Positional cloning and candidate gene analysis revealed that the EHC phenotype results from a point mutation in a splice donor site of the Myh10 gene, which encodes NMHC IIB. Complementation testing confirmed that the Myh10 mutation causes the EHC phenotype. Characterisation of the EHC cardiac defects revealed abnormalities in myocardial development, consistent with observations from previously generated NMHC IIB null mouse lines. Analysis of the EHC mutant hearts also identified defects in the formation of the coronary vasculature. We attribute the coronary vessel abnormalities to defective epicardial cell function, as the EHC epicardium displays an abnormal cell morphology, reduced capacity to undergo epithelial-mesenchymal transition (EMT), and impaired migration of epicardial-derived cells (EPDCs) into the myocardium. Our studies on the EHC mutant demonstrate a requirement for NMHC IIB in epicardial function and coronary vessel formation, highlighting the importance of this protein in cardiac development and ultimately, embryonic survival.
Author summary
In order for the heart to function properly it must have its own blood supply. Blood is delivered to the heart through a system of vessels called the coronary vasculature. During development, some of the cells that form the coronary vessels originate from the epicardium, the outer layer of the heart. These epicardial-derived cells migrate into the cardiac tissue where they contribute to the formation of the coronary vascular network. We have found that a mouse mutant containing a mutation in the gene Myh10, which encodes the cytoskeletal protein non-muscle myosin IIB, fails to form the coronary vasculature. Our work reveals defects in the epicardium, which contribute to the lack of coronary vessel development in this mutant. Surprisingly, we discovered that whilst the mutant epicardial cells are capable of movement when extracted from the embryo, these cells fail to mobilise into a vascular network in the context of the developing embryonic heart. We propose that this migration failure is due to abnormalities in the extracellular environment in the mutant heart. This work highlights the importance of the Myh10 gene in the critical developmental process of coronary vessel formation.
Introduction
A functional coronary vasculature is essential to supply the heart with oxygenated blood. Cessation of the coronary circulation deprives the working myocardium of oxygen and nutrients, leading to irreversible damage to cardiac muscle and myocardial infarction. Coronary artery disease is the main form of cardiovascular disease, and causes significant morbidity and mortality world-wide [1]. Although mammals and other higher vertebrates have insufficient capacity to restore cardiac function following ischemia, a number of studies that exogenously reactivate elements of embryonic coronary vessel formation have demonstrated neovascularisation and regeneration of the infarcted mouse heart, consequently improving cardiac function [2–5]. Moreover, experiments using thymosin β4 have revealed that the specific restoration of the quiescent adult epicardium, the outer epithelial layer of the heart, to an embryonic state, permits the activation of cardiac precursors that contribute to neovascularisation in vitro [6] and in vivo [2, 7]. However, our comprehension of both the cellular and molecular mechanisms that control this regeneration remain incomplete. Therefore, a deeper understanding of the processes that underpin coronary vessel formation may facilitate the generation of advanced and novel therapies to repair the injured heart.
During mammalian cardiac development cells from the proepicardial organ (PEO) migrate onto the surface of the heart and adhere to the nascent myocardium of the post-looped heart tube [8]. This gives rise to an outer epithelial layer, termed the epicardium, which completely envelops the developing heart. Epicardial function is critical for cardiac development, since the epicardium provides a source of paracrine signals for myocardial growth (reviewed in [9–11]). Additionally, as cardiac development progresses, a subset of epicardial cells undergo EMT and migrate through the subepicardial space to contribute to the formation of the coronary vasculature and cardiac fibroblasts (reviewed in [11–15]). While the contribution of the epicardium to the endothelial cell lining of the coronary vessels has been challenged recently [15, 16], it is clear that epicardial function is absolutely essential to establish the coronary vasculature and facilitate cardiogenesis. Furthermore, reactivating embryonic processes in the quiescent adult epicardium has been shown to facilitate the repair and regeneration of cardiac tissue in response to injury, highlighting the therapeutic potential of this tissue [5, 17, 18].
A comprehensive understanding of the critical molecular mechanisms that underpin epicardial function during mammalian cardiogenesis is needed to facilitate the development of epicardial-based therapies. To this end, we have studied mouse mutants with cardiac defects isolated from a balancer chromosome mutagenesis screen. We found that the l11Jus27 mutant [19] carried two different embryonic lethal mutations, one of which displayed a phenotype of embryonic hydrocephalus and cardiac defects (EHC). EHC mutant embryos fail to form a mature, functional coronary system, resulting in late-gestation lethality. We identified a mutation in a splice-donor site in the Myh10 gene as the cause of the EHC phenotype. Myh10 encodes Non-Muscle Myosin Heavy Chain IIB (NMHC IIB), a component of the protein hexamer Non-Muscle Myosin IIB (NMIIB). There are 3 different NMHC II isoforms (namely IIA, IIB and IIC), each displaying specific cellular expression and functionality (reviewed extensively in [20, 21]). NMHC IIB is a cytoskeletal protein with diverse functions, including: cytokinesis [22, 23], regulation of cell shape [24], adhesion [25, 26] and migration [27, 28]. Prior studies of other mutants with abnormal coronary vessel development have shown that these defects arise primarily from epicardial cell dysfunction [29–32]. Accordingly, EHC mutant epicardial cells form an abnormal epithelial layer on the surface of the heart. In addition, the migration of EHC epicardial-derived cells into the underlying myocardium is impaired. EHC epicardial cells also show decreased expression of EMT markers within the epicardium, suggesting that NMHC IIB not only plays an important role in regulating EPDC migration, but also in promoting epicardial EMT. Ultimately, NMHC IIB may therefore function in multiple processes during coronary vessel formation and cardiogenesis, which could potentially be manipulated to repair and regenerate the heart in the context of cardiovascular disease.
Results
Isolation of the EHC mutant
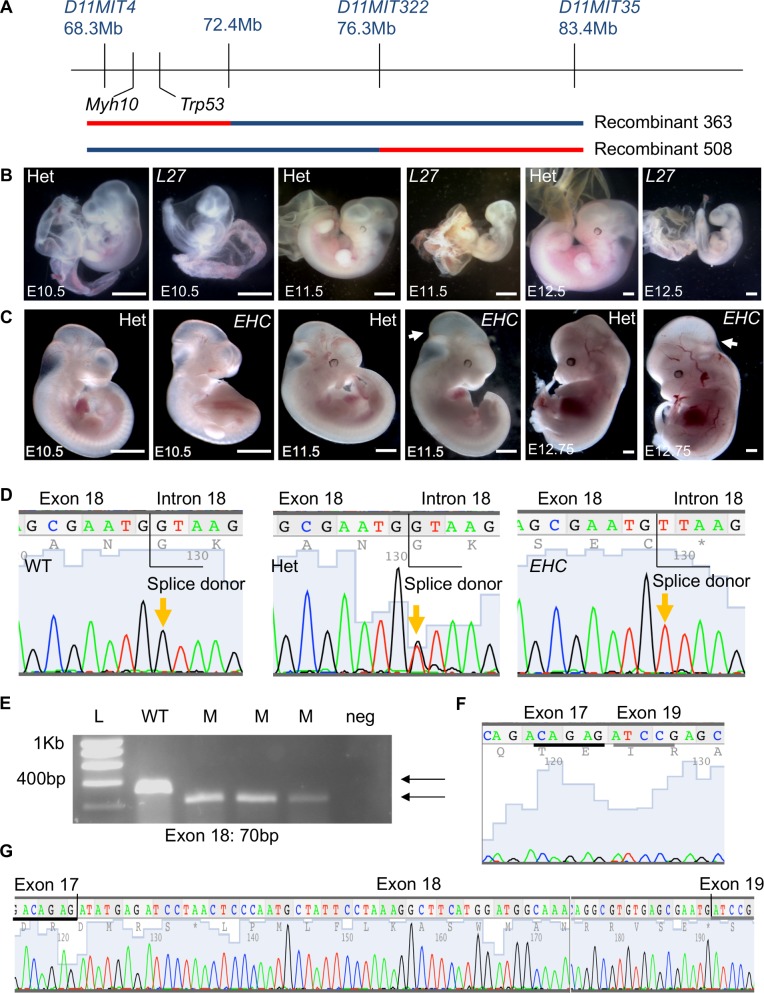
A study of ENU (N-ethyl-N-nitrosourea) mutagenised mouse strains with embryonic lethal recessive mutations revealed that homozygous mutant embryos from the l11Jus27 mouse line displayed enlarged, distended hearts with oedema [19]. A meiotic mapping approach was employed to further refine the l11Jus27 candidate region. Animals with recombination events within the balancer chromosome interval were test-crossed to known l11Jus27 carriers, and viable offspring were genotyped for several markers in the balancer chromosome region. Correlations between the inheritance of C57BL/6 genomic DNA (mutagenised strain) and the l11Jus27 phenotype (early embryonic lethality) were evaluated. The failure of a recombinant animal crossed to a known l11Jus27 carrier to produce homozygous C57BL/6 offspring would suggest that the homozygous C57BL/6 embryos died in utero, consistent with the l11Jus27 phenotype. This finding would indicate that the l11Jus27 mutation is located in the genomic region where the recombinant animal has inherited C57BL/6 DNA. Unexpectedly, two recombinant mice failed to produce homozygous C57BL/6 viable offspring when mated to known l11Jus27 heterozygotes (Fig 1A). Both of these animals carried C57BL/6 DNA in non-overlapping sub-regions of the balancer interval, suggesting that they could not carry the same embryonic lethal mutation. Recombinant 363 carried C57BL/6 genomic DNA in a 4.1Mb region extending beyond the Trp53 endpoint of the balancer chromosome, and recombinant 508 carried C57BL/6 genomic DNA in the central region of the balancer interval (Fig 1A). To determine if recombinants 363 or 508 produced mutant embryos with the l11Jus27 phenotype, timed matings were performed, and resulting embryos analysed at embryonic day (E) 10.5–12.5. At these developmental stages, the l11Jus27 phenotype was apparent, with most mutant embryos dying by E12.5 (Fig 1B). However, homozygous C57BL/6 embryos produced from the cross of recombinant 363 to l11Jus27 heterozygotes did not display the l11Jus27 phenotype (Fig 1C). Instead, mutant embryos were viable past mid-gestation, and exhibited severe hydrocephalus in the mid-brain region from E11.5 (Fig 1C, arrows). We found that embryos generated from the cross of recombinant 508 did display the l11Jus27 phenotype (Fig 1B), indicating that the l11Jus27 mutation is located in the region between the polymorphic markers D11MIT322 and D11MIT35 on mouse chromosome 11.
Fig 1. The l11Jus27 mouse line carries two embryonic lethal mutations, one of which is in Myh10.
A: Two recombinant mice show linkage of the phenotype (lethality) with distinct sub-regions of the mouse chromosome 11 balancer interval. The blue line indicates the 129S5 non-mutagenised chromosome. The C57BL/6 region (red) in recombinant mouse 363 extends beyond the balancer chromosome endpoint at Trp53. B: l11Jus27 heterozygous and homozygous mutant embryos at E10.5, E11.5, and E12.5. For this and all subsequent figures, images are representative findings from a minimum of n = 3 observations unless otherwise stated. C: Dissections of embryos from crosses of l11Jus27 heterozygotes with recombinant 363 reveal that at E10.5 –E12.75 the l11Jus27 phenotype is not apparent in homozygous C57BL/6 embryos. At E11.5 and E12.75 embryos homozygous for C57BL/6 DNA have prominent hydrocephalus (arrow), revealing a second embryonic lethal phenotype in the l11Jus27 line. D: Sequencing of the Myh10 gene reveals a ‘G’ to ‘T’ point mutation in the splice donor site of exon 18 (orange arrows). E: The Myh10 point mutation causes exon 18 to be skipped in the Myh10 mutant transcript. RT-PCR in three homozygous EHC mutant embryos (M) reveals a smaller transcript than in one wild type (WT) embryo. The reduction in size of the PCR product is consistent with skipping exon 18. F: Sequencing Myh10 transcript from mutant embryos confirms that exon 18 is missing, causing exons 17 and 19 to be joined. G: A wild type transcript that contains exons 17, 18, and 19. The black and grey lines in (F) and (G) highlight the sequence from exon 17 and sequence from exon 19 respectively shown in both the mutant and wild type. Scale bars: 1mm. Abbreviations: L27: l11Jus27; L: molecular size ladder; Het: heterozygote; WT: wild type; M: mutant; neg: negative control.
Based on the new phenotype displayed in the offspring from recombinant mouse 363, we concluded that two embryonic lethal mutations were present in the l11Jus27 mice, and that mutant embryos carrying both mutations exhibited the more severe l11Jus27 phenotype. A new line of mice displaying the hydrocephalus phenotype was generated from crossing recombinant 363 to balancer chromosome animals, so that the new mutation could be maintained in trans to the balancer. This new mutant line was named EHC. Candidate gene analysis of the C57BL/6 region inherited by EHC mice revealed Myh10 as a strong candidate gene. Myh10 encodes NMHC IIB, the heavy chain component of the NMIIB protein complex, and targeted deletion of Myh10 results in late-gestation lethality with hydrocephalus and cardiac defects [26, 33]. Therefore, due to similarities in phenotype, we sequenced Myh10 genomic DNA from EHC mutant mice. We found a ‘G’ to ‘T’ transversion mutation in the splice donor site following exon 18 (Fig 1D). This mutation causes exon 18 to be skipped from the Myh10 EHC mutant transcript, producing a smaller product from an RT-PCR reaction performed with primers in exons 17 and 19 (Fig 1E). An abnormal Myh10 transcript created from the fusion of exon 17 and exon 19 was present in EHC mutant embryos (Fig 1F), while wild type embryos contained a transcript with exons 17, 18, and 19 (Fig 1G).
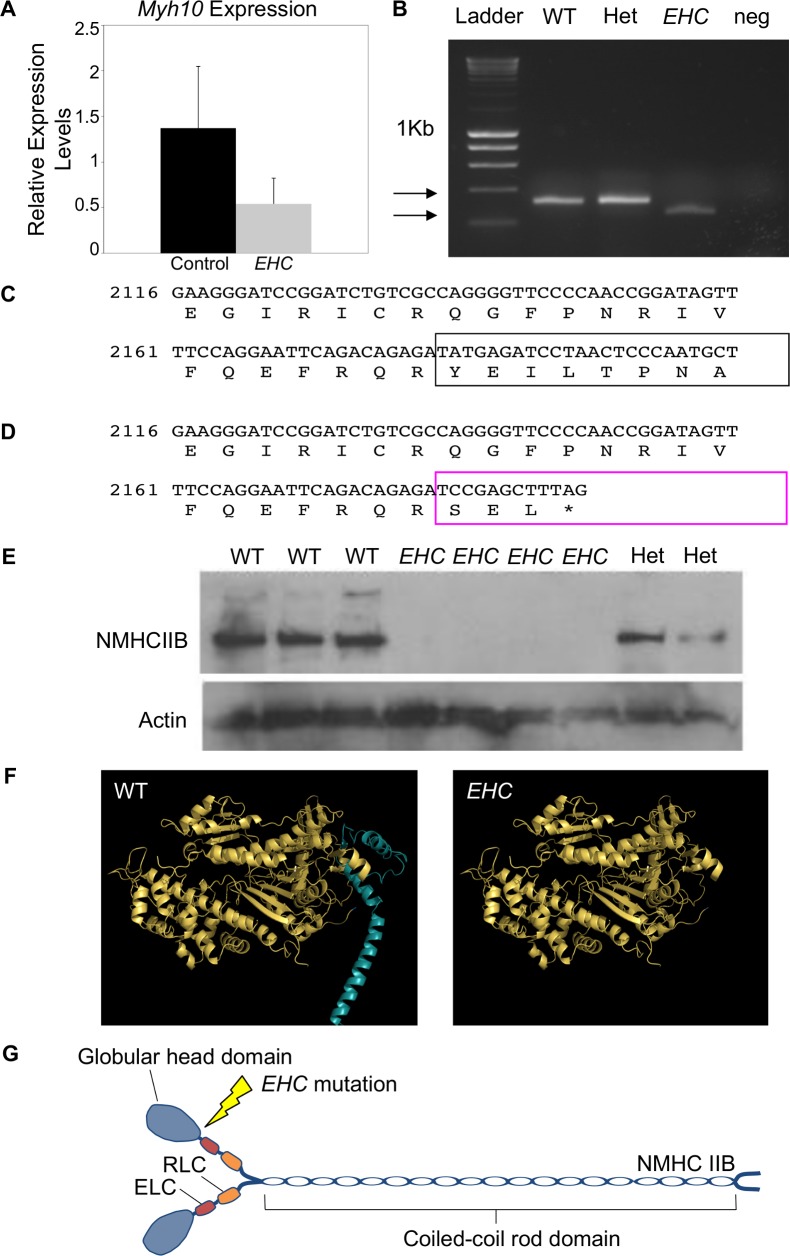
EHC mutant embryos showed a reduction in full-length Myh10 transcript levels as analysed by qPCR (Fig 2A) or RT-PCR amplifying the region from exons 17–19 (Fig 2B). The NMHC IIB protein is a 230kDa molecule comprising 1976 amino acids [20, 21, 34]. The wild type NMHC IIB protein sequence translated from exons 17 and 18 is shown in Fig 2C. The abnormal fusion of exons 17 and 19 in the EHC Myh10 transcript causes a change in the reading frame, resulting in a truncated protein (1–703 amino acids). Additionally, the final three amino acids are divergent from the wild type sequence with a YEI to SEL change (Fig 2D, boxed area). Using a C-terminal NMHC IIB antibody for western blotting, we confirmed that full-length NMHC IIB protein is not detectable in EHC mutant embryos (Fig 2E). Modelling the effect of the EHC mutation on the NMHC IIB protein demonstrated that the premature stop-codon will cause truncation of the protein in the actin binding head domain, resulting in the synthesis of NMHC IIB devoid of the coiled coil rod domain (Fig 2F). NMIIB is dependent upon interactions between the heavy chain rod domains to associate into bipolar filaments in order to exert its cellular function (Fig 2G) [20, 21]. We predict that if a truncated NMIIB protein was produced in EHC mutant embryos, this aberrant protein lacking the essential rod domain would be unable to partake in molecular interactions and therefore be unable to exert contractile force upon the actin cytoskeleton.
Fig 2. Predicted alterations to NMHC IIB protein in EHC mutants.
