Abstract
OBJECTIVES: To analyze the distant metastasis possibility based on computed tomography (CT) radiomic features in patients with lung cancer. METHODS: This was a retrospective analysis of 348 patients with lung cancer enrolled between 2014 and February 2015. A feature set containing clinical features and 485 radiomic features was extracted from the pretherapy CT images. Feature selection via concave minimization (FSV) was used to select effective features. A support vector machine (SVM) was used to evaluate the predictive ability of each feature. RESULTS: Four radiomic features and three clinical features were obtained by FSV feature selection. Classification accuracy by the proposed SVM with SGD method was 71.02%, and the area under the curve was 72.84% with only the radiomic features extracted from CT. After the addition of clinical features, 89.09% can be achieved. CONCLUSION: The radiomic features of the pretherapy CT images may be used as predictors of distant metastasis. And it also can be used in combination with the patient's gender and tumor T and N phase information to diagnose the possibility of distant metastasis in lung cancer.
Introduction
Lung cancer is one of the most common malignant tumors with the highest morbidity and mortality, which is a considerable threat to people's health and life [1]. Advanced lung cancer is likely to metastasize, which may lead to corresponding symptoms in patients with great pain, and are even life-threatening. This phenomenon is referred to as the distant metastasis of lung cancer, which is represented by the M staging in the TNM staging system [2].
Metastatic tumors are very common in the later stages of cancer. The spread of metastasis may occur through blood or lymphatic vessels or both pathways. The most common sites of metastases are the lungs, liver, brain, and bone. Because of the metastasis of cancer cells to various parts of the body, treatment becomes more difficult, surgery is almost impossible, and in most cases, only wide-range radiation therapy or chemotherapy may be used to inhibit cancer cell growth [3].
Computed tomography (CT) imaging is a widely used method for the evaluation of tumor prognosis. Based on the analysis of the CT image of the tumor, the image texture feature description can be extracted.
Doctors can diagnose the distant metastasis of the tumor using CT or positron emission tomography (PET)-CT of secondary tumors (metastatic tumors). Quantitative analysis has been employed to study the characteristics of various types of tumor metastases to predict the therapeutic effect [4], [5]. The clinical features of these cases can be used to predict the survival time of patients [6], [7]. However, there is lack of evidence regarding the prediction of distant metastasis based on the tumor. [8] In this paper, we employed radiomic methods to predict the possibility of distant metastasis of lung cancer based on the information of the tumor and the clinical features.
Radiomic refers to the comprehensive quantification of tumor phenotype using a large number of image features [9], [10], [11], [12]. It is defined as the conversion of image data to higher dimensional space and the subsequent mining of these data for improved decision support [13]. The radiomic method has been widely used in diagnosis [14], such as survival analysis [15], or lymph node metastasis prediction [16].
Diagnosis based on radiomic features has been used in the literature [16], [17], [18], but for the analysis of the prediction of M stage tumors, it remains inadequate. The aim of our study was to analyze the possibility of tumor metastasis from two aspects: clinical features and radiomic features extracted by CT, to investigate the relationship between the two kinds of features and the occurrence of distant metastasis.
Materials and Methods
Patients
The study was approved by the West China Hospital, Sichuan University. Approximately 404 patients with lung cancer (enrolled from West China Hospital, Sichuan University) were enrolled in this study. These data include small cell carcinoma and non–small cell carcinoma patients; tumor histological subtypes include squamous cell carcinoma, adenocarcinoma, and small cell carcinoma. Patient data registration time ranged from 2014 to February 2015. Patients with missing data were excluded, and the remaining 348 data sets were used in the study. The demographic and tumor characteristics of all patients are summarized in Table 1. The study design was approved by the appropriate ethics review boards.
Table 1.
