Abstract
Chemokines and chemokine receptors rapidly diversified in teleost fish but their immune functions remain unclear. We report here that Chemokine (C-C motif) ligand 19 (CCL19), a chemokine known to control lymphocyte migration and compartmentalization of lymphoid tissues in mammals, diversified in salmonids leading to the presence of six CCL19-like genes named CK10a, CK10b, CK12a, CK12b, CK13a and CK13b. Salmonid CCL19-like genes all contain the DCCL conserved motif but share low amino acid sequence identity. CK12 (but not CK10 or CK13) is constitutively expressed at high levels in all four trout MALT. Nasal vaccination with a live attenuated virus results in sustained up-regulation of CK12 (but not CK10 or CK13) expression in trout NALT. Trout recombinant CK12a (rCK12a) is not chemotactic in vitro but it increases the width of the nasal lamina propria (LP) when delivered intranasally (I.N.). rCK12a delivered I.N or i.p. stimulates the expression of CD8α, granulysin, and IFNγ in mucosal and systemic compartments and increases nasal CD8α+ cell numbers. rCK12a is able to stimulate proliferation of head kidney leucocytes (HKLs) from antigen experienced trout but not naïve controls yet it does not confer protection against viral challenge. These results show that local nasal production of CK12a contributes to antiviral immune protection both locally and systemically via stimulation of CD8 cellular immune responses and highlights a conserved role for CK12 in the orchestration of mucosal and systemic immune responses against viral pathogens in vertebrates.
Introduction
Chemokines are a family of cytokines that play important roles in homeostasis as well as immunity (1–3). As small secreted molecules, chemokines are the extracellular messengers of the immune system and largely control the extravasation of different immune cells from the bloodstream into the tissues (4). It is known that chemokine responses can be induced by a number of stimuli including viral infection and these responses are vital for control of the virus (5). Chemokines are classified in different families based on their amino acid sequence. Homeostatic chemokines such as CCL19, CCL21, CXCL12 and CXCL13, are expressed in lymphoid organs and regulate the migration and compartmentalization of lymphocytes and antigen presenting cells within lymphoid tissues (6–9). CCL19 and CCL21 are also known to orchestrate the development and organization of secondary lymphoid organs including the nasopharynx-associated lymphoid tissue (NALT) (10, 11). Viral infections trigger CCL19 responses in mammals (12), which attract naive T and B lymphocytes to lymphoid organs after 24–48 h of infection (13).
A number of viruses are known to enter the host via the olfactory route (14–18). This route of infection provides a clear advantage to neurotropic viruses since they may gain access to the central nervous system (CNS) via the olfactory bulb. Moreover, in mammals, viral infection of the upper respiratory surfaces may result in infection of the lungs (19–22). Finally, viral infection of nasal tissues, if not controlled locally, can trigger strong systemic responses once the virus enters the bloodstream via the nasal capillary beds. As a consequence, rapid onset of systemic antiviral immunity may be vital to control nasal viral infections.
Nasal vaccines are known to offer a number of advantages over other mucosal vaccines including the ability to stimulate potent systemic antibody immune responses (23–26). The profuse network of capillaries that connects the olfactory organ (OO) with the systemic bloodstream may explain this property of nasal vaccines. In addition, molecular immune mechanisms connecting the nasal and systemic lymphoid tissues must be pivotal for the orchestration and rapid communication of local and systemic immune responses. In this regard, chemokines and other cytokines may provide the required signals for this communication, yet a full understanding of these mechanisms and their evolutionary origins are still missing.
We recently discovered the presence of NALT in teleost fish (27, 28) as a diffuse network of immune cells in the nasal mucosa that shares the same canonical features of other teleost MALT. We also showed that nasal vaccination of rainbow trout with a live attenuated viral vaccine results in both local nasal and systemic immune responses (27, 29). The latter opened up many questions including what are the molecular mechanisms that explain the protective effects of nasal vaccines in fish and how are nasal and systemic immune responses orchestrated in cold-blooded vertebrates. As a preliminary effort, oligomicroarray studies following nasal vaccination with a viral vaccine revealed that early antiviral immune responses in rainbow trout NALT are characterized by dramatic increases in the expression of a CCL19-like molecule (27). Thus, we hypothesized that salmonid CCL19-like chemokines may provide the molecular link between local (nasal) and systemic immune compartments during the course of antiviral immune responses and therefore can orchestrate both types of responses following nasal vaccination.
Evolutionary speaking, chemokines and their receptors can be detected in lower deuterostome animals (30–32) and they are considered rapidly evolving immune molecules (30, 33). To date, many chemokine and chemokine receptors have been found in bony fish (34–40), a group that has more chemokines and chemokine receptors than any other vertebrate including amphibians and mammals. Previous studies have identified rapid, tandem duplications in a number of teleost species including salmonids and zebrafish (38, 41, 42). It has been suggested that the diversification observed in the numbers and sequences of chemokines in bony fish may reflect the adaptation of the individual species to their respective biological environment (42). Despite this plethora of chemokines and chemokine receptor molecules, very little information is available with regards to their biological activities in teleost fish.
Using the DCCL amino acid signature motif, CCL19-like molecules have been previously identified in a few teleost species including turbot (Scophtalmus maximus) (43, 44) and striped murrel (Channa striatus) (45). Their reported activities resemble canonical mammalian CCL19 functions including leucocyte trafficking, cell proliferation and antiviral and antibacterial properties (43, 45). Recently, a study in Atlantic salmon (Salmo salar) detected by microarray, the increased expression of CCL19-like in the HK of resistant and susceptible salmon strains challenged with infectious pancreatic necrosis virus (46). Moreover, a study on catfish showed that CCL19 gene expression is up-regulated significantly following Edwardsiella ictaluri and Flavobacterium columnare infection (41). In rainbow trout, the CCL19-like CK12 was suggested to have a role in mucosal immune responses given its constitutive expression in mucosal tissues such as gill, gut and skin (47). Furthermore, the mRNA levels of two other trout CCL19-like, CK10 and CK12 mRNA, increase following bath infection with viral hemorrhagic septicemia virus (VHSV) as well as infectious pancreatic necrosis virus (IPNV) at the fin base and ovary, respectively (40, 48–50). Trout CK12 expression also increased in HK and spleen of VHSV infected or Poly I:C injected trout and in trout HKLs after in vitro exposure to Poly I:C, IPNV and VHSV (40, 48–50). However, whether teleost CCL19-like chemokines play a role in nasal immune responses deserves further investigation (27).
Since we found a clear induction of CCL19-like by oligomicroarray in trout NALT following nasal vaccination with a live attenuated viral vaccine (27), using the DCCL amino acid signature motif, we conducted further analysis in the NCBI database, in particular searching the rainbow trout and salmon genomes, in order to identify additional CCL19-like forms. Three CCL19-like genes have been described in rainbow trout thus far: CK10, CK12a and CK12b (34, 38) based on EST libraries. However, here we report that six CCL19-like genes exist in rainbow trout and salmon (CK10a, CK10b, CK12a, CK12b, CK13a and CK13b) forming three major clusters (“CK10”, “CK12” and “CK13”). Using a number of in vitro and in vivo studies, we provide the first evidence that CCL19-like diversification resulted in the acquisition of a CCL19-like form, CK12a, that is able to induce both nasal mucosal and systemic antiviral immune responses. Our results not only highlight the conserved role of CCL19 in nasal immunity in vertebrates but also show that this chemokine facilitates the onset of systemic immune responses following nasal detection of antigens as well as the recruitment of antigen presenting cells and CD8+ T cells to the local nasal mucosa.
Materials and Methods
Molecular identification of trout CCL19-like, sequence analysis, phylogenetic analysis and 3-D structure modeling
Rainbow trout CK12a sequence was identified by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (51) using the EST sequence printed in the ‘Trout_imm_v1’ (Agilent array design 028918) (27). Rainbow trout CK12a sequence was used to identify the sequence of rainbow trout CK13 and CK10 by BLAST against the available rainbow trout genome (52). The three rainbow trout sequences possess the canonical DCCL amino acid motif typical of vertebrate CCL19 sequences. Prediction of the open reading frame (ORF) was performed with the programs BLAST and the ExPASy proteomics server (ca.expasy.org/). The ORF of each sequence was amplified by RT-PCR using specific primers (Table I) and the PCR products were cloned as previously described (53). A multiple sequence alignment was created using CLUSTALW (http://align.genome.jp/) (54). Phylogenetic tree was constructed from generated alignments using the Neighbor-Joining (NJ) method within the software MEGA 7 (55); data were analyzed using Poisson correction and gaps were removed by pairwise deletions. To evaluate the topological stability of the NJ tree, a bootstrap of 1000 replicates was applied; with only values over 50% shown. To obtain the percentages of identity and similarities of the six sequences, the software MatGat 2.02 (56) was used. 3-D prediction of protein structures for CCL19-like molecules was performed via Phyre2 (57) online tool (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) and the .pdb files were modeled using PyMOL (https://www.pymol.org).
Table 1.
Oligonucleotides used in this study.
