Conspectus
Expansion of the genetic code allows unnatural amino acids (Uaas) to be site-specifically incorporated into proteins in live biological systems, thus enabling novel properties selectively introduced into target proteins in vivo for basic biological studies and for engineering of novel biological functions. Orthogonal components including tRNA and aminoacyl-tRNA synthetase (aaRS) are expressed in live cells to decode a unique codon (often the amber stop codon UAG) as the desired Uaa. Initially developed in E. coli, this methodology has now been expanded in multiple eukaryotic cells and animals. In this account, we focus on addressing various biological challenges for rewriting the genetic code, describing impacts of code expansion on cell physiology, and discussing implications for fundamental studies of code evolution. Specifically, a general method using the type-3 polymerase III promoter was developed to efficiently express prokaryotic tRNAs as orthogonal tRNAs and a transfer strategy was devised to generate Uaa-specific aaRS for use in eukaryotic cells and animals. The aaRSs have been found to be highly amenable for engineering substrate specificity toward Uaas that are structurally far deviating from the native amino acid, dramatically increasing the stereochemical diversity of Uaas accessible. Preparation of the Uaa in ester or dipeptide format markedly increases the bioavailability of Uaas to cells and animals. Nonsense-mediated mRNA decay (NMD), an mRNA surveillance mechanism of eukaryotic cells, degrades mRNA containing a premature stop codon. Inhibition of NMD increases Uaa incorporation efficiency in yeast and C. elegans. In bacteria, release factor one (RF1) competes with the orthogonal tRNA for the amber stop codon to terminate protein translation, leading to low Uaa incorporation efficiency. Contradictory to the paradigm that RF1 is essential, it is discovered that RF1 is actually nonessential in E. coli. Knockout of RF1 dramatically increases Uaa incorporation efficiency and enables Uaa incorporation at multiple sites, making it feasible to use Uaa for directed evolution. Using these strategies, the genetic code has been effectively expanded in yeast, mammalian cells, stem cells, worms, fruit flies, zebrafish, and mice. It is also intriguing to find out that the legitimate UAG codons terminating endogenous genes are not efficiently suppressed by the orthogonal tRNA/aaRS in E. coli. Moreover, E. coli responds to amber suppression pressure promptly using transposon insertion to inactivate the introduced orthogonal aaRS. Persistent amber suppression evading transposon inactivation leads to global proteomic changes with a notable up-regulation of a previously uncharacterized protein YdiI, for which an unexpected function of expelling plasmids is discovered. Genome integration of the orthogonal tRNA/aaRS in mice results in minor changes in RNA transcripts but no significant physiological impairment. Lastly, the RF1 knockout E. coli strains afford a previously unavailable model organism for studying otherwise intractable questions on code evolution in real time in the laboratory. We expect that genetically encoding Uaas in live systems will continue to unfold new questions and directions for studying biology in vivo, investigating the code itself, and reprograming genomes for synthetic biology.
Graphical abstract
INTRODUCTION
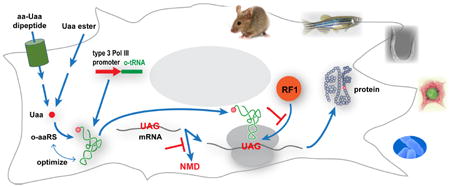
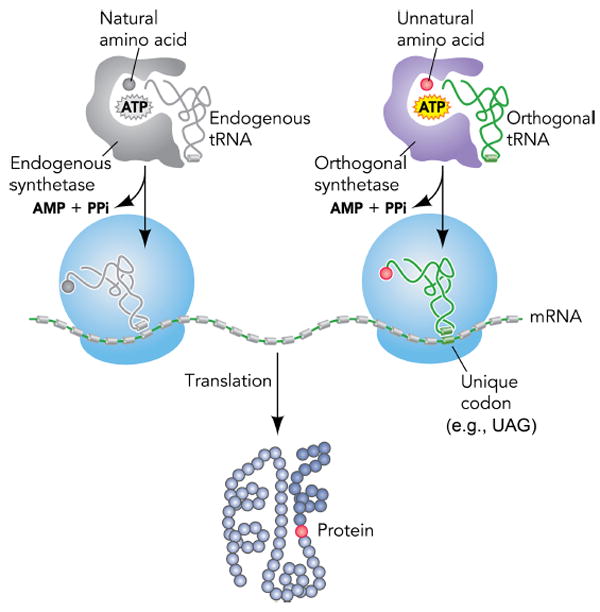
The genetic code uses 61 codons to specify 20 common amino acids and 3 stop codons for termination in protein translation. This code is preserved in virtually all life forms on earth with minor variations.1 In recent years, the genetic code has been expanded to encode unnatural amino acids (Uaas) by introducing into live cells a new tRNA/aminoacyl-tRNA synthetase (aaRS) pair,2,3 which is orthogonal to endogenous tRNA/aaRS pairs (Figure 1). The orthogonal aaRS is evolved to charge a desired Uaa onto the orthogonal tRNA, and the resultant Uaa-tRNA then incorporates the Uaa into proteins in response to a unique codon, such as the amber stop codon UAG. The first expansion of the genetic code was achieved in E. coli in 2001.4-7 Subsequently, this methodology has been proven generally applicable to various cells and organisms, and over 150 Uaas have now been incorporated using this approach.8,9 In this account, we focus on our efforts in expanding the genetic code in various cells and animals, with emphases on biological challenges of such engineering and its impacts on the biological system. For Uaa applications and other aspects, readers are referred to these papers.1,3,8-16
Figure 1.
A general method for expansion of the genetic code to incorporate Uaas into proteins in live cells and animals.6 Adapted with permission from ref.57 . Copyright 2008 American Physiological Society.
Developing a General Method for Expressing Orthogonal tRNAs in Eukaryotic Cellsand Animals
To expand the genetic code in eukaryotic cells and animals, a critical challenge is how to functionally express an orthogonal tRNA, often derived from prokaryotes, in eukaryotic cells. Prokaryotic tRNAs are transcribed through promoters upstream of the tRNA gene, whereas eukaryotic tRNAs are transcribed through promoter elements within the tRNA known as the A- and B-box (Figure 2A). The A- and B-box elements are conserved among eukaryotic tRNAs but are often absent in prokaryotic tRNAs.
