Abstract
Type 1 diabetes (T1D) results from T cell–mediated destruction of insulin-producing β-cells. Insulin represents a key self-antigen in disease pathogenesis, as recent studies identified proinsulin-responding T cells from inflamed pancreatic islets of organ donors with recent-onset T1D. These cells respond to an insulin B-chain (InsB) epitope presented by the HLA-DQ8 molecule associated with high T1D risk. Understanding insulin-specific T-cell frequency and phenotype in peripheral blood is now critical. We constructed fluorescent InsB10–23:DQ8 tetramers, stained peripheral blood lymphocytes directly ex vivo, and show DQ8+ patients with T1D have increased tetramer+ CD4+ T cells compared with HLA-matched control subjects without diabetes. Patients with a shorter disease duration had higher frequencies of insulin-reactive CD4+ T cells, with most of these cells being antigen experienced. We also demonstrate that the number of insulin tetramer+ effector memory cells is directly correlated with insulin antibody titers, suggesting insulin-specific T- and B-cell interactions. Notably, one of four control subjects with tetramer+ cells was a first-degree relative who had insulin-specific cells with an effector memory phenotype, potentially representing an early marker of T-cell autoimmunity. Our results suggest that studying InsB10–23:DQ8 reactive T-cell frequency and phenotype may provide a biomarker of disease activity in patients with T1D and those at risk.
Introduction
The strongest genetic risk factor associated with autoimmune type 1 diabetes (T1D) is genes within the HLA complex. The HLA-DR4-DQ8 haplotype in humans and MHC class II (MHCII) IAg7 in NOD mice, a spontaneous murine model of autoimmune diabetes, provide the strongest genetic risk for T1D, supporting a critical role for CD4+ T cells in disease development (1). CD4+ and CD8+ T cells, as well as B cells and dendritic cells, are important for the progression of T1D in mice and humans (2). CD8+ T cells mediate direct islet killing, whereas CD4+ T cells may play a critical role to initiate disease by providing help for CD8+ T cells and B cells (3). Interestingly, HLA-DQ8 and mouse IAg7 molecules share structural similarity and have comparable peptide binding preferences (4).
Historically, the strongest biological indicator of future T1D onset is the presence of insulin autoantibodies (IAAs), because they can appear years before the clinical onset of T1D and almost all patients diagnosed with T1D aged younger than 6 years with the DR4-DQ8 haplotype are IAA positive (5). In addition, there is considerable evidence in mouse models that insulin is a major target during the development of diabetes (6–9). Using a transgenic NOD mouse model, Nakayama et al. (6) determined that a single amino acid substitution in a T-cell receptor contact site within the insulin B-chain (InsB) conferred complete T1D protection by masking the dominant immune peptide target. In separate studies, we and others determined that T cells specific for InsB amino acids 9–23 (InsB9–23) are critical for disease development in the spontaneous diabetes NOD mouse model (6, 10). Notably, the amino acid sequence of InsB9–23 is identical in mice and humans, which has led others to investigate T-cell reactivity to this epitope in humans. In a very recent report, InsB9–23–reactive CD4+ T cells were identified from the inflamed pancreatic islets of two organ donors with recent-onset T1D, indicating that these cells are relevant to human disease (11). In separate studies, InsB-specific T cells could be detected in the peripheral blood of patients with new-onset T1D with the use of indirect cytokine ELISAs (12) and expanded from the peripheral blood of patients with T1D with established disease (13). With these discoveries, it is now critical to understand the phenotype of these cells in the peripheral circulation, how the insulin-specific T-cell response relates to disease duration, and whether monitoring insulin-specific CD4+ T-cell responses could be a useful biomarker of disease activity.
In the current study, we used peptide:HLAII tetramer staining to compare the frequency and phenotype of InsB-specific CD4+ T cells directly ex vivo within peripheral blood from HLA-DQ8+ patients with T1D and HLA-matched control subjects without diabetes. We found that 54% (20 of 37) of patients with T1D had detectable insulin tetramer+ cells compared with only 15% (4 of 26) of control subjects without diabetes. Within the patients with T1D, 64% of insulin tetramer+ cells were antigen experienced (CD45RO+). In fact, patients with the most tetramer+ effector memory cells (CD45RO+ CCR7−) had significantly higher insulin antibody titers and the shortest T1D duration. Importantly, tetramer+ cells were enumerated from several patients with new-onset T1D where insulin administration was shorter than 15 days, providing evidence that these cells are self-reactive. In one subject without diabetes, a genetically at-risk first-degree relative of a patient with T1D, we found effector memory tetramer+ cells in the absence of IAAs. Taken together, these data suggest that InsB-specific CD4+ T cells become activated in response to endogenous antigen and may be contributing to antibody production. Determining their frequency and phenotype may be useful for assessing disease activity after diagnosis and potentially during the preclinical period of T1D development.
Research Design and Methods
Mice
NOD mice were purchased from Taconic. C57Bl/6.H2g7 (B6.g7) mice were generated as described (14). NOD transgenic HLA-DQ8, H2-Ab1–deficient mice (NOD.DQ8.Ab0) (15) were purchased from The Jackson Laboratory. Mice were determined diabetic by blood glucose readings >250 mg/dL for 2 consecutive days. Animals were housed in a specific pathogen-free barrier facility. All animal procedures were approved by the University of Minnesota Institutional Animal Care and Use Committee.
Human Subjects
The research volunteers included in this specific study from the University of Minnesota Medical Center and Fairview hospital provided written informed consent for peripheral blood collection. The study was approved by the University of Minnesota Institutional Review Board. Patients with T1D, control subjects without diabetes, and first-degree relatives without diabetes were screened for the presence of the DQ8 haplotype (Table 1). Of the 26 control subjects without diabetes, we considered 2 at risk for prediabetes based on islet autoantibody positivity (16); however, DQ8 presence has potential elevated T1D genetic risk. Subjects with recent-onset T1D (<15 days) were recruited from the Barbara Davis Center for Diabetes Clinics, after approval from the University of Colorado Institutional Review Board, and provided written informed consent.
Table 1.
Patient demographics
| T1D | No T1D | |
|---|---|---|
| n = 37 | n = 26 | |
| Age (years) | 28 ± 20 | 33 ± 18 |
| Female sex (%) | 51 | 77 |
| T1D duration (years) | 16 ± 17 | — |
| Blood volume (mL) | 39 ± 14 | 45 ± 10 |
| Number of PBMCs stained (×106) | 78 ± 29 | 101 ± 29 |
Data are shown as mean ± SD or as indicated.