A: Comparison of full-length Myh10 transcript levels in control (WT or het) and mutant embryos by qPCR. B: RT-PCR amplification products from the region encompassing Myh10 exon 17–19 in wild type, het, and mutant samples. C: Myh10 wild type transcript and protein sequence is shown for exons 17 and 18. The boxed area highlights the translation of exon 18. Numbering refers to nucleotide positions of the Myh10 transcript, with the initiation codon ‘A’ considered base 1. D: Predicted protein sequence for the EHC mutant transcript. The boxed area highlights the abnormal reading frame created by the fusion of exons 17 and 19, indicating that the mutant protein is truncated. E: Western blot with a C-terminal NMHC IIB antibody reveals that EHC mutant embryos do not produce full-length non-muscle myosin IIB. Actin is used as a loading control. F: Structural model of wild type NMHC IIB protein displays the head domain (left). The region deleted in the EHC mutant is shown in blue. The NMHC IIB protein predicted to be generated from the Myh10 mutant transcript in EHC mutants (right). G: Schematic representation of wild type NMHC IIB dimers in complex with the essential light chains (ELC) and regulatory light chains (RLC) to form mature non-muscle myosin IIB. The location of the EHC mutation in the globular head domain of NMHC IIB is illustrated.
Confirmation of the EHC causative mutation by complementation test
To confirm that the EHC causative mutation had been correctly mapped to Myh10, and subsequently caused loss of NMHC IIB function, we performed a complementation assay with a known Myh10 null allele, denoted as Myh10∆. The Myh10∆ allele has a deletion of Myh10 exon 2 [33], and does not synthesise full-length NMHC IIB protein (S1A Fig). As with the EHC mutants, homozygous Myh10∆ mutant embryos demonstrated late gestation embryonic lethality and were not present at the expected Mendelian ratios at birth (S1B Fig, Chi squared test p = 0.0035). Heterozygous EHC and Myh10∆ animals were intercrossed; the resultant progeny were genotyped and analysed for embryonic lethality and developmental defects. Analysis of Mendelian frequencies of EHC/Myh10∆ embryos at birth revealed a deviation from expectations due to late gestation embryonic lethality (S1C Fig, Chi squared test, p = 0.0278). These results indicated a failure of complementation between the two lines, and provided strong support for the hypothesis that the Myh10 mutation causes the EHC phenotype by ablating NMHC IIB function.
EHC mutants display embryonic hydrocephalus and cardiac defects
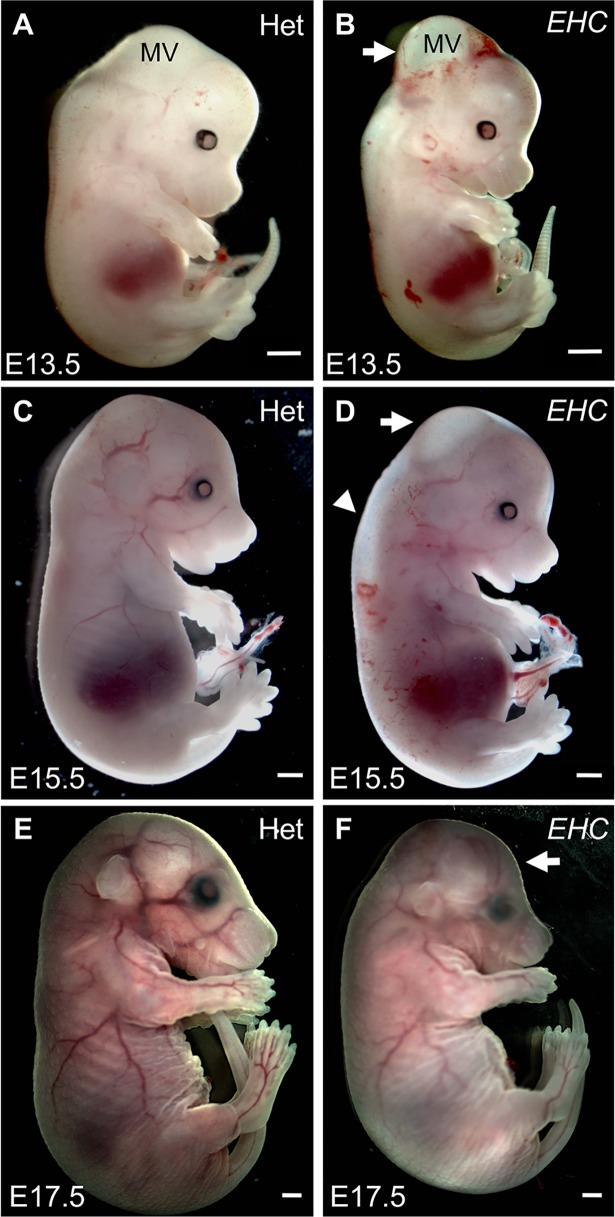
We concluded that EHC mutant embryos were alive at E12.5 since a regular heartbeat was observed at the time of dissection. We therefore examined the phenotype of EHC mutant embryos at late gestation. At E13.5, there is still prominent hydrocephalus in the mesencephalic vesicle (Fig 3A and 3B). At E15.5, EHC mutant embryos often displayed oedema in the spinal cord region (Fig 3C and 3D, arrowhead), whilst excess fluid within the mesencephalic vesicle persists (Fig 3C and 3D, arrow). It was rare to recover EHC mutant embryos past E16.5, but the embryos that did survive had abnormal dome-shaped heads, consistent with developmental hydrocephalus (Fig 3E and 3F, arrow). Based on the reduced recovery of the EHC mutants from dissections after E16.5, we conclude that the EHC phenotype causes late-gestation embryonic lethality, accompanied by defects in cranial development due to severe hydrocephalus in the early embryo.
Fig 3. EHC mutant embryo morphology.
A: Heterozygous littermate at E13.5. B: EHC mutant displays hydrocephalus in the mesencephalic vesicle (white arrow). C: Heterozygous littermate at E15.5. D: EHC mutant at E15.5 displays reduced hydrocephalus (arrow), although oedema in the spinal cord region is apparent (arrowhead). E: Heterozygous littermate at E17.5. F: EHC mutant has dome-shaped head (arrow), consistent with developmental defects arising from embryonic hydrocephalus. Scale bars: 1 mm. Abbreviations: MV: mesencephalic vesicle.
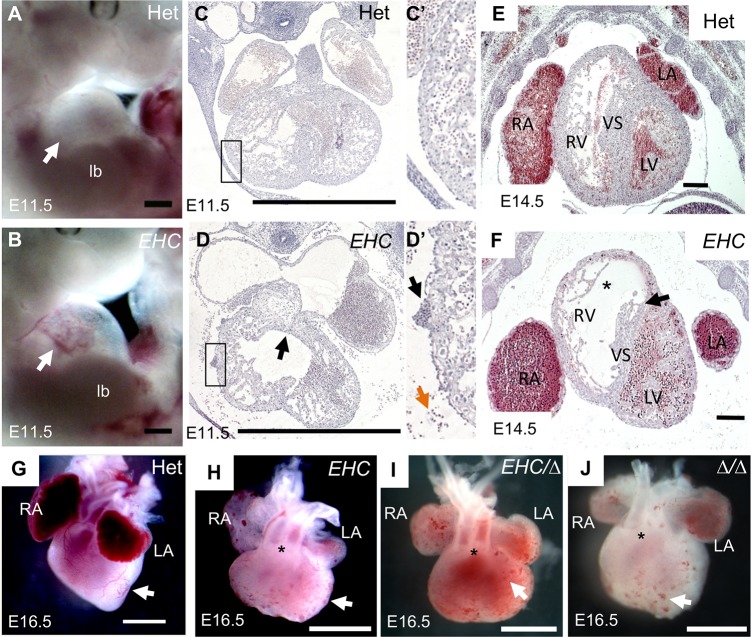
As cardiac defects have been previously described for a targeted deletion of NMHC IIB [23, 33, 35], we examined cardiac development in the EHC mutant mouse. We found several similarities between the EHC cardiac phenotype and the defects described in the NMHC IIB knock out. First, upon dissection at E11.5 we observed pericardial effusion and blood in the pericardial sac of EHC mutant embryos (Fig 4B and 4D) and on the surface of the EHC mutant heart (Fig 4D’), phenotypes consistent with cardiac developmental defects. Membranous ventricular septal defects have been reported in NMHC IIB knock out animals [33, 35, 36]. However, we found that the endocardial cushions are present and have fused in EHC mutant embryos at E11.5 (Fig 4D, arrow), indicative of initial development of the septum. Later in gestation, EHC mutants at E14.5 have a thin ventricular septum with tearing in the membranous region, suggestive of a vulnerability to septal defects (Fig 4F, arrow). We also observed that EHC mutant ventricles display reduced trabeculation, a thinner compact myocardium, and disorganisation of cells in the ventricular myocardium (Fig 4D and 4F, asterisk). In addition, we detected defects in myocardial cytokinesis (S2A–S2D Fig), comparable to previously reported data from NMHC IIB knock out mice [23, 33, 37]. We observed double-outlet right ventricle (DORV), where the aorta erroneously stems from the right ventricle, in mutant embryos at E16.5 (Fig 4H asterisk), similar to findings from NMHC IIB knock out mice [33, 36]. Interestingly, DORV was not detected when NMHC IIB was specifically deleted in cardiomyocytes [35]. However, the DORV phenotype was completely penetrant in EHC hearts at E16.5. During our morphological inspection of E16.5 hearts, we also observed that mutant hearts displayed an abnormally rounded ventricular morphology, which lacked a prominent ventricular apex (Fig 4H–4J). In addition, EHC mutants displayed distended atria, abnormally positioned in relation to the ventricles (Fig 4F and 4H), and the ventricular surface was decorated with multiple blood filled vesicle-like structures (Fig 4D’ and 4H, arrows). These vesicles can be observed as early as E11.5 and persist until embryonic lethality. Again, these malformations were fully penetrant in mutant embryos by visual inspection at E16.5.
Fig 4. EHC mutants display defects in cardiac development.
A: Heterozygous littermate control at E11.5 showing developing heart in pericardial sac. B: EHC mutant has pericardial effusion and blood visible in pericardial sac (arrow). C: Transverse section of control littermate at E11.5. The boxed region indicates the segment shown in panel C’. C’: Note the absence of blood cells on the cardiac surface and in the pericardial space. D: Transverse section of EHC mutant at E11.5 showing enlarged atria. The boxed region indicates the segment shown in panel D’. D’: Blood cells are present on the surface of the EHC heart (black arrow) and in the pericardial space (orange arrow). E: In heterozygous littermates at E14.5 the ventricular chamber is surrounded by a thick compact myocardium, which is highly organised. F: At E14.5, EHC mutants show a reduced compact ventricular myocardium (indicated by asterisk), accompanied by a thinning of the ventricular septal tissue (arrow). Note the abnormal atrial morphology and different localisation of the atria with respect to the ventricle compared to control (E). G: Heterozygous heart at E16.5 shows expected ventricular vascular connections and also coronary vessels on the cardiac surface (arrow). H: EHC mutant heart shows double-outlet right ventricle (DORV) (asterisk) as well as an absence of surface coronary vessels (arrow). Note the vesicle-like structures present on the ventricular surface. I: EHC/Myh10∆ and J: Myh10∆/Myh10∆ hearts at E16.5 similarly show DORV (asterisks) and ventricular vesicles (arrows). Scale bar = 200 μm (E, F), 1mm (A-D, G-J). Abbreviations: Het: heterozygote; lb: limb bud; LA: left atria, LV: left ventricle, RA: right atria, RV: right ventricle, VS: ventricular septum, ∆: Myh10∆.
Additionally, the morphology of EHC/Myh10∆ and Myh10∆/Myh10∆ embryonic hearts closely resembles that of the EHC mutants (Fig 4I and 4J). Together, the phenotypic similarity of EHC and Myh10∆ homozygous mutants, combined with the failure of the Myh10∆ allele to complement the EHC allele, confirms that the EHC mutation causes complete loss of Myh10 function, resulting in the observed EHC cardiac abnormalities.
Mutant hearts lack coronary vasculature
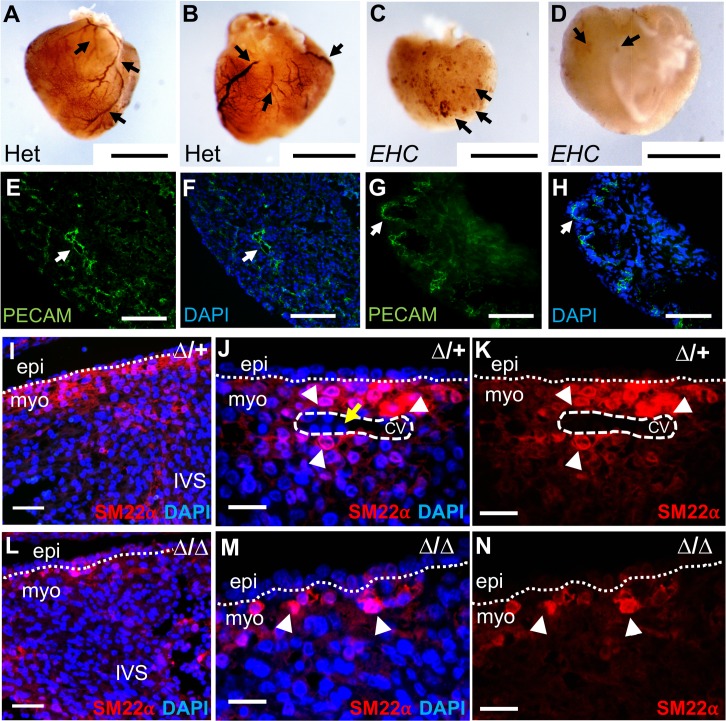
It became strikingly apparent during our dissection observations that the EHC mutants lacked blood-filled coronary vessels on their ventricular surface, in contrast to heterozygous EHC control hearts (Fig 4G compared to 4H). The Myh10∆ homozygotes and EHC/Myh10∆ compound heterozygote mutant embryos also displayed a lack of coronary vessels (Fig 4I and 4J). To confirm that EHC and Myh10∆ mutant hearts lacked a mature coronary network, we performed immunohistochemical analysis for markers of cellular components of the coronary architecture, namely vascular endothelial cells (PECAM-1/CD31), and vascular smooth muscle cells (SM22α). Heterozygous littermates at E16.5 displayed clear PECAM-1 immunoreactivity, highlighting mature coronary vasculature in which vascular endothelial cells are organised into a large and extensively branched vascular network (Fig 5A and 5B arrows). In contrast, EHC mutants at E16.5 displayed only PECAM-1 immunoreactive surface cell clusters (Fig 5C and 5D arrows). A similar staining profile was observed in EHC/Myh10∆ and Myh10∆/Myh10∆ mutant hearts (S3 Fig). To further examine the extent of coronary defects, we evaluated the localisation of vascular endothelial and smooth muscle cells (vSMCs) at E14.5. In heterozygous Myh10∆ controls, we observed intense PECAM-1 staining around the lumen of developing vessel structures, illustrating the presence of endothelial cells around the coronary vessels during maturation (Fig 5E and 5F arrow). However, Myh10∆ homozygous mutant hearts displayed PECAM-1 staining surrounding clusters of blood cells on the cardiac surface (Fig 5G and 5H arrow), consistent with results from whole mount PECAM-1 staining of EHC, EHC/Myh10∆ compound heterozygotes and Myh10∆ homozygous mutant hearts (S3 Fig). Coronary endothelial cells are present in a capillary network on the surface of EHC mutant hearts, but lack organisation into larger vessels (S5J–S5K Fig). Smooth muscle cells were present in the heart of heterozygous control embryos (Fig 5I–5K), including surrounding coronary vessels (Fig 5J arrowheads). In the interventricular septum region, SM22α distribution in Myh10∆ homozygous mutant hearts (Fig 5L) was similar to that of control hearts (Fig 5I). In the compact myocardium, Myh10∆ homozygous mutants did not display organised clusters of smooth muscle cells or vascular structures (Fig 5M and 5N). Together, these experiments demonstrate that EHC and Myh10∆ homozygous mutant hearts display similar defects in coronary vessel formation.
Fig 5. EHC mutants lack mature coronary vessels.
PECAM-1 immunohistochemistry reveals prominent coronary vessels (arrows) on the ventral (A) and dorsal (B) surface of EHC heterozygous hearts. Only clusters of PECAM-1 expressing cells (arrows) are apparent on EHC homozygous mutant ventral (C) and dorsal (D) heart surface, with no clear evidence of coronary vessels. E-F: Section immunofluorescence for PECAM-1 (green) on Myh10∆ heterozygote at E14.5, demonstrating cells in myocardial region surrounding a developing vessel (arrow). G-H: Section immunofluorescence for PECAM-1 on Myh10∆ homozygous mutant at E14.5, demonstrating staining of clusters of cells on cardiac surface (arrow), consistent with whole mount staining in panels C-D. Staining for the smooth muscle cell marker SM22α reveals few smooth muscle cells within the interventricular septum region of the heterozygote (I) and Myh10∆ homozygous mutant (L). SM22α expressing cells are localised around a developing coronary vessel (white dashed circle) in heterozygous control hearts (J-K, arrowheads), and nucleated blood cells within the vessel can be visualised with DAPI (yellow arrow). No such structures are present in Myh10∆ homozygous mutant hearts, and smooth muscle cells expressing SM22α are less clustered in mutant hearts (M-N, arrowheads). Cell nuclei were labelled with DAPI (blue). Scale bars: A-D = 1 mm, E-H = 100 μm, I and L = 50 μm, J-K and M-N = 25 μm. Abbreviations: CV: coronary vessel, epi: epicardium, IVS: interventricular septum, myo: myocardium, PECAM: platelet endothelial cell adhesion molecule-1, ∆: Myh10∆.