Figure 2.
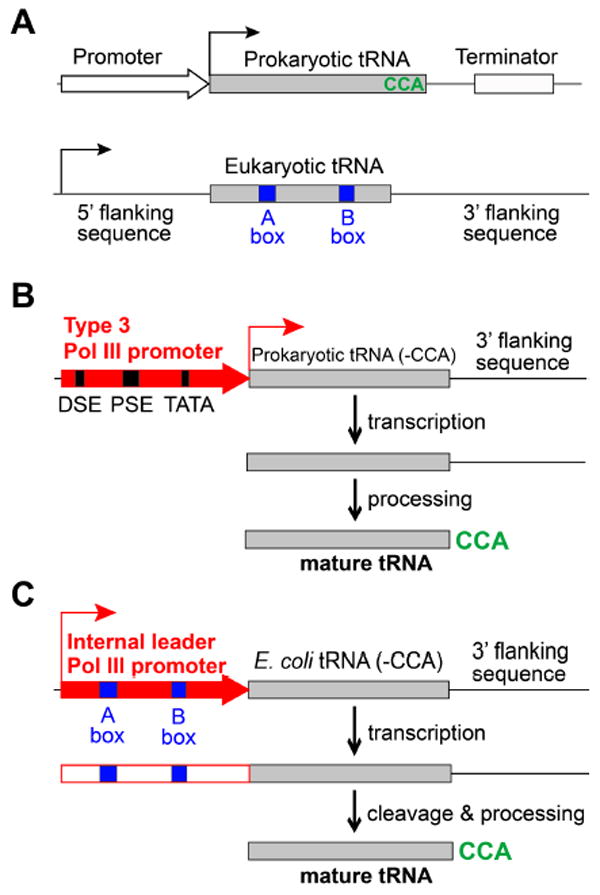
General methods for efficient expression of prokaryotic tRNA in eukaryotic cells and animals. (A) Different gene elements for tRNA transcription in prokaryotic and eukaryotic cells. (B) The type-3 Pol III promoter (e.g, H1, U6) drives functional expression of prokaryotic tRNAs in mammalian cells and animals.18 (C) The internal leader Pol III promoter (e.g., SNR52, RPR1) drives functional expression of prokaryotic tRNAs in yeast.28
Although rarely a prokaryotic tRNA, e.g., the B. stearothermophilus tRNATyr , happens to contain the consensus sequences and can be expressed in mammalian cells using multiple repeats,17 we sought for a general method to efficiently express in eukaryotic cells any tRNA regardless of its sequence. The type-3 Pol III promoter has promoter elements exclusively upstream of the coding sequence, does not require intragenic elements for transcription, and has a well-defined transcriptional initiation site. We reasoned and firstly proved that a type-3 Pol III promoter in combination with a 3’-flanking sequence can drive the functional expression of prokaryotic tRNAs in various mammalian cells (Figure 2B).18 Such a tRNA-expression cassette can also be used in multiple repeats to achieve optimal tRNA expression level.19-21 The type-3 Pol III mammalian promoter includes H1, U6 snRNA, 7SK, and MRP/7-2 promoters. We showed that this method can be used to express different tRNAs in mammalian cells, including and from E. coli 18 and from Methanosarcina.22 In addition, we applied this approach successfully in various cells and animals, including mammalian cell lines,18 primary neurons,18 stem cells,19 C. elegans,23 mouse,24,25 and zebrafish.25 Others have later applied this approach in plant26 and fruit fly.27 This general method has now been widely adopted for expressing orthogonal tRNAs in eukaryotic systems for code expansion.
Using the similar principle, we also identified two external Pol III yeast promoters, the RPR1 and SNR52 promoters, to efficiently express prokaryotic tRNAs in yeast.28,29 These promoters feature an internal leader sequence, which contains the consensus eukaryotic A- and B-box sequences (Figure 2C). When placed upstream of the E. coli tRNA, it drives the transcription of a primary RNA consisting of the promoter and the tRNA. The promoter is then cleaved yielding the mature tRNA. In comparison to the old approach of using SUP4 5’-flanking sequence, these promoters increased the activities of E. coli and in yeast by 6 to 9 fold.28
Generating Uaa-specific Synthetase for Use in Mammalian Cellsand Animals
A second challenge for code expansion in mammalian cells and animals is how to generate an orthogonal synthetase specific for the Uaa. Changing the synthetase substrate specificity from its cognate amino acid to a Uaa had been a road blocker, and the breakthrough was firstly made in E. coli by generating large mutant synthetase libraries (>109 members) followed by high-throughput selection or screen.3,6 However, it is difficult to generate huge mutant libraries in mammalian cells due to low transfection efficiency.
A transfer strategy is devised to circumvent this issue. Since the E. coli pair is orthogonal in mammalian cells as well as in yeast, and the translational machinery of yeast is homologous to that of higher eukaryotes, it should be feasible to evolve the E. coli synthetase in yeast and transfer the evolved tRNA/synthetase pairs for use in mammalian cells. This idea worked in mammalian cell lines as well as primary neurons, and other E. coli pairs such as the can also be used this way.18 In combination with the efficient tRNA expression discussed above, the transfer strategy has enabled various Uaas to be genetically encoded in mammalian cells,18 primary neurons,18 stem cells,19 and animals.23-25
Another creative solution reported by Iraha et al. is to disrupt the E. coli tRNATyr/TyrRS genes in E. coli genome and functionally replace them with the orthogonal counterpart of M. jannaschii, after which the E. coli can be expressed on a plasmid for directed evolution in the engineered E. coli.30 The evolved E. coli TyrRS mutant can then be transferred for use in mammalian cells. Although it hasn’t been widely used, evolving an E. coli aaRS in engineered E. coli is attractive, because the most efficient selection methods have been established in E. coli.3
Optimizing aaRS Recognition of tRNACUA
To decode the amber stop codon UAG, the orthogonal tRNA has to change its anticodon to CUA, which may decrease the affinity of the resultant tRNACUA toward the synthetase, since anticodon is a major recognition element of most synthetases. By mutating the anticodon-binding region of E. coli TyrRS, we identified an EwtYRS (D265R) that shows ~2 fold higher activity toward the E. coli than the WT TyrRS. This optimization can be transplanted to multiple TyrRS-derived synthetases specific for different Uaas to improve their incorporation efficiency in mammalian cells (1.6 to 5.2 fold).31
Expanding Uaa Specificity of aaRS beyond Analogs of Native Substrate
Most evolved aaRSs aminoacylate Uaas that are chemically similar to the native substrate of the WT aaRS, limiting stereochemical diversity of Uaas accessible. For instance, most Uaas incorporated by mutant pairs retain the core Lys moiety and the Nε-carbonyl group of Pyl (Figure 3A). To break this bottleneck, we found that, by mutating a conserved “gate keeper” residue (N346) of PylRS that interacts with the Nε-carbonyl and the α-amino group of amino acid substrate, PylRS can be evolved to charge Uaas structurally similar to Phe or Tyr (Figure 3B).22,32,33 A new “small-intelligent” mutagenesis approach, which uses a single codon for each amino acid, was then devised to generate mutant PylRS libraries enabling a greater number of residues simultaneously randomized, from which mutants were identified to incorporate Uaas with bulky conjugated rings34 and even Uaas containing long azobenzene side chains (Figure 3C).35,36 These results and data from other groups37 suggest unexpectedly high flexibility in engineering aaRS substrate specificity. As the can be used in both prokaryotic and eukaryotic cells, these evolved PylRS mutants enable diverse Uaas to be incorporated in various cells and organisms.
