Abstract
Recent fate mapping studies and gene expression profiles suggest that commonly used protocols to generate bone marrow-derived cultured dendritic cells yield a heterogeneous mixture, including some CD11chi cells that may not have a bona fide counterpart in vivo. In this study, we provide further evidence of the discordance between ex vivo-isolated and in vitro-cultured CD11c+ cells by analyzing an additional phenotype, the ability to support cytosolic growth of the facultative intracellular bacterial pathogen Listeria monocytogenes. Two days after foodborne infection of mice with GFP-expressing L. monocytogenes, a small percentage of both CD103neg and CD103+ conventional DC (cDC) in the intestinal lamina propria and MLN were GFP+. However, in vitro infection of the same subsets of cells harvested from naïve mice resulted in inefficient invasion by the bacteria (< 0.1 % of the inoculum). The few intracellular bacteria detected survived for only a few hours. In contrast, cultured CD103negCD11c+ cells induced by GM-CSF readily supported exponential growth of L. monocytogenes. Flt3L-induced cultures yielded CD103+CD11c+ cells that more closely resembled cDC with only a modest level of L. monocytogenes replication. For both culture protocols, the longer the cells were maintained in vitro, the more readily they supported intracellular growth. The results of this study suggest that cDC are not a niche for intracellular growth of L. monocytogenes during intestinal infection of mice.
Introduction
Conventional dendritic cells (cDC) respond to infection by secreting cytokines, migrating to draining lymph nodes, and presenting antigen to T cells. Despite the critical role of cDC in mediating both innate and adaptive immunity, direct interactions between the foodborne intracellular bacterial pathogen Listeria monocytogenes and the various subsets of DC found in vivo remain largely undefined. Most of what is known about the growth of L. monocytogenes inside dendritic cells is derived from either in vitro-generated bone marrow-derived cultured cells, or inferred from systemic infection of mice after global depletion of CD11c+ cells (1,2).
Early studies showed that GM-CSF was an important media component for generating both human and mouse DC in vitro (3–6). Bone marrow-derived cells cultured in the presence of GM-CSF are useful for in vitro studies to efficiently internalize and present antigen to CD4+ T cells or to cross-present antigen to CD8+ T cells. As a result of their frequent use in such assays, CD11c+ GM-CSF-derived cells are often thought of as prototypic DC. However, it is now appreciated that different subtypes of DC can arise from either circulating monocytes or common dendritic progenitors in the bone marrow (7–9). For example, activation of the Flt3 receptor by Flt3-L promotes the differentiation of committed DC progenitors into cDC that express CD8α+ and CD103+ in both lymphoid and non-lymphoid tissues, including the gut lamina propria (10,11). Flt3-L can also be used as a media supplement to generate CD103+ CD11chi cells from bone marrow progenitors (12,13).
cDC subsets in the intestinal lamina propria and the draining mesenteric lymph nodes (MLN) have unique characteristics compared to splenic cDC, which are likely caused by continual exposure to gut-derived microbial products and dietary antigens (14). CD103 (integrin αE chain) is expressed on the majority of DC in the intestinal lamina propria including both BATF3-dependent cDC1 and IRF4-dependent cDC2, two recently described subsets that are derived from circulating pre-DC (15). Expression of CD103 by DC in the MLN is correlated with the induction of Foxp3+ regulatory T cells and gut-homing T cells (16,17). In contrast, CD103neg DC are considered more inflammatory due to a greater ability to secrete TNF-α and IL-6 after in vitro stimulation (16), as well as efficiently inducing IFN-γ- and IL-17-producing effector T cells (11). In the intestinal LP, both CD103neg and CD103+ DC express CCR7 and migrate in lymphatic fluid (11).
L. monocytogenes are facultative intracellular pathogens, and much research effort has focused on understanding the ability of these organisms to invade a wide variety of mammalian cells, survive and replicate in the cytosol, and spread to neighboring cells. However, we recently showed that following oral transmission, a large proportion of L. monocytogenes in the gut are actually extracellular (18). Intracellular growth was not required to establish infection in either the ileum or the colon, but replication in an as yet unidentified cell type was critical for colonization of the MLN (18). Ly6Chi monocytes were the predominant cell type that associated with L. monocytogenes in the MLN, but invasion of these cells is inefficient, and they did not serve as an intracellular growth niche for L. monocytogenes (19). In this study, we examined the ability of L. monocytogenes to invade, survive, and replicate in dendritic cells. We found that cDC harvested from the MLN of naïve mice and infected directly ex vivo did not support intracellular growth of the bacteria. In contrast, L. monocytogenes replicated exponentially inside in vitro-generated, bone marrow-derived CD11c+ cells, which suggests that these cultured cells do not closely resemble intestinal cDC.
Materials and Methods
Bacteria
Mouse-adapted (InlAm-expressing) L. monocytogenes EGDe derivatives SD2000, SD2710 (constitutive GFP+) and SD2001 (GFPneg vector control) were described previously (18). All strains were grown in Brain Heart Infusion (BHI) broth shaking at 30°C to early stationary phase, aliquoted, and stored at 80°C.
Mice
Female BALBc/By/J (BALB) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) at 4 weeks of age and housed in a specific-pathogen free facility (9 AM – 7 PM dark cycle). Mice were 6–9 weeks old when used in experiments. All procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee.