NMHC IIB is not required in cardiomyocytes for coronary vessel development
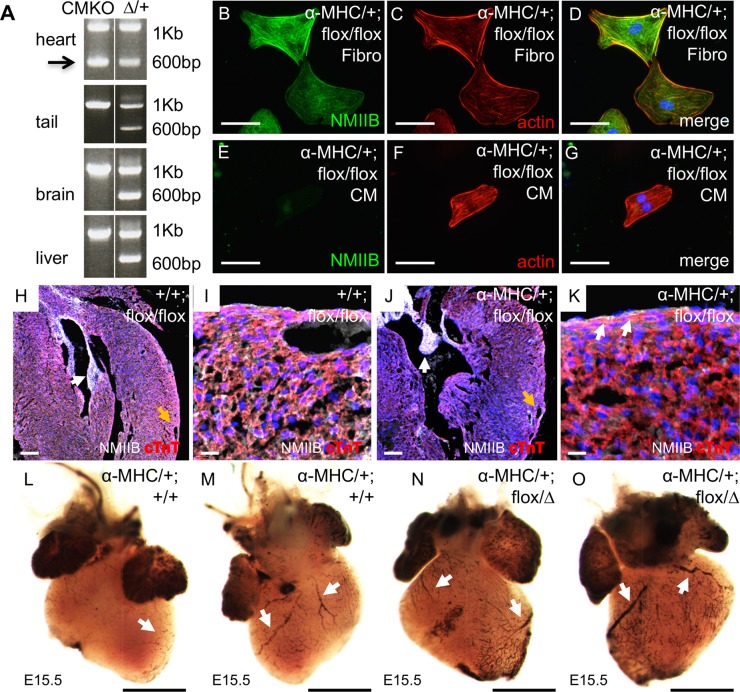
Prior research has demonstrated that mice with a cardiomyocyte-specific deletion of Myh10 are viable [35], suggesting that coronary vessel development must not be severely compromised when NMHC IIB function has been lost from cardiomyocytes. To investigate the dependence of coronary vessel development on cardiomyocytes NMHC IIB activity, we implemented the previously described strategy [35] to delete Myh10 exon 2 in cells expressing the cardiomyocyte-specific α-Myosin Heavy Chain-Cre (αMHC-Cre) transgene. Confirmation of the genomic deletion of Myh10 exon 2 in cardiac cells, but not tail, brain, or liver cells was demonstrated by genomic PCR for primers surrounding Myh10 exon 2 (Fig 6A). These primers generate a 1 Kb product when exon 2 is present in the genome, and a 600bp product after deletion of Myh10 exon 2. The 600bp deletion product is visible only in heart tissue (Fig 6A, arrow). We further demonstrated the cardiomyocyte specificity of the deletion of Myh10 by isolating cardiac cells, dissociating them in culture, and subjecting the cells to fibroblast or cardiomyocyte culture protocols [38, 39]. NMHC IIB protein persists in fibroblast cells, which show characteristic morphology (Fig 6B–6D), but not in cardiomyocytes (Fig 6E–6G), which also have distinctive morphology in culture. Histological sections of hearts at E18.5 from control and Myh10 cardiomyocyte-specific knock out embryos were examined by immunofluorescence for NMHC IIB and cardiac troponin T expression (Fig 6H–6K). Embryos inheriting the αMHC-Cre transgene and homozygous Myh10 floxed alleles had reduced NMHC IIB expression within the myocardium, although expression of NMHC IIB can be seen in non-cardiomyocyte cell populations such as the endocardial cells present in the valve leaflets (Fig 6J, white arrow). Analysis of vessel development in control embryos and cardiomyocyte knockouts reveals the presence of blood cells in organised vessels on the cardiac surface, visible directly in dissections at E18.5 (S4A–S4D Fig), with PECAM-1 staining at E16.5 (S4E and S4F Fig), and following DAB staining of blood cells within heart tissue at E15.5 (Fig 6L–6O). These results confirm that cardiomyocyte expression of NMHC IIB is not required for the development of coronary vessels.
Fig 6. NMHC IIB is not required in cardiomyocytes for coronary vessel formation.
A: Genomic PCR on tissues isolated from mice carrying the αMHC-Cre transgene and Myh10 flox/+ alleles shows a PCR product of 600 bp in cardiac tissue, consistent with deletion of Myh10 exon 2 from the genome (arrow). This product is not seen in tail, brain, or liver tissue. PCR results from a Myh10∆/+ heterozygous animal are shown for the corresponding tissues, with the 600 bp product present in all tissues. The 1 Kb product represents the Myh10 allele lacking exon 2 deletion. Products were sequenced to confirm specificity of PCR reactions. B: NMHC IIB protein (green) and C: actin (red; phalloidin stain) localisation in fibroblasts cultured from αMHC-Cre; flox/flox hearts. D: Merged image shows DAPI labeling of nuclei (blue). E: NMHC IIB protein (green) and F: actin (red; phalloidin stain) localisation in cardiomyocytes cultured from αMHC-Cre; flox/flox hearts. G: Merged image shows DAPI labelling of nuclei (blue). H: Expression patterns of NMHC IIB (white) and cardiac Troponin T (red), and nuclei stained with DAPI (blue) in αMHC-Cre; Myh10+/+ control embryo with presence of coronary vessel (orange arrow). NMHC IIB expression is seen throughout heart including valve tissue (white arrow). Co-expression of NMHC IIB and cTnT appears pink. I: Higher magnification image of cardiac tissue from αMHC-Cre; Myh10+/+ control embryo. J: Expression patterns of NMHC IIB (white) and cardiac Troponin T (red), and nuclei stained with DAPI (blue) in αMHC-Cre; Myh10 flox/flox control embryo with presence of coronary vessel (orange arrow). NMIIB expression is seen in valve tissue (white arrow) but reduced in myocardial region of heart, and is absent from cTnT positive cells, so that the pink staining indicating NMHC IIB and cTnT co-expression is reduced in the myocardium. K: Higher magnification image of cardiac tissue from αMHC-Cre; Myh10 flox/flox control embryo. Epicardial expression of NMHC IIB persists (white arrows). L: Ventral view of cardiac surface stained with DAB to identify blood cells in αMHC-Cre; Myh10+/+ heart (arrow). M: Dorsal view of cardiac surface stained with DAB to identify blood cells in αMHC-Cre; Myh10+/+ heart (arrows). N: Ventral view of cardiac surface stained with DAB to identify blood cells in αMHC-Cre; Myh10 flox/∆ heart (arrows). O: Dorsal view of cardiac surface stained with DAB to detect endogenous peroxidase activity from blood cells in αMHC-Cre; Myh10+ flox/∆ heart (arrows). Blood cells within vessels are present on the cardiac surface of cardiomyocyte-specific Myh10 mutant hearts. Similar results were seen for αMHC-Cre; Myh10 flox/flox hearts. Scale bars: B-G = 50 μm, H and J = 100 μm, I and K = 15 μm, L-O = 400 μm. Abbreviations: αMHC-Cre: Tg(Myh6-cre)2182Mds/J, CM: cardiomyocyte, cTnT: cardiac troponin T, Fibro: fibroblast, NMIIB: NMHC IIB, ∆: Myh10∆.
EHC mutants display abnormal epicardial cell morphology
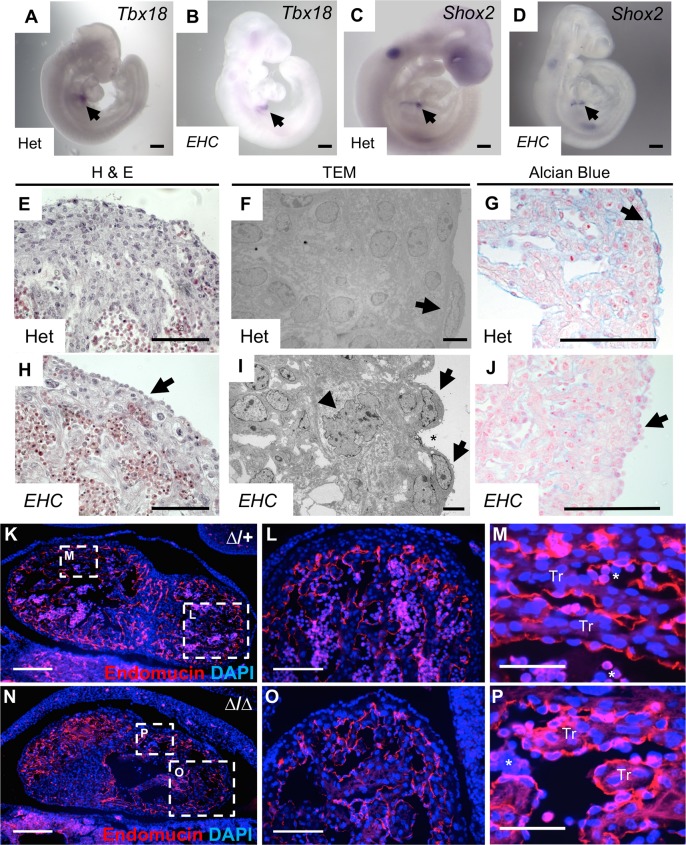
Due to the evidence that cardiomyocyte NMHC IIB is not required for coronary vessel development, we sought to determine if defects in epicardial cell function may therefore underpin the EHC mutant phenotype. A number of seminal studies have demonstrated that the epicardium plays a crucial role in the formation of the coronary vessels during mammalian development [15, 40–45]. As the coronary vessels incorporate cells and signals from the epicardium and sinus venosus during their development [13, 15], we investigated if the specification of the epicardial precursor, the proepicardial organ, and the sinus venosus occurred correctly in EHC mutant embryos. In situ hybridisation for Tbx18, a proepicardial marker [46], and Shox2, which is expressed in the sinus venosus [47, 48], showed that these molecular markers are expressed in a similar pattern in heterozygous and EHC mutant developing embryos (Fig 7A–7D, arrows), suggesting that these coronary vessel precursor tissues were present during early development in mutant embryos.
Fig 7. EHC mutants display epicardial defects.
A: In situ hybridisation demonstrating Tbx18 expression in the proepicardial organ of heterozygous and B: EHC homozygous mutant E9.5 embryos (arrows). C: Shox2 expression is detected in the sinus venosus of heterozygous and D: EHC homozygous mutant E9.5 embryos (arrows). The expression patterns of Tbx18 and Shox2 are indistinguishable between controls and EHC mutants. E: Heterozygous embryos have an epithelial layer comprised of epicardial cells coating the developing ventricle at E14.5. F: Electron microscopy showing E16.5 epicardial cells from heterozygous mice having a flattened morphology (arrow) and forming a contiguous layer with adjacent cells. G: Alcian blue staining reveals a layer of glycosaminoglycans in the extracellular matrix between the epicardium and myocardium in heterozygotes at E11.5 (arrow). H: Epicardial cells are present on the surface of the EHC homozygous mutant myocardium (arrow) at E14.5. I: Mutant epicardial cells are irregular in shape and exhibit a rounded morphology (arrows), which generates gaps between neighbouring cells (asterisk). Subepicardial cells display evidence of disrupted cellular architecture maintenance (arrowhead). J: EHC mutants lack Alcian blue staining in the subepicardial tissue domain (arrow). K-M: Endomucin immunofluorescence staining (red) of control E11.5 coronal cardiac sections delineates the ventricular endocardium. N-P: Endomucin distribution in Myh10∆ homozygous mutants is indistinguishable from controls. Areas shown at higher magnification in panels L-M and O-P are indicated with boxes on images K and N. Nucleated erythrocytes in panels M and P are indicated by asterisks. Scale bars: A-D = 250 μm, E and H = 60 μm, F and I = 5 μm, G and J = 125 μm, K and N = 200 μm, L and O = 100 μm, M and P = 50 μm. Abbreviations: H&E: haematoxylin and eosin, TEM: transmission electron micrograph Tr: trabeculae, ∆: Myh10∆.
Histology and transmission electron microscopy revealed that at E14.5, heterozygous hearts showed the formation of a characteristic epithelial layer on the surface of the myocardium (Fig 7E–7G) with flattened epicardial cells (Fig 7F, arrow). Although epicardial cells were clearly evident on the surface of the EHC mutant heart (Fig 7H, arrow), these cells had an unusual morphology compared to controls (Fig 7I, arrows). In addition, the EHC epicardium did not form a contiguous epithelial layer, and individual epicardial cells did not appear to form appropriate contacts with adjacent cells (Fig 7I, asterisk). The formation and maintenance of epicardial cell-cell gap junctions has previously been shown to be essential for correct epicardial cell function [49]. Interestingly, the ultrastructure of the subepicardial cell nucleus displayed an abnormal, multi-folded morphology, indicating defects in the maintenance of correct cellular architecture (Fig 7I, arrowhead). We calculated the percentage of epicardial cells with visually abnormal morphology in control and EHC mutant EM images, finding a statistically significant increase in the percentage of abnormal cells in EHC mutants (S2E–S2G Fig; Fisher’s exact test p<0.001).
The epicardium deposits extracellular matrix into the subepicardial space, which separates it from the underlying myocardium and is thought to play a critical role in the molecular communication between these tissues during embryonic development, homeostasis, and response to injury [13, 50, 51]. To evaluate possible defects in epicardial ECM deposition in the EHC mutants, we next analysed the subepicardial ECM by staining E11.5 cardiac sections with Alcian blue, a marker of glycosaminoglycans (GAGs) (Fig 7G and 7J). Prominent Alcian blue staining clearly delineated the subepicardial ECM in control hearts (Fig 7G, arrow). In contrast, EHC mutants lacked Alcian blue staining at the boundary between the epicardium and myocardium (Fig 7J, arrow), suggestive of abnormalities in the subepicardial ECM. Both mutant and control embryos display similar Alcian blue staining in the endocardial cushion mesenchyme (S2H–S2K Fig), illustrating that the localisation profile of GAGs within the heart is not universally disrupted in EHC mutants.
To evaluate potential requirements for NMHC IIB in other cardiac cell types with a role in coronary vessel development, we investigated whether or not NMIIB ablation disrupted the development of the endocardium by analysing the localisation of the endocardial marker endomucin, in both heterozygous control and Myh10∆ homozygous mutants E11.5 hearts (Fig 7K–7P). Myh10∆ homozygous mutants were utilised in these experiments to facilitate genotyping, as the loss of Myh10 exon 2 can be determined from a single PCR rather than requiring genomic sequencing as needed to detect the EHC point mutation. No aberrations in endomucin staining were detected in Myh10∆ homozygous mutant embryos (Fig 7N–7P), suggesting that the formation of the endocardium is not dependent upon NMIIB function. Moreover, the developing atrioventricular valves, derived from endocardial tissue, are present in heterozygous control and EHC mutant embryos at E14.5 and E16.5 (S5A–S5D Fig). As the formation of cardiac valve structures is highly dependent upon a functional endocardium [13, 52], the presence of these structures in EHC mutants supports the hypothesis that endocardial function is not significantly impaired following loss of NMHC IIB. A recently published complementary study has demonstrated that coronary vessel development is impaired when NMHC IIB is deleted specifically within the epicardium [53], supporting our findings that epicardial abnormalities in EHC mutant mice contribute to defective coronary vessel development.
NMHC IIB is the predominant form of NMII in the embryonic heart and epicardium
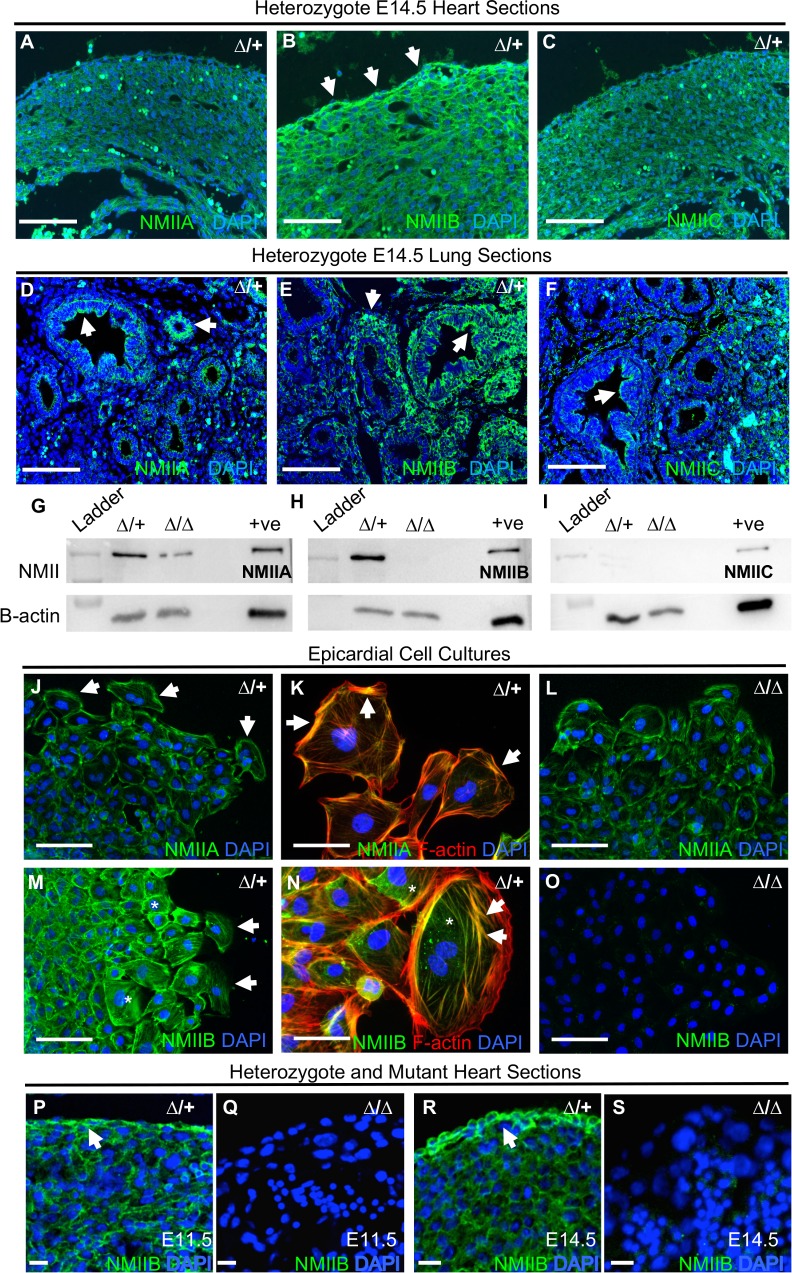
We sought to establish why the epicardium in particular demonstrates abnormalities following NMHC IIB ablation that are not exhibited in other tissue types. There are three NMHC II isoforms, IIA, IIB, and IIC, encoded by Myh9, Myh10 and Myh14 respectively in the mouse [20, 21]. The NMHC IIB null mouse displays developmental defects primarily in the brain and heart, which the authors attribute to an enrichment of NMHC IIB in these tissues [33, 37, 54]. However, the relative expression levels of individual NMHC II isoforms have not been explored specifically within the embryonic epicardium. We analysed NMHC II protein levels in control embryonic hearts using immunofluorescent microscopy. This analysis revealed that NMHC IIB is the predominant NMHC II isoform found in the E14.5 heart, and moreover, within the epicardium (Fig 8A–8C, arrows). All NMHC II isoforms were detectable in the developing lung (Fig 8D–8F, arrows), as previously described [37]. In addition, at E11.5 we found NMHC IIB together with IIA, were abundant in whole heart protein extracts probed by western blotting (Fig 8G and 8H). Somewhat surprisingly, we found that levels of NMHC IIA appeared to be diminished in NMHC IIB ablated samples (Fig 8G), whilst NMHC IIC was not detectable in either control or mutant preparations (Fig 8I). In light of this, we sought to determine the relative abundance and subcellular localisation of NMHC IIA and IIB in enriched epicardial cell cultures derived from E11.5 heart explants by immunocytochemistry. In both control and mutant cultures, NMHC IIA appeared to be primarily localised to the cell periphery in cells at both the leading edge and within the culture monolayer (Fig 8J–8L, arrows). In comparison, not only was fluorescent signal notably increased for NMHC IIB (Fig 8M and 8N), its subcellular localisation appeared to be more diffuse throughout the cell body (Fig 8N, asterisks), and frequently associated with cytoskeletal stress fibres (Fig 8N, arrows). As expected, NMHC IIB was not detectable in NMIIB ablated cultures (Fig 8O). Interestingly, mutant epicardial cells did not display either an increase in NMHC IIA signal intensity, or altered NMHC IIA subcellular distribution when compared to controls (Fig 8L compared to 8J). Higher magnification images of control hearts at E11.5 and E14.5 demonstrate NMHC IIB protein localisation to the epicardium (Fig 8P and 8R), with higher levels of epicardial expression found at E14.5 (Fig 8R) as compared to E11.5 (Fig 8P). We did not detect any immunofluorescent signal in Myh10∆ homozygous mutant hearts at either E11.5 or E14.5 using the NMHC IIB C-terminal antibody (Fig 8Q and 8S). Together, these data suggest that NMHC IIB is expressed at both a higher abundance in the epicardium, and in distinct subcellular regions to other NMHC II isoforms. Consequently, NMIIB may be serving a specialised function in the epicardial cells that cannot be compensated by other NMII isoforms when NMIIB is lost, thus exposing epicardial dysfunction.