Figure 3.
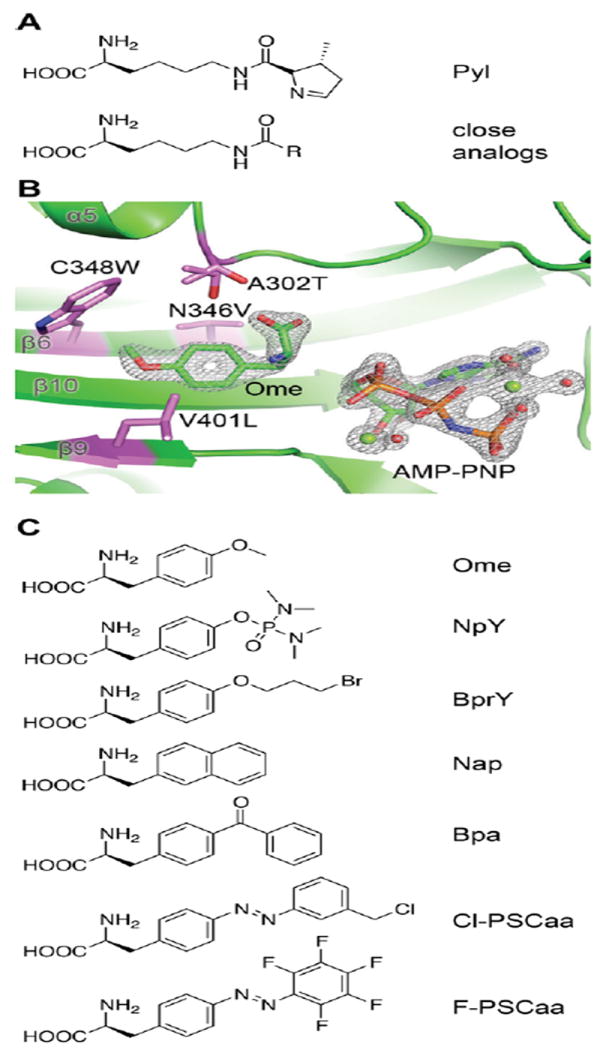
Aminoacyl-tRNA synthetase can be evolved to charge Uaas that are dramatically different from the native amino acid substrate. (A) Structure of Pyl (native substrate of PylRS) and close Uaa analogs. (B) Crystal structure of an M. mazei PylRS mutant transformed to charge Uaa o-methyl-L-tyrosine (Ome).22 Adapted with permission from ref. 22. Copyright 2011 American Chemical Society. (C) Representative Uaas that can be charged by mutant synthetases evolved from PylRS.
Besides aaRS recognition, efficient Uaa incorporation also depends on the affinity of Uaa-tRNA toward the elongation factor and ribosome. Engineering the tRNA, elongation factor and ribosome can fine tune their affinity and thus allow more diverse Uaas to be genetically encoded.38,39 To date a variety of functional groups have been installed onto the Phe/Tyr- or Lys-scaffold and incorporated into proteins as Uaas.2,3,8-10,16 Uaas with small or charged (≥2) side chains are difficult to incorporate and await more efforts.32
Increasing Bioavailability of Uaa
Uaa bioavailability inside cells is a prerequisite for evolving Uaa-specific aaRSs and Uaa incorporation. Uaas structurally close to canonical amino acids may enter cells through endogenous amino acid transporters or pathways, yet those deviating significantly or highly charged (e.g., phosphorylated Uaas) may not. We reasoned that masking the carboxyl group of Uaa with an ester would convert the Uaa zwitterion into a protonated weak base, which has a higher percentage of neutral form and increased lipophilicity for crossing the membrane (Figure 4A). Once inside the cell, intracellular esterases will cleave the ester to regenerate the Uaa. Indeed, we found that the acetoxymethyl ester of Uaa DanAla increased its intracellular concentration by 31-fold, and the Uaa-ester was hydrolyzed into Uaa inside mammalian cells within 1 h.40 Use of Uaa-ester increases the Uaa incorporation efficiency by 4-fold, and requires 75% less Uaa in growth media.
Figure 4.
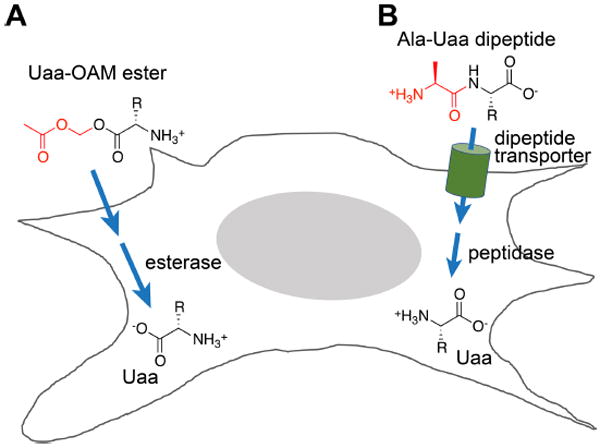
Two strategies to increase Uaa bioavailability in mammalian cells, C. elegans, and mouse: prepare the Uaa in ester (A) or in dipeptide (B) form .