HLA Haplotyping
Direct ex vivo tetramer analysis required cells to be stained fresh, so we used a rapid real-time PCR probe screening approach for the presence of DQ8 as previously described (17,18). Positive and negative DQ8 control samples were provided by Fred Hutchinson Cancer Research Center International Histocompatibility Working Group Cell and Gene Bank. Briefly, genomic DNA was isolated from 1.5 × 106 peripheral blood mononuclear cells (PBMCs) using the DNeasy Blood and Tissue Kit (Qiagen). DNA (30 ng) was mixed with primer and probes as indicated in Supplementary Table 3 in 20 μL containing 2× FastStart Universal Probe Master Mix (Roche). Conditions were 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min with 40 repeats on a 7900HT Real-Time PCR System using SDS 2.4 software (Applied Biosystems Inc.). HLA haplotyping of new-onset patients from the Barbara Davis Center was performed using linear arrays of immobilized sequence-specific oligonucleotides as previously described (19).
Peptide Sequences
Modified InsB10–23 (HLVERLYLVAGEEG) and HA306–318 (PKYVKQNTLKLAT) (Genemed Synthesis) were used in in vitro T-cell expansion and immunizations with complete Freund’s adjuvant.
Tetramers
InsB10–23:IAg7 (HLVERLYLVCGEEG), InsB10–23:DQ8 (HLVERLYLVCGEEG), and HA306–318:DR4 (PKYVKQNTLKLAT) were produced as previously described (20–22). Peptide:HLAII protein was purified by immobilized metal ion affinity chromatography using the His-Bind purification kit (Novagen-EMD Biosciences). The biotinylated peptide:HLAII protein was affinity purified using Monomeric Avidin UltraLink (Pierce). Bound peptide:HLAII molecules were eluted with 2 mmol/L biotin in PBS, and excess free biotin was removed by centrifugation and four washes with 12 mL PBS using a 30 kDa cutoff Amicon Ultra-15 filter (Millipore). Tetramers were produced by incubating peptide:HLAII monomers with streptavidin-allophycocyanin (APC; Prozyme #PJ27S) or streptavidin-phycoerythrin (PE; Prozyme #PJRS27) at a 4.5:1 molar ratio, and PBS containing 0.05% sodium azide was added to 1 µmol/L final concentration. The limit of detection for tetramer staining and enrichment was determined by a limiting dilution assay as previously described (23,24). In vitro expanded HA306–318 tetramer+ cells were diluted by twofold serial dilution and added to 30 × 106 HLA disparate cells. Tetramer+ cells were enumerated after magnetic enrichment and plotted against the theoretical frequency, yielding a limit of detection of at least 0.2 tetramer+ cells/1 million total cells.
Human PBMC Isolation and Staining
Blood was collected in BD Vacutainer tubes containing heparin, diluted 1:1 in RPMI medium, and rested overnight at room temperature. PBMCs were isolated using SepMate isolation (StemCell Technologies), and plasma was collected and frozen. We isolated an average of 87.7 × 106 PBMCs from mean blood volume of 41.9 ± 1.6 mL per patient (mean ± SEM for all patients). On the basis of the low frequency of tetramer+ cells within PBMCs, our patient inclusion criteria required a minimum of 30 mL blood. Freshly isolated PBMCs were washed and resuspended in 100 μL RPMI containing 2% FBS (Omega) and anti-CD16/32 human Fc block (BioLegend). A master mix (200 µL final volume) containing 10 nmol/L PE and 10 nmol/L APC tetramers with anti-CXCR5 (BioLegend) was added and incubated for 1 h at room temperature with gentle mixing after 30 min. Cells were washed with RPMI containing 2% FBS, resuspended in 250 μL RPMI containing 2% FBS and 25 μL anti-APC, and anti-PE magnetic beads (Miltentyi Biotec), and incubated for 25 min at 4°C. Cells were washed with PBS containing 2% FBS (FACS buffer), resuspended in 3 mL FACS buffer, and applied to a preequilibrated MACS LS column. The bound cells were washed three times with 3 mL FACS buffer, eluted by plunging 5 mL FACS buffer through the column twice, and centrifuged. The bound fraction was resuspended in 200 μL FACS buffer, and the flow through was resuspended in 500 μL FACS buffer. A 5-μL aliquot of bound fraction or flow through was removed and added to 200 μL AccuCheck counting beads to determine total CD4+ T-cell number. The remaining bound fraction or 100 μL flow through were transferred to 1.5 mL tubes, centrifuged, and resuspended in 25 μL Brilliant staining buffer (BD Biosciences). A master mix containing antibodies against CD3, CD4, CD8α, CD45RO, CCR7, CXCR3, PD-1, ICOS, CD20, CD33, and CD123 and a fixable live/dead dye was added to the cells to final volume of 100 μL (Supplementary Table 4), and the cells were incubated for 30 min at 4°C. All cells were run on an LSRII Fortessa flow cytometer equipped with 355-, 405-, 488-, 561-, and 640-nm lasers and analyzed using FlowJo 10 software (FlowJo LLC). Tetramer+ cell gates were determined by gating on CD8+ T in the bound fraction, which were always negative for dual fluorochrome–labeled tetramers (Fig. 1E, bottom). Tetramer dual staining is represented on the 45° angle because of the dual binding of each fluorophore at one-half maximal binding in each color. This dual staining allows us to gate out the nonspecific binding of the CD4+ T-cells to each individual fluorochrome-labeled tetramer reagent (22,25,26).