Foodborne infection
Aliquots of L. monocytogenes were thawed, incubated at 30°C in BHI broth without shaking for 1.5 h, washed once with PBS, and then suspended in a 2:3 mixture of PBS and salted sweet cream butter (Kroger). A 2–3 cm piece of white bread (Kroger) saturated with L. monocytogenes was fed to mice near the onset of their dark cycle as described previously (20,21). The concentration of L. monocytogenes was estimated from the known titer of the bacterial aliquot and the actual inoculum was determined by plating serial dilutions of homogenized contaminated bread pieces.
Isolation of MLN and intestinal lamina propria cells
Lymph nodes were cut into 4 pieces each, and the total MLN were placed in 4 ml of RP-5 media (RPMI 1640 (Invitrogen 21870)/20 mM HEPES/5% FBS). Collagenase type IV (300 U/ml; Worthington) and DNase I (120 U/ml; Worthington) were added and the nodes were digested for 30 min at 37°C shaking at 250 rpm in a 50 ml conical tube containing a sterile stir bar (2 cm). Large intestines (cecum and colon) were flushed with 8 ml cold CMF buffer (Ca2+/Mg2+-free HBSS/10 mM HEPES/25 mM sodium bicarbonate/2% FBS) and then everted using a sterile weaving needle with button thread (22). Mucus was removed by shaking in a 50 ml conical tube with 25 ml CMF for 1 min. Epithelial cells were removed and the LP cells were isolated from the interface of a 44%/70% Percoll gradient as described previously (20).
Flow cytometry
Antibodies specific for CD16/CD32 (93), CD45 (30-F11), CD11c (N418), CD11b (M1/70), Ly6G (1A8-Ly6g), B220 (RA3-6B2), MHC-II (M5/114.15.2), CD3 (145-2C11), F4/80 (BM8), CD19 (eBio1D3) from eBioscience; Ly6C (HK1.4) and CD103 (2E7) from BioLegend were used. Data were acquired using an iCyt Synergy and analyzed with FlowJo (Tree Star). The percentage of Listeria-associated (GFP+) cells in each population was determined by using cells from mice infected with L. monocytogenes SD2001 as a negative gating control in each experiment as described previously (18,19).
Ex vivo infection of cDC
Sorted CD103+ or CD103neg cDC isolated from the MLN were seeded in 96-well round-bottom ultra-low attachment plates (Corning) and incubated in media with 20% FBS (100 μl) for at least 30 min at 37°C in 7% CO2 prior to infection. In some experiments, cells were plated in half-area 96-well dishes to increase cell density; however, no differences were noted based on the type of dish used. Due to the relatively low yield of sorted cDC (~0.4 to 3 × 104 per mouse), cells were divided equally into two wells and infected at a normalized MOI. The cells isolated from each mouse were used to analyze infection at two different time points; no technical replicates were plated. Thus, sorted cells from individual mice served as biological replicates for each experiment. Cells were used for intracellular growth assays as indicated below except that only one wash was used to remove extracellular bacteria.
Intracellular growth assay
Gentamicin protection assays were used to identify the number of intracellular CFU. Cultured cells were seeded in either 96-well round bottom (1 × 105/well) or 24-well (2.5 × 105/well) flat-bottom ultra-low attachment dishes (Corning). Where indicated, 12 mm diameter glass coverslips were placed in the wells prior to cell seeding. Aliquots of L. monocytogenes were thawed, incubated at shaking at 37°C in BHI broth for 1.5 h, washed once with PBS, and then suspended at the appropriate concentration. The concentration of L. monocytogenes was estimated from the known titer of the bacterial aliquot and the actual MOI was determined by plating serial dilutions of the inoculum. Extracellular bacteria were removed 30–60 min after infection by washing 3× with pre-warmed HBSS and then suspended in RP-10 media containing 10 μg/ml gentamicin. The percentage of inoculum internalized was calculated by dividing the number of CFU recovered after at least 20 min. exposure to gentamicin by the total number of CFU added to each well. At each time point, cells were harvested, washed once, lysed in sterile H2O and plated on BHI agar.
In vitro generation of bone marrow-derived CD11c+ cells
Bone marrow was flushed from the femurs and tibias of uninfected mice and suspended in 10 ml RPMI 1640 (Invitrogen # 21870), L-glutamine, HEPES, 2-ME, and 10% FBS (RP-10 media) in 100 mm non-TC petri plates. For GM-CSF-induced cultures, the protocol of Lutz et al. (23) was followed using 3% J558 supernatant as a source of GM-CSF. For Flt3-L-induced cultures, CHO FLT3-L-secreting cells (provided by Thomas Mitchell, University of Louisville with permission from Nick Nicola, The Walter and Eliza Hall Institute of Medical Research) were used as a source of Flt3-L as described (24). For day 16 cultures, 1.5 × 106 bone marrow cells/ml were cultured in RP-10 with 12.5% Flt-3L supernatant plus 0.75% J558 supernatant as described (25). 5 ml of fresh media was added on day 5 and non-adherent cells were removed on day 9 and transferred to new plates in fresh media (3 × 105 cells/ml). For day 6 cultures, 1.5 × 106 bone marrow cells/ml were cultured in RP-10 supplemented with 20% Flt3-L supernatant and no J558 supernatant. 10 ml of fresh media was added on day 3.