Fig 8. NMHC IIB is the predominant form of NMII in the embryonic heart and epicardium.
A-C: Immunohistochemical comparison of the three different NMHC II isoforms (green) in the Myh10∆/+ E14.5 heart. NMHC IIB is the predominant isoform, and shows intense staining in the epicardium (B, arrows). D-F: E14.5 embryonic lung tissue was used as a control for the NMII isoform immunohistochemical analysis (arrows). Cell nuclei are stained with DAPI (blue). G-I: Western blot analysis of NMII isoforms in E11.5 Myh10∆ heterozygous control and homozygous mutant whole heart protein extracts. Protein extract from adult mouse lung was run as a positive (+ve) control. NMIIA and IIB are found in relative abundance at this developmental stage in heterozygous hearts (G and H). As expected, NMIIB is absent from mutant samples (H, lane 3), which also show a reduction in NMIIA (G, lane 3). NMIIC is not detectable in either control or mutant samples (I). Protein extracts pooled from at least n = 3 hearts for each genotype. J-O: Immunocytochemistry for the predominant IIA and IIB isoforms in Myh10∆ heterozygote (J, K, M and N) and mutant (L, O) epicardial cell cultures from E11.5 heart explants. Note localisation of NMIIA to the periphery of epicardial cells (J, K, arrows), whilst NMIIB is localised throughout the whole cell body (M, N, asterisks), and is frequently associated with actin stress fibres (M, N, arrow). Consistent with western blot analysis (H), NMHC IIB is not detectable in mutant samples (O). Images representative of cell cultures obtained from four hearts per genotype. P: NMIIB immunofluorescence (green) in E11.5 control sample. NMIIB is expressed in the epicardium (arrow). Nuclei are labeled with DAPI (blue). Q: NMIIB expression in Myh10∆ homozygous mutant heart at E11.5. R: NMIIB immunofluorescence (green) at E14.5 in control sample. NMIIB is highly expressed in the epicardium (arrow). Nuclei are labeled with DAPI (blue). S: NMIIB expression in Myh10∆ homozygous mutant heart at E14.5. NMIIB is not detectable in Myh10∆ homozygous mutant samples, consistent with western blot (H) and cell culture results (O). Scale bars: A-F, J, L, M and O = 100 μm, K, N, P-S = 50 μm. Images in A-C and D-F obtained at same magnification. Abbreviations: ∆: Myh10∆.
EHC epicardial cells do not exhibit migration defects in vitro
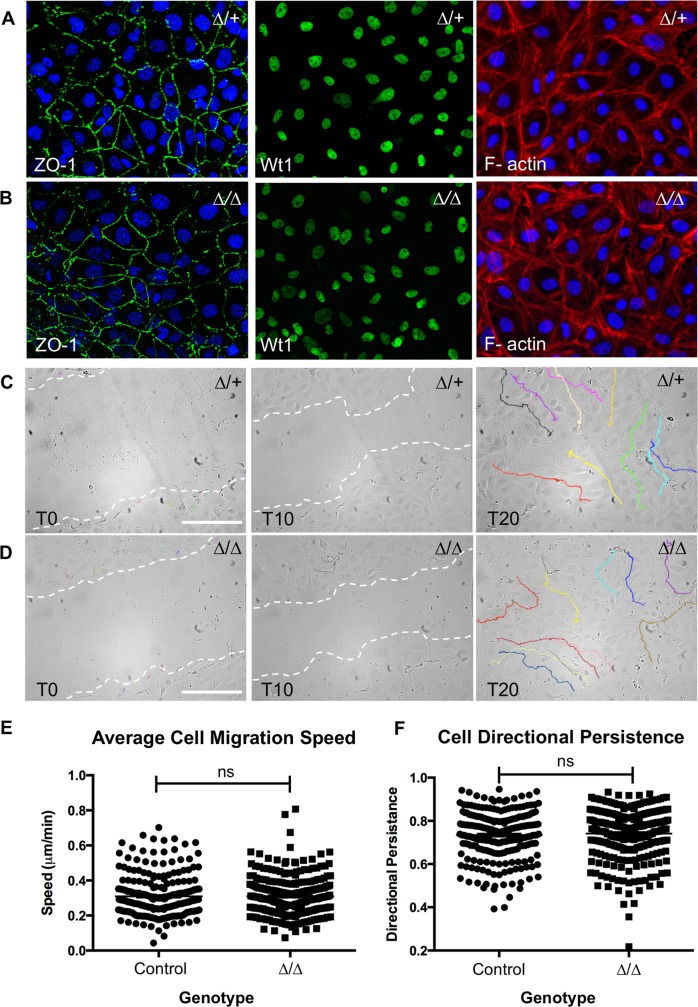
The formation of the epicardium is wholly reliant upon the migration of cells from the PEO to the surface of the developing myocardium [8]. NMHC IIB has been shown to play an important role in cell migration, through the generation of traction forces, and guidance of directional persistence [20, 21, 28, 36]. In light of this, we sought to investigate whether NMHC IIB null epicardial cells demonstrated motility defects in vitro by performing a scratch wound assay on epicardial cells cultured from embryonic heart explants as previously described [55]. Epicardial cells from control (+/+ or Myh10∆/+) and NMHC IIB null (Myh10∆/Myh10∆) E11.5 hearts were enriched and cultured for 48 hours to confluence on gelatin-coated 24-well plates. The Myh10∆ line was used for these experiments to expedite genotyping. The epicardial nature of the resultant cell populations was confirmed by immunostaining for the epithelial marker, ZO1, epicardial marker, Wt1 (Wilms Tumour 1), and the epithelial ‘cobble-stone’ morphology of filamentous actin staining with rhodamine phalloidin (Fig 9A and 9B). These epicardial monolayers were scratched with a P10 pipette tip and wound closure was imaged over a 20-hour period (S1 and S2 Movies). For each image series, we identified 10 cells at the leading edge of the wound at T0, and manually tracked their migration (Fig 9C and 9D). Cell migration speed and directional persistence were subsequently analysed using ImageJ. Somewhat surprisingly, we found that NMHC IIB null epicardial cells exhibited normal migratory behaviour when compared to controls, with no significant difference in either the average migration speed (Fig 9E, Mann-Whitney, p = 0.6717), or directional persistence (Fig 9F, Mann-Whitney, p = 0.2494). This finding suggests that epicardial cells do not require NMHC IIB to exhibit normal migratory behaviour in vitro.
Fig 9. NMHC IIB ablated epicardial cells do not show migration defects in vitro.
A: Cells cultured from E11.5 Myh10∆/+ heterozygote and B: Myh10∆ homozygous mutant heart explants form an epicardial monolayer. Epicardial status was confirmed by positive staining for the epithelial marker ZO-1 (left panels), and the epicardial marker, Wt1 (centre panels). Rhodamine-phalloidin staining of the actin cytoskeleton also revealed characteristic ‘cobblestone’ morphology, indicative of an epithelial cell population (right panels). C: Control and D: mutant epicardial monolayers were scratched at T0 (left panels) and imaged at 10-minute intervals for 20 hours. The migration of ten cells per field of view was tracked using ImageJ (right panels, coloured lines). E: Graph showing the subsequent comparison of mean cell migration speed (control = 0.3099μm/min, mutant = 0.3098μm/min, Mann-Whitney U-test p = 0.6717). F: Graph showing the comparison of cell migration directional persistence (control = 0.7342, mutant = 0.7411, Mann-Whitney U-test p = 0.2494). Total tracked cells = 240 control and 270 mutant. Cultures were generated from at least four hearts for each genotype. Control refers to data compiled from both wild type and ∆/+ genotypes. Scale bars: 250 μm. Abbreviations: ∆: Myh10∆.
EHC epicardial-derived cells demonstrate disrupted functionality in vivo
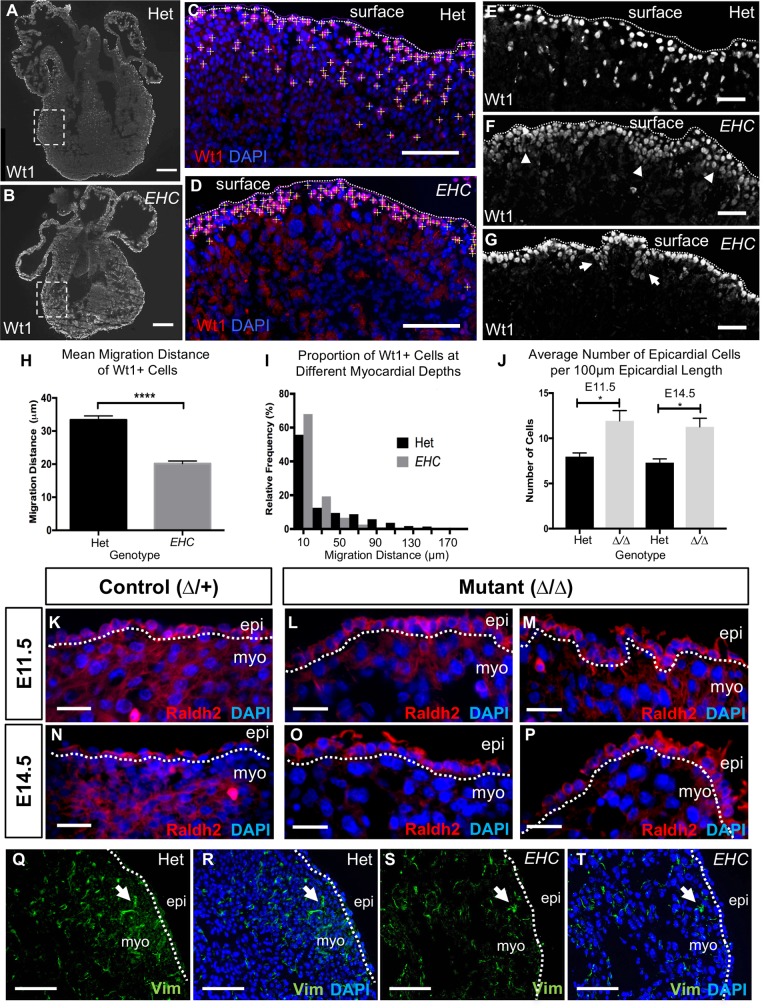
It is well documented that during development, a sub-population of epicardial cells undergo EMT and give rise to epicardial-derived cells that have acquired the ability to invade the underlying myocardium, where they differentiate into multiple cell lineages, including interstitial and perivascular fibroblasts and vascular smooth muscle cells [15, 45]. In addition to this cellular contribution, it is suspected that EPDCs regulate myocardial development through expression of paracrine signalling molecules [56–60]. It is clear that the correct execution of epicardial EMT is essential for both coronary and myocardial development. Since we did not detect epicardial motility defects in vitro, we examined epicardial cell migration into the underlying cardiac tissues in vivo by evaluating the localisation of cells expressing the epicardial marker Wt1 in EHC mutant and control embryos (Fig 10A–10G). Measurements of the distance between Wt1 positive cells in the ventricular myocardium of E14.5 hearts (Fig 10C and 10D, yellow crosshairs) and the cardiac surface indicated that EHC EPDCs had not penetrated as deeply into the myocardium as EPDCs in control hearts (Fig 10H, Mann Whitney, p<0.0001). Consequently, the majority of EHC EPDCs resided in a tightly restricted region of the subepicardial space (Fig 10F and 10I) or specifically around the blood-filled ventricular vesicles apparent in mutant hearts (Fig 10G). To confirm that the cells we detected at the cardiac surface were epicardial cells, we used the marker Raldh2. We found that Raldh2 expressing cells are located at the cardiac surface at E11.5 and E14.5 in both heterozygous control and Myh10∆ homozygous mutants (Fig 10K–10P). Quantification of the number of Raldh2 positive cells per length of epicardium reveals a significant increase in epicardial cell number in Myh10∆ homozygous mutants at both E11.5 and E14.5 when compared to control littermates (Fig 10J). We also evaluated vimentin expression at the cardiac surface in EHC mutant embryos and heterozygote controls (Fig 10Q–10T). We did not detect increased vimentin staining in EHC mutant embryos, indicating that there is not an increase in mesenchymal cell populations at the surface of the EHC heart. These in vivo results mirror the defects exhibited in other mouse models with compromised epicardial cell function [32, 61–63]. In addition, an in vivo cell migration defect may well be predicted from previous studies in which NMHC IIB is ablated, or its activation inhibited [28, 32]. Together, these data provide a strong evidence base to support our hypothesis that epicardial cell abnormalities contribute to the coronary vessel defects displayed by EHC mutant mice.
Fig 10. EHC epicardial-derived cells show defective migration in vivo.
A: Representative images of Wt1 immuno-stained coronal cryo-sections of EHC heterozygous and B: homozygous mutant E14.5 hearts. The dashed box indicates the area of ventricular wall used for measurement analysis in (C) and (D). C: Wt1 positive epicardial-derived cells were marked with yellow crosshairs in heterozygous and D: mutant hearts. The distance between these cells and the cardiac surface (C-G: dashed line) was manually measured using ImageJ. E: Wt1 positive cells (white) in the heterozygous heart can be seen at the cardiac surface (dashed line) indicating localization in the epicardium, as well as in deeper cardiac tissue below the dashed line. F: EHC mutants have Wt1 positive cells (white) primarily at the cardiac surface (dashed line), but these cells are organised into abnormal clusters (arrowheads), with few cells at deeper positions in the underlying myocardium. G: Wt1 positive cells surround the ventricular surface blisters (arrows) present in EHC mutant hearts. H: Graph comparing the mean migration distance of Wt1 positive cells: Het = 33.37 μm (+/-1.197 μm SEM, n = 975), mutant = 20.14 μM (+/-0.8136 μm SEM, n = 1180), Mann-Whitney U-test p = <0.0001). I: Histogram showing the relative frequency of Wt1 positive cells at increasing depths within the myocardial wall. A higher proportion of mutant Wt1 positive cells reside in the subepicardial region (<50 μm from the apical epicardial boundary) compared to controls. J. Comparison of the mean number of Raldh2 expressing cells found on the cardiac surface of hearts per unit of epicardial length for each genotype at E11.5 and E14.5. Mean number of epicardial cells/100μm in E11.5 controls = 7.975 (+/- 0.4008, n = 4) and mutants = 11.93 (+/- 1.145, n = 7); Mean number of epicardial cells/100μm in E14.5 controls = 7.3 (+/- 0.4243, n = 4) and mutants = 11.27 (+/- 0.9502, n = 10). A significant difference is detected between genotypes at E11.5 (2-tailed unpaired t test, p = 0.0335) and E14.5 (2-tailed unpaired t test, p = 0.0257). Error bars represent standard error of the mean. K: Raldh2 protein localisation (red) in Myh10∆ heterozygous control and L-M: Myh10∆ homozygous mutant heart at E11.5. Sections from two different embryos are shown. N: Raldh2 protein localisation (red) in Myh10∆ control and O-P: Myh10∆ homozygous mutant heart at E14.5. Sections from one embryo at different cardiac depths are shown (O-P). Q: Vimentin immunofluorescence in E14.5 control heart. Spindle-shaped mesenchymal cells are present (arrow). R: Merged image showing nuclei (blue). S: Vimentin immunofluorescence in E14.5 EHC mutant heart. Spindle-shaped mesenchymal cells are present (arrow). T: Merged image showing nuclei (blue). The epicardial boundary with the myocardium is labeled with a dashed line. Scale bars: A and B = 250 μm, C, D, Q-T = 100 μm, E-G = 50 μm, K and N = 25 μm. Abbreviations: epi: epicardium, myo: myocardium.