Patient Demographics and Clinic Pathological Characteristics of the Training and Validation Set for the Metastasis Analysis
| Demographic or Clinic pathological Characteristics | Training Set |
Validation Set |
||||
|---|---|---|---|---|---|---|
| Without Metastasis | Metastasis | Without Metastasis | Metastasis | |||
| Total | 149 | 92 | 66 | 41 | ||
| Gender, no. (%) | Male | 105 (70.5) | 59 (64.1) | 45 (68.2) | 22 (53.7) | |
| Female | 44 (29.5) | 33 (35.9) | 21 (31.8) | 19 (46.3) | ||
| Age (years), no. (%) | ≤60 | 64 (43.0) | 38 (41.3) | 19 (28.8) | 10 (24.4) | |
| >60 | 85 (57.0) | 54 (58.7) | 47 (71.2) | 31 (75.6) | ||
| Smoking status | Smoker | 82 | 41 | 34 | 16 | |
| Stage, no. (%) | I | 46 (30.9) | 19 (28.8) | |||
| II | 32 (21.5) | 11 (16.7) | ||||
| III | 71 (47.6) | 36 (54.5) | ||||
| IV | 92 (1) | 41 (1) | ||||
| Histological subtype | Squamous cell carcinoma | 75 | 30 | 31 | 2 | |
| Adenocarcinoma | 66 | 50 | 34 | 33 | ||
| Small cell carcinoma | 12 | 8 | 1 | 6 | ||
| TNM no. (%) | T | T1 | 33 (22.1) | 1 (1.0) | 10 (15.2) | 2 (4.9) |
| T2 | 61 (41.0) | 31 (33.7) | 28 (42.4) | 11 (26.8) | ||
| T3 | 29 (19.5) | 11 (12.0) | 12 (18.2) | 7 (17.1) | ||
| T4 | 26 (17.4) | 49 (53.3) | 16 (24.2) | 21 (51.2) | ||
| N | N0 | 73 (49.0) | 11 (12.0) | 31 (47.0) | 1 (2.4) | |
| N1 | 21 (14.1) | 3 (3.3) | 8 (12.1) | 5 (12.2) | ||
| N2 | 48 (32.2) | 54 (58.7) | 18 (27.3) | 27 (65.9) | ||
| N3 | 7 (4.7) | 24 (26.0) | 9 (13.6) | 8 (19.5) | ||
CT Images and Tumor Segmentation
Tumor segmentation and CT images were provided by the hospital (West China Hospital, Sichuan University). CT images were loaded into the ITK-SNAP software (version 2.2.0; www.itksnap.org) for three-dimensional manual segmentation. A radiologist with 8 years of experience with CT performed the tumor segmentations in all patients. The following parameters were used to obtain the CT images: collimator with 64 × 0.6 mm, voxel spacing 0.638672 × 0.638672 × 5 mm, and tube voltage of 100 kV. The tube current is calculated according to the individual's weight, height, and body mass index. The tube current was 220 mA for body mass index <25 kg/m2 and 330 mA for body mass index >25 kg/m2. The region of interest in the CT images can be extracted based on the segmentation.
Feature Extraction and Analysis
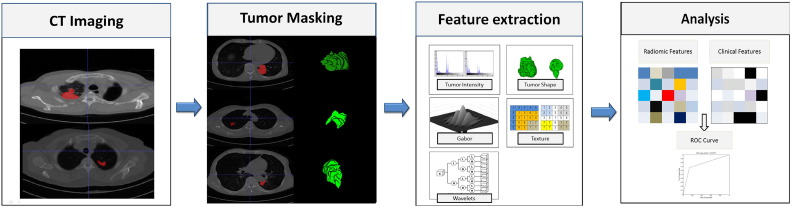
A complete lung cancer tumor radiomic features set included its volume, texture [19], [20], and Gabor and wavelet features. According to a previous study [9], the 485 radiomic features could be divided into four groups based on the tumor's intensity, shape, texture, and wavelet. The first group's quantified tumor intensity characteristics were calculated from the histogram of tumor voxel intensity values by using first-order statistics. Group 2 consisted of features based on the shape of the tumor, such as volume, surface area, and sphericity. Group 3 was composed of intratumor texture features, using “gray-level nonuniformity” to measure intratumor heterogeneity. Group 4 included the calculated intensity and textural features from the wavelet decomposition of the original image. We used the gray-level co-occurrence matrix and gray-level run length matrix (GLRLM) features as used in the study [9], and added the Gabor descriptor (we use six different angles of Gabor function for convolution of the image to obtain the features) to represent it in the decomposed images. The workflow of feature extraction is shown in Figure 1.