Microscopy
For Diff-Quik® (Dade-Behring) staining, cells were spun onto Superfrost slides (VWR) for 6 min. at 600 rpm using a Cytospin. Cells were then fixed in methanol 5 sec, stained in solution I for 10 sec, and stained in solution II 5 sec. Cells were dried and mounted with Permount®under glass coverslips. For F-actin staining, cells were spun onto slides, air-dried, fixed with formalin for 10 min, washed 3 times with PBS, and then permeabilized in TBS-T (TBS/0.1% Triton X-100/1% BSA, pH=8.8) for 15 min at room temp. Texas Red®-X Phalloidin (ThermoFisher) was added for 20 min at room temp. Slides were then washed 8 times in TBS-T, and 8 times with TBS alone. For differential “in/out” staining of L. monocytogenes, cells were washed 3 times with cold buffer (Ca2+/Mg2+-free HBSS/1% FBS/1 mM EDTA) and then incubated with Difco Listeria O Antiserum Poly (BD Biosciences cat. # 223021) (1:10) in PBS with 3% BSA for 20 min on ice. Washed cells were then incubated with goat anti-rabbit IgG-Texas Red® (ThermoFisher) for 20 min on ice. Stained cells were spun onto poly-L-lysine-coated Superfrost slides (VWR), formalin fixed at 4°C for 10 min, washed with PBS, and mounted under coverslips with ProLong® Diamond antifade (Molecular Probes). All cells were visualized using a Zeiss Axio Imager.Z1 with a 100×/1.4NA PlanApo oil immersion objective and analyzed with AxioVision software.
Statistics
Statistical analysis was performed using Prism for Macintosh (version 6; Graph Pad). P values of <0.05 were considered significant and are indicated as follows: *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
Results
L. monocytogenes associate with both CD103+ and CD103neg DC in the gut
Ly6Chi monocytes were previously identified as the primary cell type associated with GFP-expressing L. monocytogenes in the gut-draining MLN two days after foodborne infection with either wild type or murinized (InlAm-expressing) bacteria (19). CD11c+ cells represented only a minor fraction of the total GFP+ cells at that early time point, but the number of L. monocytogenes-associated CD11c+ cells did increase slightly by 72 hpi. In this study, we used a similar flow cytometric approach to confirm that oral infection also resulted in an association of the bacteria with CD11c+ cells in the intestinal lamina propria (Fig. S1). Only 1.8% of all GFP+ cells were Ly6G−Ly6C−/+CD11chi (Fig. S1D), similar to what was observed previously in the MLN (19). The key difference noted in the lamina propria was that the majority of L. monocytogenes-associated cells were Ly6Ghi neutrophils, rather than Ly6Chi monocytes.
CD103+ cDC can express E-cadherin (26), and binding of the L. monocytogenes surface protein InlA to E-cadherin is known to induce a “zipper-like” mechanism of bacterial uptake (27). Therefore, we postulated that L. monocytogenes would associate primarily with CD103+ subsets of DC in the gut. To test this, female BALB/c/By/J mice were fed 108 CFU of a mouse-adapted L. monocytogenes strain that constitutively expressed GFP. Two days post-infection (dpi), mononuclear cells from the large intestine lamina propria were analyzed by flow cytometry (Fig. 1A) and the percentage of L. monocytogenes-associated (GFP+) cells in each cDC subset was determined using the approach outlined in Fig. S1C. Approximately equal numbers of CD103neg and CD103+ cDC were isolated from the large intestine lamina propria of each infected mouse (Fig. 1B). As expected, only a minor fraction of the cDC was GFP+. However, there was no preferential association of L. monocytogenes with the CD103+ cells. Thus, within 48 hours of foodborne transmission, L. monocytogenes were directly interacting with both CD103+ and CD103neg cDC in the intestinal lamina propria.
FIGURE 1.
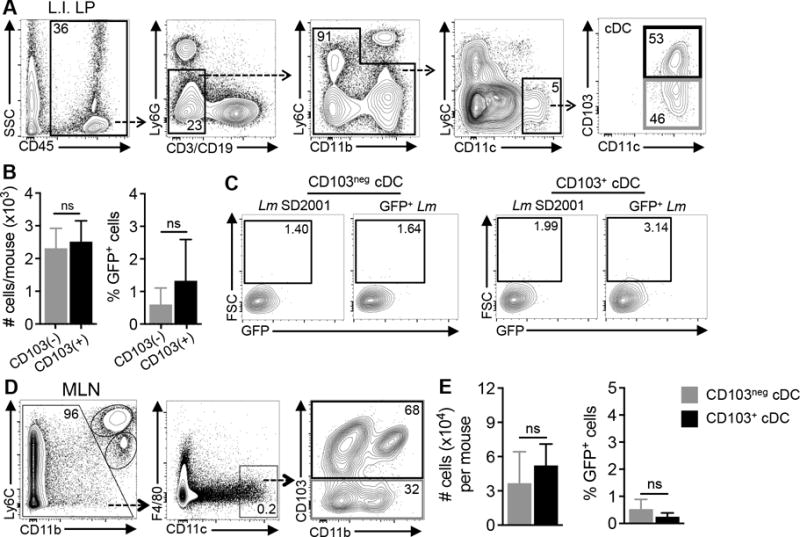
L. monocytogenes do not preferentially associate with CD103+ cDC in the gut. BALB mice were fed 1–6 × 108 CFU of either Lm SD2710 (GFP+) or Lm SD2001 (vector control). (A and C) Gating schemes used to identify CD103neg and CD103+ cDC isolated from the large intestine lamina propria (LI-LP) 48 hpi or MLN 72 hpi. (B and D) Mean (±SD) total number of cDC (left graph) and mean (±SD) percentage of GFP+ (Lm-associated) cDC in each subset (right graph). Pooled data for n=6 mice (LI-LP) or n=4 mice (MLN) from at least two separate experiments are shown. Statistical significance was determined using Mann-Whitney analysis.