The ECM environment is altered in NMHC IIB mutant hearts
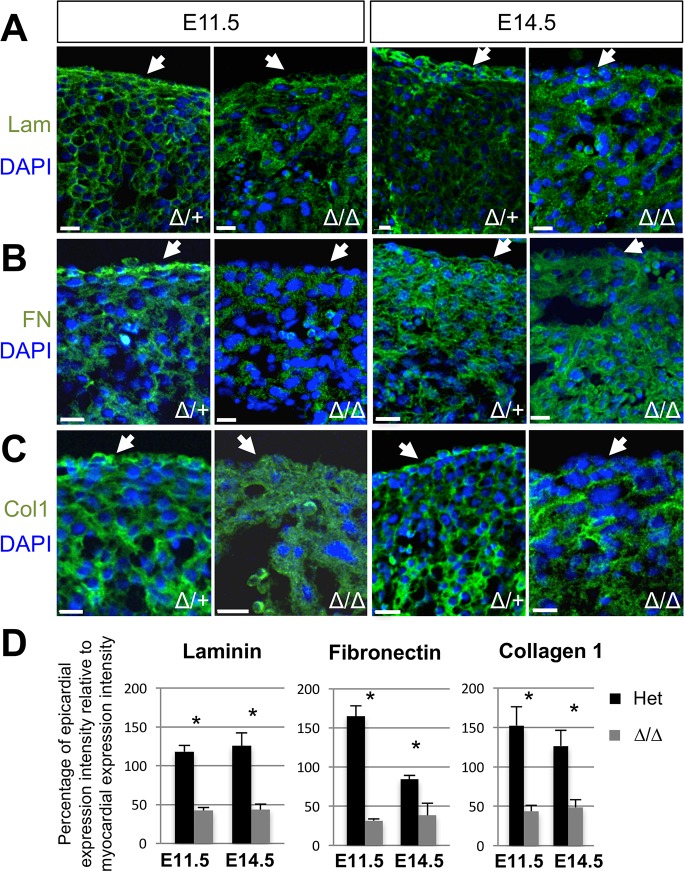
Following the observation that Myh10∆ homozygous mutant epicardial cells display altered migration in vivo, but not in scratch wound assays in vitro, we hypothesised that the coronary vessel development defects in the EHC and Myh10∆ mutant embryos might be due to an altered in vivo environment affecting epicardial derived cell migration. We have shown that Alcian blue staining for GAGs was reduced in the EHC subepicardial ECM (Fig 7G and 7J). Since GAGs constitute a major molecular component of the extracellular matrix, we hypothesised that the loss of NMIIB disrupts ECM protein distribution in the developing heart, thus hindering epicardial cell motility or migration in vivo. We therefore evaluated the expression of laminin and fibronectin, ECM components documented to be expressed in the human embryonic heart [64]. Fibronectin is of interest particularly in the developing epicardium, since it has been shown to be required for directional persistence during epicardial cell migration [65]. In the mouse heart, fibronectin has been reported to localise to the epicardium from E12.5 to E16.5, where it co-localises with collagen I [66]. To evaluate ECM distribution we performed immunofluorescence for laminin, fibronectin, and collagen I in Myh10∆/+ control and Myh10∆ homozygous mutant hearts at E11.5 and E14.5. We found that laminin, fibronectin and collagen I were abundantly present in the subepicardial ECM of control hearts, and indeed, throughout the ventricular myocardium (Fig 11A–11C, arrows). The distribution of these ECM components was altered in Myh10∆ homozygous mutant hearts, with little detectable staining in the epicardial layer and generally reduced levels throughout the heart (Fig 11A–11C, arrows). Quantification of the staining intensity in the epicardial region relative to staining intensity in the myocardial region on the same tissue section was performed. All ECM components analysed demonstrated a significantly reduced ratio of expression in Myh10∆ homozygous mutants at E11.5 and E14.5 (Fig 11D), suggesting that the extracellular environment of the mutant heart has been altered. Furthermore, Myh10∆ homozygous mutant epicardial explants cultured independently from myocardial cells display an altered pattern of fibronectin distribution, with poor organisation and a reduced network of fibrils (S6 Fig), confirming that mutant epicardial cells have an impaired ability to produce fibronectin.
Fig 11. Examination of ECM composition in Myh10∆ control and mutant embryos.
A: Laminin immunofluorescence (green) is present at E11.5 and E14.5 in the Myh10∆ heterozygous ventricular myocardium and is continuously distributed in the epicardium (arrow). Myh10∆ homozygous mutants show reduced laminin immunoreactivity in the epicardium (arrow) at both E11.5 and E14.5. Nuclei are marked with DAPI (blue). B: Fibronectin immunofluorescence (green) is present at E11.5 and E14.5 in the Myh10∆ heterozygous ventricular myocardium and is continuously distributed in the epicardium (arrow). Myh10∆ homozygous mutants appear to have reduced signal, especially in the epicardium (arrow) at E11.5 and E14.5. Nuclei are marked with DAPI (blue). C: Collagen 1 immunofluorescence (green) is present at E11.5 and E14.5 in the Myh10∆ heterozygous ventricular myocardium and is continuously distributed throughout the epicardium (arrow). Myh10∆ homozygous mutants have reduced collagen 1 signal in the epicardium (arrow) at E11.5 and E14.5. Nuclei are marked with DAPI (blue). Genotypes and developmental stages are labeled on the image. D: Quantification of the ratios of expression levels of epicardial immunofluorescence intensity to myocardial immunofluorescence intensity for each protein. Three embryos of each genotype were analysed, with measurements taken from five different areas of three different sections from each embryo. All markers and time points showed a statistically significant reduction (*) in immunofluorescence intensity of the epicardium relative to the myocardium in Myh10∆ homozygous mutants as compared to controls (two-tailed t-test, p<0.05). Ratios of expression for each genotype were compared to each other for each marker and developmental stage. Error bars represent standard deviation. Genotypes, markers and stages are labeled on the graphs. Scale bars: 15 μm. Abbreviations: Lam: laminin, FN: fibronectin, Col1: Collagen I.
Analysis of epicardial cell apoptosis, proliferation, and signaling in NMHC IIB mutants
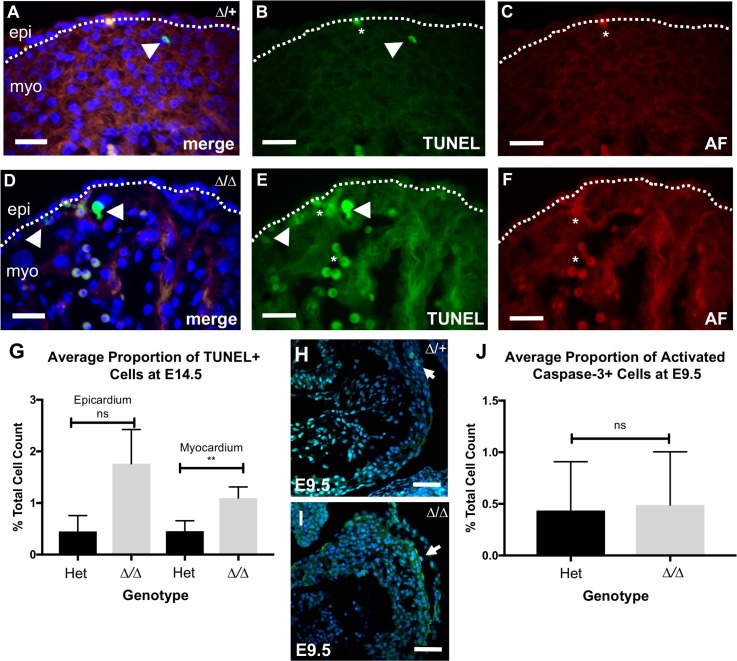
Correct deposition of the cardiac ECM is necessary for appropriate cardiac development [67]. In the adult heart, alterations in ECM composition are associated with apoptosis [68]. After detecting ECM defects in NMHC IIB ablated mutants, we therefore assessed apoptosis in the Myh10∆ homozygous mutant heart at E14.5 using a TUNEL assay, and compared our findings to control samples (Fig 12A–12G). At this developmental stage, control hearts displayed low levels of apoptosis in both the epicardium and underlying myocardium (Fig 12A and 12B). We found a small but statistically significant increase in the average number of apoptotic cells within the mutant myocardium, compared to control littermates (Fig 12G; Mann Whitney U test, p<0.006). In contrast, whilst the mutant epicardium also displayed elevated apoptosis rates when compared to controls, this finding was not statistically significant (Fig 12G; Mann Whitney U test p = 0.07). Similarly, no statistically significant difference was observed between the number of apoptotic cells in activated caspase-3 stained control (Fig 12H) and mutant (Fig 12I) hearts during early development at E9.5 (Fig 12J; unpaired 2-tailed Mann-Whitney U-test, p = 0.9292). These data suggest that loss of NMHC IIB does not cause significant epicardial apoptosis.
Fig 12. Apoptosis in Myh10∆ mutant hearts.
A: TUNEL analysis (green) of Myh10∆ heterozygous heart at E14.5 shown with nuclei stained with DAPI (blue). B: Single channel of TUNEL staining from (A). Apoptotic cells (arrowhead) are present along with autofluorescent erythrocytes (asterisk). C: Autofluorescence of erythrocytes (red) from (A). D: TUNEL analysis (green) of Myh10∆ homozygous mutant heart at E14.5 shown with nuclei stained for DAPI (blue). E: Single channel of TUNEL staining from (D). Apoptotic cells (arrowhead) are present along with clusters of erythrocytes (asterisks). F: Autofluorescence of erythrocytes (red) from (D). G: Graph comparing mean number of TUNEL positive cells as a proportion of total cells in either the epicardium, or myocardial wall. Mean percentage of apoptosis in control = 0.4484% (+/- 0.3082%), and mutant epicardium = 1.762% (+/- 0.662%) (unpaired 2-tailed Mann-Whitney U-test, p = 0.0744). Mean percentage of apoptosis in control = 0.4532% (+/- 0.204%), and mutant myocardial wall = 1.097% (+/- 0.2132%) (unpaired 2-tailed Mann-Whitney U-test, p = 0.0053). Three hearts per genotype and six non-consecutive sections per heart were analysed. H: Sagittal section of control and I: Myh10∆ homozygous mutant hearts at E9.5 showing immunofluorescence for activated Caspase-3 (turquoise, arrows), with nuclei visualised with DAPI (blue). J: Graph comparing mean number of activated caspase-3 positive cells as a percentage of total cells in cardiac sections of control and mutant E9.5 hearts. Mean percentage of apoptosis in control = 0.435% (+/- 0.0.1579%), and mutant hearts = 0.4885% (+/- 0.1722%). Three hearts were examined per genotype, and three sections were analysed for each heart. There is no significant difference in apoptosis between the two genotypes (unpaired 2-tailed Mann-Whitney U-test, p = 0.9292). Error bars in G and J represent standard error of the mean. Scale bars: A-F = 25 μm, H-I = 50 μm.
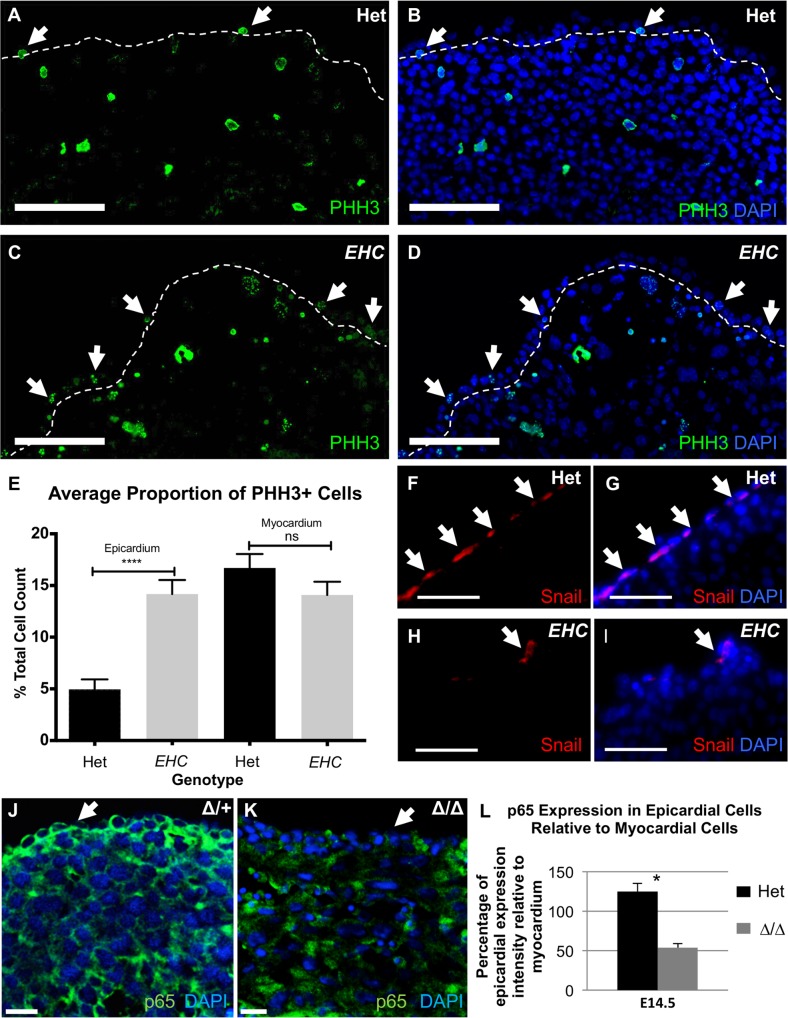
We next sought to establish whether the function of EHC epicardial cells was compromised prior to EPDC migration, specifically focusing on the process of epicardial EMT. Analysis of the proliferation marker phosphohistone H3 (PHH3) revealed a distinct increase in proliferation within the epicardium of EHC mutants at E14.5 as compared to controls (Fig 13A–13D, arrows; Fig 13E, unpaired t-test, p<0.0001). We found no significant difference in cell proliferation in the underlying cardiac tissue between controls and EHC mutant hearts (Fig 13E, unpaired t-test, p = 0.1684), nor in total cardiac tissue between control and Myh10∆ homozygous mutant hearts at E9.5 (S6C Fig, unpaired t-test, p = 0.57). This finding is consistent with a reduced incidence of EMT induction, as cells undergoing EMT attenuate cell division in favour of changes to cell morphology [69]. Correspondingly, we examined the localisation of the EMT marker, Snail, within the epicardium at E14.5. Similarly, we found that the EHC mutants demonstrated a reduction in the number of Snail positive epicardial cells (Fig 13F–13I, arrowheads). Recently it has been reported that NF-κB signaling is required downstream of TGFβ and PDGF inputs to mediate epicardial cellular changes associated with EMT [70]. The NF-κB component p65 is expressed in the mouse epicardium during EMT [70]. We examined p65 expression at E14.5 in heterozygote control and Myh10∆ homozygous mutant hearts (Fig 13J–13K) and calculated the ratio of epicardial staining intensity compared to myocardial staining intensity. We found a statistically significant reduction in the ratio in Myh10∆ mutant embryos, indicating reduced p65 expression in the mutant epicardium (Fig 13L). Together, this evidence is highly suggestive of epicardial EMT dysregulation specifically in the mutant epicardium. This result suggests an important role for NMHC IIB in the promotion or execution of EMT, via NF-κB signaling, that to our knowledge has not been reported previously.
Fig 13. EHC epicardial cells show defects in EMT.
A-B: EHC Heterozygous and C-D: EHC homozygous mutant E14.5 heart cryo-sections were stained for the proliferation marker, PHH3 (green). Proliferating epicardial cells are indicated by arrowheads. The dashed line indicates the boundary between the epicardium and myocardium. Cell nuclei were stained with DAPI (blue). E: Graph comparing the proportions of proliferating cells (as a percentage of total cells) in both the epicardium and myocardium between controls and homozygous mutants. Average proportion of proliferating cells in control = 4.953% (+/- 0.9674%) and EHC mutant epicardium = 14.17% (+/- 1.356%) (unpaired 2-tailed t-test, p = <0.0001). Average proportion of proliferating cells in control = 16.70% (+/- 1.348%), and mutant myocardium = 14.09% (+/- 1.275%) (unpaired 2-tailed t-test, p = 0.1684). Error bars represent standard error of the mean. F-I: Localisation of the EMT marker, Snail in the control and mutant epicardium (arrowheads). Cell nuclei are indicated by DAPI staining (blue). The EHC epicardium shows a reduction in the number of cells in which EMT is activated. J: p65 immunofluorescence (green) in Myh10∆ heterozygous heart showing expression in the epicardium (arrow). Nuclei are labeled with DAPI (blue). K: p65 immunofluorescence (green) in Myh10∆ homozygous mutant heart showing diminished immunoreactivity in the epicardium (arrow). L: Quantification of p65 immunofluorescence intensity in the epicardium relative to the myocardium. Genotypes are labeled on the graph. A statistically significant reduction in p65 epicardial expression is seen in Myh10∆ homozygous mutants (t-test, p = 0.003). Error bars represent standard deviation. Images were analysed following the same methodology as reported for Fig 11D. Scale bars: A-D = 100μm, F-I = 50μm, J-K = 15 μm. Abbreviations: Wt1: Wilms tumour 1, PHH3: phosphohistone H3.
Discussion
The present study demonstrates a requirement for NMHC IIB during the formation of the mammalian coronary vasculature. Here, we report the characterisation of a mutant mouse line, generated from a balancer chromosome ENU mutagenesis screen [19], that displays embryonic hydrocephalus and cardiac defects. The EHC point mutation in Myh10 results in a global loss of NMIIB function, as confirmed by genetic complementation of the EHC allele in trans to the Myh10∆ allele. Notably, the Myh10 gene is located approximately 1 Mb outside the Trp53 endpoint of the balancer chromosome interval [19]. In breeding the EHC mutants we have only had 1 animal out of more than 1180 in total that showed recombination between the Myh10 mutation and the balancer chromosome end point, indicating that the balancer chromosome can be used to maintain balanced heterozygous stocks for embryonic lethal mutations in genes located outside the balancer interval.
Our studies of the EHC mouse have made advances in understanding the essential role undertaken by NMHC IIB during cardiogenesis. The EHC mutant coronary vessel abnormalities are markedly similar to phenotypic observations of mice in which the epicardium has been specifically disrupted [29, 31, 63], and are consistent with a primary defect in epicardial cell function. We have shown that EHC mutant epicardial cells have a highly perturbed morphology, impaired epicardial EMT, and disrupted subepicardial ECM composition, in addition to reduced migration of EPDCs into the myocardium. The absence of mature coronary vessels in EHC mutants suggests that in vivo these dysfunctional EPDCs are incapable of contributing to the vascular network. A complementary study has demonstrated that coronary vessel development is impaired when Myh10 is deleted specifically in the epicardium [53], consistent with our conclusion that epicardial defects underpin disrupted coronary vessel development in EHC mutant mice.
NMHC IIB plays a key role in a broad variety of fundamental cellular processes [20, 21]. Similarly, the defects we have documented encompass a range of physiological events, including, but not limited to: cell migration, adhesion, proliferation and apoptosis. Our finding that EHC mutants display defects in epicardial EMT is of particular interest, as other fundamental developmental processes that depend upon the correct initiation and execution of EMT (e.g. gastrulation, craniofacial development) occur in the EHC embryo. Indeed, formation of the atrioventricular valves, a process dependent upon endocardial cushion EMT [13], is evident in EHC mutant hearts (S5 Fig). We have demonstrated that NMHC IIB is the predominant NMHC II isoform in epicardial cells both in vivo and in vitro, and that NMHC IIB is localised to distinct subcellular regions (in agreement with the findings of Lo et al., [28], and Ma et al., [53]). Therefore, it may be hypothesised that NMHC IIB plays critical roles in epicardial and cardiac development, which cannot be replaced by other NMHC II isoforms when NMHC IIB function is globally ablated. The observation that loss of NMHC IIB leads to decreased expression of NMIIA presents the possibility of altered transcriptional regulation in EHC mutants, which needs to be further explored to understand the roles of NMIIB in epicardial cell function, and embryonic development as a whole. Notably, unlike NMHC IIA or IIC, NMHC IIB displays cell type and cell-cycle specific mechanosenstivity [71], which may be specifically altered in mutant epicardial cells during development.