| Gene | Primer name | Primer sequence (5′→3′) | Application |
|---|---|---|---|
| EF-1a | EF-1a F EF-1a R |
CAACGATATCCGTCGTGGCA ACAGCGAAACGACCAAGAGG |
qPCR |
| CK12 | CK12 F CK12 R |
CTCTGAGGTACCCGTGGATTGC CCTTAGGGACTATTGTTCTTCAGC |
qPCR |
| CK13 | CK13 F CK13 R |
CGACCGATACCAAGCTTCCC CCTTATGCGATTTCCTCTTCAG |
qPCR |
| CK10 | CK10 F CK10 R |
GGCCAGATGGTGATGGACTGTG GGTAGTGAAGACCACAGCGCTG |
qPCR |
| CD8α | CD8α F CD8α R |
ATGAAAATGGTCCAAAAGTGGATGC GGTTAGAAAAGTCTGTTGTTGGCTATAGG |
qPCR |
| CD8β | CD8β F CD8β R |
CAACGGTGTGCTTGTGGAAAAC ACACTTTTTGGGTAGTCGGCTGAA |
qPCR |
| Granulysin | Granulysin F Granulysin R |
GCCTACTGGCTTGTTCAGTTTGG TGGTCCTCACGTCATCACTGG |
qPCR |
| IFNγ | IFNγ F IFNγ R |
GCTGTTCAACGGAAAACCTGTTT TCACTGTCCTCAAACGTG |
qPCR |
| CCR7 | CCR7 F CCR7 R |
TTCACTGATTACCCCACAGACAATA AAGCAGATGAGGGAGTAAAAGGTG |
qPCR |
| MHC II | MHC II F MHC II R |
CATATTCTCTGGAACAGATGGATA GCTCAACTGTCTTGTCCAGTATGGCGC |
qPCR |
| CXCR3 | CXCR3 F CXCR3 R |
GGACATCGCCTTTAGACAGGTG GTAGCAGTAGAGCATGACCAGC |
qPCR |
| IL-7R | IL-7R F IL-7R R |
GTGGAGAAGAATTGGTTGAC CCTCCATTTCATCATCGGTGTC |
qPCR |
| CK12 probe | 5′ → 3′ | GGTACCCGTGGATTGCTGTCTCCTCACCA--CTGAGACACGTTTCCCT | FISH |
Constitutive expression of CK12, CK13 and CK10 by RT-qPCR
RNA was extracted by homogenization in 1ml TRIZol (Invitrogen) using tungsten carbide beads (3 mm, Qiagen) and shaking (300 times per min) following the manufacturer’s instructions. The RNA pellet was washed in 500 ml 80% ethanol, air dried and resuspended in RNase-free H2O. The RNA concentration was determined by spectrophotometry (Nanodrop ND1000, LabTech) and the integrity of the RNA was determined by electrophoresis (Agilent Bioanalyser, 2100). cDNA synthesis was performed using 1 μg of total RNA, which was denatured (65°C, 5 min) in the presence of 1 μl of oligo-dT17, 1 μl dNTP (deoxynucleoside triphosphate mix 10 mM each (Promega) and RNA/DNA free water (Sigma) in a volume of 13 μl. Synthesis was carried out using 1 μl Superscript III enzyme reverse transcriptase (Invitrogen) in the presence of 5 μl of 5x first strand buffer, 1 μl 0.1 M DTT, made up to a final volume of 25 μl with water, and incubated at 55°C for 1 h. The resultant cDNA was stored at −20°C. The expression of CK12, CK13 and CK10 was measured by RT-qPCR using specific primers (Table I). Due to the sequence similarities between CK12a and b and between CK13a and b, we were only able to design primers sets that would amplify all “CK12” genes, all “CK13” genes. Although it may be possible to design primers to analyze gene expression of CK10a and b separately, for consistency purposes, we designed primers that would amplify both “CK10” genes. The qPCR was performed using 3 μl of a diluted cDNA template as described in Tacchi et al. 2013 (53). Trout elongation factor EF-1α was used as control gene for normalization of expression.
Animals and nasal vaccination with viral vaccine
Triploid female adult rainbow trout (mean weight 200 g) were obtained from the Lisboa Springs Hatchery, Pecos, New Mexico. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of New Mexico, protocol number 16-200384-MC. For nasal vaccination studies, adult rainbow trout received live attenuated infectious hematopoietic necrosis virus (IHNV) vaccine or phosphate buffer saline (PBS) intranasally (I.N.) as described in (27). The OOs (N=5) from each group were sampled 1 day, 4 days and 7 days post-vaccination and placed in RNAlater for gene expression studies
Recombinant protein production, SDS-PAGE and western blot
Recombinant His-tagged trout CK12a (rCK12a) (aa sequence: MHHHHHHFSEVPVDCCLLTTETRFPRHFKMVSYLLQTTEKGCDIDATVFITKTGVRLCTPHPTKSKWVADYIKRLERTISL) was produced in a bacterial expression system (E. coli, expression vector E3, GenScript USA, Inc) with a theoretical molecular weight of the recombinant protein of 9.4 KDa. Endotoxin level was <0.01 EU/μg. The recombinant protein was then refolded using gel filtration as previously described (58) and filtered sterilized sing a 0.22 μm filter. The obtained protein was analyzed by SDS-PAGE under non-reducing and reducing conditions by loading 2 μg of recombinant protein onto a 4%-2-% SDS-PAGE gel followed by Coomassie Blue staining to confirm the presence of a band of the expected m.w. (~12 kDa) (Supplemental Fig. 1A). We only observed one band after refolding. Moreover, no aggregates were observed in the gel. The gel was transferred to a PVDF membrane and incubated with anti-histag antibody (1:1000; Pierce) followed by incubation with HRP-labeled anti-histag mouse IgG1 (1:7500; ThermoScientific) (Supplemental Fig. 1B). Recombinant protein was then refolded using gel filtration as explained elsewhere (58).
Head kidney leucocyte isolation and chemotaxis assays
After bleeding from the caudal vein, HKLs, spleen leucocytes (SLs) and gut leucocytes (GLs) were isolated by Percoll density gradients as described in Salinas (2007 and 2011) (59, 60). Cells were then washed twice, counted in a Neubauer chamber and adjusted to 5×106 cells/ml. Chemotaxis assays were carried out in Transwell plates (pore size 3 μm, Costar, Corning). To test the capacity of rainbow trout rCK12a to attract HKLs and SLs from naive and vaccinated trout, 400 μl of culture media containing different concentrations of rCK12a (10, 100 or 1000 ng/ml) were placed at the bottom of the wells and 5×105 HKLs, SLs or GLs were carefully loaded onto the upper chamber. After 90 min at 18°C in a CO2 chamber, cells were collected from the bottom of the plate and adherent cells detached by trypsinization. The total number of cells that had migrated to the bottom of the wells was quantified by flow cytometry in an Attune Flow Cytometer (Applied Biosystems) by counting the number of events in 2 min. Positive controls consisted of freshly isolated HKLs and SLs, HKLs and SLs directly placed at the bottom of the well so test the maximum collection capacity and wells to where zymosan (ZAS) activated trout serum (1:30) was added to the medium. Chemorepellent activity of rCK12a was tested by placing both zymosan and rCK19a (10, 100, 1000 ng/ml) at the bottom of the Transwell plates. Negative controls consisted of medium to which the same volume of PBS or an irrelevant protein (a histag Drosophila melanogaster recombinant protein refolded as rCK12a in PBS) rather than rCK12a was added.
CFSE proliferation assay and flow cytometry
Freshly isolated rainbow trout HKLs cells (106 cells/ml) from control or IHNV I.N. vaccinated fish (n=5) were labeled with 1 μM CFSE (Molecular Probes, Thermofisher) according to manufacturer’s instruction. Cells were then stimulated with rCK12a (100 ng/ml or 1000 ng/ml) for 7 days. Unstimulated labeled HKLs were used as negative controls. Positive controls consisted of HKLs incubated with Vibrio anguillarum lysate. Cell division was measured as the percentage of cells where the CFSE intensity was lower than the unstimulated control in an Attune Flow Cytometer (Applied Biosystems). A total of 20,000 events were collected per sample.
Fluorescence in situ hybridization (FISH)
OO cryosections from adult control or nasally vaccinated IHNV rainbow trout were stained with rainbow trout CK12 or NON-CK12 (negative control) oligonucleotide probes labeled at their 5′ ends with indodicarbocyanine (Eurofins MWG Operon). The probe sequence used is shown in Table I. Hybridizations were performed at 37°C overnight with hybridization buffer (2X SSC/50% formamide) containing 1 μg/ml of the labeled probe. Slides were then washed with hybridization buffer without probes followed by two more washes in washing buffer (0.1X SSC) and two washes in PBS at 37°C. Nuclei were stained with DAPI (5 ng/ml) solution for 25 min at 37°C. Slides were mounted with fluorescent mounting media and images were acquired and analyzed with a Nikon Ti fluorescence microscope and the Elements Advanced Research Software (version 4.2).
Immunofluorescence microscopy
OO cryosections from control, I.N. rCK12a treated and i.p. rCK12a treated (n = 3) rainbow trout were stained with anti-trout CD8α+ and MHC-II antibody as explained elsewhere (29, 61, 62). Nuclei were stained with DAPI DNA stain and slides were observed under a Nikon Ti fluorescence microscope. A total number of ten images per sample from six different cryosections were captured and the number of CD8α+ cells and MHC-II+ cells were visually counted by two different researchers. The distance from 0 to 100 μm was considered the apical LP and between 100 and 250 μm was considered the mid LP. The width of LP in apical and mid points of the lamella was measured in ten different lamellae per sample. All analyses were carried out with the Nikon Elements Advanced Research Software v. 4.2.
In vitro effects of rCK12a on immune gene expression
A total of 106 HKLs (n=6) in DMEM + 10% FBS per well were pipetted onto flat-bottom 24 well plates. After 4 h, rCK12a (100 ng/ml) or PBS (control) were added to the cell cultures for 6, 24 and 48 h. Cells were collected and placed in TRIZol (Invitrogen). Total RNA was extracted and cDNA produced as explained elsewhere (63). Expression levels of CD8α, CD8β, granulysin, IFNγ, IL7-R, CCR7, MHC-IIb and CXCR3 was measured by RT-qPCR using specific primers (Table I). Trout elongation factor EF-1α was used as control gene for normalization of expression. The relative expression level of the genes was compared to the unstimulated control and determined using the Pfaffl method (64).
In vivo delivery of rCK12a, effects on gene expression and challenge experiments
Rainbow trout (mean weight 3 g) were divided into three experimental groups. The first group was a control group that received PBS I.N. and i.p., the second group received 3 μg of refolded rCK12a in PBS I.N.; the third group received 3 μg of refolded rCK12a in PBS i.p. OO and HKs (n=5) were sampled at days 1, 5 and 8 after rCK12a administration. Expression of CD8α, granulysin, IFNγ, CCR7 and CK12 was measured by RT-qPCR and analyzed as previously described. Eight days after rCK12a treatment, trout were challenged with virulent IHNV as explained elsewhere (27). A total of 25 fish per tank in duplicate tanks were used for the challenge experiment. Controls consisted of mock-vaccinated fish (received PBS I.N. and i.p.) and an unchallenged group. Mortalities were recorded for 28 days post-challenge.
Statistical analyses
Data are expressed as the mean ± sem. Gene expression data was analyzed by t test to identify statistically significant differences between groups. Data analysis was performed in GraphPad Prism version 5.0. One-way ANOVA and a Tukey post hoc analysis test were performed to identify statistically significant differences among groups. For the proliferation assay a multivariate ANOVA test followed by a Fisher’s least significant difference post hoc test for multiple comparisons was performed. Survival proportions among experimental groups in the challenge experiment were compared using the logrank test within Prism. The p values < 0.05 were considered statistically significant.