CD103+ cDC are migratory cells that traffic from the gut to the MLN (28). To find out if L. monocytogenes preferentially associated with the CD103+ cells that disseminated to the draining lymph nodes, we also analyzed total cells from the MLN at 72 hpi (Fig. 1C). Similar numbers of CD103+ and CD103neg cDC were found in the MLN and there was still no preferential association of L. monocytogenes with either cDC subset (Fig. 1D). Similar numbers of GFP+ cells were found in the CD11b+ and CD11bneg subsets (data not shown), suggesting that L. monocytogenes did not preferentially associate with any of the four subsets of intestinal DC. To find out if the cells contained live, replicating intracellular bacteria, sorted cells were lysed and plated on BHI agar. In the majority of cases, no CFU were detected; the cells from one mouse yielded one CFU (data not shown). This was not a surprising result given the small number of cDC that were isolated and the low percentage of cells that were GFP+ two days post-infection; the expected number of CFU would be just at or below the limit of detection using this approach. However, it was also possible that 48 hours after infection, most of the cDC present in gut had been exposed to pro-inflammatory cytokines that activated the cells to restrict intracellular growth of L. monocytogenes.
L. monocytogenes inefficiently invade cDC and cannot sustain intracellular survival
To find out if either CD103neg or CD103+ cDC were capable of supporting intracellular growth of L. monocytogenes, the cells were isolated from the MLN of uninfected mice and then infected ex vivo (Fig. 2A). L. monocytogenes was added to approximately 1 × 104 sorted cells per well in low attachment plates at an MOI of 10. One hour after infection, only 0.1% of the inoculum added to either CD103neg or CD103+ DC was protected from gentamicin (Fig. 2B). By comparison, adherent phagocytic cells such as bone marrow-derived macrophages can take up nearly all of an inoculum within 1 hour, and phagocytose about 1–5 % of the bacteria if the cells are cultured in low adherence dishes (19). Non-phagocytic cells such as Caco-2 intestinal epithelial cells also typically internalize 1–3% of L. monocytogenes when infected at high MOI (19,29). No gentamicin resistant bacteria were recovered when the cells were infected at low dose (MOI=0.1–0.5) or at an intermediate dose (MOI=1–5) (data not shown). Therefore, the cDC infected directly ex vivo did not efficiently internalize mouse-adapted L. monocytogenes.
FIGURE 2.
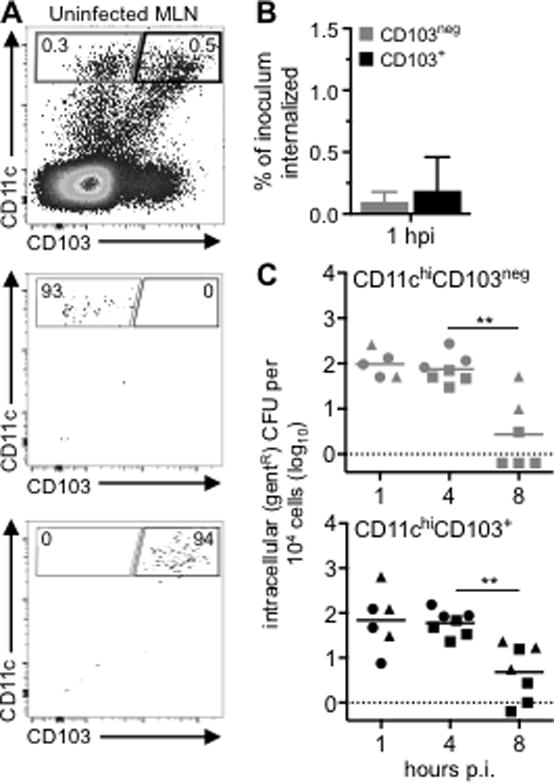
cDC isolated from the MLN of naïve mice do not support intracellular growth of L. monocytogenes. (A) CD11chiCD103neg or CD11chiCD103+ cells were sorted from the MLN of uninfected BALB mice. Sorted cells were infected with Lm SD2000 (MOI=10–14) directly ex vivo. (B) Mean percentage (±SD) of the Lm inoculum that was resistant to 10 μg/ml gentamicin (gentR) after 1 h. (C) Intracellular growth assay for CD103neg (grey) and CD103+ (black) cDC infected directly ex vivo. Pooled data from three separate experiments (designated by circles, squares, & triangles) are shown. Cells sorted from a single mouse were used at two time points. Statistical significance was determined using Mann-Whitney analysis.