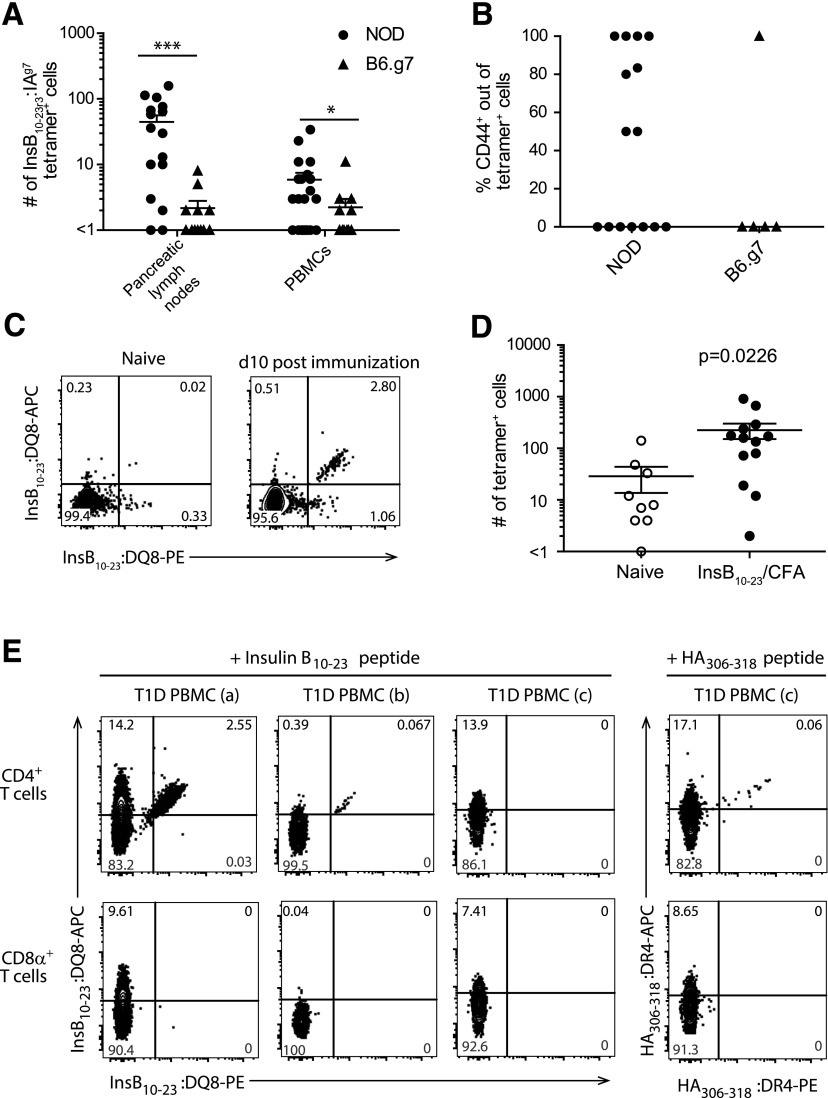
Figure 1.
Identification of InsB:IAg7 tetramer–binding cells in PBMCs from NOD and B6.g7 mice, with in vitro and in vivo validation of the InsB:DQ8 tetramer reagent. A: Quantification of InsB:IAg7 tetramer–binding cells from pancreatic lymph nodes and PBMCs isolated from 0.5 mL blood per mouse of 4- to 15-week-old NOD and age-matched B6.g7 mice. B: The percentage of CD44+ cells of InsB:IAg7 tetramer–binding cells within PBMCs from panel A (n = 6–11 mice/group [biological replicate], data are pooled from three independent experiments [technical replicate] for panels A and B). C: Representative flow cytometry plots showing dual InsB:DQ8 tetramer staining of CD4+ T cells from the secondary lymphoid organs of naïve (left panel) and InsB10–23 immunized (right panels) NOD.DQ8.Ab0 transgenic mice (data are representative of three independent experiments [technical replicates]). D: Quantification of tetramer+ cells shown in panel C from the secondary lymphoid organs of NOD.DQ8.Ab0 transgenic naïve mice (n = 4–6 [biological replicates]) and mice immunized (n = 9–13) with InsB10–23 peptide emulsified in complete Freund’s adjuvant (CFA). Cells were stained with InsB10–23:DQ8 tetramers in APC and PE (data are pooled from three independent experiments [technical replicates]). E: Representative flow cytometry plots showing dual tetramer staining within peripheral blood CD4+ T-cell (top) and no dual staining of CD8α+ T-cell compartments (bottom) from DR4+/DQ8+ human subjects after 14 days of in vitro stimulation with modified InsB10–23 or HA306–318 (data are representative of four independent experiments [technical replicate], n = 8 T1D patients for each tetramer [biological replicate]). Three patients are shown: T1D PBMC (a)—male, age 14, T1D duration of 3.5 years; T1D PBMC (b)—female, age 6, T1D duration of 2.6 years; and T1D PBMC (c)—male, age 34, T1D duration of 32 years. The solid lines in panels A, B, and D represent the mean ± SEM. The Mann-Whitney U test was used to determine statistical significance for nonparametric data. *P < 0.05, ***P < 0.0001. PE, R-phycoerythrin.
In Vitro Peptide Stimulation of PBMCs
Frozen PBMCs were washed and resuspended at 4–6 × 106 cells/mL in RPMI containing 10% pooled human serum, penicillin/streptomycin, sodium pyruvate, and 2-mercaptoethanol. Modified InsB10–23 (HLVERLYLVAGEEG) or HA306–318 (PKYVKQNTLKLAT) peptide was added to a final concentration of 10 μmol/L, and the cells were cultured in 24-well plates for 10–14 days at 37°C with 5% CO2. Recombinant human interleukin 2 (PeproTech) was added on day 5 at a final concentration of 10 units/mL.
Mouse Cell Isolation and Tetramer Staining
Lymphocytes were isolated in D10 media (DMEM containing 10% FBS, HEPES, 2- mercaptoethanol, nonessential amino acids, and penicillin/streptomycin) into a single cell suspension. PBMCs were isolated from 0.5 mL blood obtained by retro-orbital puncture. After centrifugation, the plasma was collected, and the pellet was resuspended in 1 mL PBS, layered onto 1.5 mL Ficoll-Paque in 5 mL tubes, and centrifuged at 400g for 10 min at room temperature. The interface was collected and washed twice with FACS buffer. Cells were resuspended in anti-CD16/32 Fc-Block and incubated for 10 min at 4°C. Tetramer staining and magnetic enrichment was performed as previously described (22). Cells were stained with surface antibodies against CD3ε, CD4, CD8α, PD-1, CD44, B220, CD11b, CD11c, and a fixable live/dead dye for 30 min at 4°C, washed with FACS buffer, and run on a BD LSRII Fortessa or BD Fortessa X-20 flow cytometer and analyzed using FlowJo software.
Autoantibody Analysis
Autoantibodies in blood plasma were determined by radiobinding assay at the University of Colorado, Denver Autoantibody Service Center.
Statistics
Statistical analyses were performed using GraphPad Prism 7.0 software or R 3.2.4 software. Unsupervised hierarchical clustering and heat map generation was done using the “Hierarchical Clustering” modules from GenePattern (27) (http://genepattern.broadinstitute.org) and JColorGrid (28) (www.jcolorgrid.sourceforge.net), respectively. The statistical tests used for each analysis are described in the figure legends.
