(
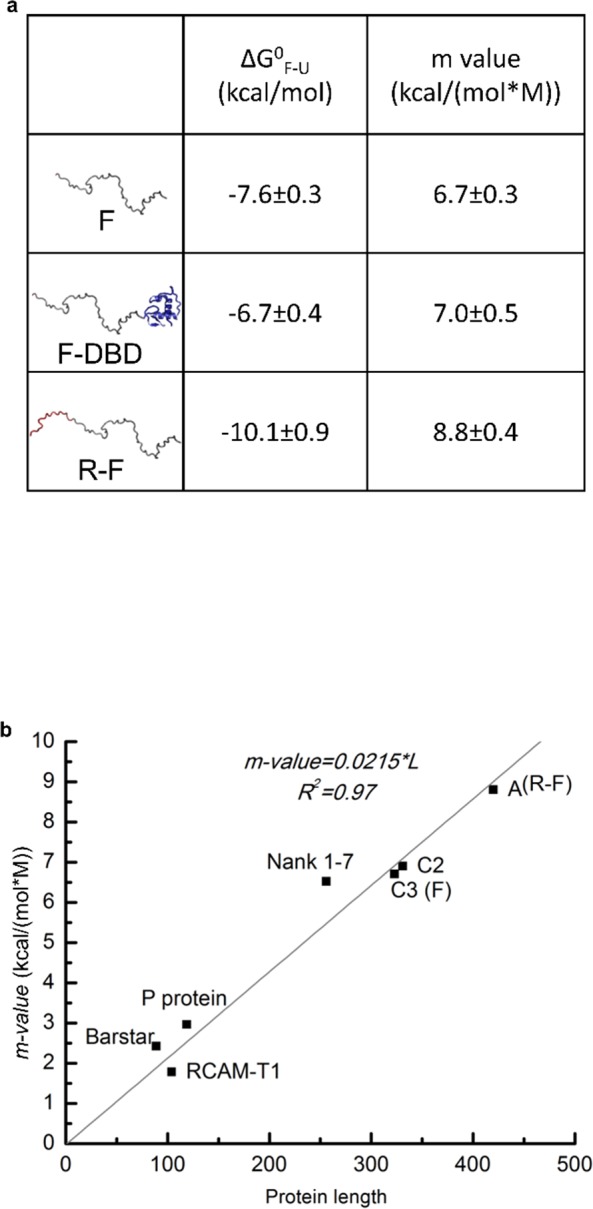
a) Thermodynamic parameters obtained from TMAO-induced folding experiments demonstrate that DBD stabilizes a folded conformation of the F-domain while the R-domain destabilizes that conformation. Parameter values obtained from the fits of the TMAO-induced folding data for the constructs shown in
Figure 2a. See previous publication (
Li et al., 2012) for details. Because the thermodynamic analyses report on the free energy differences and not the mechanistic bases of the energy differences, the reported free energies of folding (
) may not necessarily reflect the stability of unique conformations. Instead, they may be reporting on the stability of a conformationally heterogeneous ensemble. Toward this end, it has been reported that ID proteins may adopt poly-beta sheet formations (
Han et al., 2012) as part of their functional states. As the effect of TMAO on structure is almost entirely defined by the influence on backbone atoms (i.e. excluding backbone hydrogen bond donors and acceptors from solvent) (
Auton and Bolen, 2005), any state or ensemble of states that facilitates removal of backbone atoms from solvent will be stabilized by TMAO. As such, the current analysis provides evidence that a functional state exists whose probability is proportional to transcriptional activation. However, the structural properties of that state are not known. (
b) Comparison of the
m-values fitted from TMAO induced folding of F-domain and R-F with the published
m-values for known representative globular proteins (see previous publication (
Li et al., 2012) for details). The
m-value comparison suggests that the F-domain and R-domain can adopt folded conformations similar to globular proteins in terms of surface area buried upon folding. We note that the data in (
b) are the same as presented in
Li et al., 2012 (
Figure 4a), according to the current nomenclature, wherein the R-F construct is equivalent to the A isoform of NTD and the F construct is equivalent to the C3 isoform of NTD.