After incubating for three additional hours (4 hpi), the number of gentamicin-resistant CFU recovered from the cDC remained the same (Fig. 2C). This suggested that intracellular L. monocytogenes survived, but did not replicate inside the cDC. To find out if the intracellular bacteria had escaped from the phagocytic vacuole into the cytosol, we stained F-actin and examined the cells microscopically for the presence of actin tails (Fig. 3A). Of 137 infected cells recovered from three different mice, only three cells harbored bacteria with actin tails (Fig. 3B). The vast majority of GFP+ cells 4 hpi did not co-localize with actin, and most cells were associated with only one to three bacilli per cell (Fig. 3C). By eight hours post-infection, the number of intracellular L. monocytogenes had decreased significantly in both CD103neg and CD103+ cDC, and in some cases, no gentamicin-resistant bacteria were recovered (Fig. 2C). Together, these results indicate that L. monocytogenes are inefficiently taken up by cDC, and those few intracellular bacteria that survive have a truncated life cycle compared with L. monocytogenes intracellular growth previously observed in macrophages.
FIGURE 3.
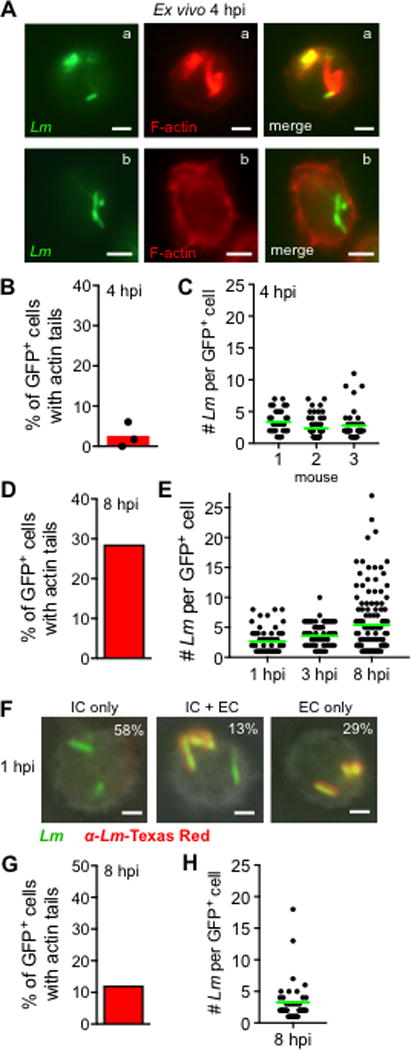
Microscopic analysis of L. monocytogenes-infected CD11c+ cells. (A-C) CD11chi cells were sorted from the MLN of uninfected BALB mice and infected with Lm SD2710 (MOI=8) directly ex vivo for 1 h. Cells were spun onto slides and stained with phalloidin-Texas Red 4 hpi. (A) Representative images: cell in “a” contains Lm associated with actin tails and cell in “b” contains Lm not associated with actin. (B) Bar indicates mean percentage of GFP+ cells that had Lm associated with actin tails from each mouse (n=3). (C) Dots represent the number of Lm associated with each infected cell; horizontal green lines indicate mean values. (D-E) Bone marrow-derived CD11c+MHC-IIhi cells sorted after 6 d of culture in GM-CSF were infected for 30 min with Lm SD2710 (MOI = 0.2). A total of 300 cells were examined for each time point. (D) Percentage of Lm-infected cells that contained bacteria with actin tails. (E) Dots indicate the number of bacilli associated with each infected cell. (F-H) Bone marrow-derived cells cultured in FLT3-L for 6 d were infected with Lm SD2710 (MOI=1). (F) Representative images for differential “in/out” staining of unpermebilized cells spun onto slides and stained with Lm-specific antibodies 1 hpi. The percentage of cells with intracellular bacteria (IC only), both intracellular and extracellular (IC + EC), or only extracellular (EC only) are given in the upper right corner of each image. A total of 400 cells were counted. (G) Percentage of Lm-infected cells that contained bacteria with actin tails. (H) Dots indicate the number of Lm associated with each infected cell 8 hpi. Scale bars, 2 μm.
In vitro-generated, bone marrow-derived CD103negCD11c+ cells readily support intracellular growth of L. monocytogenes
Westcott et al. previously reported that bone marrow-derived cultured CD11chi cells were a suboptimal niche for intracellular growth of L. monocytogenes when compared to bone marrow-derived macrophages (30). However, in contrast to our results with ex vivo infections, in that study, L. monocytogenes survived inside the CD11c+ cells for at least 8 hours and the number of gentamicin-resistant bacteria grew 30-fold during that time period. To explain this discrepancy and to better understand the culture conditions that lead to an intracellular growth phenotype, we re-examined the ability of L. monocytogenes to replicate in cultured CD11c+ cells.