Results
The goal of this study was to determine whether the frequency and activation profile of InsB-specific CD4+ T cells analyzed directly ex vivo from human peripheral blood could provide insight into their role in T1D pathogenesis. We first tested whether insulin-specific CD4+ T cells from PBMCs could be used as a readout of pancreas-specific autoimmunity in mice. Using a well-described InsB10–23:IAg7 tetramer, we compared insulin-specific CD4+ T cells in PBMCs and pancreatic lymph node cells from diabetes-susceptible NOD to MHCII-matched diabetes-resistant C57Bl/6.H2g7 (B6.g7) mice (7,14,22,29). We determined that NOD mice had significantly more insulin-specific CD4+ T cells in the pancreatic lymph nodes and pancreas compared with B6.g7 mice (P < 0.0001) (Fig. 1A) (22). Importantly, the number of insulin-specific CD4+ T cells was also significantly higher in the peripheral blood of nondiabetic NOD mice compared with B6.g7 mice (P < 0.0478) (Fig. 1A). Activated insulin-specific cells were present in the peripheral blood of 54.4% (8 of 15) of NOD mice compared with 20% (1 of 5) of B6.g7 mice, but this was not statistically significant (Fig. 1B). Insulin-specific CD4+ T cells did not stain with an irrelevant hen-egg lysozyme tetramer from PBMCs or the pancreatic lymph nodes in NOD or B6.g7 mice (data not shown) and as previously reported (22). We therefore conclude that peripheral blood lymphocytes are reflective of diabetes progression in mice.
To investigate insulin-reactive CD4+ T-cell frequency and phenotype in peripheral blood in humans, we first generated an InsB10–23:DQ8 tetramer (InsB-tetramer), analogous to the InsB10–23:IAg7 reagent used previously (4,30,31). This reagent contains a modified InsB10–23 peptide, which has a substitution of B22R→E (HLVERLYLVCGEEG) bound to the HLA-DQ8 class II molecule, allowing for improved presentation in a position or register favorable for T-cell binding to InsB10–23:DQ8 (22). To validate this InsB-tetramer, we immunized NOD transgenic HLA-DQ8, H2-Ab1–deficient mice (NOD.DQ8.Ab0) (15) with peptide emulsified in complete Freund’s adjuvant, and harvested the spleen and draining lymph nodes after immunization. Control NOD.DQ8.Ab0 mice had an average of 29 cells/mouse and NOD.DQ8.Ab0 mice immunized with modified InsB10–23 peptide had an average of 225 tetramer binding cells/mouse, reflecting a 7.75-fold expansion (Fig. 1C and D).
To further validate our InsB-tetramer, we used modified InsB10–23 peptide to expand insulin-reactive T cells in vitro to generate T-cell lines from PBMCs isolated from DR4/DQ8+ patients with T1D with established disease. Using dual-color tetramer labeling, we determined that five of eight patients with T1D had insulin-reactive T cells that expanded after in vitro stimulation in agreement with a previous report (13). As a control, we used HA306–318 peptide to expand influenza-specific CD4+ T cells from PBMCs of all eight patients, including those who did not have detectable InsB-tetramer–specific cells, thus validating our approach and sample viability (Fig. 1E).
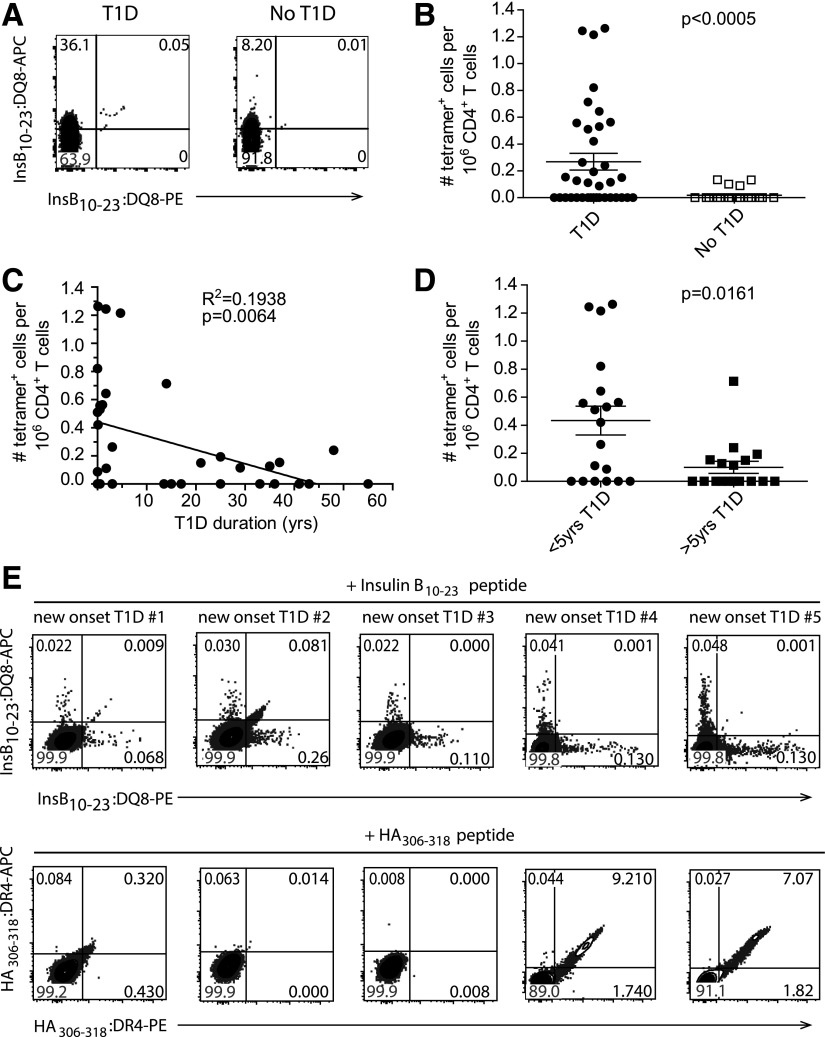
Yang et al. (13) previously demonstrated that InsB-specific cells could be expanded from PBMCs from patients with T1D but not from control subjects without diabetes. However, because these cells were cultured in vitro, the initial frequency and phenotype of the cells could not be determined. To overcome these limitations, we directly determined the frequency of InsB-tetramer–binding cells ex vivo from freshly isolated PBMCs using magnetic bead enrichment of tetramer-stained cells (20,24). Figure 2A shows representative staining of tetramer+ CD4+ T cells from DQ8+ patients with T1D and control subjects without diabetes (Supplementary Table 1). We identified insulin-specific cells in 54% (20 of 37) of patients with T1D, whereas only 15% (4 of 26) of individuals without diabetes had detectable tetramer+ cells (Fig. 2B). Interestingly, of the individuals that had InsB-tetramer+ cells, patients with T1D had a significantly higher frequency of tetramer+ cells out of the total CD4+ T-cell compartment compared with control subjects without diabetes (1 in 4 million CD4+ T cells vs. 1 in 9 million CD4+ T cells; P < 0.0005) (Fig. 2B). Moreover, we also found a significant inverse relationship between the frequency of InsB-tetramer+ cells and T1D disease duration (R2 = 0.1938, P = 0.0064) (Fig. 2C). In fact, patients with T1D duration of less than 5 years had a significantly higher frequency of tetramer+ cells/1 million CD4+ T cells compared with patients with T1D duration of greater than 5 years (P = 0.0161) (Fig. 2D). To confirm that these InsB-tetramer+ cells are autoreactive and not expanded as a result of exogenous insulin therapy, we assessed whether InsB-tetramer+ cells were present in PBMCs isolated from patients with T1D within 15 days of the T1D diagnosis (Supplementary Table 2). As a result of sample volume limitations, PBMCs were stimulated in vitro with register 3 modified InsB peptide to expand any peptide-specific cells. After 10 days of stimulation, we detected InsB-tetramer+ cells in PBMCs derived from two of five patients with new-onset T1D (Fig. 2E), which is a slightly lower frequency of patients compared with our established cohort with T1D but similar to what others have reported using ex vivo peptide stimulation (13). As a control, we also expanded HA-specific cells in three of five patients shown in Fig. 2E. These results suggest that InsB-tetramer+ cells become activated in response to endogenous antigen and that their frequency is associated with recent-onset T1D, presumably because recent-onset patients have residual endogenous insulin production (32).