Figure 1.
Extracting radiomic data from images. At the left are the example CT images of patients with lung cancer. CT images with tumor segmentation left; three-dimensional visualizations right. The following pictures show the strategy for extracting radiomic data from images. (I) Experienced physicians contour the tumor areas on all CT slices (red section in the picture). (II) Features are extracted from within the defined tumor contours on the CT images, quantifying tumor intensity, shape, texture, and Gabor and wavelet texture. (III) For the analysis, the radiomic features are combined with clinical data.
On this basis, the patient's clinical characteristics were added as reference features, including the patient's age, smoking status, tumor classification, and T and N staging. TNM and final staging was performed as per the American Joint Committee on Cancer guidelines [2].
Machine Learning Methods
In this paper, we used machine learning methods for feature selection and classification. A key feature selection method can improve the utilization rate of data and features and attain better performance [21]. FSV [22] was used in this process to locate the features that performed best by using the selected features and then through SVM classification to make the decision.
A support vector machine (SVM) was used as the classifier in this group classification model [23]. As used in previous studies [24], [25], the SVM used the nearest point (support vector) to adjust the classification line so that the distance between the line and support vectors was as similar as possible, and the final classification line was only related to the support vectors. By using the SVM method, we can get a good classification result by using only a few data points. This method has been applied in classification based on the radiomic method [26].
Feature selection via convex minimization (FSV) [22] can extract the features by minimizing the subspace of the misclassified points. The FSV method is as follows: in the sample space, attain the subspace made of the misclassified samples with a linear classifier. In the convex optimization misclassified sample space minimization process, the most effective features can be discovered. By using the program in a previous study [27], we used the FSV method to select features and used the SVM classifier with a 10-fold cross-validation for evaluation.
Stochastic gradient descent (SGD), also known as incremental gradient descent, is a stochastic approximation of the gradient descent optimization method for minimizing an objective function that is written as a sum of differentiable functions. In other words, SGD was a method trying to find the minima or maxima by iteration.
The SVM and SGD methods used in this paper are derived from Sklearn package in Python3.5.
Statistical Analysis
The classification results were evaluated by both accuracy (ACC) and area under the curve (AUC). The ACC can reflect the intuitive results, and the receiver operating characteristic (ROC) curves combined with a true-positive rate and false-positive rate can be observed with a more comprehensive result. AUC signifies the area under the ROC curve. In this paper, ROC and AUC were calculated based on Sklearn package in Python3.5.
We generated a 10-fold cross-validation process to evaluate the feature selection. By this method, the features selected were more universal, and we could attain stable results in most cases. In the performance validation section of the model, we selected 241 (before October 2014) participants from the earlier dates of admission for the training set and the remaining 107 participants for the validation set for the final model test.
Path Analysis
Path analysis is a method of estimating and testing the internal coherence of the factors that assume causal structures [28]. By decomposing the relevant structure into the form of a path, the contribution between the factors is decomposed into direct contribution and indirect contribution [29]. In this paper, path analysis was used to detect the contribution of radiomic and clinical features for the prediction of metastasis in lung cancer. Path analysis was conducted with SPSS18. The total contribution relationship is found by the partial correlation coefficient, and the standard coefficient (path coefficient) is obtained using stepwise linear regression. The path coefficient represents the direct contribution of the factor, and the partial correlation coefficient is equivalent to the total contribution.
Results
Feature Selection
Results from the feature selection section showed that the radiomic features from the pretherapy CT images could be combined with the clinical features to achieve a better prediction of tumor metastasis. In the case of only considering the radiomic features from the pretherapy CT images, we selected four features with the FSV method. The resulting ACC and AUC were 71.02% and 72.84%. In the case of clinical features, FSV selected features as patient's' gender, T stage, and the N stage state. The resulting ACC and AUC were 79.44% and 84.09%. In the synthesis of the two kinds of features, seven features were selected by the FSV method, which included four radiomic features, such as texture and wavelet, and three clinical features such as the patient's' gender, T stage, and N stage (the statistical information of clinical features is described in Figure 2).