The myocardium also plays a role in epicardial cell behaviour, through the secretion of paracrine signalling molecules which traverse the subepicardial ECM and communicate with the epicardium to orchestrate correct epicardial function [13, 15, 50, 60]. The requirement for NMHC IIB in myocardial development is well documented, with previous reports observing myocardial disorganisation, cardiomyocyte cytokinesis defects, and a reduction of the myocyte population in NMHC IIB null hearts [23, 33, 36, 37]. However, cardiomyocyte-specific Myh10 ablated mice are viable, and importantly, demonstrate a reduced instance of VSD and the complete absence of DORV [35]. Here we report that NMHC IIB is not required within the cardiomyocyte population for coronary vessel development. This finding suggests that the severe morphological defects present in NMHC IIB null hearts are caused by loss of NMHC IIB from other cardiac cell populations. It has recently been demonstrated that epicardial-specific deletion of Myh10 does impair coronary vessel development [53], although the reported defects are not as severe as we have documented here for the EHC mutant, EHC/Myh10∆ compound heterozygote, or Myh10∆ homozygous mutant embryos. Further investigation is required to determine if these phenotypic differences are due to differences in assays used or due to requirements for NMHC IIB in multiple cell types during coronary vessel development.
NMHC IIB has been directly shown to be important in cell migration as a key component of the actin-myosin cytoskeletal machinery [20, 21, 28, 36]. Our use of the epicardial cell culture model shows that the migration of EHC epicardial cells progresses unimpeded in vitro, both in the context of epicardial cell outgrowth from embryonic heart explants, and in a wound-healing assay. Moreover, cultured primary epicardial cells have been previously shown to express Snail [72], indicating that the process of epicardial cell migration from explant heart tissue in vitro involves EMT activation. A key difference between the explant model and the EHC mutant is the context of the extracellular environment. We performed our explant assays on gelatin-coated plates, and it has been demonstrated that the provision of exogenous ECM substrate can compensate for migration defects in cells with ECM production deficiencies [73]. It is known that the subepicardial ECM plays an important role in epicardial function, in both the adhesion of the epicardial monolayer to the myocardium and in facilitating molecular communication through the subepicardial space [13, 50, 60, 74]. During EMT, TGFβ signaling (a key input for epicardial EMT [60]) is known to be affected by ECM substrate rigidity [75]. Additionally, alterations in cell tension, provoked by changes in the ECM, can disrupt nuclear architecture and chromatin structure, with subsequent effects on transcriptional regulation [76]. Disruption of the subepicardial ECM as seen in EHC and Myh10∆ mutants may therefore alter the ability of EHC EPDCs to migrate into the myocardium.
The detection of abnormalities in EMT signaling, indicated by increased epicardial cell proliferation and reduced Snail expression in EHC mutants, is surprising, as NMHC IIB would be expected to be a downstream effector of cell motility in EMT. Although EMT defects have been detected in the epicardial-specific Myh10 knock out [53], comparison of the results of that study with ours is complicated by the use of different assay methods and the potential for requirements for NMHC IIB in non-epicardial cells to influence our findings. However, our results suggest that the loss of NMHC IIB disrupts processes required for EMT signaling, through the NF-κB pathway acting downstream of TGFβ and PDGF inputs [70]. PDGF is produced in the myocardium and serves as a paracrine signal to promote epicardial EMT [60]. Phenotypically, the epicardial cell morphology defects we note from EM studies are similar to those reported for a PDGFRβ knock out [63]. Interestingly, it has previously been shown that changes in ECM composition, particularly collagen, can alter PDGF responsive gene activation during wound healing [77]. We have detected reduced p65 in Myh10∆ mutants, indicative of impaired NF-κB pathway activation. Since NF-κB pathway activation occurs in response to PDGFBB ligand inputs [70], we speculate that the observed alterations in the subepicardial ECM of mutant embryos may impede PDGF signalling, which subsequently hinders NF-κB pathway activation, thus contributing to epicardial EMT dysregulation in EHC and Myh10∆ homozygous mutant embryos.
In summary we have demonstrated a requirement for NMHC IIB to generate the appropriate cardiac extracellular matrix environment at the subepicardial space. We also demonstrate that signals promoting epicardial cell EMT are deficient in EHC and Myh10∆ mutant embryos, and that migration of EPDCs into the myocardium is impaired. Together, these data indicate that the coronary defects observed in the EHC embryos are underpinned by compromised epicardial function, and suggest that NMHC IIB plays a crucial role in normal epicardial biology. Confirmation of our findings is provided from a recent study demonstrating that mice with an epicardial-specific knock out of Myh10 display defects in coronary vessel development [53]. Myh10 is a pleiotropic gene that performs multiple roles in different developmental processes including tension generation [78], growth factor receptor internalisation [79], cell adhesion [20], and extracellular matrix protein secretion [80]. Moreover, the different NMHC II isoforms have functionally distinct roles [24]. Further investigation of the molecular functions of NMHC IIB in the epicardium and other cardiac cell types may inform therapeutic strategies to reactivate epicardial processes in injured cardiac tissue and enhance coronary vessel repair and regeneration.
Materials and methods
Ethics statement
Experiments using animals were performed in accordance with legislation in the UK Animals (Scientific Procedures) Act of 1986 (PPL 70/8858 to Graham Morrissey). Experiments were approved by the University of Manchester Animal Welfare and Ethical Review Body.
Mouse strains and genotyping
The l11Jus27 mouse line was generated from a balancer chromosome mutagenesis screen. The two mutations carried in the l11Jus27 mouse line were maintained in trans to 129S5.Inv(11)8BrdTrp53-Wnt3 [19]. Genomic DNA was prepared from ear punches of adult mice and yolk sacs (<E11.5) or tails (≥E11.5) of embryos. Genotypes were determined by PCR analysis with microsatellite marker D11MIT327, D11MIT35, D11MIT31 or D11MIT322 to differentiate between C57BL/6 (mutant) and 129S5 (wild type) strains of mice. Myh10tm7Rsad mice were obtained from the MMRRC, and were crossed to Tg(Nes-cre)Wme mice to generate global deletion of Myh10 exon 2. Tg(Myh6-cre)2182Mds/J mice were bred to Myh10tm7Rsad mice to generate cardiomyocyte-specific deletion of Myh10 exon 2. Genotyping primers are listed in S1 Table. PCR products were sequenced to confirm specificity of genotyping PCR reactions.
Meiotic mapping and NMHC IIB sequencing
For mutation mapping, mice were bred to 129S5 wild type mice. Animals inheriting the l11Jus27 mutation but not the balancer chromosome were selected for further breeding. l11Jus27 mice without the balancer were crossed to 129S5 wild type animals and progeny examined for recombination events. Recombination events were identified using microsatellite and SNP polymorphic markers between C57BL/6 and 129S5 mouse strains (primer sequences in S1 Table). Recombinant mice were intercrossed to l11Jus27 mice and progeny analysed. An absence of homozygous C57BL/6 pups indicated that the mutation was present in the recombinant animal. A timed mating was performed to confirm the mutant phenotype. Recombinant mouse 363, which only carried the ‘EHC’ point mutation, was bred with 129S5.Inv(11)8BrdTrp53-Wnt3 to generate the EHC mouse line. The EHC point mutation was identified by genomic sequencing of all annotated Myh10 exons (UCSC genome Browser mm9 assembly). PCR amplification was performed on each exon, products precipitated, and cycle sequenced using Big Dye v1.1 reaction mix (ABI) according to manufacturer’s instructions (primer sequences in S1 Table).
Complementation assay
To confirm genomic deletion of Myh10 exon 2, PCR was performed for primers flanking exon 2 (Fig 6) and products were sequenced to confirm specificity to Myh10. Matings of Myh10∆/+ and EHC/+ male and female adult mice were set up and pregnancies were allowed to proceed to term. Neonate litters were culled and tail biopsies from euthanised mice were used to genotype each animal for the Myh10∆ and EHC mutations. The observed genotypes were compared to expected Mendelian ratios and data sets were analysed using a Chi-squared test with 2 degrees of freedom (http://graphpad.com/quickcalcs/chisquared1.cfm).
Embryo dissection and analysis
Mice were set up for timed matings and the morning of the vaginal plug was defined as day E0.5. Mice were sacrificed according to Home Office Schedule 1 methods. Embryos were then dissected from decidua at the desired time point and subsequently imaged in PBS using a Leica MZ6 microscope and DFC420 camera.
Protein structure predictive modelling
The three dimensional structure of residues 1–815 of mouse NMHC IIB were predicted using homology modelling. The sequences of mouse NMHC IIB and chicken smooth muscle myosin were aligned using ClustalW [81], and structure predicted by Modeller [82] using the known chicken myosin structure (PDB id 1BR1) [83] as a template. The sequence identity between the two proteins was 83% over the aligned region. Twenty models were built, and the one with the best score was used for further analysis.
SDS-PAGE and Western blotting
Protein was extracted from E11.5 embryos by homogenisation in RIPA buffer. Protein concentrations were determined according to the manufacturer’s protocol (Biorad Protein Assay). Protein lysate (30 μg) was loaded onto a 10% polyacrylamide gel and separated by electrophoresis before being transferred to a PVDF membrane. Membranes were blocked overnight in 5% milk and incubated in anti-nonmuscle myosin II primary antibodies (NMIIA, Covance, PRB-440P, NMIIB Covance, PRB-445P, NMIIC, Covance, PRB-444P; 1:1000) or anti-beta-actin HRP conjugate (Sigma, A3854; 1:200,000) for 1 hour at room temperature. Membranes were subsequently incubated in HRP conjugated donkey anti-rabbit secondary antibody (Santa Cruz, SC-2313; 1:1000) for 1 hour at room temperature. Protein detection was performed using the ECL Plus Western Blotting detection system (GE Healthcare) according to manufacturer’s instructions.
Quantitative real-time PCR
RNA was prepared from E12.5 wild type, heterozygous and mutant embryos using Tri reagent (Sigma). RNA (5 μg) was treated with RNase-free DNase 1 (Promega) and cDNA generated using random primers (Promega) and Bioscript reverse transcriptase (Bioline). Primers for Myh10 qPCR are listed in S1 Table.
Histology
Embryos were fixed in Bouin’s fixative or 4% paraformaldehyde (PFA), dehydrated and then cleared in xylene or histoclear. Embryos were embedded in paraffin and sectioned at 7 μm. After sectioning paraffin was removed from sections by washing 2 x 10 minutes in xylene or histoclear. Sections were then rehydrated and stained in haemotoxylin and eosin or Alcian blue and/or nuclear fast red before being mounted in depex mounting medium for analysis.
Immunohistochemistry and immunofluorescence
Embryos and embryonic hearts were fixed overnight in 4% PFA and stored in 70% ethanol. For immunohistochemistry, whole hearts were incubated with primary antibodies anti-PECAM-1 (BD Biosciences, 550274; 1:100) for 1 hour at room temperature. For immunohistochemistry on tissue sections, samples were incubated in anti-smooth muscle alpha actin (Sigma, A5228; 1:400) for 1 hour at room temperature. Staining was developed using the Vectastain Elite ABC Kit (Vector, PK-6100) and visualised using the DAB substrate kit (Vector, SK-4100). For immunofluorescence on tissue sections hearts were dehydrated, embedded in paraffin and sectioned at 7 μm. Tissue sections were subjected to antigen retrieval by heating in citrate buffer (DAKO, S1699) as per the manufacturer’s instructions. Tissue was blocked prior to incubation in primary antibodies with either 10% (v/v) serum in PBS, or DAKO serum free protein block (DAKO, X0909). Heart sections were stained with primary antibodies directed against the following proteins: laminin (custom-made antibody, kind gift from Ray Boot-Handford, University of Manchester; 1:400); fibronectin (Santa Cruz, SC-6952; 1:50 dilution), collagen 1 (Gentaur, OARA02579; 1:400), fibronectin (Santa Cruz, sc-6952, 1:50), p65 (Santa Cruz, SC-372; 1:200), cardiac Troponin T (Abcam, ab106076; 1:400), NMHC IIB (Eurogentec, PRB-445P-050; 1:400), Raldh2 (Abcam, ab75674: 1:200), SM22α (Abcam, ab14106; 1:250), endomucin (Santa Cruz, SC-65495; 1:50), sarcomeric alpha-actinin (Sigma, EA-53; 1:500), and beta-catenin (Sigma, C2206, 1:250) overnight at 4° C. Embryos were incubated in species-specific fluorescent secondary antibodies (Jackson Immunochemicals or Invitrogen; 1:500 dilution) and slides mounted in DAPI mounting media (Vector).
Blood cell peroxidase staining
Whole hearts were fixed for two hours at room temperature in 4% PFA. Hearts were washed 2X5 min in PBS, prior to incubation in the DAB substrate kit with added Nickel solution (Vector, SK-4100) according to manufacturer’s instructions.
Cryo-immunohistochemistry
E14.5 embryonic hearts were dissected into ice cold PBS, embedded in OCT (R.A. Lamb) and snap frozen in liquid nitrogen. Cryosections (14 μm thick) were fixed in 4% PFA for 15 minutes, permeabilised in PBS + 0.1% (v/v) Triton X-100 for 15 minutes and subsequently blocked with PBS + 1% (w/v) BSA + 10% (v/v) normal goat or horse serum (Vector) for 1 hour. Sections were then incubated with the following primary antibodies diluted in PBS + 0.1% (v/v) Triton X-100 for 24 hours at 4°C: rabbit polyclonal anti-mouse Wt-1 (Calbiochem, CA1026, 1:300), goat anti-Snail (Abcam, ab53519, 1:100), rabbit anti-PHH3 (Millipore, 06–570, 1:300), anti-vimentin (Proteintech, 10366-1-AP, 1:50). Sections were then incubated with species-specific biotinylated secondary antibodies (Vector) at 1:500, for 2 hours, or FITC conjugated goat anti-rabbit secondary antibody (Sigma, F9887, 1:160) (detection of PHH3). Sections were incubated with Cy3 conjugated streptavidin (GE Healthcare, PA43001), diluted 1:3000 or 1:1000 (detection of Snail) for 30 minutes. Coverslips were mounted with Vectashield with DAPI (Vector, H-1200) and sealed with nail varnish. Slides were stored in the dark at 4°C and imaged within 48 hours.
Epicardial cell culture
Cardiac fibroblast and cardiomyocyte cell populations were generated from E15.5 embryonic hearts according to previously described protocols [38, 39]. For the generation of epicardial cell cultures, embryonic hearts were dissected from E10.5–12.5 embryos and the atria and outflow tract were removed. Ventricular tissue was then carefully dissected into four pieces of comparable size, and each piece was placed onto a coverslip pre-coated with 0.1% gelatin (Sigma, G2500) (1 hour incubation at 37°C) in a 24 well tissue culture plate (Corning). Explants were cultured in 500μl DMEM (Sigma, D5796) supplemented with 15% (v/v) heat inactivated FBS (Gibco, 10500064) and 1% (v/v) Penicillin/Streptomycin (Sigma, P0781). Explants were incubated at 37°C with 5% CO2 and the media was replaced every 3 days until the cultures were required for experiments.
Immunocytochemistry
Epicardial cells cultured for 72 hours were washed with tissue culture grade PBS + MgCl2 and CaCl2 (Sigma, D8662) and then fixed in 4% PFA for 10 minutes on an orbital shaker. The coverslips were then washed and cell monolayers were permeabilised by incubating in PBS + 0.1% (v/v) Triton X-100 for 15 minutes. Cultures were then blocked in either PBS + 10% (v/v) goat serum, or PBS + 1% (w/v) BSA, for at least 1 hour before addition of relevant primary antibody diluted in PBS + 0.1% (v/v) Triton X-100 for 1 hour: rabbit anti-ZO-1 (Invitrogen, 40–2300, 1:100), rabbit anti-Wt1 (1:300, SantaCruz, SC-192), goat anti-fibronectin (Santa Cruz, sc-6952, 1:100) or rabbit anti-NMHC IIB (Biolegend, PRB445P, 1:500). Unbound antibody was removed by washing the cultures with PBS before addition of appropriate biotinylated secondary antibody (Vector) diluted 1:500 in PBS + 0.1% (v/v) Triton X-100 for 1 hour. For visualisation of fibronectin, cultures were incubated with FITC conjugated secondary antibody (Sigma, F9887, 1:160) or developed using the Vectastain Elite ABC Kit (Vector, PK-6100) and visualised using the DAB substrate kit (Vector, SK-4100) as per manufacturer’s instructions. Cultures were washed and then incubated in Cy5 conjugated streptavidin (GE Healthcare, PA45001) diluted 1:500 in PBS + 0.1% (v/v) Triton X-100 for 30 minutes. Coverslips were washed and then treated with 100nM rhodamine-phalloidin (Cytoskeleton, PHDR1) for 30 minutes to allow visualisation of the actin cytoskeleton. Following a final wash, coverslips were mounted onto microscope slides using Vectashield with DAPI mounting media (Vector, H-1200), and stored at 4°C in the dark until imaging within 48 hours. The same protocol was employed for immunofluorescence staining of NMHC IIB in cultured cardiac fibroblasts and myocytes populations on gelatin coated glass coverslips.
TUNEL assay
Paraffin sections were rehydrated and subjected to TUNEL staining (Promega DeadEnd Fluorometric TUNEL System, G3250) as per the manufacturer’s instructions.
Fluorescent microscopy image collection and analysis
Images were collected on an Olympus BX51 upright microscope and captured using a Coolsnap ES camera (Photometrics) through MetaVue Software (Molecular Devices). Specific band pass filter sets for DAPI, FITC, Cy3 and Cy5 were used to prevent bleed through from one channel to the next. Images were then processed and analysed using ImageJ software (Wayne Rasband, NIH, USA). (http://rsb.info.nih.gov/ij). IMARIS (Bitplane) 7.3.4 software was used to analyse the fluorescence intensity in epicardial and myocardial regions for ECM and p65 quantification. For each individual cell in the epicardial and myocardial layers, mean fluorescence intensities (MFI), Alexa488, and Alexa 647 were measured in pixels. Twenty randomly selected areas of the epicardium and myocardium were measured per tissue section. Five sections per embryo, and three embryos of each genotype, were measured. Myocardial measurements were taken from the compact myocardium as defined by morphology rather than trabeculae or endocardium. Two-tailed student t-test was used to test the significance of differences between two sets of data. Further details of data analysis techniques, statistical tests and numbers of samples used are provided in figure legends.