Results
Salmonids possess six CCL19-like genes
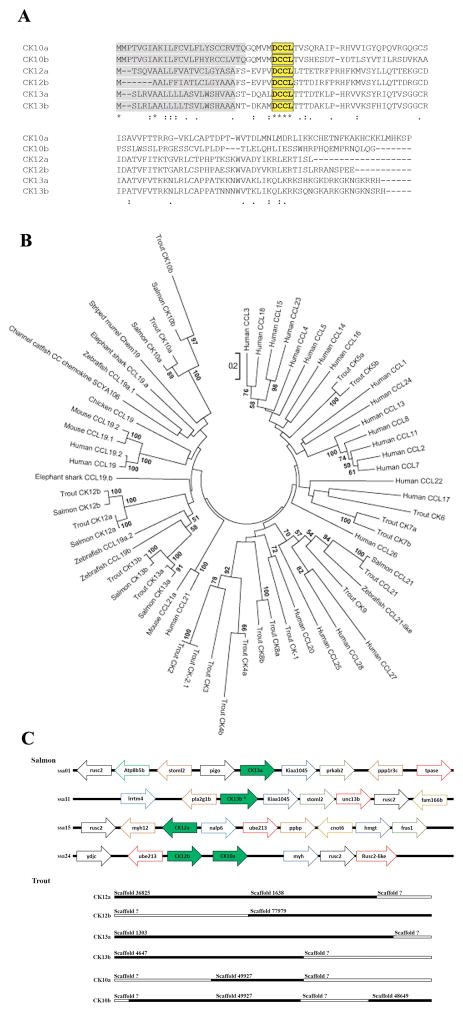
Searching for the CCL19-like motif in the rainbow trout genome revealed that salmonids possess six different genes that encode CCL19-like chemokines. We named them CK12a, CK12b, CK13a, CK13b, CK10a and CK10b (Fig. 1). Out of all the isoforms, CK12a had been identified as one of the most important immune genes up-regulated in trout NALT following nasal vaccination with a viral vaccine (27). The coding regions for CK12a, CK12b, CK13a, CK13b, CK10a and CK10b were 285, 297, 321, 324, 342 and 318 bp long, respectively. They encoded for a 95, 99, 107, 108, 114 and 106 aa long protein, respectively (Fig. 1A). Alignment of all six rainbow trout CCL19-like sequences showed a highly variable level of conservation among them with CK13a and b sharing 82.4% identity while CK13b and CK10b are the most dissimilar with only 17% identity (Supplemental Table I). Phylogenetic analyses comparing salmonid CCL19-like molecules with other available teleost and mammalian CCL19 sequences showed that there is a clear relationship between CK12 and CK13 genes (with a bootstrap over 50%) (Fig. 1B). Meanwhile, CK10a and b, appear to be phylogenetically distant from the other teleost CCL19-like genes. As expected, mammalian CCL19 sequences cluster in a separated clade from the fish homologues. When comparing all the trout and human chemokines available to date, phylogenetic analysis also shows that teleost CCL19-like genes are more closely related to mammalian CCL19 genes than to other chemokine genes. Finally, screening of the Atlantic salmon and rainbow trout genomes revealed that the six CCL19–like genes are positioned in four different chromosomes in the Atlantic salmon genome and in six different scaffolds in the rainbow trout genome (Fig. 1C). However, we could not locate CK10b in the current versions of the Atlantic salmon or trout genomes. It is possible that future versions of these genomes may reveal CK10b genomic position.
Figure 1.
Salmonids possess six CCL19-like genes. (A) Multiple sequence alignment (performed with CLUSTALW http://align.genome.jp/) of Trout CCL19-like isoforms with the conserved DCCL motif boxed in bold. (B) Phylogenetic tree showing the evolutionary relationship of CCL19-like genes among fish and other vertebrates. Tree was constructed using the Neighbor-Joining method in MEGA 7. The tree was bootstrapped 1000 times, and only values over 50% are shown. Accession numbers used to construct the tree were obtained from https://www.ncbi.nlm.nih.gov: trout CK1 (AF093802); trout CK2 (AF418561); trout CK4a (CA371157); trout CK4b (CA352593); trout CK5a (CA383670); trout CK5b (CA374135); trout CK6 (CA355962); trout CK7a (CA343117); trout CK7b (CA346976); trout CK8a (CB494647);trout CK8b (CA353159); trout CK9 (CA378686); trout CK12a (KF683302.1); trout CK12b (CA346383.1); trout CK13a (DV196492.3); trout CK13b (CO471983.1); CK10a (CA361535.1); trout CK10b (DV194065); trout CCL21 (CDQ86623.1 (unnamed product)); salmon CK12a (XM_014143452.1, named Salmo salar C-C motif chemokine 4-like (LOC106570886); salmon CK12b (XP014028076.1; named SsCCL4l12); salmon CK13a (BT125229.1); salmon CK13b (XP013984336.1); salmon CK10a (ACI69602.1); salmon CK10b (XM_014172597.1, named Salmo salar C-C motif chemokine 19-like (LOC106585878)); Salmon CCL21 (NP001134739.1); zebrafish CCL19b_F1QHU2; zebrafish CCL19a.2_B3DHA6; zebrafish CCL19a.1_A2BIR2; channel catfish chemokine SCYA106 (ABA54953.1); elephant shark CCL19.a (XP_007910129.1); Elephant shark CCL19.b (XP_007910128.1); stripped murrel CCL19 (AGN52674.1); human CCL1 (P22362); human CCL2 (P13500); human CCL3 (P10147); human CCL4 (P13236); human CCL5 (P13501); human CCL7 (P80098); human CCL8 (P80075); human CCL11 (P51671); human CCL13 (Q99616); human CCL14 (Q16627); human CCL15 (Q16663); human CCL16 (O15467); human CCL17 (Q92583); human CCL18 (P55774); human CCL19 (Q99731); human CCL19.2 (Q5VZ75) human CCL21 (Q6ICR7); mouse CCL19.1 (Q548P0); mouse CCL19.2 (A0A0N4SUZ8); mouse CCL21a (NP_035254.1); chicken CCL19 (R4GM50). (C) Genomic configuration of CCL19-like genes in the Atlantic salmon genome (top) and rainbow trout genome (bottom). Salmon CCL19-like genes are represented with green arrows and they are located in 4 chromosomes: ssa01 for CK13a, ssa11 for CK13b, ssa15 for CK12a and ssa24 for CK12b and CK10. Asterisk on CK13b, highlight the presence of 3 different variants. We were not able to locate the chromosome containing salmon CCL19c-like. In the lower part of the figure, trout CCL19-like genes are represented within several scaffolds (in black); while the missing part of the sequences appear in white.
3-D protein structures of all six rainbow trout CCL19-like molecules (CK12a, CK12b, CK13a, CK13b, CK10a and CK10b) are shown in Supplemental Fig. 2. Mammalian CCL19 has a canonical chemokine fold consisting of a flexible N-terminus and N-loop followed by an antiparallel three stranded β-sheet, a C-terminal α-helix, and a short flexible C-terminus (65). Predicted rainbow trout CCL19-like structures show some commonalities but also important differences compared to mammalian CCL19. All trout CCL19-like forms have a short flexible C-terminus, a C-terminal α-helix, a three stranded β-sheet and an N-loop. However, the N-terminus differs significantly from the resolved mammalian CCL19 structure. Moreover, consistent with the low sequence identity among some of the trout CCL19-like molecules, their N-termini differ considerably. Whereas CK12a has a very short flexible N-terminus followed by a N-terminus α-helix, CK13a, b and CK10a all have long flexible N-termini. CK12b and CK10b, in turn, had intermediate length flexible N-termini. These data indicate some degree of molecular structural conservation among CK12, CK13 and CK10 molecules in salmonids.
CK12, CK13 and CK10 show distinct constitutive tissue distribution in rainbow trout
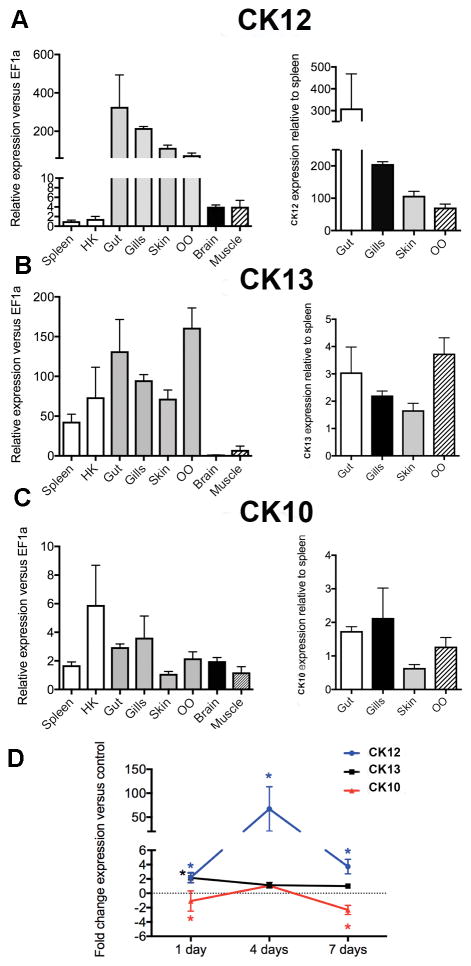
In order to gain some understanding on the physiological functions of the three CCL19-like genes found in rainbow trout, we measured their constitutive expression in main lymphoid organs (HK and spleen), mucosal lymphoid tissues (gut, gill, skin and OO), neuronal tissues (brain) as well as muscle. We normalized the expression in every tissue to that of the brain. As shown in Fig. 2A–C, each CCL19-like gene displays a unique constitutive expression pattern. In agreement with a previous report (47), CK12 is primarily expressed in mucosal lymphoid tissues including the OO, with levels between 100–300 fold greater than those recorded in main lymphoid organs such as the spleen (Fig. 2A). CK13 was expressed in all immune tissues examined with no clear difference between mucosal and non-mucosal immune sites although it was expressed between 1.5–4 times more in mucosal tissues than in the spleen (Fig. 2B). Finally, CK10 was expressed in every tissue examined, including non-immune tissues such as the brain and the muscle (Fig. 2C). At mucosal sites, expression of CK10 was comparable to that measured in the spleen. This expression pattern may indicate a more homeostatic role for CK10 compared to the other two (Fig. 2C). Combined, these results suggested that CK12 plays a more important role in trout mucosal immunity than CK13 and CK10.
Figure 2.
CK12a is highly expressed in mucosal tissues and is up-regulated following nasal IHNV vaccination. Tissue distribution of major isoforms of CCL19-like (CK12, CK13 and CK10) in different tissues of control trout and following IHNV vaccination was measured by RT-qPCR. Constitutive expression of CK12 (A) CK13 (B) CK10 (C) in seven different trout tissues. Expression levels were normalized to the expression in the brain. (D) Gene expression levels of CK12, CK13 and CK10 were measured in the olfactory organ of rainbow trout 1 day, 4 days and 7 days following I.N. IHNV vaccination (2*107 PFU). Gene expression levels were normalized to the housekeeping gene EF-1a and are expressed as the fold change compared with the expression levels in mock-vaccinated controls using the Pfaffl method. Results are representative of two different experiments (n = 6). *p< 0.05.
Kinetics of trout CCL19-like expression in trout NALT following nasal viral vaccination
We previously identified CCL19-like (CK12a in this study) as one of the major innate immune genes that is up-regulated in NALT following nasal vaccination with an IHNV vaccine (27). In order to know if this response is specific of this CCL19-like form, we measured CK12, CK13 and CK10 expression in the OO of trout 1 day, 4 days and 7 days post-nasal vaccination using the same IHNV vaccine model (Fig. 2D). CK12 was up-regulated 2-fold on day 1, 67-fold on day 4 and 4-fold on day 7 in the OO of vaccinated trout compared to controls. The strong up-regulation of CK12 expression on day 4 was confirmed by FISH (Supplemental Fig. 3). FISH staining showed that the increase in CK12 expression was due to a few high expressing cells rather than an increase throughout the tissue.