Figure 2.
Enumeration of InsB:DQ8 tetramer+ cells from HLA-DQ8+ patients with T1D and individuals without diabetes. A: Representative flow cytometry plots showing dual InsB:DQ8 tetramer+ stained cells concatenated from six patients with T1D patients (left) and four concatenated DQ8+ individuals without diabetes (right). Cells were gated on size, single cells, live, CD20−, CD33−, CD123−, CD8α−, CD3ε+, CD4+. B: Frequency of insulin-tetramer+ cells/1 million CD4+ T cells from all patients with T1D (n = 37) and control subjects without diabetes (n = 26; biological replicates). C: Correlation of the frequency of tetramer+ cells with disease duration for all patients with T1D. D: Comparison of the frequency of tetramer+ cells in individuals with T1D duration shorter and longer than 5 years. E: Flow cytometry plots showing dual InsB:DQ8 (top) and HA:DR4 (bottom) tetramer staining within peripheral blood CD4+ T cells from five DR4+/DQ8+ subjects with new-onset after 10 days of in vitro stimulation with modified InsB10–23 or HA306–318 (data are from two independent experiments). Bars represent the mean. The Mann-Whitney U test for nonparametric data (B and D) or linear regression (C) was used to determine significance. PE, R-phycoerythrin.
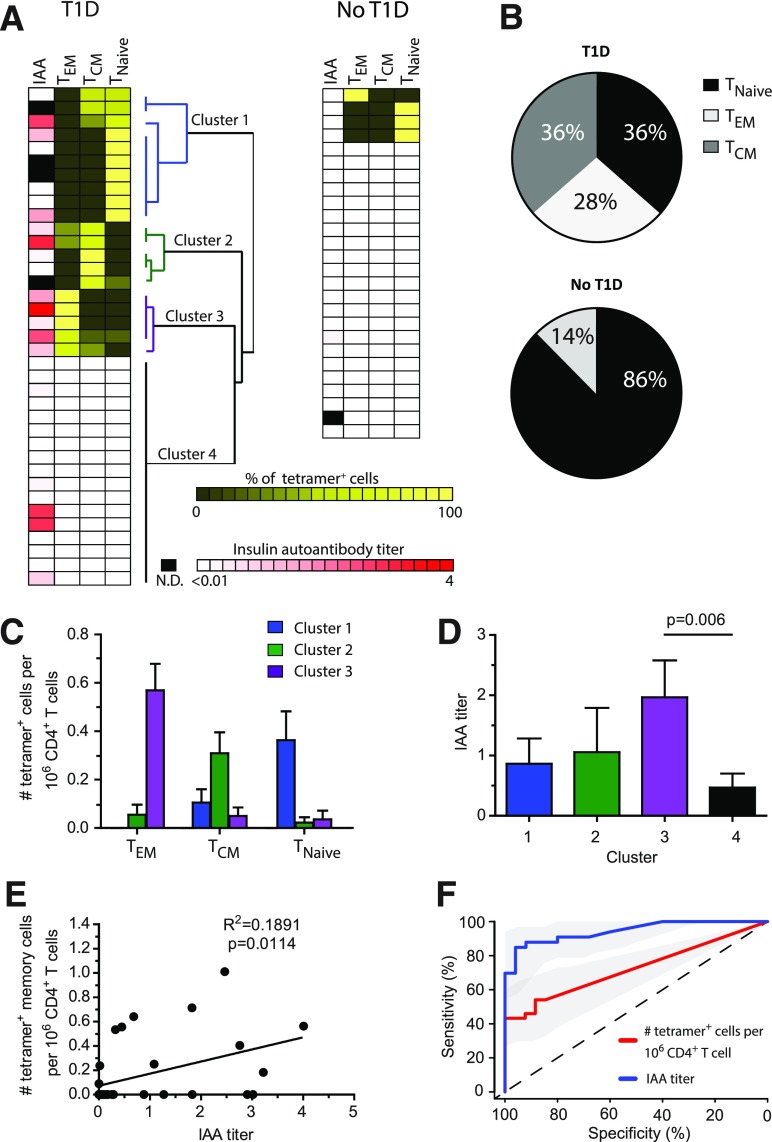
We next examined the activation phenotype of insulin-specific T cells. We used unbiased hierarchical clustering analysis to group individual patients with T1D into four clusters based on the presence or absence of InsB-tetramer+ cells and the phenotype of InsB-tetramer+ cells (CD4+ T effector memory [TEM] CD4+ T central memory [TCM], and naïve CD4+ T cell [TNaïve]) (Fig. 3A). TNaïve CD4+ T cells were defined as CD45RO−CCR7+, TEM CD4+ T cells were CD45RO+CCR7−, and TCM CD4+ T cells were CD45RO+CCR7+ (33,34). Of the insulin-specific cells from patients with T1D, 64% were antigen-experienced memory cells, with 28% TEM (CD45RO+CCR7−) and 36% TCM (CD45RO+CCR7+) phenotype (33,34) (Fig. 3A and B). To verify that these InsB-tetramer+ T cells were antigen experienced, we also evaluated CXCR3 and CXCR5 expression and determined that 73.5% expressed either chemokine receptor compared with 6% in the naïve InsB-tetramer+ populations. Cluster 1 from the hierarchical analysis included patients with the highest frequency of naïve InsB-tetramer+ cells (0.363/1 million CD4+ T cells), whereas clusters 2 and 3 contained patients with predominantly TCM (0.309/1 million CD4+ T cells) or TEM (0.568/1 million CD4+ T cells), respectively (Fig. 3C). In contrast, of the 26 control subjects without diabetes, 22 (84%) did not have any InsB-tetramer+ cells, 1 had TEM cells, and 3 had detectable TNaïve InsB-tetramer+ T cells (Fig. 3A). Interestingly, the control subject with an InsB-tetramer+ cell effector memory phenotype was a 3-year-old first-degree relative with the HLA-DQ8/DR4 haplotype associated with highest T1D risk.