Figure 2.
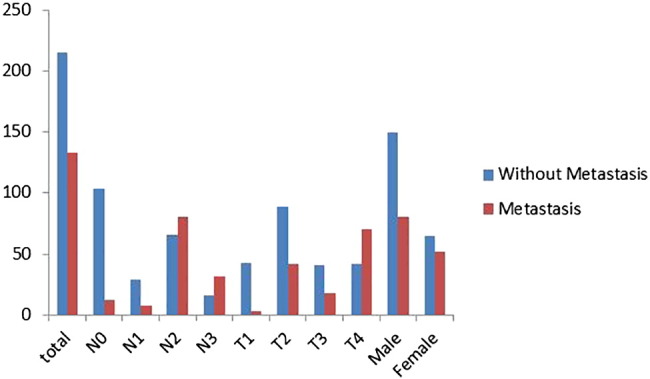
Histogram statistics of clinical features.
Classification Results
Based on the seven features selected by FSV, we employed the SVM with the SGD method (penalty = ‘l2’, alpha = 0.001) to construct the M staging group classification model. The resulting ACC and AUC were 85.98% and 89.09%. The ROC curve of the classification model was plotted (as shown in Figure 3).
Figure 3.
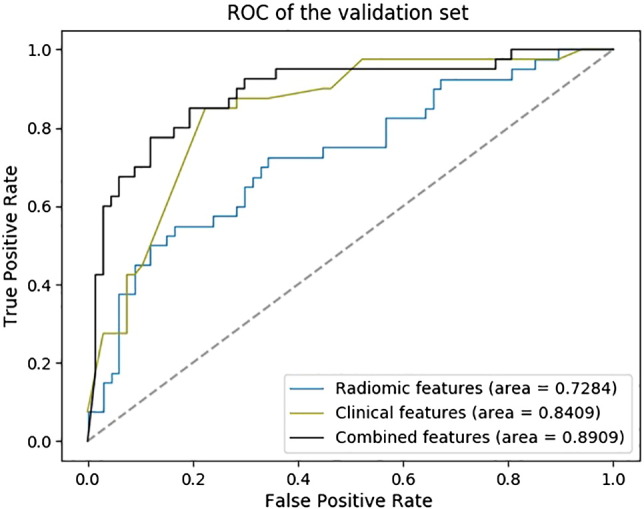
The ROC curve of the M stage group classification model (241 patients from the earlier date of admission were used for the training set, and the following 107 patients were used for the validation set).
Representative Phenotypic Features
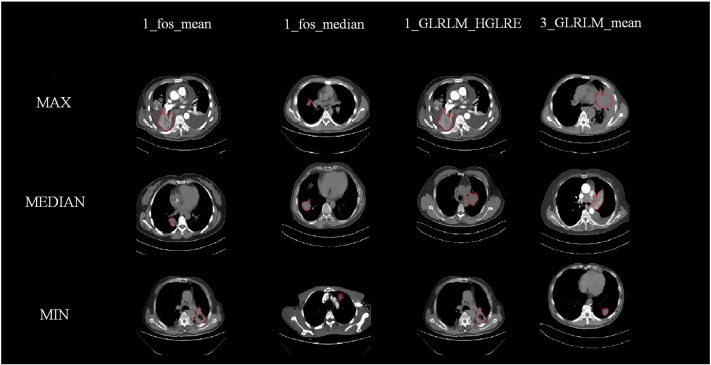
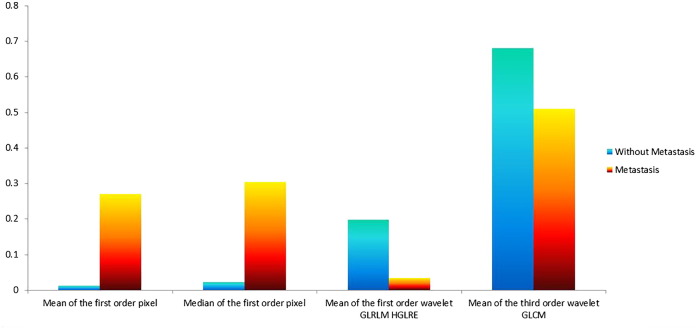
The seven features that were selected included three clinical features: patient's age, tumor's T stage, and N stage, and four radiomic texture features: including the mean and median of the first -order wavelet pixel, the mean of the first-order wavelet high gray-level run emphasis, and the third-order wavelet GLRLM. The selected radiation characteristics are mainly composed of first-order wavelet characteristics and auxiliary classification, which show that the statistical features decomposed by the first-order wavelet can effectively assist in the diagnosis of distant metastasis [9]. We selected the most prominent examples of these features in Figure 4 for comparison. In Figure 5, we compare the mean values of the four features.