CK13 only showed a moderate up-regulation on day 1 (2-fold) and no-significant changes in expression at any other time points. Interestingly, CK10 responded very differently to the other two CCL19-like genes since it was significantly down-regulated (~3-fold) on day 1 and back to basal levels after 4 days and 7 days. These expression patterns indicated that CK12 is the main CCL19-like gene involved in trout nasal immune responses in vivo.
rCK12a is not chemotactic in vitro
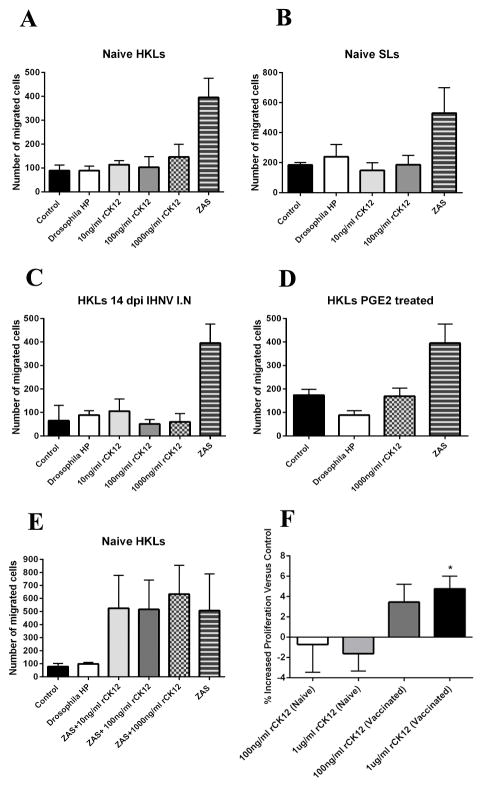
Previous studies have shown that teleost CCL19 has a canonical chemotactic activity similar to that reported in mammals (43, 45). It is important to note that those studies albeit calling the molecule of interest CCL19, they used recombinant proteins that do not cluster with trout CK12 based on phylogenetic analysis (Fig. 1B). Moreover, a study in rainbow trout tested chemotactic activities of recombinant protein CK12b (acc. number CA346383) in HKLs, PBLs and SLs of rainbow trout (47). They showed no chemotactic activity of 0.1, 1 and 10 ng/ml of recombinant protein CK12b toward HKLs and PBLs cells in vitro using the Transwell system. Nevertheless, they observed moderate chemotactic activity in SLs exposed to 10 ng/ml of trout recombinant CK12b (47). We also tested the chemotactic ability of rCK12a in vitro using a Transwell assay system (Fig. 3A–E). We could not detect any significant migration of naïve HKLs, SLs (Fig. 3A and B) or GLs (data not shown) towards a wide range of concentrations of rCK12a. We attempted to pre-activate trout leucocytes with LPS (data not shown), IHNV (Fig. 3C) or prostaglandin E2 (PGE2) (Fig. 3 =D), yet no chemotaxis towards rCK12a was observed in vitro. To test whether rCK12a has chemorepellent activities, we added rCK12a to our positive control (ZAS) at different concentrations. However, we did not detect any significant difference in HKLs migration between treatments (Fig. 3E) indicating that rCK12a has no chemorepellent activity.
Figure 3.
rCK12a is not chemotactic in vitro but stimulates proliferation of HKLs from IHNV vaccinated fish. Chemotaxis assays were carried out in Transwell plates and migrated cells were measured by FACS. Chemotactic activity of rCK12a on naïve HKL cells (A) SLs (B) HK cells from IHNV vaccinated fish (C) Prostaglandin E2 (PGE2) treated HK cells (D) are shown. (E) Chemorepellent activity of rCK12a on naïve HKL cells was shown. Positive controls consisted of cells migrated towards zymosan (ZAS) activated trout serum. HKLs cells from control or IHNV I.N. vaccinated fish (N=5) were labeled with CFSE and treated with rCK12a then proliferation was measured by FACS. (F) Increased proliferation (%) of HKLs from naïve and IHNV vaccinated fish incubated with or without rCK12a is shown (comparisons were carried out in cells from the same individual). Transwell experiments were conducted three independent times with N=3. Proliferation experiments were conducted twice with N=5. *p < 0.05.
rCK12a stimulates proliferation of HKLs from IHNV vaccinated fish but not naive fish
In order to know if trout rCK12a is able to induce proliferation of trout immune cells, we used CFSE to label HKLs isolated from control and IHNV vaccinated fish (14 days post-immunization, dpi) followed by incubation with rCK12a (100 ng or 1 μg) or PBS for 7 days. The FACS analyses showed a significant proliferation rate in HKLs from IHNV vaccinated fish that were treated with 1 μg rCK12a, compared to the control (Fig. 3F). However, we did not observe any proliferation in naïve HKLs that were treated with rCK12a at both doses compared to the control. Thus, the results suggest that CK12 plays a major role in cell proliferation in the antigen-experienced leucocytes.
Nasal delivery of rCK12a results in infiltration of MHC-II+ cells into the nasal mucosa and in enlargement of the olfactory LP in trout
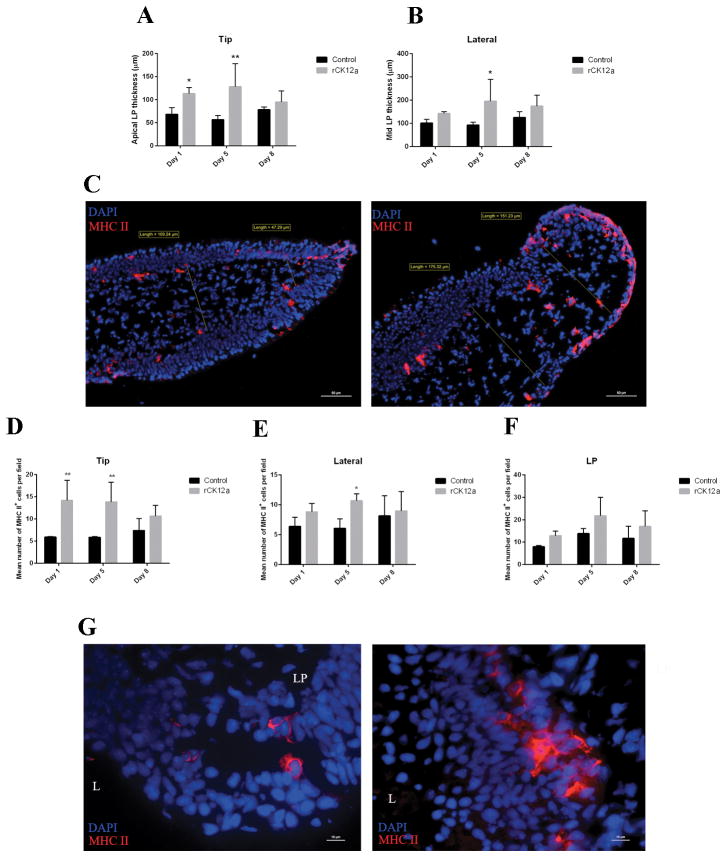
Nasal vaccination of rainbow trout with live attenuated IHNV vaccine results in the recruitment of myeloid and lymphoid cells to the local nasal environment (27). Leucocyte recruitment is easily visualized by histological examination due to the presence of an enlarged LP in the olfactory lamellae of vaccinated animals compared to controls (27). Furthermore, we have shown in the past that nasal delivery of IHNV vaccine results in MHC-II+ responses in the OO of trout (29). Since in vitro Transwell assays may not mimic in vivo immune responses, we delivered rCK12a I.N. to rainbow trout and measured the effects on MHC-II+ cells as well as on the width of the olfactory LP. As shown in Fig. 4, rCK12a delivery results in changes in the morphology and localization of MHC-II+ cells in trout NALT especially on day 5. Moreover, the olfactory LP of rCK12a treated fish was expanded significantly compared to PBS controls in the tip of the lamella on day 1 (from ~70 to ~110 μm) and in both the tip (from ~60 to ~130 μm) and medial regions (from ~100 to ~200 μm) of the LP on day 5. We observed LP expansions in rCK12a treated group at other time points in both the apical and mid lamella regions but the increase was not statistically significant (Fig. 4A–C). Our analyses showed that I.N. delivery of rCK12a led to a significant (~3-fold) increase in the number of MHC-II+ cells in the tip on day 1 and a ~2.5-fold increase in both the tip and medial regions of the lamella on day 5 (Fig. 4D and E). In addition, we observed that rCK12a increased MHC-II intensity staining in OO which may be due to stimulation of antigen presenting process (Fig. 4G). Treatment with rCK12a also resulted in higher numbers of MHC-II+ cells in LP in all time points compared to controls, however, the increases were not significant (Fig. 4F). These results indicate that rCK12a has inflammatory, chemotactic properties and suggest that it can stimulate antigen presentation in vivo.
Figure 4.
In vivo nasal delivery of rCK12a (I.N.) results in enlargement of trout olfactory organ lamina propria (LP). Trout were delivered rCK12a (3μg/fish) intranasally (I.N.) and the olfactory organs were sampled 1, 5 and 8 days later and cryosections obtained. (A) and (B) The width of LP at the tip (100 μm away from the lamellar tip) and medial (250 μm away from the lamellar tip) regions of the olfactory lamella were measured by image analysis of ten individual lamellae from three different fish per treatment. The mean distance ± SE is shown. (C) Immunofluorescence staining of control (left) and rCK12a treated (I.N.) rainbow trout olfactory organ stained with anti-trout MHC-II antibody (red) showing our image analysis strategy and the enlargement in the apical and medial regions of the LP in the rCK12a treated fish (right). For (C) and (G), cell nuclei were stained with DAPI DNA stain (blue). Results are representative of two different experiments (n = 3). Scale bars: 50 μm. (D), (E) and (F) Quantification of the number of MHC-II+ cells present in the tip, lateral and LP regions of control and rCK12a treated (I.N.) rainbow trout olfactory organ (n = 3). *p < 0.05 and **p < 0.01. (G) Magnified view of an immunofluorescence staining of control (left) and rCK12a treated (right) rainbow trout olfactory organ labeled with anti-trout MHC-II antibody (red) showing higher expression of MHC-II in the rCK12a treated fish (right) compared to control (left). LP: lamina propria; L: Lumen. Scale bars: 10 μm.
rCK12a modulates CD8+ T cell related genes in rainbow trout in vitro and in vivo
Since CK12 appeared to be critical in the local nasal anti-viral immune response of rainbow trout, we next asked whether locally produced CK12a may modulate immune genes that may contribute to defense against viruses in the systemic compartment. We incubated HKLs with rCK12a in vitro for 6, 24 and 48 h and measured by RT-qPCR the expression change of CD8+ T cells related immune genes (Fig. 5A and B). At 6 h, CD8α, IFNγ and CCR7 expression was significantly up-regulated whereas after 24 h, rCK12a resulted in an up-regulation of CD8α expression; however, the change observed was not significant. No significant changes were recorded at 48 h (data not shown).