C. elegans has a protective cuticle that excludes many compounds from internalization. We reasoned that Uaas might be taken in as food via the intestinal route and trafficked to multiple tissues, although intestinal transporters may not recognize Uaas with drastically different structures. Indeed, we found that O-methyl-L-tyrosine (OmeY), structurally similar to Tyr, could be incorporated into proteins in muscle by feeding worms with OmeY, whereas similarly fed DanAla was sequestered in intestinal cells.23 We then developed a dipeptide strategy to deliver the Uaa (Figure 4B).23 PEPT-1 and PEPT-2 are dipeptide transporters present on many C. elegans cell surface. We reasoned that an Ala-Uaa dipeptide should enter cells through dipeptide transporters and subsequently be hydrolyzed by cellular peptidases to generate the Uaa. Indeed, when Ala-DanAla dipeptide was fed to worms, DanAla was found to be incorporated into proteins in various tissues of C. elegans.23
When incorporating Uaas in mouse brain in vivo, we also found that the use of Uaa-Ala dipeptide greatly increases Uaa bioavailability in brain cells,24 as PEPT-2 is highly expressed in rodent brain. The Uaa-Ala dipeptide was injected into the lateral or third ventricle of the mouse brain, and the corresponding Uaa was successfully incorporated into proteins in neocortex, thalamus and hypothalamus, with efficiencies dramatically higher than injecting the Uaa.24,41
Inactivating NMD to Stabilize UAG-containing mRNA
Eukaryotic cells have an mRNA surveillance mechanism, nonsense-mediated mRNA decay (NMD), to identify mRNAs containing premature stop codons and target the mRNA for rapid degradation. Use of the amber stop codon for encoding Uaa will subject the mRNA to NMD, thus decreasing protein yield (Figure 5). We were the first to investigate whether inactivation of NMD would preserve the stability of UAG-containing mRNA and thus enhance Uaa incorporation efficiency.28 An NMD-deficient yeast strain was successfully generated by knocking out UPF1 gene, an essential component for NMD.28,29 This upf1∆ strain increased Uaa incorporation more than 2-fold than the WT yeast.
Figure 5.
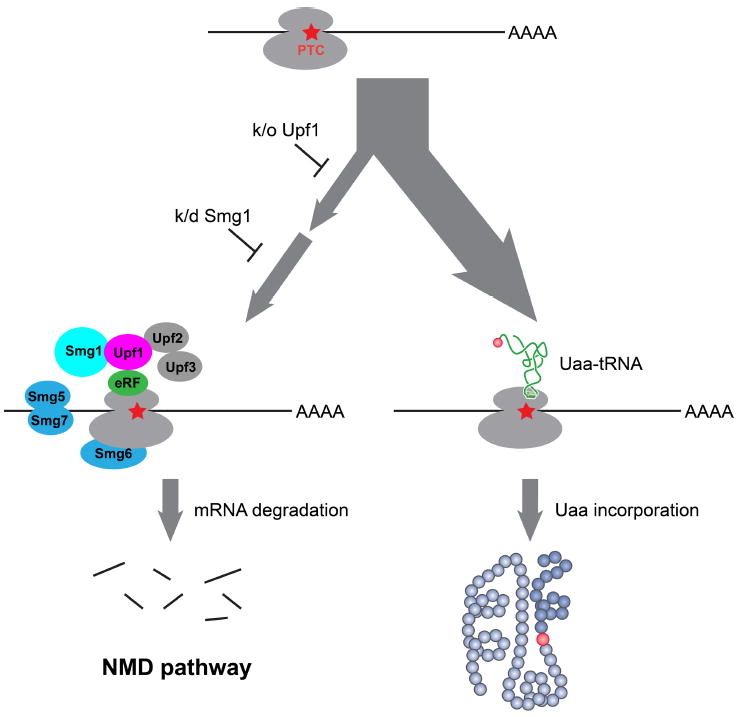
Inhibition of NMD surveillance increases Uaa incorporation efficiency in eukaryotic cells and animals. When a premature termination codon (PTC) is encountered during translation, eRF can recruit Upf and Smg proteins leading to mRNA degradation. Knock out or knock down of essential NMD factors enhances mRNA stability and Uaa incorporation efficiency.
We also demonstrated that NMD has a similar effect on Uaa incorporation in C. elegans.23 After knocking down the NMD component smg-1 in C. elegans using RNA interference, 5.6 fold more Uaa-containing protein was purified than from control worms.
Knocking out RF1 to Increase Uaa Incorporation Efficiency and Enable Multiple Site Incorporation
Class I release factors (RFs) recognize stop codons to terminate translation. While eukaryotes and archaea use a single RF to recognize all three stop codons, bacteria use two: RF1 for UAA/UAG and RF2 for UAA/UGA (Figure 6A). Synthetically recoding a genome may afford new properties through encoding Uaas. Although Uaas can be specified with the UAG codon, RF1 makes UAG ambiguous, being a stop signal and a Uaa simultaneously. RF1 competition for UAG also limits Uaa incorporation at a single site with low efficiency (10-30%); additional UAG codons decrease protein yields precipitously, preventing effective Uaa use at multiple sites.
Figure 6.