Figure 4.
The contrast of different radiomic features.
Figure 5.
The comparison of normalized mean of radiomic features.
Path Analysis Results
Through path analysis, we computed the direct and indirect contribution of the feature to the prediction of distant metastasis of the tumor. From the results, the total contribution of radiomic features was smaller than their indirect contribution, indicating the need for complex relationships to characterize their contribution. The total contribution of clinical features was greater than their direct contribution, indicating that these features can express the association of distant metastases intuitively. The detailed results are shown in Table 2.
Table 2.
The Path Analysis Results of the Selected Features
| Features | Contribution |
|
|---|---|---|
| Direct Contribution | Total Contribution | |
| Mean of the first-order pixel | 0.054 | 0.13 |
| Median of the first-order pixel | −0.1 | −0.04 |
| Mean of the first-order wavelet GLRLM HGLRE | 0.222 | 0.191 |
| Mean of the third-order wavelet GLCM | 0.133 | 0.131 |
| Gender | 0.153 | 0.176 |
| T stage | 0.227 | 0.240 |
| N stage | 0.387 | 0.389 |
Features that do not pass the correlation test will be filtered out.
HGLRE, high gray-level run emphasis, GLCM, gray-level co-occurrence matrix.
Discussion
From the Results section, we can summarize that if only the features extracted from the CT image are used, the predictive effect is not significant, but it can be used as a reference to judge M as staging before operation. If combined with information such as pathological T/N staging, diagnostic effect will be more significant.
From the section of feature selection, we determined some relationships between the features and their ability to predict distant metastasis. In the clinical features, we can find that the status of N staging is very important for distant metastasis; in addition, the T stage and gender of the patients will have an impact on the prediction of the M staging of the tumor. To wit, there appears to be an association between lung cancer metastasis and gender of the patient, the size of the tumor, and the differentiation of the tumor. Regarding the radiomic features, it can be seen that the texture feature based on the first-order wavelet has a good auxiliary classification effect, accounting for three out of four of the selected radiomic features.
The study provided evidence that machine learning methods can be used to predict the metastasis of lung cancer. In this study, a certain degree of diagnostic accuracy could be attained from combining radiomic features obtained from the pretherapy CT images with the patients' clinical features. We also showed that the texture features play a role in the prognosis of distant metastasis in lung cancer and can be helpful in predicting metastasis.
At present, it is difficult to detect and predict the metastasis of lung cancer. It is usually necessary to sample and analyze all kinds of information in the cell [30], [31], [32]. These methods need to be analyzed after the tumor metastasized, which is usually detected only after it has already developed to a certain stage. The method employed in this study can be used to predict or diagnose the metastasis of tumors to some extent. Most of the clinical information needed can be obtained from CT. In addition, numerous methods have been reported on the prediction of tumor N staging [33], [34], which can be concurrently employed to augment the prognosis prediction. It can be used in conjunction with the N-stage prediagnostic method.
This study had several limitations. First, the patients were all from Asia, which may lead to biased results. Second, the use of the CT image is nonenhanced; there is no enhancement of the image or any obvious features. Moreover, it was not feasible to compare CT images of patients acquired at different time points. Besides, the CT images all have a slice thickness of 5 mm. Finally, there may be more accurate description of radiomic features that remains to be discovered. These issues will be addressed in future research.