Figure 5.
rCK12a modulates CD8+ T cell related genes in rainbow trout in vitro and in vivo. Control rainbow trout HKLs were cultured in the presence of rCK12a for 6 or 24 h and expression of immune genes was measured by RT-qPCR. (A) and (B) Gene expression levels of CD8α, CD8β, granulysin, IFN-γ, IL7-R, chemokine receptor CCR7, MHC-II and CXCR3 are shown. Gene expression levels were normalized to the housekeeping gene EF-1a and expressed as the fold change compared with the untreated HKLs expression levels using the Pfaffl method. Results are representative of three different experiments (n=5). *p<0.05, **p<0.01. For in vivo experiments, 3 μg of rCK12a were delivered I.N. or i.p. in rainbow trout (n = 5) and olfactory organ (OO) and head kidney (HK) were sampled 1, 5 and 8 days later. Gene expression levels of CD8α (C), granulysin (D), IFNγ (E), chemokine receptor CCR7 (F), CK12 (G) and MHC-II (H) are shown. Results are representative of four different experiments. *p<0.05, **p<0.01 compared with the HK.
Administration of rCK12a i.p. or I.N. led to a down-regulation of CD8α, granulysin, and IFNγ expression in the HK and OO 1 day post-administration (Fig. 5C–E). In contrast, on day 5, all cell-mediated immune genes were found to be up-regulated in the HK and OO as a result of rCK12a administration both i.p. and I.N (Fig. 5C–E). Similarly, on day 8 cell-mediated response genes such as CD8α, granulysin and IFNγ expression levels remained significantly higher than those observed in control OO (Fig. 5C–E).
In mammals, CCR7 is known to be the receptor for CCL19 and CCL21 (66) and a CCR7 homolog has been reported in rainbow trout (37). Our RT-qPCR data show that CCR7 was significantly up-regulated in the HK 1 day post-administration of rCK12a both i.p. and I.N. However, a significant down-regulation in CCR7 expression was observed in the OO at this time point. On days 5 and 8, CCR7 expression was significantly higher in the OO in both the I.N. and i.p. administered groups compared to the control (Fig. 5F). Administration of rCK12a led to increased CK12 transcript levels in the HK 5 days after I.N delivery and decreased expression levels on day 8 (Fig. 5G). In the OO, only I.N administration resulted in significant increases in CK12 expression on day 8 (Fig. 5G). In agreement with our microscopy results (Fig. 4C–F), we observed a significant up-regulation in MHC-II expression in the OO 5 days post-administration (I.N) of rCK12a (Fig. 5H) with no other changes in any other time points or tissues. Our results indicate that presence of CK12 either in the local nasal environment or systemically results in activation of CD8+ T cell related genes as well as CCR7 in rainbow trout.
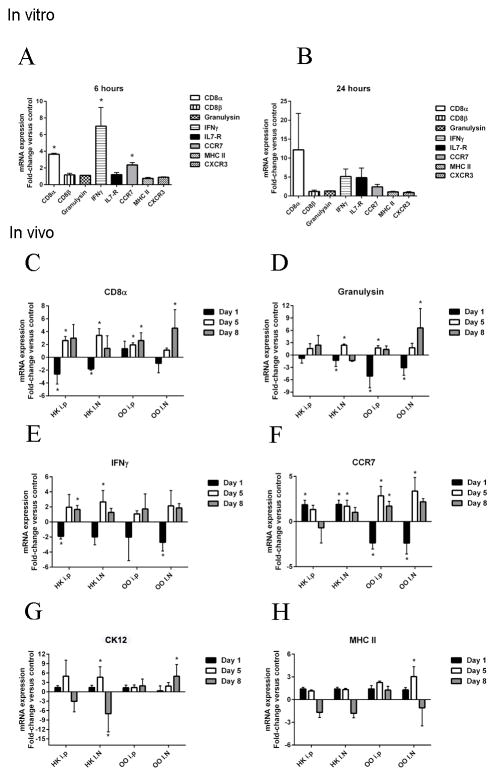
In vivo delivery of rCK12a increases NALT CD8α+ cell numbers in rainbow trout
We showed that i.p. and I.N. delivery of rCK12a modulates CD8α+ T cell related genes especially in OO both in vitro and in vivo. To test the number of CD8α+ T cells in OO changes following the administration of rCK12a, we next measured the total numbers of CD8α+ cells in the OO of rainbow trout using IF microscopy on days 1, 5 and 8 after rCK12a delivery (i.p. or I.N.). We observed a non-significant increase in the number of CD8α+ cells on day 5 and a significant increase on day 8 (Fig. 6A and B). This increase was due to increase numbers in the neuroepithelial compartment whereas no changes were observed in the mucosal tip (data not shown). This result suggests that rCK12a may stimulate local proliferation of CD8α+ cells or their migration from lymphoid organs to the nasal mucosa.
Figure 6.
In vivo delivery of rCK12a increases NALT CD8α+ T cell numbers in rainbow trout. rCK12a was delivered I.N. or i.p. (n = 5) and the olfactory organ (OO) was sampled 1, 5 and 8 days later. Control fish received the same volume of PBS I.N. and i.p. (A) Quantification of the number of CD8α+ T cells present in neuroepithelial region of the olfactory organ of control and rCK12a treated (I.N. or i.p.) rainbow trout (n = 3). *p < 0.05 and **p < 0.01. (B) Representative immunofluorescence staining of control (left) and rCK12a treated (right) rainbow trout olfactory organ labeled with anti-trout CD8α (green) monoclonal antibody showing higher number of CD8α+ cells in the rCK12a treated fish (right) compared to control (left). DAPI was used to stain cell nuclei (blue). Scale bars: 10 μm.
I.N. or i.p. delivery of rCK12a does not protect rainbow trout against viral challenge
In order to investigate if rCK12a is sufficient to confer protection against IHNV, we administered rCK12a I.N. or i.p. to trout and challenged them eight days later with live IHNV. No significant levels of protection were observed between rCK12a treated and untreated (control) groups at the tested rCK12a dose and administration regime following viral challenge (Fig. 7). Thus, rCK12a when delivered alone does not lead to protection against viral pathogen.
Figure 7.
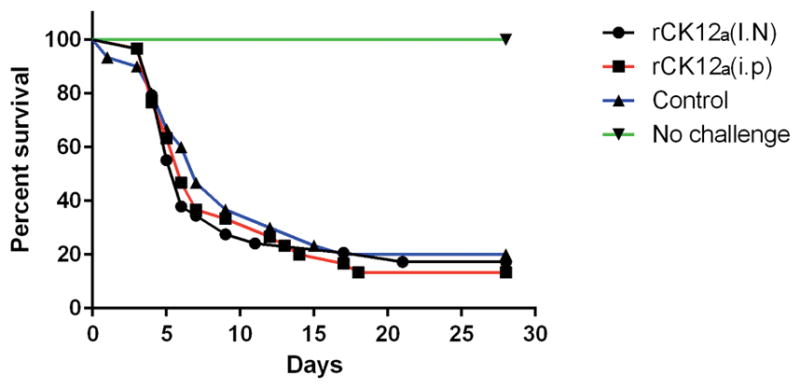
I.N. or i.p. delivery of rCK12a does not protect rainbow trout against viral challenge with IHNV. Kaplan-Meier curves showing the percent survival of rainbow trout 28 days after challenge with virulent IHNV. Eight days after rCK12a treatment, trout were challenged with virulent IHNV by injection. A total of 25 fish per tank in duplicate tanks were used for the challenge experiment. Controls consisted of mock-vaccinated fish (received PBS I.N. and i.p.) and an unchallenged group.
Discussion
Chemokines are small proteins that act as extracellular messengers of the immune system (3, 67, 68). CCL19 plays many biological roles in mammals including homing of T cells and DCs to T cell areas of lymphoid tissues, secondary lymphoid tissue organogenesis, antibody responses, and regulatory and memory T cell function (69–71). CCL19 is also one of the chemokines whose expression is induced by viral infection in a number of host-viral models (13, 71) and it contributes to inflammatory responses against viruses (71–73).
It is believed that the chemokine system appeared around 650 mya (31). Chemokines are present in both jawless and jawed vertebrates but not in invertebrates (31, 33). Chemokine and chemokine receptor genes have undergone several gene duplications in teleosts (31), however, the functional consequences of this molecular diversification are largely unknown. The goal of the present study was to investigate the functional role of CCL19-like as a messenger between the nasal and the systemic immune compartments during antiviral immune responses in rainbow trout.
Although previous studies had reported the presence of some CCL19-like genes in salmonids, the present study provides an updated and comprehensive analysis of this chemokine family using recently released genomes. Interestingly, we found an unprecedented diversity of CCL19-like genes in salmonids, with six genes in rainbow trout forming three major clusters (“CK12”, “CK13” and “CK10”). This finding raised the question, are all these CCL19-like forms involved in nasal immune responses? We first observed unique tissue distribution patterns of each CCL19-like gene, CK12 showing a more “mucosal-like” profile than the other two. These results are consistent with previous studies in trout were CK12 was mainly expressed in mucosal tissues (47). It is also worth mentioning that the CK12 primers in previous studies (47) and the current study amplify both CK12a and CK12b genes due to sequence similarities. Moreover, we observed that only CK12 expression was significantly up-regulated in a sustained manner over a period of 7 days in the local nasal environment, supporting the idea that CK12 is specialized in nasal mucosal immune responses against viruses. These findings are in agreement with previous studies in trout where CK12 expression increased in response to experimental rhabdoviral infection (49). Combined, the unique tissue distributions as well as the unique transcriptional responses of trout CK12, CK13 and CK10 suggest specialized functions for each of these molecules. Further studies should address the specific immune roles of CK13 and CK10 in salmonids.
Along with the local induction of CK12 in the nasal mucosa, we had previously recorded strong immune responses both in the nasal mucosa and in the main systemic lymphoid tissue of teleosts (the HK) in response to nasal vaccination with IHNV vaccine especially 4 days post-vaccination (27). These findings led us to hypothesize this CCL19-like chemokine coordinates both local and systemic antiviral immune responses in rainbow trout.