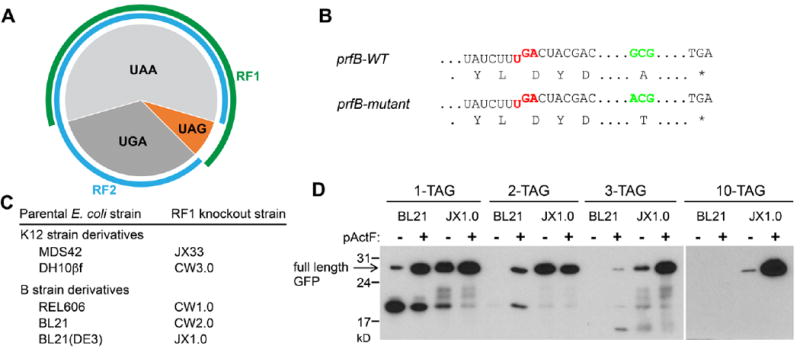
RF1 knockout enables multisite incorporation of Uaa. (A) Bacteria use RF1 to recognize UAG/UAA and RF2 to recognize UGA/UAA. UAA is the dominant while UAG is the least used stop codon in E. coli. (B) Features of the prfB gene encoding E. coli RF2. K12 strains have the mutant prfB. (C) List of RF1 knockout strains. (D) Incorporation of Uaa p-acetyl-L-phenylalanine (pActF) into GFP at multiple sites in the RF1 knockout strain JX1.0. Adapted with permission from ref. 44. Copyright 2012 American Chemical Society.
To fully reassign UAG to a sense codon, it is imperative to knock out RF1. However, no free-living bacterium has been found lacking either RF1 or RF2. RF1 has been reported essential for E. coli since 1980s.42 Contradictory to this paradigm, we discovered that the apparent essentiality of RF1 was caused by an RF2 mutation (A264T) in K12 strains, which makes RF2 inefficient in terminating the UAA stop codon (Figure 6B).43 As UAA is the dominant stop codon in E. coli, inefficient termination at UAA may result in lethality, preventing RF1 knockout. We thus fixed the A246T mutation and removed an in-frame UGA stop codon in RF2 gene that autoregulates RF2 expression level. After such fixation, RF1 was readily knocked out from E. coli.43 In addition, we further found that, in all E. coli B strains that contain the WT RF2 gene, RF1 can be knocked out unconditionally (Figure 6C).44 Moreover, when the A246T mutation was reverted in K12 strains, RF1 can be knocked out as well.
These results demonstrate that RF1 is nonessential in E. coli for the first time. A series of RF1-knockout E. coli strains were generated.44 These strains allow Uaa incorporation at the UAG codon with efficiency close to those of natural amino acids, and more importantly, enable simultaneous Uaa incorporation at multiple sites (Figure 6D). As many as 10 UAG sites, either spreading out or in tandem, have been shown to be decoded as Uaas in such RF1-knockout strains. These unprecedented properties make the RF1-knockout bacterium a unique host to efficiently harness Uaa in the same manner as natural amino acids for evolving new protein properties and biological activities.
Expanding the Genetic Code in Various Cellsand Animals
Mammalian cells
In 2001, the principle of genetically encoding Uaas using an orthogonal tRNA/synthetase pair was firstly demonstrated in E. coli (Figure 7).6 For biological and biomedical research, mammalian cells and animal models are desired to effectively study physiological and pathological processes in vivo. Two impediments had stymied intensive efforts to incorporate Uaas in mammalian cells for many years: the difficulty in expressing prokaryotic tRNAs and the infeasibility of evolving Uaa-specific synthetases in mammalian cells. We solved these problems by harnessing the type-3 polymerase III promoter and a transfer strategy, respectively. These new approaches enabled us to genetically encode different Uaas in various mammalian cell lines and primary cells such as neurons.18 Furthermore, we markedly increased Uaa incorporation efficiency by optimizing tRNA/synthetase affinity31 and enhancing cellular uptake of Uaas with chemical modifications.40
Figure 7.
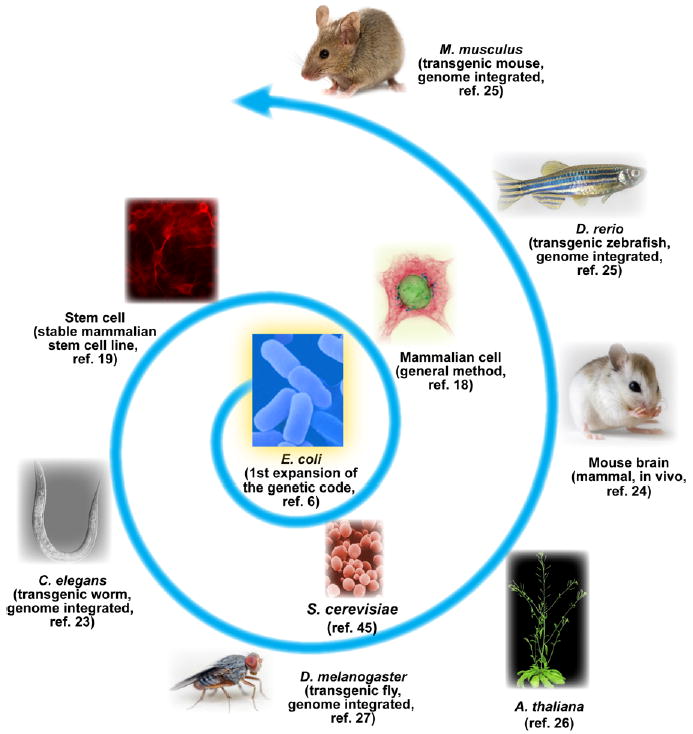
Expansion of the genetic code in various cells and organisms.
Yeast
The low Uaa incorporation efficiency in yeast had prevented effective applications. We found that orthogonal E. coli tRNAs expressed in yeast using the conventional method are not competent in translation. To solve this problem we developed a new expression method for tRNA by using internal leader Pol III yeast promoters (RPR1 and SNR52 promoters).28 In addition, we demonstrated for the first time that disabling NMD markedly increases Uaa incorporation in eukaryotic cells. These new strategies increased the yield of Uaa-containing proteins from tens of micrograms45 to tens of milligrams per liter in yeast.28 This dramatic improvement has enabled the community to start using Uaas in yeast for research.46
Stem cells
Stem cell lines stably incorporating Uaas provide not only novel means for studying stem cell biology but also an attractive source of Uaa-encoding mature cells (e.g., neurons) that are otherwise difficult or expensive to procure. Uaa-incorporation methods for mature cells lack sufficient temporal resolution for studying the long-term differentiation process of stem cells. In addition, it was unknown whether Uaa incorporation would perturb stem cell differentiation. We developed a lentiviral-based method and achieved stable Uaa incorporation in neural stem cells.19 These stable stem cell lines maintain the ability to incorporate Uaa throughout differentiation and in the differentiated neurons. No notable interference with differentiation by incorporating Uaas was observed. This work reports the first stable mammalian stem cell line for genetically incorporating Uaas.19
Invertebrate animals
Research into development, intercellular communication, differentiation, cancerous transformation, and various other signaling processes necessitates multicellular organisms. We developed new strategies to address every challenging aspect of Uaa incorporation in C. elegans, a multicellular model organism extensively used for studying biology and human diseases.23 The type-3 Pol III C. elegans promoter, rpr-1 promoter, was identified to efficiently express orthogonal E. coli tRNAs in worms, and worms were fed with either Uaa or Ala-Uaa dipeptide for various tissue bioavailability. We have generated a series of stable transgenic worms capable of genetically encoding Uaas using different tRNA/aaRS pairs.