Scratch wound assay
Epicardial cells were cultured from ventricular explants as described above. After 48 hours in culture, explants were removed and the cell monolayer was briefly washed twice with complete media and returned to the incubator for 24 hours. Cell monolayers were scratched using a P10 pipette tip, rinsed twice in complete media, and photographed at 10 minute intervals for 20 hours using an AS MDW live cell imaging system, maintained at 37°C and 5% CO2. Ten cells at the leading edge of the denuded area were tracked per field of view, using MTrackJ in ImageJ. Total tracked cells = 240 control and 270 mutant. Cultures were generated from at least four hearts for each genotype. Data sets for directional persistence and migration speed measurements were subjected to Mann-Whitney U test to assess statistical significance.
Whole mount in situ hybridisation
Embryos were fixed overnight in 4% PFA and in situ hybridisation was performed as previously described [84, 85]. Tbx18 and Shox2 plasmids were obtained from A. Kispert (Institute of Molecular Biology Hannover, Germany). Anti-sense probes were synthesized from linear template DNA using RNA polymerase, digoxygenin nucleotide mix and transcription buffer (Roche). RNA probes were purified by precipitation and added to hybridisation mix at a concentration of 0.5 μg/μl.
Transmission electron microscopy
Samples were prepared for electron microscopy as described previously [86]. Sections (50–70 nm-thick) were generated using a Reichert-Jung Ultracut (Leica Microsystems, UK) and stained using 2% uranyl acetate and 0.3% lead citrate. Sections were examined with an FEI Tecnai 12 Biotwin transmission electron microscope. Images were recorded using a Gatan Orius SC1000 camera (11 Mpixels, 4008 x 2672).
Supporting information
A: Western blot analysis of E11.5 wild type, Myh10∆ heterozygous control and Myh10∆ homozygous mutant heart protein extracts for NMHC IIB expression using a C-terminal antibody. NMHC IIB is abundant in controls but not detected in homozygous mutant samples. B: Table showing expected Mendelian frequencies vs observed frequencies of wild type, Myh10∆ heterozygous and Myh10∆ homozygous mutant progeny from intercrossed Myh10∆ heterozygous animals (Chi squared test, p = 0.0035). C: Table showing expected Mendelian frequencies vs observed frequencies of wild type, EHC/+ or Myh10∆/+ heterozygous and EHC/Myh10∆ compound heterozygous mutant progeny from complementation test of intercrossed Myh10∆/+ and EHC/+ heterozygous animals (Chi squared test, p = 0.0278). The Myh10∆ allele does not rescue the EHC embryonic lethality phenotype, and therefore fails to complement the EHC allele.
(TIFF)
Comparison of ventricular cells in heterozygous control embryos at E12.5 (A) and EHC mutant hearts (B) at E12.5 reveals mutant cells have large nuclei (B, arrows), consistent with prior descriptions of cytokinesis defects in Myh10 knockout embryos. Immunofluorescent staining at E10.5 of heterozygous control (C) and EHC mutant hearts (D). Cell-cell contacts are stained with β-catenin (green) and myofibrils are stained with α-actinin (red), DAPI labels the nuclei (blue). Large nuclei are also present in EHC mutant embryos at E10.5 (D, asterisks). EM imaging shows epicardial cells in control embryos (E) and EHC mutant embryos (F-G). Multiple epicardial cells in EHC mutants display abnormal morphology (F-G, white arrows), and large nuclei are present in cells within the underlying myocardial tissue (F, black arrow). The percentage of epicardial cells with abnormal morphology compared to total epicardial cell number was calculated from images of 5 individual control embryos and 4 individual EHC mutant embryos. Overall there was a statistically significant difference in the percentage of abnormal epicardial cells in control embryos (11% abnormal) as compared to EHC mutants (71% abnormal; Fishers exact text p<0.0001). Alcian blue staining indicates the presence of glycosaminoglycans in endocardial cushion tissue in control littermate (H, J) and EHC mutant heart (I, K). The areas magnified in panels J and K are indicated with boxes on panels H and I. Scale bars: A-D: 25 μm, E-F: 5 μm, G = 17 μm, H-I: 1mm.
(TIF)
PECAM-1 staining at E16.5 reveals prominent vessels on the dorsal surface of the Myh10∆/+ control heart (A). The Myh10∆/∆ (B) and Myh10∆/EHC (C) mutant hearts do not have surface vessels, but rather clusters of PECAM-1 immunoreactive cells (arrows). Section immunohistochemistry for SMαA on D: EHC heterozygote, E: EHC homozygous mutant, or F: EHC/Myh10∆ compound heterozygote at E16.5. Higher magnifications of the boxed areas in sections D-F reveals vessels ringed with SMαA cells in G: EHC heterozygote, but no such structures are present in H: EHC homozygous mutant or I: EHC/Myh10∆ compound heterozygote. Similar results were observed for Myh10∆ homozygous mutant embryos. J: High magnification view of PECAM-1 immunostaining on control heart at E16.5 reveals large coronary vessels. K: EHC mutant heart at E16.5 immunostained for PECAM-1 shows some evidence of a capillary network, along with surface blisters, but no large vessels. Scale bars: A-C = 1mm, D-F and J-K = 0.25mm; G-I = 0.05mm. Abbreviations: PECAM: platelet endothelial cell adhesion molecule-1, SMαA: smooth muscle alpha-actin, ∆: Myh10∆.
(TIF)
A-B: Ventral and dorsal views, respectively, of control littermate showing prominent blood-filled coronary vessels (arrows) at dissection at E18.5. C-D: Ventral and dorsal views, respectively, of Myh10 cardiomyocyte-specific knock out heart showing prominent blood-filled coronary vessels (arrows) at dissection at E18.5. E: PECAM-1 staining reveals prominent vessels (arrows) on the surface of the control heart at E16.5. F: PECAM-1 staining reveals prominent vessels (arrows) on the surface of the Myh10 cardiomyocyte-specific knock out heart at E16.5. Scale bars: 1 mm (A-F). Each pair of images is taken at the same magnification. Genotypes are labeled on the image. Abbreviations: α-MHC: α-myosin heavy chain encoded by Myh6, PECAM: platelet endothelial cell adhesion molecule-1.
(TIF)
Nuclear fast red staining of E14.5 (A-B) and E16.5 (C-D) EHC heterozygote control and EHC homozygous mutant hearts demonstrating the presence of both the mitral and tricuspid atrioventricular valve leaflets (arrows). Scale bars: 1 mm.
(TIF)
Immunohistochemistry on epicardial explant cultures for control (A) and Myh10∆ homozygous mutant (E) to detect fibronectin (arrows). Higher power images show a well-organised fibronectin fibril network in control sample (B, arrows), but not in Myh10∆ homozygous mutant (F). Immunofluorescence staining confirms that control samples have prominent localization of fibronectin (C, green) while staining levels are reduced in mutants (G). Nuclei are visualised with DAPI (blue). The underlying ECM in the explant culture dish was assayed for the presence of fibronectin following removal of explant cells with 20mM ammonium hydroxide, revealing greater prominence of fibronectin in the control sample (D, arrows) as compared to the Myh10∆ homozygous mutant sample (H, arrows). Explants were cultured on gelatin-coated plates. Scale bars: A and E = 100 μm, B-C and F-G = 200 μm.
(TIF)
Phosphohistone H3 immunofluorescence (green) labels proliferating cells (arrows) in control (A) and Myh10∆ homozygous mutant (B) cardiac sagittal sections at E9.5. No statistically significant difference (C) was detected in the percentage of proliferating cells in mutant samples as compared to controls, (two-tailed t-test. p = 0.57). Three sections each from three embryos of each genotype were counted. PHH3 stained cells in cardiac tissue only (identified by morphology) were counted. DAPI positive nuclei (blue) in cardiac tissue only were counted to determine total cell number in each sample. Abbreviations: PHH3: phosphohistone H3, ∆/∆ = Myh10∆homozygous mutant. Scale bars: 50 μm.
(TIF)
Time-lapse microscopy imaging of wound healing in control (+/+) epicardial cell cultures. A ‘denuded’ area was created by scratching D3 epicardial cell culture monolayers with a P10 pipette tip. Point visiting was used to image the same field of view at 10-minute intervals for a period of 20 hours. Ten cells per field of view were tracked over the duration of the experiment using the MTrackJ plugin in ImageJ (as indicated by multicoloured lines). Lines indicate the tracking of cells that were measured for speed and directional persistence.
(AVI)
Lines indicate the tracking of cells that were measured for speed and directional persistence, using the same methodology as reported for S1 Movie.
(AVI)
(DOCX)
Acknowledgments
We thank Ray Boot-Handford for providing the custom-made laminin antibody. We thank Nicoletta Bobola, Kimberly Mace, Berenika Plusa, and Peter Scambler for providing aliquots of various commercially available antibodies. We thank Richa Garva for providing the collagen I staining protocol. We thank Andreas Kispert for providing in situ probe plasmids. The l11Jus27 mouse line was provided by Monica Justice. The Tg(Nes-cre)Wme mice were provided by Michael Briggs and the Tg(Myh6-cre)2182Mds/J were provided by Elizabeth Cartwright. We thank Nicola Smart and Paul Riley for advice on epicardial cell cultures. We thank Patrick Caswell for advice on scratch wound assays and cell tracking. We thank Nancy Papalopulu and Christopher Thompson for microscope access and Steven Marsden for assistance with microscopy. We thank Nathan Hunter for performing immunohistochemistry experiments. We thank Graham Bell and Michael Harris for performing genotyping and histology.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by British Heart Foundation (www.bhf.org.uk) grants PG/06/144/21898 and PG/10/87/28624 to KEH and Wellcome Trust (wellcome.ac.uk/funding/) grant 110126/Z/15/Z to KEK. BK is supported by a British Heart Foundation Personal Chair (CH/13/2/30154). LAR was supported by BBSRC (http://www.bbsrc.ac.uk/) Industrial CASE studentship BB/J012866/1 awarded to KEH. KM and LAS were supported by BBSRC Doctoral Training Account Studentships awarded to the University of Manchester. CC was supported by a Wellcome Trust VIP award awarded as core funding to the University of Manchester Faculty of Life Sciences [grant 092403/Z/10/Z]. The Bioimaging Facility microscopes used in this study were purchased with grants from BBSRC, Wellcome Trust and the University of Manchester Strategic Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brown JR, O'Connor GT. Coronary heart disease and prevention in the United States. The New England journal of medicine. 2010;362(23):2150–3. doi: 10.1056/NEJMp1003880 . [DOI] [PubMed] [Google Scholar]
- 2.Rui L, Yu N, Hong L, Feng H, Chunyong H, Jian M, et al. Extending the time window of mammalian heart regeneration by thymosin beta 4. Journal of cellular and molecular medicine. 2014;18(12):2417–24. doi: 10.1111/jcmm.12421 ; PubMed Central PMCID: PMC4302647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winter EM, Grauss RW, Hogers B, van Tuyn J, van der Geest R, Lie-Venema H, et al. Preservation of left ventricular function and attenuation of remodeling after transplantation of human epicardium-derived cells into the infarcted mouse heart. Circulation. 2007;116(8):917–27. doi: 10.1161/CIRCULATIONAHA.106.668178 . [DOI] [PubMed] [Google Scholar]
- 4.Xiang FL, Liu Y, Lu X, Jones DL, Feng Q. Cardiac-specific overexpression of human stem cell factor promotes epicardial activation and arteriogenesis after myocardial infarction. Circulation Heart failure. 2014;7(5):831–42. doi: 10.1161/CIRCHEARTFAILURE.114.001423 . [DOI] [PubMed] [Google Scholar]
- 5.Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474(7353):640–4. doi: 10.1038/nature10188 ; PubMed Central PMCID: PMC3696525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riley PR, Smart N. Thymosin beta4 induces epicardium-derived neovascularization in the adult heart. Biochemical Society transactions. 2009;37(Pt 6):1218–20. doi: 10.1042/BST0371218 . [DOI] [PubMed] [Google Scholar]
- 7.Bock-Marquette I, Shrivastava S, Pipes GC, Thatcher JE, Blystone A, Shelton JM, et al. Thymosin beta4 mediated PKC activation is essential to initiate the embryonic coronary developmental program and epicardial progenitor cell activation in adult mice in vivo. Journal of molecular and cellular cardiology. 2009;46(5):728–38. doi: 10.1016/j.yjmcc.2009.01.017 ; PubMed Central PMCID: PMC2768533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. The Anatomical record. 1981;201(1):157–68. doi: 10.1002/ar.1092010117 . [DOI] [PubMed] [Google Scholar]
- 9.Masters M, Riley PR. The epicardium signals the way towards heart regeneration. Stem Cell Res. 2014;13(3 Pt B):683–92. doi: 10.1016/j.scr.2014.04.007 ; PubMed Central PMCID: PMCPMC4241487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy-Lydon T, Rosenthal N. Cardiac regeneration: epicardial mediated repair. Proc Biol Sci. 2015;282(1821):20152147 doi: 10.1098/rspb.2015.2147 ; PubMed Central PMCID: PMCPMC4707759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miquerol L, Kelly RG. Organogenesis of the vertebrate heart. Wiley Interdiscip Rev Dev Biol. 2013;2(1):17–29. doi: 10.1002/wdev.68 . [DOI] [PubMed] [Google Scholar]
- 12.Gittenberger-de Groot AC, Winter EM, Bartelings MM, Goumans MJ, DeRuiter MC, Poelmann RE. The arterial and cardiac epicardium in development, disease and repair. Differentiation. 2012;84(1):41–53. doi: 10.1016/j.diff.2012.05.002 . [DOI] [PubMed] [Google Scholar]
- 13.von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circulation research. 2012;110(12):1628–45. doi: 10.1161/CIRCRESAHA.111.259960 ; PubMed Central PMCID: PMC3427736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakajima Y, Imanaka-Yoshida K. New insights into the developmental mechanisms of coronary vessels and epicardium. Int Rev Cell Mol Biol. 2013;303:263–317. doi: 10.1016/B978-0-12-407697-6.00007-6 . [DOI] [PubMed] [Google Scholar]
- 15.Tian X, Pu WT, Zhou B. Cellular origin and developmental program of coronary angiogenesis. Circulation research. 2015;116(3):515–30. doi: 10.1161/CIRCRESAHA.116.305097 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, et al. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Developmental cell. 2012;22(3):639–50. doi: 10.1016/j.devcel.2012.01.012 ; PubMed Central PMCID: PMC3306604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. The Journal of clinical investigation. 2011;121(5):1894–904. doi: 10.1172/JCI45529 ; PubMed Central PMCID: PMC3083761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell JL, Goetsch SC, Gaiano NR, Hill JA, Olson EN, Schneider JW. A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circulation research. 2011;108(1):51–9. doi: 10.1161/CIRCRESAHA.110.233262 ; PubMed Central PMCID: PMC3042596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Liu B, et al. Functional genetic analysis of mouse chromosome 11. Nature. 2003;425(6953):81–6. doi: 10.1038/nature01865 . [DOI] [PubMed] [Google Scholar]
- 20.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nature reviews Molecular cell biology. 2009;10(11):778–90. doi: 10.1038/nrm2786 ; PubMed Central PMCID: PMC2834236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conti MA, Adelstein RS. Nonmuscle myosin II moves in new directions. Journal of cell science. 2008;121(Pt 1):11–8. doi: 10.1242/jcs.007112 . [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Wei Q, Adelstein RS, Wang PJ. Non-muscle myosin IIB is essential for cytokinesis during male meiotic cell divisions. Developmental biology. 2012;369(2):356–61. doi: 10.1016/j.ydbio.2012.07.011 ; PubMed Central PMCID: PMC3584343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeda K, Kishi H, Ma X, Yu ZX, Adelstein RS. Ablation and mutation of nonmuscle myosin heavy chain II-B results in a defect in cardiac myocyte cytokinesis. Circulation research. 2003;93(4):330–7. doi: 10.1161/01.RES.0000089256.00309.CB . [DOI] [PubMed] [Google Scholar]
- 24.Gutzman JH, Sahu SU, Kwas C. Non-muscle myosin IIA and IIB differentially regulate cell shape changes during zebrafish brain morphogenesis. Developmental biology. 2015;397(1):103–15. doi: 10.1016/j.ydbio.2014.10.017 . [DOI] [PubMed] [Google Scholar]
- 25.Huang IH, Hsiao CT, Wu JC, Shen RF, Liu CY, Wang YK, et al. GEF-H1 controls focal adhesion signaling that regulates mesenchymal stem cell lineage commitment. Journal of cell science. 2014;127(Pt 19):4186–200. doi: 10.1242/jcs.150227 ; PubMed Central PMCID: PMC4179489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X, Bao J, Adelstein RS. Loss of cell adhesion causes hydrocephalus in nonmuscle myosin II-B-ablated and mutated mice. Molecular biology of the cell. 2007;18(6):2305–12. doi: 10.1091/mbc.E07-01-0073 ; PubMed Central PMCID: PMC1877086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, Adelstein RS. A point mutation in Myh10 causes major defects in heart development and body wall closure. Circulation Cardiovascular genetics. 2014;7(3):257–65. doi: 10.1161/CIRCGENETICS.113.000455 ; PubMed Central PMCID: PMC4106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo CM, Buxton DB, Chua GC, Dembo M, Adelstein RS, Wang YL. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Molecular biology of the cell. 2004;15(3):982–9. doi: 10.1091/mbc.E03-06-0359 ; PubMed Central PMCID: PMC363055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(46):18109–14. doi: 10.1073/pnas.0702415104 ; PubMed Central PMCID: PMC2084304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trembley MA, Velasquez LS, de Mesy Bentley KL, Small EM. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development. 2015;142(1):21–30. doi: 10.1242/dev.116418 ; PubMed Central PMCID: PMC4299137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu SP, Dong XR, Regan JN, Su C, Majesky MW. Tbx18 regulates development of the epicardium and coronary vessels. Developmental biology. 2013;383(2):307–20. doi: 10.1016/j.ydbio.2013.08.019 ; PubMed Central PMCID: PMC4172450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips HM, Hildreth V, Peat JD, Murdoch JN, Kobayashi K, Chaudhry B, et al. Non-cell-autonomous roles for the planar cell polarity gene Vangl2 in development of the coronary circulation. Circulation research. 2008;102(5):615–23. doi: 10.1161/CIRCRESAHA.107.160861 . [DOI] [PubMed] [Google Scholar]