Several studies in mammals have shown that CCL19 is chemotactic for the immune cells that express its receptor CCR7 such as DCs (74, 75), B cells (76–78), CD4+ and CD8+ T cells (77–79). Since migration rates of human cells increase with increasing CCR7 ligands doses up to 1000 ng/ml in vitro (80–82), we tested different concentrations of rCK12a up to 1000 ng/ml but we did not observe any chemotactic effects regardless of the concentration tested. These results were consistent regardless of the source of cells used (HKLs, SLs or GLs) in vitro even when we pre-activated leucocytes were used. We have previously shown that HK and gut CD8α+ cells express CCR7 (29) indicating that the source of cells used in our studies should not be reason why chemotaxis results were negative. Some reports have shown the importance of PGE2 on the migratory ability of CCL19 in mammalian monocytes (83–86). Thus, we also pre-activated HKLs with PGE2 but still failed to observe any chemotactic effects in vitro. Testing the bioactivity of mammalian chemokines such as CCR7 ligands in vitro requires gradients generated by sustained release from microparticles (81), the presence of immobilized but not soluble chemokines (87) or the use of three dimensional collagen gel assays where soluble CCL19 gradients are formed (87, 88). Thus, further studies may evaluate the chemoattractive activity of trout CCL19-like molecules in vitro using assays other than Transwell assays. In any case, our data highlight that trout CK12a and CK12b have distinct biological functions since CK12b has some chemotactic ability in vitro (47) whereas CK12a does not.
In vivo delivery of the rCK12a (I.N.) led to expansion of LP in both the apical and mid regions of the olfactory lamella as well as infiltration of MHC-II+ cells 5 days after delivery. These results are consistent with our previous work showing significant enlargement of the olfactory LP and infiltration of immune cells 4 days after nasal vaccination with IHNV vaccine (27). This data indicates that trout CK12a promotes the recruitment and extravasation of leucocytes into the nasal mucosa in vivo.
We have previously shown that I.N. delivery of IHNV vaccine changes the morphology of the MHC-II+ cells dramatically (29). The present study revealed that delivery of rCK12a (I.N.) increases MHC-II+ expression and changes in MHC-II+ cell morphology 5 days after the delivery. In line with our findings, mammalian studies had showed that CCL19 induces changes in morphology, activation and maturation of MHC-II+ DCs in mice (71, 89). Combined, these results suggest that trout CK12a is responsible for the activation of antigen presentation in the nasal mucosa of rainbow trout in response to nasal viral antigen delivery.
CCL19 is known to modulate a myriad of adaptive immune responses in mammals including memory B and T cell function (69–71). Thus, we investigated the possible role of trout CCL19-like in the onset of secondary immune responses. We observed no increased proliferation in naïve HKLs incubated with rCK12a. However, HKLs extracted from IHNV vaccinated fish responded to rCK12a stimulation with increasing proliferation rates. In support, Marsland et al; 2005 (71) reported that CCL19 did not induce any proliferation to murine naïve T cells. Yet, the same study observed that CCL19 enhanced T cell proliferation in the presence of antigens (71). Overall, our findings support the idea that CCL19-like molecules have adopted multiple innate and adaptive immunological functions during evolution and that these functions are largely conserved from teleosts to mammals.
CD8+ T cells play an important role in adaptive immune responses especially against viral pathogens in vertebrates. Some reports have indicated the importance of CD8+ T cell responses at the transcript level in salmonids during IHNV infection (90–92). It is also shown that CCL19 stimulates Th1 responses rather than Th2 responses by induction of DCs in mice (71). We hypothesized that rCK12a plays an important role in the stimulation of CD8+ T cell responses. Interestingly, we observed a significant up-regulation of CD8+ T cells related genes in HKLs that were treated with rCK12a in vitro. Similarly, in vivo delivery of rCK12a (i.p. or I.N.) modulated CD8+ T cells related genes in HK and OO especially 5 and 8 days post-delivery. These results indicate that rCK12a is a major factor for CD8α+ T cell activation in trout. Importantly, we detected significant increases in the total number of CD8α+ cells present in the nasal mucosa 8 days after rCK12a delivery regardless of the route (i.p. or I.N.) suggesting that CK12a mobilizes CD8α+ cells from systemic to nasal tissues whether produced locally or systemically. Although the effects of CCL19 on nasal CD8 responses have not been studied in mammalian models to date, our results are in line with previous work in mice where delivery of plasmid DNA encoding for CCL19 either I.N. or i.m. resulted in enhanced mucosal (vaginal) and systemic antibody responses across isotypes (93). Similarly, although in a different mucosal site, in this case the sublingual mucosa, the CCR7-CCL19/CCL21 pathway has been shown to regulate efficient Ag-specific systemic and mucosal T and B cell immune responses (94).
We recently showed that NALT CD8α+ cells mostly express T cell markers but not NK or DC markers and that both nasal (but not gut) and systemic CD8α+ cells express similar levels of CCR7 in trout (29). Thus, direct ligand-receptor interactions may have driven the observed effects of CK12a on CD8α+ cells. The increased numbers of CD8α+ cells may have been the result of local proliferation or migration from other lymphoid sites into the nasal mucosa. Given that nasal vaccination of trout with IHNV vaccine results in negligible local proliferation (29), we believe that infiltration of the local olfactory epithelium is the main contributing mechanism by which CK12a stimulates nasal CD8 cellular responses.
Teleost MALT lack organized lymphoid structures such as LNs as well as GC formation (27, 28, 95). However, we recently characterized two unique tissue microenvironments with unique immune cell populations in the trout olfactory organ (29). Specifically, the apical mucosal portion of the olfactory lamella harbors clusters of CD8+ T cells and this region also has the highest expression of CK12. This is particularly interesting since the CCL19/CCR7 axis appears to be vital for the homing of naïve and regulatory T lymphocytes to LNs and the strategic positioning of these cells within LNs (66, 70, 96, 97). Thus, in the absence of these organized lymphoid structures in teleosts, CCL19-like chemokines may still have evolved a role in the strategic positioning of T cells within mucosal lymphoid tissues.
Since rCK12a was able to recapitulate some of the main characteristic immune responses elicited by entire nasal vaccine formulations, we hypothesized that delivery of rCK12a in the absence of antigen may lead to protection against IHNV in naïve rainbow trout. Fish were challenged 8 days after rCK12a administration since we had observed significant increases in CD8+ T cell numbers in NALT following rCK12a delivery at this time point. However, we were not able to observe any protection against live viral challenge following administration of rCK12a (i.p./I.N.). One explanation may be that the dose we used was too low to afford protection. In this regard, murine studies have shown no adjuvant effects of CCL19 when delivered at 50 μg/mouse but immunostimulatory effects when delivered at a dose of 100 μg/mouse (93). Our results may also be interpreted as follows: the high levels of protection against IHNV following I.N. IHNV vaccination require triggering a combination of immune factors and cells beyond CCL19 responses. Alternatively, rCK12a delivery may not result in sustained enough immune responses. Nasal vaccination with IHNV vaccine results in presence of the antigen for 4 days in the local environment (98). Thus, a single delivery of rCK12a may not have achieved the levels of this chemokine required for protection on the day of the challenge. In addition, the half-life of the recombinant protein may require multiple administrations of rCK12a in order to confer protection. Finally, since we did not co-administered rCK12a with a vaccine, our results may simply indicate that rCK12a alone did not stimulate specific immune responses in naïve rainbow trout. In other words, the increased CD8+ cell numbers we observed in the nasal mucosa are not protective unless those lymphocytes are antigen specific. Future studies should address whether co-delivery of rCK12a with a vaccine enhances vaccine efficacy and/or protection.
In summary, the present study demonstrates that the diversification of CCL19-like genes in salmonids resulted in the acquisition of one CCL19-like molecule, CK12, that plays a cardinal role in antiviral immune responses following nasal immunization. The ability of CCL19 to not only regulate local mucosal immune responses against viruses but also systemic ones underscores the biological role of chemokines as messengers between nasal and systemic lymphoid tissues of vertebrates. The presence of this CCL19-like driven immune mechanism in teleost fish suggests that this chemokine has evolutionary conserved roles in vertebrate immune systems.
Supplementary Material
Acknowledgments
The authors thank Dr. U. Fisher for the anti-trout CD8α antibody, Dr. C. Tafalla for the anti-trout MHC-II antibody, Dr. C. A. Johnston for the D. melanogaster recombinant protein and Michael Burnett for technical assistance with microscopy. We thank Dr. G. Wiens for insightful discussions on the trout genome and trout CC19 genes. The authors thank Lisboa Springs Hatchery for kindly providing animals and the staff at Clear Springs Foods Inc. for helping with sample collection.
This work was funded by USDA AFRI Grant # 2DN70-2RDN7 to IS, NIH COBRE grant # P20GM103452 and NIH 2R01GM085207-05 to JOS.