The results of our study suggest that further studies of CT image phenotypic features are necessary [35], [36]. These can effectively improve the performance of the auxiliary diagnosis of the possibility of metastasis of lung cancer in order to increase the potential of offering timely and reliable treatment.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81227901, 81771924, 81501616, 61231004, 81671851, and 81527805); National Key R&D Program of China (2017YFA0205200, 2017YFC1308700, 2017YFC1308701, 2017YFC1309100); the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (KFJ-SW-STS-160); the Instrument Developing Project of the Chinese Academy of Sciences (YZ201502); the Beijing Municipal Science and Technology Commission (Z161100002616022); the Key Program from the Department of Science and Technology, Sichuan Province, China (2017SZ0052); and the Youth Innovation Promotion Association CAS.
Contributor Information
Hairong Zheng, Email: hr.zheng@siat.ac.cn.
Weimin Li, Email: weimin003@163.com.
Jie Tian, Email: tian@ieee.org.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Edge S, Byrd D, Compton C. Cancer Staging Manual. 2010. AJCC (American Joint Committee on Cancer) [DOI] [PubMed] [Google Scholar]
- 3.Inamura K, Ishikawa Y. Lung cancer progression and metastasis from the prognostic point of view. Clin Exp Metastasis. 2010;27(6):389–397. doi: 10.1007/s10585-010-9313-4. [DOI] [PubMed] [Google Scholar]
- 4.Tamura A, Matsubara O, Yoshimura N, Kasuga T, Akagawa S, Aoki N. Cardiac metastasis of lung cancer. A study of metastatic pathways and clinical manifestations. Cancer. 1992;70(2):437–442. doi: 10.1002/1097-0142(19920715)70:2<437::aid-cncr2820700211>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Hidaka T, Ishii Y, Kitamura S. Clinical features of skin metastasis from lung cancer. Intern Med. 1996;35(6):459. doi: 10.2169/internalmedicine.35.459. [DOI] [PubMed] [Google Scholar]
- 6.Coleman RE. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin Cancer Res. 2006;12(20 Pt 2):6243s. doi: 10.1158/1078-0432.CCR-06-0931. [DOI] [PubMed] [Google Scholar]
- 7.Sugiura Hideshi, Yamada Kenji. Predictors of survival in patients with bone metastasis of lung cancer. Clin Orthop Relat Res. 2008;466(3):729–736. doi: 10.1007/s11999-007-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LB, Gui Yuan LI, Wang JH, Zhou XJ, Zang J, Zhang Q, Chu XY, Geng HC. Study on prediction of metastasis in human non-small cell lung cancer. J Med Postgrad. 2003:168–171. [Google Scholar]
- 9.Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, Bussink J, Monshouwer R, Haibekains B, Rietveld D. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Z, Fang M, Huang Y, He L, Chen X, Liang C, Huang X, Cheng Z, Dong D, Liang C. CT-based radiomics signature for differentiating Borrmann type IV gastric cancer from primary gastric lymphoma. Eur J Radiol. 2017;91:142–147. doi: 10.1016/j.ejrad.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y, Tian J. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recogn. 2017;61(61):663–673. [Google Scholar]
- 13.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278(2):563. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW, Mak RH, Aerts HJ. Radiomic phenotype features predict pathological response in non–small cell lung cancer. Radiother Oncol. 2016;119(3):480–486. doi: 10.1016/j.radonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non–small cell lung cancer. Radiology. 2016;281(3):947. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 16.Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 17.Song JD, Liu ZY, Zhong WZ, Huang YQ, Ma ZL, Dong D, Liang CH, Tian J. Non–small cell lung cancer: quantitative phenotypic analysis of CT images as a potential marker of prognosis. Sci Rep. 2016;6:9. doi: 10.1038/srep38282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balaji G, Elleny P, Kate B, Sabina D, Ken M. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur Radiol. 2012;4:796–802. doi: 10.1007/s00330-011-2319-8. [DOI] [PubMed] [Google Scholar]