In the study by Westcott et al., bone marrow cells were cultured with rGM-CSF for six days before the loosely adherent CD11c+ cells were magnetically enriched, allowed to adhere to glass coverslips, and then infected. In contrast, the ex vivo cDC infections shown in Fig. 2 were conducted in round-bottom low-attachment plates. Since in vitro culture of bone marrow cells with GM-CSF also generates adherent cells with more macrophage-like features (3), it was possible that allowing the cells to attach was promoting efficient invasion and replication. To test this, bone marrow cells were cultured for eight days in the presence of GM-CSF containing cell supernatants and then the loosely adherent cells were transferred to low-attachment 24-well plates with or without glass coverslips and cultured for an additional 24 hours. At this time point, approximately 80% of the cells expressed intermediate to high levels of CD11c and lacked CD103 expression regardless of whether or not the cells were transferred to glass coverslips during the last day of culture (Fig. 4A and data not shown). Attachment to glass also did not alter the expression levels of MHC-II or B220 (Fig. 4B). One hour after infection with L. monocytogenes, similar numbers of gentamicin-resistant bacteria were recovered, which suggested that adherence to glass did not significantly alter initial invasion of L. monocytogenes (Fig. 4C). The number of intracellular bacteria increased 32-fold over eight hours for the cells cultured on glass coverslips (Fig. 4D). However, only a 6-fold increase in CFU was observed for cells infected in suspension in low attachment plates. Thus, adherence to glass accounted for at least some of the ability of L. monocytogenes to replicate inside in vitro-generated CD103negCD11c+ cells.
FIGURE 4.
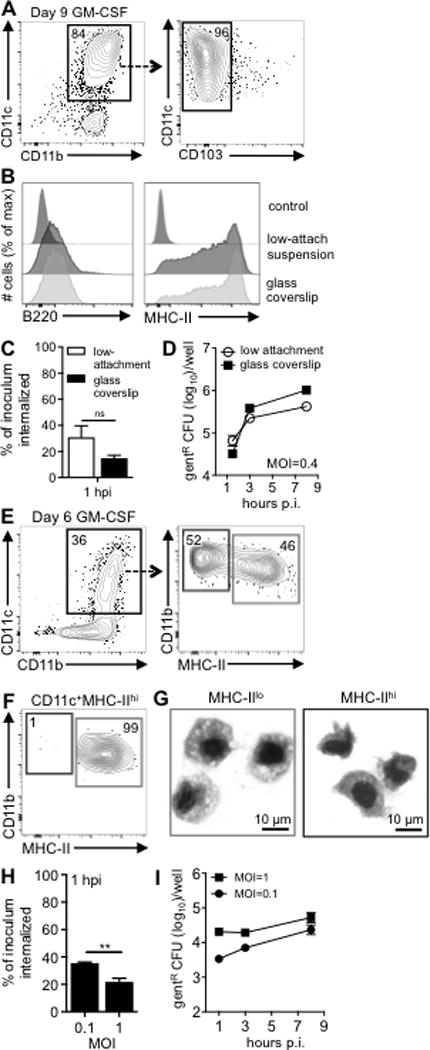
In vitro generated, bone marrow-derived CD103negCD11c+ cells readily support exponential growth of L. monocytogenes. (A-D) Bone marrow cells were cultured in GM-CSF for 8 days seeded in low-attachment dishes with or without glass coverslips and analyzed on day 9. (A) The majority of cultured CD11c+ cells lacked CD103. (B) Overnight incubation on glass did not alter expression of MHC-II or B220. (C) Mean percentage (±SD) of the Lm SD2000 inoculum that was gentR after 30 min. infection (MOI=0.4). (D) Intracellular growth assay indicates mean values (±SD) for triplicate samples. (E) Gating scheme used to identify CD11c+MHC-IIlo and CD11c+MHC-IIhi cells after 6 d of culture in GM-CSF. (F) Post sort analysis of sorted CD11c+MHC-IIhi cells. (G) Diff-Quik staining of sorted CD11c+MHC-IIlo and CD11c+MHC-IIhi cells. (H) Mean percentage (±SD) of the Lm SD2000 inoculum that was gentR after 30 min. infection at the indicated MOI. (I) Intracellular growth assay showing mean values (±SD) for triplicate samples. For all panels, data from one of two separate experiments is shown.
GM-CSF can promote the development of macrophages as well as neutrophils (31), and ~15–20% of the cells in our cultures were typically CD11cneg (Fig. 4A). Thus, it was possible that a minor proportion of heavily infected CD11cneg cells in the bulk cultures could account for the intracellular growth we observed. To test this, CD11c+ and CD11cneg cells were sorted on day eight and then cultured overnight in low-attachment plates prior to infection. As shown in Fig. S2A, the CD11c+ cells had a round mononuclear morphology with a relatively large cytoplasmic to nucleus ratio. The CD11cneg cells had characteristic monocytic kidney-shaped nuclei, and a minor proportion of the cells had lobular nuclei characteristic of granulocytes. Gentamicin-resistant L. monocytogenes were recovered from the CD11c+, but not the CD11cneg cells (Fig S2B). The sorted CD11c+ cells had a slight reduction in total CFU compared to unsorted bulk cultures, but exponential growth of intracellular bacteria was still observed. Thus, the enhanced ability of in vitro-generated CD11c+ cells to support intracellular growth of L. monocytogenes was not explained by the presence of other CD11cneg cells in the culture.