Figure 3.
Phenotype and correlation analysis of InsB-tetramer+ cells with insulin antibody. A: IAA titer along with unbiased hierarchical clustering analysis based on tetramer+ cell frequency and phenotype. Each row represents an individual patient (n = 37) or sample from a control subject without diabetes (n = 26), biological replicates. N.D., not determined. B: Combined phenotype of all tetramer+ cells from all patients shown in panel A. C: Frequency of insulin-tetramer+ cells within TEM, TCM, and TNaïve phenotypes from patients within each cluster as determined in panel A. D: Plasma IAA titer among patients within each cluster as defined in panel A. E: Correlation of the frequency of tetramer+ memory (TEM and TCM) cells identified in each patient with their IAA titer. F: Receiver operating characteristic curve analysis using R 3.2.4 software to measure the sensitivity, specificity, and overall performance of tetramer+ cell frequency or IAA to identify individuals with T1D. Bars represent the mean ± SEM. A one-way ANOVA test (D) or linear regression (E) was used to determine significance.
We next determined whether a correlation existed between the InsB-tetramer+ cell phenotype and IAA titers. We compared the tetramer+ cell frequency and activation phenotype for each cluster (Fig. 3A) with the serum titer of IAA. We found that patients in cluster 3 (i.e., those with the highest frequency of TEM CD4+ T cells) had significantly higher IAA titers than patients in the other clusters (P = 0.006) (Fig. 3D). We extended this analysis to evaluate the relationship between the IAA titer and the frequency of all memory (TCM and TEM) InsB-tetramer+ T cells and detected a weak but statistically significant positive correlation (P = 0.0114) (Fig. 3E).
The correlation of InsB-tetramer+ cells with IAA titers suggests that determining the frequency and activation phenotype of InsB-tetramer+ cells may have additional value in understanding diabetes progression. Receiver operating characteristic curve analysis was used to measure the sensitivity, specificity, and overall performance of tetramer+ cell frequency compared with the IAA titer to identify individuals with T1D. We determined that InsB-tetramer+ cell frequency was correlative and successfully discriminated patients with and without T1D with an area under the curve of 0.705 (Fig. 3F). As predicted, the IAA titer was also highly correlative and successfully predicted T1D with an area under the curve of 0.892 (Fig. 3F). These results indicate that InsB-tetramer+ frequency and the IAA titer both correlate with T1D and achieve specificity, whereas IAA titers achieved higher sensitivity, indicating a higher rate of true positives. As T1D duration progressed, the frequency of InsB-tetramer+ cells diminished faster than the IAA titers, resulting in lower sensitivity (Fig. 3F).
Discussion
In the current study, we examined the frequency and phenotype of insulin-reactive CD4+ T cells directly ex vivo, without expansion, from HLA-DQ8+ patients with T1D and HLA-matched control subjects without diabetes. Using our approach, we were able to identify InsB-tetramer+ T cells in 54% (20 of 37) of patients with T1D and in 15% (4 of 26) of HLA-DQ8+ control subjects. A similar reagent recently developed by Yang et al. (13) was used to demonstrate that insulin-specific cells could be expanded in vitro from the peripheral blood of 37% of patients with T1D but not from control subjects without diabetes, highlighting the differential sensitivity of the two approaches. Our analysis determined that the average frequency of self-reactive T cells was 1 in 4 million CD4+ T cells from patients with T1D and 1 in 9 million CD4+ T cells from HLA-DQ8+ control individuals without diabetes (Fig. 2B). This frequency of self-reactive T cells is similar to recent reports on DR4+ healthy donors (24) but lower than those reported for a GAD250–266 epitope (∼1 in 0.2 million) where all tested patients with T1D had tetramer+ cells (23). It is important to note that we did not detect InsB-specific CD4+ T cells in all patients with T1D, and only 15% of the HLA-DQ8+ control subjects without diabetes had InsB-tetramer+ T cells. This could be the result of differences in T-cell trafficking, antigen targets, epitope spreading, disease duration, or complex peptide-binding registers for HLA haplotypes. Furthermore, we did detect InsB-specific T cells in patients with recent-onset T1D (within 15 days), indicating that these cells are unlikely to be induced from exogenous insulin administration. These results also indicate that analysis of self-reactive T cells from peripheral blood can be used to measure T1D disease activity even though the pancreas is the organ targeted for immune attack and would be the preferred site for measuring disease activity, but not clinically feasible.
The work showed here supports previous reports of effector memory and central memory skewing of self-antigen specific T cells in patients with T1D but not in HLA-matched control subjects without diabetes (Fig. 3) (12,35–37). This is not surprising, however, because previous reports have used in vitro expansion, and thus, the memory cells would be favored over naïve T-cells. Human PBMCs stimulated in vitro with insulin peptide predominantly produce interferon-γ, interleukin 2, and tumor necrosis factor-α (11–13), consistent with inflammatory cytokines interferon-γ and tumor necrosis factor-α produced by insulin-specific T cells in NOD mice after challenge (22). We extended these results to show a positive correlation between the frequency of insulin-specific memory cells (TCM or TEM) and IAA titers at the same time of T-cell analysis (Fig. 3E). This result is consistent with the published studies correlating the IAA titer as NOD mice age (6) and InsB-tetramer+ CD4 T-cell abundance and activation as NOD mice age (22). We did not find a correlation of the number of tetramer+ cells and their cell phenotype with the duration of T1D. However, we found that patients with T1D who were sampled within 5 years of disease onset were more likely to harbor insulin-specific CD4+ T cells in peripheral blood. We speculate that the presence of residual β-cell function drives the autoimmune T-cell response and that some insulin-specific CD4+ T cells may provide help to insulin-specific B cells as previously identified in mouse models of T1D (38). In support of this, we found that two of five IAA+ patients with new-onset T1D (within 15 days of T1D diagnosis) had detectable InsB-tetramer+ cells at a time when the immunological effects from exogenous insulin therapy were minimal. We detected insulin-specific T cells in four individuals without diabetes and without IAA, one of whom had a first-degree relative with T1D and had TEM tetramer+ cells. This is an interesting result, given that T-cell activation generally precedes B-cell antibody responses, because B cells require T-cell help. Critically, our strategy allows direct enumeration and examination of T-cell activation and effector profiling that is not possible after in vitro expansion, thus increasing the utility of tetramer-based approaches to studying CD4+ T-cell responses in T1D.