The transgenic C. elegans we generated have the orthogonal tRNA/aaRS and reporter genes all stably integrated into the chromosome,23 whereas in the study by Greiss et al. these genes were transiently expressed on extrachromosomal arrays to be maintained by antibiotics.47 Extrachromosomal arrays overexpress reporter genes, result in nonspecific and Uaa-independent amber codon readthrough, and show expression inconsistency between animals and within a single animal. The rate of generating transgenic worms using extrachromosomal array is very low (1–5 per hundreds of worms), and only 5% of the animals in the established line express the reporter. In contrast, genome-integration generates transgenic worms with a success rate of 25–50%, and the transgenes are transmitted in 100% efficiency with clean genetic uniformity. Each of these transgenic worms showed Uaa-dependent responses reproducibly. Therefore, a transgenic animal effective for genetic code expansion should have the orthogonal tRNA/aaRS pair stably integrated into the genome with genetic inheritance and uniformity,23 rather than transiently present in extrachromosome showing genetic instability and mosaicism.47
Following this principle, a second transgenic invertebrate animal, the fruit fly Drosophila melanogaster was later generated by Bianco et al., in which a type-3 pol III promoter U6 was similarly used to express .27 The model plant Arabidopsis thaliana has also been transformed by Li et al. to incorporate Uaas, wherein a type-3 Pol III Arabidopsis promoter 7SL4 was used to express .26
Vertebrate animals
Vertebrate animals share a higher degree of genetic homology with human than invertebrates. In particular, mice are widely used experimental mammals for developmental, physiological, neurological, and pathological studies. To explore the possibility of genetically encoding Uaas in mouse in vivo, we began by introducing the orthogonal tRNA/aaRS genes into embryonic mouse brain through in utero electroporation.24 Genes containing the orthogonal E. coli (driven by type-3 Pol III promoter H1) and the CmnRS were delivered and electroporated into brain cells of live embryonic mouse. The photocaged Uaa Cmn was made bioavailable by injecting the Cmn-Ala dipeptide into the brain ventricle. Cmn was site-specifically incorporated into the K+ channel Kv1.2 in neurons of live mouse brain. Upon light stimulation, Cmn was photolyzed into Cys and Kv1.2 channel was selectively activated, immediately silencing neuron firing in brain tissues.24,41 This work represents the first success of Uaa incorporation in mammals in vivo, which also enables optical regulation of neuronal protein function selectively in its native habitat.
An exciting stride is the recent generation of transgenic vertebrates capable of encoding Uaas. Chen et al. reported the success in creating the first transgenic zebrafish.25 Zebrafish is a non-mammalian species that has conserved cellular mechanisms with mammals, and is a convenient vertebrate model for live imaging. A type-3 Pol III promoter, the human U6 promoter, was used to drive the expression of bacterial B. stearothermophilus , and the AzFRS specific for Uaa AzF was driven by the zebrafish ubiquitin promoter. This construct and mRNA of Tol2 transposase were co-injected into embryos to generate the transgenic zebrafish, which have stable integration of transgene in germline. By incubating the embryos in water containing 2.5 mM AzF, AzF was incorporated into various cell types of the fish in vivo, including mesenchymal, notochord and muscle cells.
The ultimate challenge is whether a transgenic mouse can be generated to have code expansion system-wide. Although transient mouse transgenesis is feasible and tolerated as initially demonstrated in mouse brain,24 it remained unclear whether the biological complexity of mouse allows the introduction, maintenance, and transmission of the genetic material for code expansion. The breakthrough was recently achieved:25 Again, the orthogonal B. stearothermophilus was driven by the type-3 Pol III U6 promoter, and AzFRS driven by the ubiquitous human EF1α promoter. The pronuclei injection approach was used to allow random insertion of the transgene into mouse genome. The transgene was transmitted stably and efficiently from founders to subsequent generations, and an average of 15 copies of transgenes were found in the transgenic line. Uaa incorporation was verified in primary cells derived from the adult transgenic mice, including neurons and bone marrow cells.
What happens to Legitimate UAG codons?
Although UAG is the least-used stop codon in E. coli, it still terminates ~7% of the total genes. A long-standing question is how endogenous genes ending with the UAG are affected by code expansion. After examining the expression of endogenous UAG-ending E. coli genes, we found that the amber suppressor does not efficiently incorporate its cognate Tyr at the TAG sites in the presence of RF1.43 This surprising finding suggests there may be unknown mechanisms preventing legitimate stop codons from being suppressed. It also explains why E. coli can tolerate the orthogonal amber suppressor tRNA/synthetase for Uaa incorporation without causing notable adverse effect. However, we also discovered that, upon RF1 knockout in E. coli, the UAG codon of endogenous genes is then efficiently suppressed by the orthogonal tRNA/synthetase pair, and such suppression leads to a slower growth phenotype.43,44
Organism Response to Amber Suppression
How would suppression of the stop codon affect cellular physiology? And how would the host cell and organism respond to such perturbation? To address these questions, we introduced into E. coli the orthogonal amber suppressor pair,5 which decodes the UAG codon as Tyr in high efficiency, and monitored cell response. We found that E. coli promptly counteracts the strong amber suppression pressure through transposon inactivation.48 Within three passages in liquid culture, most of the cells have the orthogonal TyrRS gene disrupted by E. coli transposons, losing the ability to suppress the UAG codon.