- 19.Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA. Non–small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology. 2013;266(1):326–336. doi: 10.1148/radiol.12112428. [DOI] [PubMed] [Google Scholar]
- 20.Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging. 2013;40(1):133. doi: 10.1007/s00259-012-2247-0. [DOI] [PubMed] [Google Scholar]
- 21.Chang CY, Chen SJ, Tsai MF. Application of support-vector-machine-based method for feature selection and classification of thyroid nodules in ultrasound images. Pattern Recogn. 2010;43(10):3494–3506. [Google Scholar]
- 22.Bradley PS, Mangasarian OL. Fifteenth International Conference on Machine Learning. 1999. Feature Selection via Concave Minimization and Support Vector Machines; pp. 82–90. [Google Scholar]
- 23.Thorsten Joachims. Learning to classify text using support vector machines: methods, theory and algorithms. Comput Linguist. 2002;29(4):655–661. [Google Scholar]
- 24.Mangasarian OL. Springer; US: 2003. Data Mining via Support Vector Machines; pp. 91–112. [Google Scholar]
- 25.Wahba G. 1998. Support vector machines, reproducing kernel Hilbert spaces, and randomized GACV. Advances in kernel methods; pp. 69–88. [Google Scholar]
- 26.Gao X, Chu C, Li Y, Lu P, Wang W, Liu W, Yu L. The method and efficacy of support vector machine classifiers based on texture features and multi-resolution histogram from 18 F-FDG PET-CT images for the evaluation of mediastinal lymph nodes in patients with lung cancer ✩. Eur J Radiol. 2015;84(2):312–317. doi: 10.1016/j.ejrad.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Roffo G, Melzi S, Cristani M. Infinite Feature Selection. IEEE Int Conf Comput Vis. 2015:4202–4210. [Google Scholar]
- 28.Tunali I. Palgrave Macmillan; UK: 1990. Path Analysis. [Google Scholar]
- 29.Alwin DF, Hauser RM. The decomposition of effects in path analysis. Am Sociol Rev. 1975;40(1):37–47. [Google Scholar]
- 30.Li Q, Han Y, Fei G, Guo Z, Ren T, Liu Z. IL-17 promoted metastasis of non-small-cell lung cancer cells. Immunol Lett. 2012;148(2):144. doi: 10.1016/j.imlet.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Bai J, Guo C, Sun W, Li M, Meng X, Yu Y, Jin Y, Tong D, Geng J, Huang Q. DJ-1 may contribute to metastasis of non–small cell lung cancer. Mol Biol Rep. 2012;39(3):2697–2703. doi: 10.1007/s11033-011-1024-7. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen BD, Fletcher GP, Patel AC. PET/CT imaging of conus medullaris metastasis from lung cancer. Clin Nucl Med. 2005;30(4):253–256. doi: 10.1097/01.rlu.0000156376.63311.99. [DOI] [PubMed] [Google Scholar]
- 33.Takenaka T, Yano T, Morodomi Y, Ito K, Miura N, Kawano D, Shoji F, Baba S, Abe K, Honda H. Prediction of true-negative lymph node metastasis in clinical IA non–small cell lung cancer by measuring standardized uptake values on positron emission tomography. Surg Today. 2012;42(10):934. doi: 10.1007/s00595-012-0277-7. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Ma C, Yin H, Zhang J. Prediction of ipsilateral mediastinal lymph node metastasis (N2 disease) in patients with lung cancer. Chin J Lung Cancer. 2001;4(2):105–108. doi: 10.3779/j.issn.1009-3419.2001.02.08. [DOI] [PubMed] [Google Scholar]
- 35.Wang Shuo, Zhou Mu, Liu Zaiyi, Liu Zhenyu, Gu Dongsheng, Zang Yali, Dong Di, Gevaert Olivier, Tian Jie. Medical Image Analysis. 2017. Central Focused Convolutional Neural Networks: Developing a Data-driven Model for Lung Nodule Segmentation. (accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B., Tian J., Dong D., Gu D., Dong Y., Zhang L., Lian Z., Liu J., Luo X., Pei S. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2017;23(15):4259–4269. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]