We recognize that our analysis was limited to a single peptide, and recent reports have identified additional CD4+ T-cell targets. Several antigens have been implicated in T1D pathogenesis, including proinsulin, GAD65, islet-specific glucose-6-phosphatase catalytic subunit related protein, insulinoma antigen-2, phogrin, chromogranin A, and islet zinc transporter (3,5,11,39–45). Recent reports indicate that novel peptides generated through a fusion process, termed transpeptidation, may also be involved in T1D pathogenesis as targets of islet-specific CD4+ T cells in both mice and humans (40,45–47). We predict that the ability to build additional peptide:HLAII tetramer reagents that incorporate different epitopes within the suspected protein targets will enhance our ability to 1) characterize the autoimmune T-cell repertoire in patients with T1D, 2) define a peripheral T-cell biomarker of disease activity within the islet, and 3) potentially allow timing of immune interventions such as islet antigen-specific tolerance (48–50).
Potential therapeutic approaches could include deletion of InsB-tetramer+–specific T cells using toxin-coupled tetramers, as previously reported for CD8+ T cells in mouse models of T1D (51), or modulation of the InsB-tetramer+ T-cell specific response using monoclonal antibodies targeting InsB peptide in the context of MHCII. This approach has recently been shown to delay diabetes in NOD mice (10,52). Finally, antigen-specific tolerance is a third possibility to modulate the InsB-tetramer+ T cells using peptide-pulsed nanoparticles or antigen-coupled cells, as previously described (53,54).
Although this study significantly advances our knowledge of the frequency and effector memory phenotype of insulin-reactive T cells from patients with T1D, it has some limitations. First, the linear regression analysis demonstrated a positive relationship between InsB-tetramer+ cell frequency and IAA that was statistically significant but not overwhelmingly predictive.
Second, our analysis was limited to a single diabetes-associated antigen. Although this choice is based on animal models and human pathogenesis demonstrating InsB-specific T cells infiltrate islets, future studies will include additional antigens in the context of diabetes-associated HLAII molecules that may encompass a broader spectrum of autoreactive T-cell specificities. We recognize that 30 mL blood for a single analysis is not clinically viable for large-scale screening. Multiplexing several additional T1D target peptides by using additional tetramers would provide a significant advantage for increased sensitivity.
Third, T-cell characterization was limited to naïve and memory phenotypes. Quantification of additional T-cell subsets (i.e., regulatory T-cells, T helper 1, or T follicular helper cells) might increase our understanding of disease pathogenesis and potential tolerance mechanisms in those without T1D (12).
Fourth, tetramer measurements were performed at a static time point. A logical next step is to expand the analysis to include longitudinal assessments in the same subjects, which may identify T-cell expansion and contraction phases (i.e., relapse and remittance of T-cell–mediated disease activity) with temporal associations before disease onset. These current limitations provide future areas of research strategies. Our intent is not to replace autoantibody screening for T1D risk assessment but to add T-cell evaluation together with B-cell activity to strengthen risk assessment and identify therapeutic opportunities.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Vivian Gersuk, from the Benaroya Research Institute, for assistance and advice on HLA haplotyping, and Trevor Ormann, Anne Street, and Beth Pappenfus, from the University of Minnesota, for help with patient recruitment. The HLA control samples were kindly provided by the Fred Hutchinson Cancer Research Center International Histocompatibility Working Group Cell and Gene Bank.
Funding. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases DK-108868 (A.W.M.); National Institute of Allergy and Infectious Diseases P01-AI-35296 (M.K.J. and B.T.F.), R01-AI-106791 (B.T.F.), and U24-AI-118635 (B.T.F.); Regenerative Medicine Minnesota RMM-11215-TR002 (B.T.F.); and JDRF 3-2014-215 (J.A.S.) and 17-2014-3 (B.T.F.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.A.S., N.L.S., J.C.W., T.M., and B.T.F. designed and performed the experiments and analyzed the data. J.A.S., T.D., M.K.J., and B.T.F. designed, generated, and validated the Ins:DQ8 tetramer. J.A.S., A.W.M., and B.T.F. directed the studies, prepared the figures, and wrote the manuscript. A.L.B., E.B.F., A.W.M., and A.M. procured the human samples. E.B.F. and B.R.B. provided critical feedback on the manuscript. All authors reviewed the manuscript. B.T.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-0666/-/DC1.
See accompanying article, p. 2940.
References
- 1.Pociot F, Lernmark Å. Genetic risk factors for type 1 diabetes. Lancet 2016;387:2331–2339 [DOI] [PubMed] [Google Scholar]
- 2.Anderson MS, Bluestone JA. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol 2005;23:447–485 [DOI] [PubMed] [Google Scholar]
- 3.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med 2012;2:a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suri A, Walters JJ, Gross ML, Unanue ER. Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J Clin Invest 2005;115:2268–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steck AK, Dong F, Waugh K, et al. Predictors of slow progression to diabetes in children with multiple islet autoantibodies. J Autoimmun 2016;72:113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama M, Abiru N, Moriyama H, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 2005;435:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci U S A 2010;107:10978–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naik RG, Beckers C, Wentwoord R, et al. Precursor frequencies of T-cells reactive to insulin in recent onset type 1 diabetes mellitus. J Autoimmun 2004;23:55–61 [DOI] [PubMed] [Google Scholar]
- 9.Standifer NE, Burwell EA, Gersuk VH, Greenbaum CJ, Nepom GT. Changes in autoreactive T cell avidity during type 1 diabetes development. Clin Immunol 2009;132:312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Crawford F, Yu L, et al. Monoclonal antibody blocking the recognition of an insulin peptide-MHC complex modulates type 1 diabetes. Proc Natl Acad Sci U S A 2014;111:2656–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michels AW, Landry LG, McDaniel KA, et al. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes 2017;66:722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama M, McDaniel K, Fitzgerald-Miller L, et al. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc Natl Acad Sci U S A 2015;112:4429–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Chow IT, Sosinowski T, et al. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci U S A 2014;111:14840–14845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zucchelli S, Holler P, Yamagata T, Roy M, Benoist C, Mathis D. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity 2005;22:385–396 [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Purdy LE, Rabinovitch S, Jevnikar AM, Elliott JF. Major DQ8-restricted T-cell epitopes for human GAD65 mapped using human CD4, DQA1*0301, DQB1*0302 transgenic IA(null) NOD mice. Diabetes 1999;48:469–477 [DOI] [PubMed] [Google Scholar]