To investigate the effect of long-term amber suppression on cells, we grew E. coli cells harboring the pair on plate and continued to passage cells that evade transposon inactivation. After ~500 generations, proteomic change of cells was characterized quantitatively. Over 30 proteins were downregulated and 21 proteins up-regulated. The most remarkable change identified was a hypothetic protein, YdiI, showing 16-fold increase. Intriguingly, we discovered that this uncharacterized YdiI protein has an unexpected function of expelling plasmids from E. coli, helping cells to eliminate amber suppression pressure.48 These results reveal that E. coli has multiple creative ways to respond and adapt to amber suppression.
The impact of the orthogonal amber suppressor tRNA/synthetase gene in live transgenic mice has also been evaluated.25 Various tissues including brain, heart, liver, colon, kidney, skeletal muscle and lung were analyzed but showed no detectable morphological changes. The liver transcriptomes of the transgenic mice showed minor changes in 97 upregulated and 47 downregulated transcripts, mostly related to metabolism, but these changes did not impair the liver function. The transgenic mice were then fed with 30 mg/mL of Uaa AzF for 10 days, and no obvious defects were observed. These results suggest that genome integration of the orthogonal tRNA/synthetase genes in mice did not cause significant physiological impairment; whether long-term Uaa feeding would cause problems awaitsfurther study.
CONCLUSIONS AND PERSPECTIVES
Since the first expansion of the genetic code to include Uaas in E. coli in 2001,6,7 the methodology has been proven generally applicable in various cells and animals. It has been pleasantly surprising to witness the flexibility of aminoacyl-tRNA synthetases, after directed evolution, to charge a wide range of Uaas, and of various cells and organisms to tolerate artificial code expansion and rewriting. Genetically encoding Uaas in live systems is providing novel avenues for studying biology both in vitro and in vivo.
Progress made in the past 16 years keeps unfolding exciting questions and new directions. Engineering the genetic code may afford ample opportunities to investigate the genetic code itself, from the perspective of fundamental code evolution to recoding for synthetic biology. For instance, the RF1 knockout strains now afford a previously unavailable model organism for studying otherwise intractable questions on code evolution in real time in the laboratory,43,44,49 such as whether the altered code can eventually be fixed, how long this process would take, and what physiological changes will accompany such adaptations. Answers to these questions would also provide rare empirical data to guide genome recoding for specific synthetic purposes and generation of completely orthogonal genetic systems. In addition, we shall be able to set aside additional triplet codons through genome editing or to use unnatural base pairs ultimately for simultaneous coding of multiple different Uaas.50,51 Once we are able to harness the Uaa in live systems as nature does to the common amino acids, i.e., in highly efficient, diverse, and autonomous (genetically stable and Uaa self-sustained) manner, we will then be poised to evolving desired new biological functions and even systems. Moreover, the concept of code expansion and the recently introduced bioreactive Uaas52 may afford novel approaches for biotherapeutical applications.53-56
Acknowledgments
Funding
Grants from the National Institutes of Health (1DP2OD004744, 1R01GM118384, 1RF1MH114079) and California Institute for Regenerative Medicine (RN1-00577) are acknowledged.
Biography
Lei Wang graduated from Peking University with B.S. and M.S. and received Ph.D. from UC Berkeley in chemistry. After postdoctoral research at UCSD, he joined the faculty of the Salk Institute for Biological Studies in 2005 and moved to UCSF in 2014.
References
- 1.Ling J, O–Donoghue P, Soll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat Rev Microbiol. 2015;13:707–721. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Schultz PG. Expanding the genetic code. Chem Commun. 2002:1–11. doi: 10.1039/b108185n. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Magliery TJ, Liu DR, Schultz PG. A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J Am Chem Soc. 2000;122:5010–5011. [Google Scholar]
- 5.Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 7.Wang L. Expanding the genetic code. Science. 2003;302:584–585. doi: 10.1126/science.302.5645.584. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Chen PR. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat Chem Biol. 2016;12:129–137. doi: 10.1038/nchembio.2024. [DOI] [PubMed] [Google Scholar]
- 12.Nikic I, Lemke EA. Genetic code expansion enabled site-specific dual-color protein labeling: superresolution microscopy and beyond. Curr Opin Chem Biol. 2015;28:164–173. doi: 10.1016/j.cbpa.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 13.Huber T, Sakmar TP. Chemical biology methods for investigating G protein-coupled receptor signaling. Chem Biol. 2014;21:1224–1237. doi: 10.1016/j.chembiol.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr Opin Chem Biol. 2010;14:774–780. doi: 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanagisawa T, Umehara T, Sakamoto K, Yokoyama S. Expanded genetic code technologies for incorporating modified lysine at multiple sites. ChemBioChem. 2014;15:2181–2187. doi: 10.1002/cbic.201402266. [DOI] [PubMed] [Google Scholar]
- 16.Wang L. Genetically encoding new bioreactivity. N Biotechnol. 2017;38:16–25. doi: 10.1016/j.nbt.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio K, Saito K, Shirouzu M, Hirao I, Yokoyama S. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen B, Xiang Z, Miller B, Louie G, Wang W, Noel JP, Gage FH, Wang L. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cells. 2011;29:1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coin I, Katritch V, Sun T, Xiang Z, Siu FY, Beyermann M, Stevens RC, Wang L. Genetically encoded chemical probes in cells reveal the binding path of Urocortin-I to CRF class B GPCR. Cell. 2013;155:1258–1269. doi: 10.1016/j.cell.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coin I, Perrin MH, Vale WW, Wang L. Photo-cross-linkers incorporated into G-protein-coupled receptors in mammalian cells: a ligand comparison. Angew Chem Int Ed Engl. 2011;50:8077–8081. doi: 10.1002/anie.201102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takimoto JK, Dellas N, Noel JP, Wang L. Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parrish AR, She X, Xiang Z, Coin I, Shen Z, Briggs SP, Dillin A, Wang L. Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem Biol. 2012;7:1292–1302. doi: 10.1021/cb200542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang JY, Kawaguchi D, Coin I, Xiang Z, O–Leary DD, Slesinger PA, Wang L. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 2013;80:358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Ma J, Lu W, Tian M, Thauvin M, Yuan C, Volovitch M, Wang Q, Holst J, Liu M, Vriz S, Ye S, Wang L, Li D. Heritable expansion of the genetic code in mouse and zebrafish. Cell Res. 2017;27:294–297. doi: 10.1038/cr.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Zhang H, Sun Y, Pan Y, Zhou J, Wang J. Expanding the genetic code for photoclick chemistry in E. coli, mammalian cells, and A. thaliana. Angew Chem Int Ed Engl. 2013;52:9700–9704. doi: 10.1002/anie.201303477. [DOI] [PubMed] [Google Scholar]