Footnotes
Abbreviations: NALT: nasopharynx-associated lymphoid tissue; OO: olfactory organ; LP: lamina propria; HKLs: head-kidney leucocytes; IHNV: infectious hematopoietic necrosis virus; I.N.: intranasal
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Le Y, Zhou Y, Iribarren P, Wang J. Chemokines and chemokine receptors: their manifold roles in homeostasis and disease. Cell Mol Immunol. 2004;1:95–104. [PubMed] [Google Scholar]
- 2.Viola A, Luster AD. Chemokines and their receptors: drug targets in immunity and inflammation. Annu Rev Pharmacol Toxicol. 2008;48:171–197. doi: 10.1146/annurev.pharmtox.48.121806.154841. [DOI] [PubMed] [Google Scholar]
- 3.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 4.Gale LM, McColl SR. Chemokines: extracellular messengers for all occasions? BioEssays. 1999;21:17–28. doi: 10.1002/(SICI)1521-1878(199901)21:1<17::AID-BIES3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 5.Melchjorsen J, Sørensen LN, Paludan SR. Expression and function of chemokines during viral infections: from molecular mechanisms to in vivo function. J Leukoc Biol. 2003;74:331–343. doi: 10.1189/jlb.1102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Takamura K, Fukuyama S, Nagatake T, Kim DY, Kawamura A, Kawauchi H, Kiyono H. Regulatory role of lymphoid chemokine CCL19 and CCL21 in the control of allergic rhinitis. J Immunol. 2007;179:5897–5906. doi: 10.4049/jimmunol.179.9.5897. [DOI] [PubMed] [Google Scholar]
- 8.Yoshie O, Imai T, Nomiyama H. Novel lymphocyte-specific CC chemokines and their receptors. J Leukoc Biol. 1997;62:634–644. doi: 10.1002/jlb.62.5.634. [DOI] [PubMed] [Google Scholar]
- 9.Zlotnik A, Morales J, Hedrick J. Recent advances in chemokines and chemokine receptors. Crit Rev Immunol. 1999;19:1. [PubMed] [Google Scholar]
- 10.Fukuyama S, Nagatake T, Kim DY, Takamura K, Park EJ, Kaisho T, Tanaka N, Kurono Y, Kiyono H. Cutting edge: uniqueness of lymphoid chemokine requirement for the initiation and maturation of nasopharynx-associated lymphoid tissue organogenesis. J Immunol. 2006;177:4276–4280. doi: 10.4049/jimmunol.177.7.4276. [DOI] [PubMed] [Google Scholar]
- 11.Rangel-Moreno J, Moyron-Quiroz J, Kusser K, Hartson L, Nakano H, Randall TD. Role of CXC chemokine ligand 13, CC chemokine ligand (CCL) 19, and CCL21 in the organization and function of nasal-associated lymphoid tissue. J Immunol. 2005;175:4904–4913. doi: 10.4049/jimmunol.175.8.4904. [DOI] [PubMed] [Google Scholar]
- 12.Choi YK, Fallert BA, Murphey-Corb MA, Reinhart TA. Simian immunodeficiency virus dramatically alters expression of homeostatic chemokines and dendritic cell markers during infection in vivo. Blood. 2003;101:1684–1691. doi: 10.1182/blood-2002-08-2653. [DOI] [PubMed] [Google Scholar]
- 13.Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood. 2006;107:2613–2618. doi: 10.1182/blood-2005-07-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker H, Beth Genter M. The olfactory system and the nasal mucosa as portals of entry of viruses, drugs, and other exogenous agents into the brain. In: Doty R, editor. Handbook of olfaction and gustation. Marcel Dekker; New York: 2003. pp. 549–574. [Google Scholar]
- 15.Mori I, Nishiyama Y, Yokochi T, Kimura Y. Olfactory transmission of neurotropic viruses. J Neurovirol. 2005;11:129–137. doi: 10.1080/13550280590922793. [DOI] [PubMed] [Google Scholar]
- 16.Munster VJ, Prescott JB, Bushmaker T, Long D, Rosenke R, Thomas T, Scott D, Fischer ER, Feldmann H, de Wit E. Rapid Nipah virus entry into the central nervous system of hamsters via the olfactory route. Scientific reports. 2012;2:736. doi: 10.1038/srep00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morales J, Herzog S, Kompter C, Frese K, Rott R. Axonal transport of Borna disease virus along olfactory pathways in spontaneously and experimentally infected rats. Med Microbiol Immunol. 1988;177:51–68. doi: 10.1007/BF00189527. [DOI] [PubMed] [Google Scholar]
- 18.Owen SJ, Batzloff M, Chehrehasa F, Meedeniya A, Casart Y, Logue CA, Hirst RG, Peak IR, Mackay-Sim A, Beacham IR. Nasal-associated lymphoid tissue and olfactory epithelium as portals of entry for Burkholderia pseudomallei in murine melioidosis. J Infect Dis. 2009;199:1761–1770. doi: 10.1086/599210. [DOI] [PubMed] [Google Scholar]
- 19.Mizgerd JP. Acute lower respiratory tract infection. N Engl J Med. 2008;358:716–727. doi: 10.1056/NEJMra074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Riel D, V, Munster J, De Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5N1 virus attachment to lower respiratory tract. Science. 2006;312:399–399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 21.Simoes EA. Respiratory syncytial virus infection. The Lancet. 1999;354:847–852. doi: 10.1016/S0140-6736(99)80040-3. [DOI] [PubMed] [Google Scholar]
- 22.Hament JM, Kimpen JL, Fleer A, Wolfs TF. Respiratory viral infection predisposing for bacterial disease: a concise review. FEMS Immunol Med Microbiol. 1999;26:189–195. doi: 10.1111/j.1574-695X.1999.tb01389.x. [DOI] [PubMed] [Google Scholar]
- 23.Davis S. Nasal vaccines. Adv Drug Delivery Rev. 2001;51:21–42. doi: 10.1016/s0169-409x(01)00162-4. [DOI] [PubMed] [Google Scholar]
- 24.Jabbal-Gill I. Nasal vaccine innovation. J Drug Target. 2010;18:771–786. doi: 10.3109/1061186X.2010.523790. [DOI] [PubMed] [Google Scholar]
- 25.Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med. 2005;11:S45–S53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 26.Neutra MR, Kozlowski PA. Mucosal vaccines: the promise and the challenge. Nat Rev Immunol. 2006;6:148–158. doi: 10.1038/nri1777. [DOI] [PubMed] [Google Scholar]
- 27.Tacchi L, Musharrafieh R, Larragoite ET, Crossey K, Erhardt EB, Martin SA, LaPatra SE, Salinas I. Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat Commun. 2014;5:6205. doi: 10.1038/ncomms6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sepahi A, Salinas I. The evolution of nasal immune systems in vertebrates. Mol Immunol. 2016;69:131–138. doi: 10.1016/j.molimm.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sepahi A, Casadei E, Tacchi L, Muñoz P, LaPatra SE, Salinas I. Tissue microenvironments in the nasal epithelium of rainbow trout (Oncorhynchus mykiss) define two distinct CD8α+ cell populations and establish regional immunity. J Immunol. 2016;197:4453–4463. doi: 10.4049/jimmunol.1600678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomiyama H, Osada N, Yoshie O. A family tree of vertebrate chemokine receptors for a unified nomenclature. Dev Comp Immunol. 2011;35:705–715. doi: 10.1016/j.dci.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 31.DeVries ME, Kelvin AA, Xu L, Ran L, Robinson J, Kelvin DJ. Defining the origins and evolution of the chemokine/chemokine receptor system. J Immunol. 2006;176:401–415. doi: 10.4049/jimmunol.176.1.401. [DOI] [PubMed] [Google Scholar]
- 32.Flajnik M, Du Pasquier L. Evolution of the immune system. In: Paul W, editor. Fundamental Immunology. 7. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 67–128. [Google Scholar]
- 33.Nomiyama H, Osada N, Yoshie O. The evolution of mammalian chemokine genes. Cytokine Growth Factor Rev. 2010;21:253–262. doi: 10.1016/j.cytogfr.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Laing KJ, Secombes CJ. Trout CC chemokines: comparison of their sequences and expression patterns. Mol Immunol. 2004;41:793–808. doi: 10.1016/j.molimm.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 35.Dixon B, Shum B, Adams EJ, Magor K, Hedrick RP, Muir DG, Parham P. CK-1, a putative chemokine of rainbow trout (Oncorhynchus mykiss) Immunol Rev. 1998;166:341–348. doi: 10.1111/j.1600-065x.1998.tb01274.x. [DOI] [PubMed] [Google Scholar]
- 36.Dixon B, Luque A, Abós B, Castro R, González-Torres L, Tafalla C. Molecular characterization of three novel chemokine receptors in rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2013;34:641–651. doi: 10.1016/j.fsi.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Ordás MC, Castro R, Dixon B, Sunyer JO, Bjork S, Bartholomew J, Korytar T, Köllner B, Cuesta A, Tafalla C. Identification of a novel CCR7 gene in rainbow trout with differential expression in the context of mucosal or systemic infection. Dev Comp Immunol. 2012;38:302–311. doi: 10.1016/j.dci.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peatman E, Liu Z. Evolution of CC chemokines in teleost fish: a case study in gene duplication and implications for immune diversity. Immunogenetics. 2007;59:613–623. doi: 10.1007/s00251-007-0228-4. [DOI] [PubMed] [Google Scholar]
- 39.Lally J, Al-Anouti F, Bols N, Dixon B. The functional characterisation of CK-1, a putative CC chemokine from rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2003;15:411–424. doi: 10.1016/s1050-4648(03)00008-1. [DOI] [PubMed] [Google Scholar]
- 40.Bird S, Tafalla C. Teleost chemokines and their receptors. Biology. 2015;4:756–784. doi: 10.3390/biology4040756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Q, Yang Y, Li C, Zeng Q, Zhou T, Li N, Liu Y, Li Y, Wang X, Liu S. The chemokinome superfamily: II. The 64 CC chemokines in channel catfish and their involvement in disease and hypoxia responses. Dev Comp Immunol. 2017;73:97–108. doi: 10.1016/j.dci.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Nomiyama H, Hieshima K, Osada N, Kato-Unoki Y, Otsuka-Ono K, Takegawa S, Izawa T, Yoshizawa A, Kikuchi Y, Tanase S. Extensive expansion and diversification of the chemokine gene family in zebrafish: identification of a novel chemokine subfamily CX. BMC Genomics. 2008;9:222. doi: 10.1186/1471-2164-9-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Hu Y-h, Xiao Z-z, Sun L. SmCCL19, a CC chemokine of turbot Scophthalmus maximus, induces leukocyte trafficking and promotes anti-viral and anti-bacterial defense. Fish Shellfish Immunol. 2013;35:1677–1682. doi: 10.1016/j.fsi.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 44.Chen S, Liu Y, Dong X, Meng L. Cloning, characterization, and expression analysis of a CC chemokine gene from turbot (Scophthalmus maximus) Fish Physiol Biochem. 2010;36:147–155. doi: 10.1007/s10695-008-9218-1. [DOI] [PubMed] [Google Scholar]