- 33.Tullio AN, Accili D, Ferrans VJ, Yu ZX, Takeda K, Grinberg A, et al. Nonmuscle myosin II-B is required for normal development of the mouse heart. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(23):12407–12. ; PubMed Central PMCID: PMC24969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuzovic L, Yu L, Zeng W, Li X, Lu H, Lu HM, et al. A human de novo mutation in MYH10 phenocopies the loss of function mutation in mice. Rare diseases. 2013;1:e26144 doi: 10.4161/rdis.26144 ; PubMed Central PMCID: PMC3927488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma X, Takeda K, Singh A, Yu ZX, Zerfas P, Blount A, et al. Conditional ablation of nonmuscle myosin II-B delineates heart defects in adult mice. Circulation research. 2009;105(11):1102–9. doi: 10.1161/CIRCRESAHA.109.200303 ; PubMed Central PMCID: PMC2792753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao J, Ma X, Liu C, Adelstein RS. Replacement of nonmuscle myosin II-B with II-A rescues brain but not cardiac defects in mice. The Journal of biological chemistry. 2007;282(30):22102–11. doi: 10.1074/jbc.M702731200 . [DOI] [PubMed] [Google Scholar]
- 37.Ma X, Jana SS, Conti MA, Kawamoto S, Claycomb WC, Adelstein RS. Ablation of nonmuscle myosin II-B and II-C reveals a role for nonmuscle myosin II in cardiac myocyte karyokinesis. Molecular biology of the cell. 2010;21(22):3952–62. doi: 10.1091/mbc.E10-04-0293 ; PubMed Central PMCID: PMC2982113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louch WE, Sheehan KA, Wolska BM. Methods in cardiomyocyte isolation, culture, and gene transfer. Journal of molecular and cellular cardiology. 2011;51(3):288–98. doi: 10.1016/j.yjmcc.2011.06.012 ; PubMed Central PMCID: PMCPMC3164875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sreejit P, Kumar S, Verma RS. An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell Dev Biol Anim. 2008;44(3–4):45–50. doi: 10.1007/s11626-007-9079-4 . [DOI] [PubMed] [Google Scholar]
- 40.Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Developmental biology. 2001;234(1):204–15. doi: 10.1006/dbio.2001.0254 . [DOI] [PubMed] [Google Scholar]
- 41.Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. The International journal of developmental biology. 2002;46(8):1005–13. . [PubMed] [Google Scholar]
- 42.Dettman RW, Denetclaw W Jr., Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Developmental biology. 1998;193(2):169–81. doi: 10.1006/dbio.1997.8801 . [DOI] [PubMed] [Google Scholar]
- 43.Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(20):9504–8. ; PubMed Central PMCID: PMC50160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circulation research. 1998;82(10):1043–52. . [DOI] [PubMed] [Google Scholar]
- 45.Zhou B, Pu WT. Genetic Cre-loxP assessment of epicardial cell fate using Wt1-driven Cre alleles. Circulation research. 2012;111(11):e276–80. doi: 10.1161/CIRCRESAHA.112.275784 ; PubMed Central PMCID: PMC3532881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100(1):83–6. . [DOI] [PubMed] [Google Scholar]
- 47.Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115(14):1830–8. doi: 10.1161/CIRCULATIONAHA.106.637819 . [DOI] [PubMed] [Google Scholar]
- 48.Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Developmental biology. 2009;327(2):376–85. doi: 10.1016/j.ydbio.2008.12.028 ; PubMed Central PMCID: PMCPMC2694185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li WE, Waldo K, Linask KL, Chen T, Wessels A, Parmacek MS, et al. An essential role for connexin43 gap junctions in mouse coronary artery development. Development. 2002;129(8):2031–42. . [DOI] [PubMed] [Google Scholar]
- 50.Perez-Pomares JM, de la Pompa JL. Signaling during epicardium and coronary vessel development. Circulation research. 2011;109(12):1429–42. doi: 10.1161/CIRCRESAHA.111.245589 . [DOI] [PubMed] [Google Scholar]
- 51.Balmer GM, Bollini S, Dube KN, Martinez-Barbera JP, Williams O, Riley PR. Dynamic haematopoietic cell contribution to the developing and adult epicardium. Nature communications. 2014;5:4054 doi: 10.1038/ncomms5054 ; PubMed Central PMCID: PMC4059938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circulation research. 2004;95(5):459–70. doi: 10.1161/01.RES.0000141146.95728.da ; PubMed Central PMCID: PMCPMC2810618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma X, Sung DC, Yang Y, Wakabayashi Y, Adelstein RS. Nonmuscle myosin IIB regulates epicardial integrity and epicardium-derived mesenchymal cell maturation. Journal of cell science. 2017;130(16):2696–706. doi: 10.1242/jcs.202564 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma X, Adelstein RS. In vivo studies on nonmuscle myosin II expression and function in heart development. Frontiers in bioscience. 2012;17:545–55. ; PubMed Central PMCID: PMC3476727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen T, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, et al. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Developmental biology. 2002;250(1):198–207. . [DOI] [PubMed] [Google Scholar]
- 56.Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Developmental biology. 2003;255(2):334–49. . [DOI] [PubMed] [Google Scholar]
- 57.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, et al. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes & development. 2006;20(12):1651–66. doi: 10.1101/gad.1411406 ; PubMed Central PMCID: PMC1482484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, et al. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs). Developmental biology. 2002;247(2):307–26. . [DOI] [PubMed] [Google Scholar]
- 59.Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dolle P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128(7):1019–31. . [DOI] [PubMed] [Google Scholar]
- 60.Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circulation research. 2010;106(5):818–32. doi: 10.1161/CIRCRESAHA.109.209197 ; PubMed Central PMCID: PMC2843003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, et al. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101(7):729–39. . [DOI] [PubMed] [Google Scholar]
- 62.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126(9):1845–57. . [DOI] [PubMed] [Google Scholar]
- 63.Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, et al. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circulation research. 2008;103(12):1393–401. doi: 10.1161/CIRCRESAHA.108.176768 ; PubMed Central PMCID: PMC2757035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim H, Yoon CS, Kim H, Rah B. Expression of extracellular matrix components fibronectin and laminin in the human fetal heart. Cell Struct Funct. 1999;24(1):19–26. . [DOI] [PubMed] [Google Scholar]
- 65.Benesh EC, Miller PM, Pfaltzgraff ER, Grega-Larson NE, Hager HA, Sung BH, et al. Bves and NDRG4 regulate directional epicardial cell migration through autocrine extracellular matrix deposition. Molecular biology of the cell. 2013;24(22):3496–510. doi: 10.1091/mbc.E12-07-0539 ; PubMed Central PMCID: PMCPMC3826988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanson KP, Jung JP, Tran QA, Hsu SP, Iida R, Ajeti V, et al. Spatial and temporal analysis of extracellular matrix proteins in the developing murine heart: a blueprint for regeneration. Tissue Eng Part A. 2013;19(9–10):1132–43. doi: 10.1089/ten.TEA.2012.0316 ; PubMed Central PMCID: PMCPMC3609645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bowers SL, Baudino TA. Laying the groundwork for growth: Cell-cell and cell-ECM interactions in cardiovascular development. Birth Defects Res C Embryo Today. 2010;90(1):1–7. doi: 10.1002/bdrc.20168 . [DOI] [PubMed] [Google Scholar]
- 68.Richter M, Kostin S. The failing human heart is characterized by decreased numbers of telocytes as result of apoptosis and altered extracellular matrix composition. Journal of cellular and molecular medicine. 2015;19(11):2597–606. doi: 10.1111/jcmm.12664 ; PubMed Central PMCID: PMCPMC4627565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes & development. 2004;18(10):1131–43. doi: 10.1101/gad.294104 ; PubMed Central PMCID: PMC415638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, Urban A, Midura D, Simon HG, Wang QT. Proteomic characterization of epicardial-myocardial signaling reveals novel regulatory networks including a role for NF-kappaB in epicardial EMT. PloS one. 2017;12(3):e0174563 doi: 10.1371/journal.pone.0174563 ; PubMed Central PMCID: PMCPMC5373538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schiffhauer ES, Luo T, Mohan K, Srivastava V, Qian X, Griffis ER, et al. Mechanoaccumulative Elements of the Mammalian Actin Cytoskeleton. Curr Biol. 2016;26(11):1473–9. doi: 10.1016/j.cub.2016.04.007 ; PubMed Central PMCID: PMCPMC4899209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeichi M, Nimura K, Mori M, Nakagami H, Kaneda Y. The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PloS one. 2013;8(2):e57829 doi: 10.1371/journal.pone.0057829 ; PubMed Central PMCID: PMCPMC3585213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sung BH, Zhu X, Kaverina I, Weaver AM. Cortactin controls cell motility and lamellipodial dynamics by regulating ECM secretion. Curr Biol. 2011;21(17):1460–9. doi: 10.1016/j.cub.2011.06.065 ; PubMed Central PMCID: PMCPMC3175319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riley PR, Smart N. Vascularizing the heart. Cardiovascular research. 2011;91(2):260–8. doi: 10.1093/cvr/cvr035 . [DOI] [PubMed] [Google Scholar]
- 75.O'Connor JW, Riley PN, Nalluri SM, Ashar PK, Gomez EW. Matrix Rigidity Mediates TGFbeta1-Induced Epithelial-Myofibroblast Transition by Controlling Cytoskeletal Organization and MRTF-A Localization. J Cell Physiol. 2015;230(8):1829–39. doi: 10.1002/jcp.24895 . [DOI] [PubMed] [Google Scholar]
- 76.Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, et al. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater. 2016;15(12):1287–96. doi: 10.1038/nmat4729 ; PubMed Central PMCID: PMCPMC5121013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu J, Clark RA. Extracellular matrix alters PDGF regulation of fibroblast integrins. J Cell Biol. 1996;132(1–2):239–49. ; PubMed Central PMCID: PMCPMC2120701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Norstrom MF, Smithback PA, Rock RS. Unconventional processive mechanics of non-muscle myosin IIB. The Journal of biological chemistry. 2010;285(34):26326–34. doi: 10.1074/jbc.M110.123851 ; PubMed Central PMCID: PMCPMC2924056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim JH, Wang A, Conti MA, Adelstein RS. Nonmuscle myosin II is required for internalization of the epidermal growth factor receptor and modulation of downstream signaling. The Journal of biological chemistry. 2012;287(33):27345–58. doi: 10.1074/jbc.M111.304824 ; PubMed Central PMCID: PMCPMC3431632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalson NS, Starborg T, Lu Y, Mironov A, Humphries SM, Holmes DF, et al. Nonmuscle myosin II powered transport of newly formed collagen fibrils at the plasma membrane. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(49):E4743–52. doi: 10.1073/pnas.1314348110 ; PubMed Central PMCID: PMCPMC3856828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2:Unit 2 3. doi: 10.1002/0471250953.bi0203s00 . [DOI] [PubMed] [Google Scholar]
- 82.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626 . [DOI] [PubMed] [Google Scholar]
- 83.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94(5):559–71. . [DOI] [PubMed] [Google Scholar]
- 84.Risley MD, Clowes C, Yu M, Mitchell K, Hentges KE. The Mediator complex protein Med31 is required for embryonic growth and cell proliferation during mammalian development. Developmental biology. 2010;342(2):146–56. doi: 10.1016/j.ydbio.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 85.Tenin G, Clowes C, Wolton K, Krejci E, Wright JA, Lovell SC, et al. Erbb2 Is Required for Cardiac Atrial Electrical Activity during Development. PloS one. 2014;9(9). Artn E107041 doi: 10.1371/Journal.Pone.0107041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Canty EG, Lu Y, Meadows RS, Shaw MK, Holmes DF, Kadler KE. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J Cell Biol. 2004;165(4):553–63. doi: 10.1083/jcb.200312071 ; PubMed Central PMCID: PMCPMC2172348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Western blot analysis of E11.5 wild type, Myh10∆ heterozygous control and Myh10∆ homozygous mutant heart protein extracts for NMHC IIB expression using a C-terminal antibody. NMHC IIB is abundant in controls but not detected in homozygous mutant samples. B: Table showing expected Mendelian frequencies vs observed frequencies of wild type, Myh10∆ heterozygous and Myh10∆ homozygous mutant progeny from intercrossed Myh10∆ heterozygous animals (Chi squared test, p = 0.0035). C: Table showing expected Mendelian frequencies vs observed frequencies of wild type, EHC/+ or Myh10∆/+ heterozygous and EHC/Myh10∆ compound heterozygous mutant progeny from complementation test of intercrossed Myh10∆/+ and EHC/+ heterozygous animals (Chi squared test, p = 0.0278). The Myh10∆ allele does not rescue the EHC embryonic lethality phenotype, and therefore fails to complement the EHC allele.
(TIFF)
Comparison of ventricular cells in heterozygous control embryos at E12.5 (A) and EHC mutant hearts (B) at E12.5 reveals mutant cells have large nuclei (B, arrows), consistent with prior descriptions of cytokinesis defects in Myh10 knockout embryos. Immunofluorescent staining at E10.5 of heterozygous control (C) and EHC mutant hearts (D). Cell-cell contacts are stained with β-catenin (green) and myofibrils are stained with α-actinin (red), DAPI labels the nuclei (blue). Large nuclei are also present in EHC mutant embryos at E10.5 (D, asterisks). EM imaging shows epicardial cells in control embryos (E) and EHC mutant embryos (F-G). Multiple epicardial cells in EHC mutants display abnormal morphology (F-G, white arrows), and large nuclei are present in cells within the underlying myocardial tissue (F, black arrow). The percentage of epicardial cells with abnormal morphology compared to total epicardial cell number was calculated from images of 5 individual control embryos and 4 individual EHC mutant embryos. Overall there was a statistically significant difference in the percentage of abnormal epicardial cells in control embryos (11% abnormal) as compared to EHC mutants (71% abnormal; Fishers exact text p<0.0001). Alcian blue staining indicates the presence of glycosaminoglycans in endocardial cushion tissue in control littermate (H, J) and EHC mutant heart (I, K). The areas magnified in panels J and K are indicated with boxes on panels H and I. Scale bars: A-D: 25 μm, E-F: 5 μm, G = 17 μm, H-I: 1mm.
(TIF)
PECAM-1 staining at E16.5 reveals prominent vessels on the dorsal surface of the Myh10∆/+ control heart (A). The Myh10∆/∆ (B) and Myh10∆/EHC (C) mutant hearts do not have surface vessels, but rather clusters of PECAM-1 immunoreactive cells (arrows). Section immunohistochemistry for SMαA on D: EHC heterozygote, E: EHC homozygous mutant, or F: EHC/Myh10∆ compound heterozygote at E16.5. Higher magnifications of the boxed areas in sections D-F reveals vessels ringed with SMαA cells in G: EHC heterozygote, but no such structures are present in H: EHC homozygous mutant or I: EHC/Myh10∆ compound heterozygote. Similar results were observed for Myh10∆ homozygous mutant embryos. J: High magnification view of PECAM-1 immunostaining on control heart at E16.5 reveals large coronary vessels. K: EHC mutant heart at E16.5 immunostained for PECAM-1 shows some evidence of a capillary network, along with surface blisters, but no large vessels. Scale bars: A-C = 1mm, D-F and J-K = 0.25mm; G-I = 0.05mm. Abbreviations: PECAM: platelet endothelial cell adhesion molecule-1, SMαA: smooth muscle alpha-actin, ∆: Myh10∆.
(TIF)
A-B: Ventral and dorsal views, respectively, of control littermate showing prominent blood-filled coronary vessels (arrows) at dissection at E18.5. C-D: Ventral and dorsal views, respectively, of Myh10 cardiomyocyte-specific knock out heart showing prominent blood-filled coronary vessels (arrows) at dissection at E18.5. E: PECAM-1 staining reveals prominent vessels (arrows) on the surface of the control heart at E16.5. F: PECAM-1 staining reveals prominent vessels (arrows) on the surface of the Myh10 cardiomyocyte-specific knock out heart at E16.5. Scale bars: 1 mm (A-F). Each pair of images is taken at the same magnification. Genotypes are labeled on the image. Abbreviations: α-MHC: α-myosin heavy chain encoded by Myh6, PECAM: platelet endothelial cell adhesion molecule-1.
(TIF)
Nuclear fast red staining of E14.5 (A-B) and E16.5 (C-D) EHC heterozygote control and EHC homozygous mutant hearts demonstrating the presence of both the mitral and tricuspid atrioventricular valve leaflets (arrows). Scale bars: 1 mm.
(TIF)
Immunohistochemistry on epicardial explant cultures for control (A) and Myh10∆ homozygous mutant (E) to detect fibronectin (arrows). Higher power images show a well-organised fibronectin fibril network in control sample (B, arrows), but not in Myh10∆ homozygous mutant (F). Immunofluorescence staining confirms that control samples have prominent localization of fibronectin (C, green) while staining levels are reduced in mutants (G). Nuclei are visualised with DAPI (blue). The underlying ECM in the explant culture dish was assayed for the presence of fibronectin following removal of explant cells with 20mM ammonium hydroxide, revealing greater prominence of fibronectin in the control sample (D, arrows) as compared to the Myh10∆ homozygous mutant sample (H, arrows). Explants were cultured on gelatin-coated plates. Scale bars: A and E = 100 μm, B-C and F-G = 200 μm.
(TIF)
Phosphohistone H3 immunofluorescence (green) labels proliferating cells (arrows) in control (A) and Myh10∆ homozygous mutant (B) cardiac sagittal sections at E9.5. No statistically significant difference (C) was detected in the percentage of proliferating cells in mutant samples as compared to controls, (two-tailed t-test. p = 0.57). Three sections each from three embryos of each genotype were counted. PHH3 stained cells in cardiac tissue only (identified by morphology) were counted. DAPI positive nuclei (blue) in cardiac tissue only were counted to determine total cell number in each sample. Abbreviations: PHH3: phosphohistone H3, ∆/∆ = Myh10∆homozygous mutant. Scale bars: 50 μm.
(TIF)
Time-lapse microscopy imaging of wound healing in control (+/+) epicardial cell cultures. A ‘denuded’ area was created by scratching D3 epicardial cell culture monolayers with a P10 pipette tip. Point visiting was used to image the same field of view at 10-minute intervals for a period of 20 hours. Ten cells per field of view were tracked over the duration of the experiment using the MTrackJ plugin in ImageJ (as indicated by multicoloured lines). Lines indicate the tracking of cells that were measured for speed and directional persistence.
(AVI)
Lines indicate the tracking of cells that were measured for speed and directional persistence, using the same methodology as reported for S1 Movie.
(AVI)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.