- 16.Orban T, Sosenko JM, Cuthbertson D, et al.; Diabetes Prevention Trial-Type 1 Study Group . Pancreatic islet autoantibodies as predictors of type 1 diabetes in the Diabetes Prevention Trial-Type 1. Diabetes Care 2009;32:2269–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gersuk VH, Nepom GT. A real-time PCR approach for rapid high resolution subtyping of HLA-DRB1*04. J Immunol Methods 2006;317:64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gersuk VH, Nepom GT. A real-time polymerase chain reaction assay for the rapid identification of the autoimmune disease-associated allele HLA-DQB1*0602. Tissue Antigens 2009;73:335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bugawan TL, Erlich HA. Rapid typing of HLA-DQB1 DNA polymorphism using nonradioactive oligonucleotide probes and amplified DNA. Immunogenetics 1991;33:163–170 [DOI] [PubMed] [Google Scholar]
- 20.Moon JJ, Chu HH, Pepper M, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 2007;27:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CP, Crawford F, Marrack P, Kappler J. T cell positive selection by a high density, low affinity ligand. Proc Natl Acad Sci U S A 1998;95:4522–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauken KE, Linehan JL, Spanier JA, et al. Cutting edge: type 1 diabetes occurs despite robust anergy among endogenous insulin-specific CD4 T cells in NOD mice. J Immunol 2013;191:4913–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok WW, Liu AW, Novak EJ, et al. HLA-DQ tetramers identify epitope-specific T cells in peripheral blood of herpes simplex virus type 2-infected individuals: direct detection of immunodominant antigen-responsive cells. J Immunol 2000;164:4244–4249 [DOI] [PubMed] [Google Scholar]
- 24.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 2013;38:373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tubo NJ, Pagán AJ, Taylor JJ, et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 2013;153:785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwok WW, Tan V, Gillette L, et al. Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire. J Immunol 2012;188:2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet 2006;38:500–501 [DOI] [PubMed] [Google Scholar]
- 28.Joachimiak MP, Weisman JL, May BC. JColorGrid: software for the visualization of biological measurements. BMC Bioinformatics 2006;7:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford F, Stadinski B, Jin N, et al. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc Natl Acad Sci U S A 2011;108:16729–16734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corper AL, Stratmann T, Apostolopoulos V, et al. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science 2000;288:505–511 [DOI] [PubMed] [Google Scholar]
- 31.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol 2001;2:501–507 [DOI] [PubMed] [Google Scholar]
- 32.The Diabetes Control and Complications Trial Research Group Effect of intensive therapy on residual beta-cell function in patients with type 1 diabetes in the diabetes control and complications trial. A randomized, controlled trial. Ann Intern Med 1998;128:517–523 [DOI] [PubMed] [Google Scholar]
- 33.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol 2004;22:745–763 [DOI] [PubMed] [Google Scholar]
- 34.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–712 [DOI] [PubMed] [Google Scholar]
- 35.Danke NA, Yang J, Greenbaum C, Kwok WW. Comparative study of GAD65-specific CD4+ T cells in healthy and type 1 diabetic subjects. J Autoimmun 2005;25:303–311 [DOI] [PubMed] [Google Scholar]
- 36.Monti P, Scirpoli M, Rigamonti A, et al. Evidence for in vivo primed and expanded autoreactive T cells as a specific feature of patients with type 1 diabetes. J Immunol 2007;179:5785–5792 [DOI] [PubMed] [Google Scholar]
- 37.Chow IT, Yang J, Gates TJ, et al. Assessment of CD4+ T cell responses to glutamic acid decarboxylase 65 using DQ8 tetramers reveals a pathogenic role of GAD65 121-140 and GAD65 250-266 in T1D development. PLoS One 2014;9:e112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan X, Thomas JW, Unanue ER. Class-switched anti-insulin antibodies originate from unconventional antigen presentation in multiple lymphoid sites. J Exp Med 2016;213:967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kracht MJ, van Lummel M, Nikolic T, et al. Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat Med 2017;23:501–507 [DOI] [PubMed] [Google Scholar]
- 40.Delong T, Wiles TA, Baker RL, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 2016;351:711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadinski BD, Delong T, Reisdorph N, et al. Chromogranin A is an autoantigen in type 1 diabetes. Nat Immunol 2010;11:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman SM, DiLorenzo TP. A comprehensive guide to antibody and T-cell responses in type 1 diabetes. Tissue Antigens 2003;62:359–377 [DOI] [PubMed] [Google Scholar]
- 43.Arvan P, Pietropaolo M, Ostrov D, Rhodes CJ. Islet autoantigens: structure, function, localization, and regulation. Cold Spring Harb Perspect Med 2012;2:a007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pietropaolo M, Towns R, Eisenbarth GS. Humoral autoimmunity in type 1 diabetes: prediction, significance, and detection of distinct disease subtypes. Cold Spring Harb Perspect Med 2012;2:a012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin N, Wang Y, Crawford F, et al. N-terminal additions to the WE14 peptide of chromogranin A create strong autoantigen agonists in type 1 diabetes. Proc Natl Acad Sci U S A 2015;112:13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiles TA, Delong T, Baker RL, et al. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J Autoimmun 2017;78:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cresswell P. Cell biology. Cutting and pasting antigenic peptides. Science 2004;304:525–527 [DOI] [PubMed] [Google Scholar]
- 48.Diabetes Prevention Trial–Type 1 Diabetes Study Group Effects of insulin in relatives of patients with type 1 diabetes mellitus. N Engl J Med 2002;346:1685–1691 [DOI] [PubMed] [Google Scholar]
- 49.Skyler JS, Krischer JP, Wolfsdorf J, et al. Effects of oral insulin in relatives of patients with type 1 diabetes: the Diabetes Prevention Trial–Type 1. Diabetes Care 2005;28:1068–1076 [DOI] [PubMed] [Google Scholar]
- 50.Bonifacio E, Ziegler AG, Klingensmith G, et al.; Pre-POINT Study Group . Effects of high-dose oral insulin on immune responses in children at high risk for type 1 diabetes: the Pre-POINT randomized clinical trial. JAMA 2015;313:1541–1549 [DOI] [PubMed] [Google Scholar]
- 51.Vincent BG, Young EF, Buntzman AS, et al. Toxin-coupled MHC class I tetramers can specifically ablate autoreactive CD8+ T cells and delay diabetes in nonobese diabetic mice. J Immunol 2010;184:4196–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spanier JA, Frederick DR, Taylor JJ, et al. Efficient generation of monoclonal antibodies against peptide in the context of MHCII using magnetic enrichment. Nat Commun 2016;7:11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singha S, Shao K, Yang Y, et al. Peptide-MHC-based nanomedicines for autoimmunity function as T-cell receptor microclustering devices. Nat Nanotechnol 2017;12:701–710 [DOI] [PubMed] [Google Scholar]
- 54.Fife BT, Guleria I, Gubbels Bupp M, et al. Insulin-induced remission in new-onset NOD mice is maintained by the PD-1-PD-L1 pathway. J Exp Med 2006;203:2737–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.