L. monocytogenes survive, but do not replicate exponentially, inside bone marrow-derived CD103negCD11c+MHC-IIhi cells that are cultured for only six days
Helft et al. recently demonstrated that CD11c+ cells generated in vitro with GM-CSF contain two populations of cells: MHC-IIhi cells that expressed DC surface markers such as CCR7 and CD135 and MHC-IIint cells that express markers of the monocyte/macrophage lineage such as CD115, F4/80 and CD64 (32). The MHC-IIhi cells were derived from both common dendritic precursors (CDP) and monocytes. In an attempt to isolate CD11c+ cells that more closely resembled cDC that recently differentiated from CDP, we harvested cells from an earlier time point in the culture (Fig. 4E) and sorted the MHC-IIhi cells (Fig. 4F). After 6 days, the CD11c+MHC-IIlo cells had a macrophage-like morphology with a large cytoplasm and phagocytic vesicles, whereas, the CD11c+MHC-IIhi cells had relatively larger nuclei with a more dendritic-like morphology (Fig. 4G).
L. monocytogenes were efficiently taken up in a dose-dependent manner by the sorted MHC-IIhi cells within one hour of infection (Fig. 4H). However, there was only a 3 to 7-fold increase in gentamicin-resistant bacteria in these cells over eight hours (Fig. 4I). Many of the intracellular bacteria were localized to the cytosol and associated with actin tails 8 hpi (Fig. 3D). The average number of bacilli associated with each infected cell increased over time with about 8% of the cells containing 15 or more L. monocytogenes by 8 hpi (Fig. 3E). Together, these results indicated that the efficiency of L. monocytogenes growth inside in vitro-generated CD103negCD11c+ cells was as least partially dependent on length of time in culture, and that GM-CSF-derived cells did not closely resemble cDC isolated from the MLN in regards to intracellular growth phenotype.
CD103+CD11c+ cells generated using FLT3-L do not efficiently support intracellular replication of L. monocytogenes
Activation of CD135 (FLT-3) is critical for differentiation of DC progenitors into cDC that express CD103, and a recent report described a longer culture method to selectively generate CD103+ DC in vitro using FLT3-L (25). Nearly all of the cells generated using this approach were CD11c+MHC-II+ after 16 days in culture, and approximately two-thirds of the cells were CD103+ (Fig. 5A). Microscopic examination of these cells revealed a unique morphology in regards to overall diameter and nuclear shape relative to the GM-CSF-cultured CD11c+ cells (Fig. 5B).
FIGURE 5.
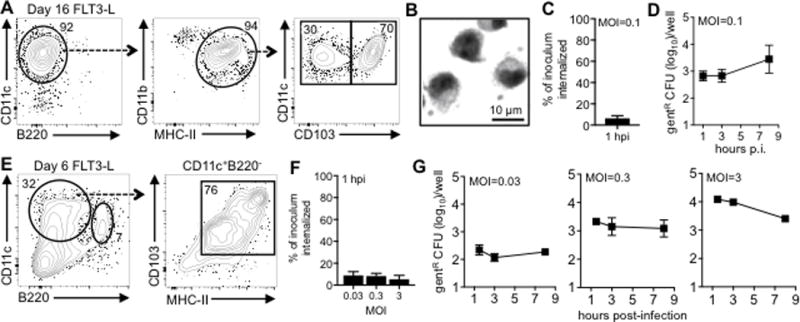
Flt3-L-cultured cells do not efficiently support the intracellular replication of L. monocytogenes. Gating scheme (A) and Diff-Quik staining (B) of bone marrow cells cultured for 16 days in 12.5% Flt3-L sup plus 0.75% GM-CSF sup. (C) Mean percentage (±SD) of Lm SD2000 inoculum that was gentR 1 h after infection of day 16 cells. (D) Intracellular growth assay using day 16 cells showing mean values (±SD) for triplicate samples. (E) Surface phenotype of bone marrow cells cultured for 6 days in 20% FLT3-L supernatant alone. (F) Mean percentage (±SD) of Lm SD2000 inoculum that was gentR 1 h after infection of day 6 cells at various MOI. (G) Intracellular growth assay using day 6 cells showing mean values (±SD) for triplicate samples. In panels C, D, F, and G data are representative of at least two separate experiments.
One hour after infection of CD103+CD11c+ cells in suspension at low MOI, ~ 6 % of the inoculum was gentamicin-resistant (Fig. 5C). Thus, the FLT3L-induced cultured CD103+ cells had an invasion rate that was less than the GM-CSF cultured CD103neg cells (Fig. 4C), but more efficient than the cDC harvested from MLN and infected directly ex vivo (Fig. 2C). As shown in Fig. 5D, there was no change in intracellular CFU recovered at 3 hpi, but by 8 hours, the number of gentamicin-resistant CFU increased 4-fold. Reducing the FLT3-L culture period to just six days resulted in a mixture of CD103+CD11chi cells as well as CD11cintB220+ plasmacytoid DC (Fig. 5E). The majority of the cells that expressed a cDC phenotype (CD11chiB220−) co-expressed both MHC-II and CD103. To determine if any of these cells could support intracellular growth of L. monocytogenes, the cells were infected in bulk at various MOI. As shown in Fig. 5F, L. monocytogenes readily invaded these cells within 30 minutes in a dose-independent manner however, the number of gentamicin-resistant CFU did not increase over time (Fig. 5G).