- 45.Arockiaraj J, Bhatt P, Harikrishnan R, Arasu MV, Al-Dhabi NA. Molecular and functional roles of 6C CC chemokine 19 in defense system of striped murrel Channa striatus. Fish Shellfish Immunol. 2015;45:817–827. doi: 10.1016/j.fsi.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Reyes-López FE, Romeo JS, Vallejos-Vidal E, Reyes-Cerpa S, Sandino AM, Tort L, Mackenzie S, Imarai M. Differential immune gene expression profiles in susceptible and resistant full-sibling families of Atlantic salmon (Salmo salar) challenged with infectious pancreatic necrosis virus (IPNV) Dev Comp Immunol. 2015;53:210–221. doi: 10.1016/j.dci.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 47.Montero J, Ordas MC, Alejo A, Gonzalez-Torres L, Sevilla N, Tafalla C. CK12, a rainbow trout chemokine with lymphocyte chemo-attractant capacity associated to mucosal tissues. Mol Immunol. 2011;48:1102–1113. doi: 10.1016/j.molimm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Montero J, Garcia J, Ordas MC, Casanova I, Gonzalez A, Villena A, Coll J, Tafalla C. Specific regulation of the chemokine response to viral hemorrhagic septicemia virus at the entry site. J Virol. 2011;85:4046–4056. doi: 10.1128/JVI.02519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaves-Pozo E, Montero J, Cuesta A, Tafalla C. Viral hemorrhagic septicemia and infectious pancreatic necrosis viruses replicate differently in rainbow trout gonad and induce different chemokine transcription profiles. Dev Comp Immunol. 2010;34:648–658. doi: 10.1016/j.dci.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 50.Montero J, Chaves-Pozo E, Cuesta A, Tafalla C. Chemokine transcription in rainbow trout (Oncorhynchus mykiss) is differently modulated in response to viral hemorrhagic septicaemia virus (VHSV) or infectious pancreatic necrosis virus (IPNV) Fish Shellfish Immunol. 2009;27:661–669. doi: 10.1016/j.fsi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, Bento P, Da Silva C, Labadie K, Alberti A. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014:5. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tacchi L, Larragoite E, Salinas I. Discovery of J Chain in African Lungfish (Protopterus dolloi, Sarcopterygii) Using High Throughput Transcriptome Sequencing: Implications in Mucosal Immunity. PLoS One. 2013;8:e70650. doi: 10.1371/journal.pone.0070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016:msw054. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campanella JJ, Bitincka L, Smalley J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics. 2003;4:1. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y-A, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salinas I, Maas E, Muñoz P. Characterization of acid phosphatases from marine scuticociliate parasites and their activation by host’s factors. Parasitology. 2011;138:836–847. doi: 10.1017/S0031182011000527. [DOI] [PubMed] [Google Scholar]
- 60.Salinas I, Meseguer J, Esteban MÁ. Assessment of different protocols for the isolation and purification of gut associated lymphoid cells from the gilthead seabream (Sparus aurata L.) Biol Proced Online. 2007;9:43–55. doi: 10.1251/bpo132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takizawa F, Dijkstra JM, Kotterba P, Korytář T, Kock H, Köllner B, Jaureguiberry B, Nakanishi T, Fischer U. The expression of CD8α discriminates distinct T cell subsets in teleost fish. Dev Comp Immunol. 2011;35:752–763. doi: 10.1016/j.dci.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 62.Granja AG, Leal E, Pignatelli J, Castro R, Abós B, Kato G, Fischer U, Tafalla C. Identification of Teleost Skin CD8α+ Dendritic-like Cells, Representing a Potential Common Ancestor for Mammalian Cross-Presenting Dendritic Cells. J Immunol. 2015;195:1825–1837. doi: 10.4049/jimmunol.1500322. [DOI] [PubMed] [Google Scholar]
- 63.Sepahi A, Cordero H, Goldfine H, Esteban MÁ, Salinas I. Symbiont-derived sphingolipids modulate mucosal homeostasis and B cells in teleost fish. Scientific reports. 2016;6:39054. doi: 10.1038/srep39054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veldkamp CT, Kiermaier E, Gabel-Eissens SJ, Gillitzer ML, Lippner DR, DiSilvio FA, Mueller CJ, Wantuch PL, Chaffee GR, Famiglietti MW. Solution structure of CCL19 and identification of overlapping CCR7 and PSGL-1 binding sites. Biochemistry. 2015;54:4163–4166. doi: 10.1021/acs.biochem.5b00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 67.Parkin J, Cohen B. An overview of the immune system. The Lancet. 2001;357:1777–1789. doi: 10.1016/S0140-6736(00)04904-7. [DOI] [PubMed] [Google Scholar]
- 68.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 69.Randall TD, Carragher DM, Rangel-Moreno J. Development of secondary lymphoid organs. Annu Rev Immunol. 2008;26:627. doi: 10.1146/annurev.immunol.26.021607.090257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Comerford I, Harata-Lee Y, Bunting MD, Gregor C, Kara EE, McColl SR. A myriad of functions and complex regulation of the CCR7/CCL19/CCL21 chemokine axis in the adaptive immune system. Cytokine Growth Factor Rev. 2013;24:269–283. doi: 10.1016/j.cytogfr.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 71.Marsland BJ, Bättig P, Bauer M, Ruedl C, Lässing U, Beerli RR, Dietmeier K, Ivanova L, Pfister T, Vogt L. CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity. 2005;22:493–505. doi: 10.1016/j.immuni.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 72.Bachmann MF, Kopf M, Marsland BJ. Chemokines: more than just road signs. Nat Rev Immunol. 2006;6:159–164. doi: 10.1038/nri1776. [DOI] [PubMed] [Google Scholar]
- 73.Luther SA, Cyster JG. Chemokines as regulators of T cell differentiation. Nat Immunol. 2001;2:102–107. doi: 10.1038/84205. [DOI] [PubMed] [Google Scholar]
- 74.Haessler U, Pisano M, Wu M, Swartz MA. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc Natl Acad Sci U S A. 2011;108:5614–5619. doi: 10.1073/pnas.1014920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Randolph GJ. Semin Immunol. Elsevier; 2001. Dendritic cell migration to lymph nodes: cytokines, chemokines, and lipid mediators; pp. 267–274. [DOI] [PubMed] [Google Scholar]
- 76.Corcione A, Arduino N, Ferretti E, Raffaghello L, Roncella S, Rossi D, Fedeli F, Ottonello L, Trentin L, Dallegri F. CCL19 and CXCL12 trigger in vitro chemotaxis of human mantle cell lymphoma B cells. Clin Cancer Res. 2004;10:964–971. doi: 10.1158/1078-0432.ccr-1182-3. [DOI] [PubMed] [Google Scholar]
- 77.Reif K, Okkenhaug K, Sasaki T, Penninger JM, Vanhaesebroeck B, Cyster JG. Cutting edge: differential roles for phosphoinositide 3-kinases, p110γ and p110δ, in lymphocyte chemotaxis and homing. J Immunol. 2004;173:2236–2240. doi: 10.4049/jimmunol.173.4.2236. [DOI] [PubMed] [Google Scholar]
- 78.Okada T, Ngo VN, Ekland EH, Förster R, Lipp M, Littman DR, Cyster JG. Chemokine requirements for B cell entry to lymph nodes and Peyer’s patches. J Exp Med. 2002;196:65–75. doi: 10.1084/jem.20020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bromley SK, Thomas SY, Luster AD. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol. 2005;6:895–901. doi: 10.1038/ni1240. [DOI] [PubMed] [Google Scholar]
- 80.Riol-Blanco L, Sánchez-Sánchez N, Torres A, Tejedor A, Narumiya S, Corbí AL, Sánchez-Mateos P, Rodríguez-Fernández JL. The chemokine receptor CCR7 activates in dendritic cells two signaling modules that independently regulate chemotaxis and migratory speed. J Immunol. 2005;174:4070–4080. doi: 10.4049/jimmunol.174.7.4070. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, Irvine DJ. Engineering chemoattractant gradients using chemokine-releasing polysaccharide microspheres. Biomaterials. 2011;32:4903–4913. doi: 10.1016/j.biomaterials.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campbell JJ, Murphy KE, Kunkel EJ, Brightling CE, Soler D, Shen Z, Boisvert J, Greenberg HB, Vierra MA, Goodman SB. CCR7 expression and memory T cell diversity in humans. J Immunol. 2001;166:877–884. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- 83.Muthuswamy R, Mueller-Berghaus J, Haberkorn U, Reinhart TA, Schadendorf D, Kalinski P. PGE2 transiently enhances DC expression of CCR7 but inhibits the ability of DCs to produce CCL19 and attract naive T cells. Blood. 2010;116:1454–1459. doi: 10.1182/blood-2009-12-258038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scandella E, Men Y, Gillessen S, Förster R, Groettrup M. Prostaglandin E2 is a key factor for CCR7 surface expression and migration of monocyte-derived dendritic cells. Blood. 2002;100:1354–1361. doi: 10.1182/blood-2001-11-0017. [DOI] [PubMed] [Google Scholar]
- 85.Scandella E, Men Y, Legler DF, Gillessen S, Prikler L, Ludewig B, Groettrup M. CCL19/CCL21-triggered signal transduction and migration of dendritic cells requires prostaglandin E2. Blood. 2004;103:1595–1601. doi: 10.1182/blood-2003-05-1643. [DOI] [PubMed] [Google Scholar]
- 86.Côté SC, Pasvanis S, Bounou S, Dumais N. CCR7-specific migration to CCL19 and CCL21 is induced by PGE 2 stimulation in human monocytes: involvement of EP 2/EP 4 receptors activation. Mol Immunol. 2009;46:2682–2693. doi: 10.1016/j.molimm.2008.08.269. [DOI] [PubMed] [Google Scholar]
- 87.Schumann K, Lämmermann T, Bruckner M, Legler DF, Polleux J, Spatz JP, Schuler G, Förster R, Lutz MB, Sorokin L. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity. 2010;32:703–713. doi: 10.1016/j.immuni.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 88.Lämmermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Söldner R, Hirsch K, Keller M, Förster R, Critchley DR, Fässler R. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 89.Yanagawa Y, Onoé K. CCL19 induces rapid dendritic extension of murine dendritic cells. Blood. 2002;100:1948–1956. doi: 10.1182/blood-2002-01-0260. [DOI] [PubMed] [Google Scholar]
- 90.Purcell MK, Nichols KM, Winton JR, Kurath G, Thorgaard GH, Wheeler P, Hansen JD, Herwig RP, Park LK. Comprehensive gene expression profiling following DNA vaccination of rainbow trout against infectious hematopoietic necrosis virus. Mol Immunol. 2006;43:2089–2106. doi: 10.1016/j.molimm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 91.Hansen JD, La Patra S. Induction of the rainbow trout MHC class I pathway during acute IHNV infection. Immunogenetics. 2002;54:654–661. doi: 10.1007/s00251-002-0509-x. [DOI] [PubMed] [Google Scholar]
- 92.Ballesteros NA, Alonso M, Saint-Jean SR, Perez-Prieto SI. An oral DNA vaccine against infectious haematopoietic necrosis virus (IHNV) encapsulated in alginate microspheres induces dose-dependent immune responses and significant protection in rainbow trout (Oncorrhynchus mykiss) Fish Shellfish Immunol. 2015;45:877–888. doi: 10.1016/j.fsi.2015.05.045. [DOI] [PubMed] [Google Scholar]
- 93.Hu K, Luo S, Tong L, Huang X, Jin W, Huang W, Du T, Yan Y, He S, Griffin GE. CCL19 and CCL28 augment mucosal and systemic immune responses to HIV-1 gp140 by mobilizing responsive immunocytes into secondary lymph nodes and mucosal tissue. J Immunol. 2013;191:1935–1947. doi: 10.4049/jimmunol.1300120. [DOI] [PubMed] [Google Scholar]
- 94.Song J-H, Kim J-I, Kwon H-J, Shim D-H, Parajuli N, Cuburu N, Czerkinsky C, Kweon M-N. CCR7-CCL19/CCL21-regulated dendritic cells are responsible for effectiveness of sublingual vaccination. J Immunol. 2009;182:6851–6860. doi: 10.4049/jimmunol.0803568. [DOI] [PubMed] [Google Scholar]
- 95.Salinas I. The mucosal immune system of teleost fish. Biology. 2015;4:525–539. doi: 10.3390/biology4030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Groom JR. Moving to the suburbs: T-cell positioning within lymph nodes during activation and memory. Immunol Cell Biol. 2015;93:330. doi: 10.1038/icb.2015.29. [DOI] [PubMed] [Google Scholar]
- 97.von Andrian UH, Mempel TR. Homing and cellular traffic in lymph nodes. Nat Rev Immunol. 2003;3:867. doi: 10.1038/nri1222. [DOI] [PubMed] [Google Scholar]
- 98.Larragoite ET, Tacchi L, LaPatra SE, Salinas I. An attenuated virus vaccine appears safe to the central nervous system of rainbow trout (Oncorhynchus mykiss) after intranasal delivery. Fish Shellfish Immunol. 2016;49:351–354. doi: 10.1016/j.fsi.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.