- 27.Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Expanding the genetic code of Drosophila melanogaster. Nat Chem Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Wang L. Genetic incorporation of unnatural amino acids into proteins in yeast. Methods Mol Biol. 2012;794:199–213. doi: 10.1007/978-1-61779-331-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iraha F, Oki K, Kobayashi T, Ohno S, Yokogawa T, Nishikawa K, Yokoyama S, Sakamoto K. Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion. Nucleic Acids Res. 2010;38:3682–3691. doi: 10.1093/nar/gkq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takimoto JK, Adams KL, Xiang Z, Wang L. Improving orthogonal tRNA-synthetase recognition for efficient unnatural amino acid incorporation and application in mammalian cells. Mol Biosyst. 2009;5:931–934. doi: 10.1039/b904228h. [DOI] [PubMed] [Google Scholar]
- 32.Hoppmann C, Wong A, Yang B, Li S, Hunter T, Shokat KM, Wang L. Site-specific incorporation of phosphotyrosine using an expanded genetic code. Nat Chem Biol. 2017;13:842–844. doi: 10.1038/nchembio.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Z, Lacey VK, Ren H, Xu J, Burban DJ, Jennings PA, Wang L. Proximity-enabled protein crosslinking through genetically encoding haloalkane unnatural amino acids. Angew Chem Int Ed Engl. 2014;53:2190–2193. doi: 10.1002/anie.201308794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacey VK, Louie GV, Noel JP, Wang L. Expanding the library and substrate diversity of the pyrrolysyl-tRNA synthetase to incorporate unnatural amino acids containing conjugated rings. ChemBioChem. 2013;14:2100–2105. doi: 10.1002/cbic.201300400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoppmann C, Lacey VK, Louie GV, Wei J, Noel JP, Wang L. Genetically encoding photoswitchable click amino acids in Escherichia coli and mammalian cells. Angew Chem Int Ed Engl. 2014;53:3932–3936. doi: 10.1002/anie.201400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoppmann C, Maslennikov I, Choe S, Wang L. In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. J Am Chem Soc. 2015;137:11218–11221. doi: 10.1021/jacs.5b06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang YS, Fang X, Wallace AL, Wu B, Liu WR. A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J Am Chem Soc. 2012;134:2950–2953. doi: 10.1021/ja211972x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrader JM, Chapman SJ, Uhlenbeck OC. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci U S A. 2011;108:5215–5220. doi: 10.1073/pnas.1102128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, Soll D. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takimoto JK, Xiang Z, Kang JY, Wang L. Esterification of an unnatural amino acid structurally deviating from canonical amino acids promotes its uptake and incorporation into proteins in mammalian cells. ChemBioChem. 2010;11:2268–2272. doi: 10.1002/cbic.201000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang JY, Kawaguchi D, Wang L. Optical control of a neuronal protein using a genetically encoded unnatural amino acid in neurons. J Vis Exp. 2016 doi: 10.3791/53818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rydén S, Isaksson L. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 43.Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson DB, Wang C, Xu J, Schultz MD, Schmitz RJ, Ecker JR, Wang L. Release factor one is nonessential in Escherichia coli. ACS Chem Biol. 2012;7:1337–1344. doi: 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 46.Majmudar CY, Lee LW, Lancia JK, Nwokoye A, Wang Q, Wands AM, Wang L, Mapp AK. Impact of nonnatural amino acid mutagenesis on the in vivo function and binding modes of a transcriptional activator. J Am Chem Soc. 2009;131:14240–14242. doi: 10.1021/ja904378z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greiss S, Chin JW. Expanding the genetic code of an animal. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, Sun T, Xu J, Shen Z, Briggs SP, Zhou D, Wang L. Response and adaptation of Escherichia coli to suppression of the amber stop codon. ChemBioChem. 2014;15:1744–1749. doi: 10.1002/cbic.201402235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson DB, Wang L. Imprints of the genetic code in the ribosome. Proc Natl Acad Sci U S A. 2010;107:8298–8303. doi: 10.1073/pnas.1000704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Jr, Romesberg FE. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, Zhou M, Singh K, Napolitano MG, Moosburner M, Shrock E, Pruitt BW, Conway N, Goodman DB, Gardner CL, Tyree G, Gonzales A, Wanner BL, Norville JE, Lajoie MJ, Church GM. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 52.Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods. 2013;10:885–888. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Si L, Xu H, Zhou X, Zhang Z, Tian Z, Wang Y, Wu Y, Zhang B, Niu Z, Zhang C, Fu G, Xiao S, Xia Q, Zhang L, Zhou D. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science. 2016;354:1170–1173. doi: 10.1126/science.aah5869. [DOI] [PubMed] [Google Scholar]
- 54.Chen XH, Xiang Z, Hu YS, Lacey VK, Cang H, Wang L. Genetically encoding an electrophilic amino Acid for protein stapling and covalent binding to native receptors. ACS Chem Biol. 2014;9:1956–1961. doi: 10.1021/cb500453a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoppmann C, Wang L. Proximity-enabled bioreactivity to generate covalent peptide inhibitors of p53-Mdm4. Chem Commun. 2016;52:5140–5143. doi: 10.1039/c6cc01226d. [DOI] [PubMed] [Google Scholar]
- 56.Yuan Z, Wang N, Kang G, Niu W, Li Q, Guo J. Controlling multicycle replication of live-attenuated HIV-1 using an unnatural genetic switch. ACS Synth Biol. 2017;6:721–731. doi: 10.1021/acssynbio.6b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin MZ, Wang L. Selective labeling of proteins with chemical probes in living cells. Physiology. 2008;23:131–141. doi: 10.1152/physiol.00007.2008. [DOI] [PubMed] [Google Scholar]