To confirm that L. monocytogenes were able to invade these cells, we performed differential “in/out” staining one hour after infection with GFP+ L. monocytogenes. As shown in the representative images in Fig. 3F, approximately 70% of the cells had at least one internalized (green) bacterium. Furthermore, actin staining of cells infected for 8 h indicated that about 10% of the cells counted harbored L. monocytogenes associated with actin tails (Fig. 3G). Consistent with the lack of exponential growth observed in the gentamicin protection assay, only 2 of the infected cells were associated with more than ten bacilli per cell (Fig. 3H). Together, these data indicate that L. monocytogenes can efficiently invade in vitro-generated CD103+CD11c+ cells, and survive in the cytosol, but intracellular growth is either inhibited or delayed compared to either macrophages or GM-CSF-induced cultured cells.
Discussion
In this study, we demonstrated that L. monocytogenes associated with a minor fraction of cDC in the intestinal LP and MLN during foodborne infection. However, L. monocytogenes inefficiently invaded these cells, and did not survive for more than a few hours. In striking contrast, in vitro-generated, bone marrow-derived-derived CD11c+ cells that phenotypically resembled cDC readily supported intracellular growth of L. monocytogenes. Interestingly, the length of time cells were maintained in monoculture with growth factors such as GM-CSF and Flt-3L correlated with increased replication of intracellular bacteria (Fig. 6). Our data suggest that Flt3-L induced cultured cells most closely resemble cDC in the gut, but still differ significantly in their ability to restrict long term survival of intracellular L. monocytogenes.
FIGURE 6.
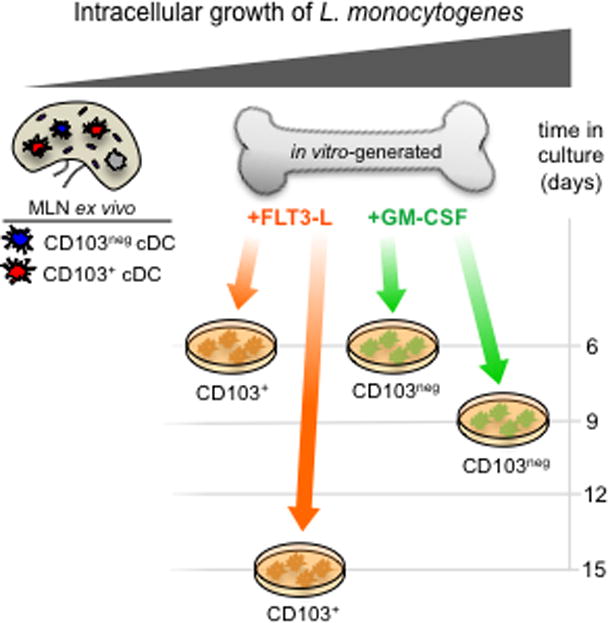
Intracellular growth of L. monocytogenes in CD11c+ cells is ontogeny-dependent. Cartoon depicts the relative ability of the cells used in this assay to support intracellular growth of L. monocytogenes. Cells on the left are most restrictive and cells on the right are most permissive for exponential replication.
Pron et al. previously used a microscopy approach to show that cells that expressed OX-62 (CD103) were the first cell type to associate with L. monocytogenes near the Peyer’s Patches and in the draining MLN of rats infected via a ligated intestinal loop (33). In the mouse, interactions between L. monocytogenes and DC have been primarily studied in the spleen following intravenous infection. The migratory CD8α+ DC found in the spleen are thought to be equivalent to the CD103+ DC found in non-lymphoid tissues, based on transcriptional network analysis and the sharing of key transcriptional factors required for development, (34,35). Although some investigators have reported that L. monocytogenes preferentially interacted with CD8α+ DC in the spleen (1,2), another study found more CD8α− DC associated with L. monocytogenes (36). Similar to our results, these earlier studies found only a small number of live, replicating L. monocytogenes associated with DC in the spleen (1–13 CFU per 1000 cells) (1,2,37,38). Despite this association of live CFU with splenic DC, it is still unclear if L. monocytogenes actively replicated inside those cells over time. In fact, it has been suggested that the key role for CD8α+ DC in the spleen may actually be to translocate bacteria from the red pulp into the periarteriolar lymphoid sheath of the white pulp, allowing for subsequent replication in other permissive cell types such as macrophages (1,36,39). In that case, there would be no requirement for intracellular survival or replication in DC, and transport of extracellular, adherent bacteria would be most efficient.
The exact mechanism by which DC limit intracellular growth of L. monocytogenes remains unclear. As we previously proposed in monocytes (19), it is possible that a reduced acidification of phagosomes in cDC or monocyte-derived DC can delay bacterial escape by inhibiting optimal activity of the pore-forming toxin listeriolysin O. Once in the cytosol of DC, L. monocytogenes may be killed by autophagy. Matsumura et al. recently suggested that the actin-bundling protein Fascin-1 may promote an association between the autophagosomal marker LC3 and L. monocytogenes in GM-CSF-induced cultured CD11c+ cells (40). L. monocytogenes normally avoid being trapped in autophagosomal membranes by expressing ActA at one pole to mediate actin-based motility in the cell cytosol (41). Expression of ActA is typically induced at least 200-fold after L. monocytogenes gain to the host cell cytosol (42), and this has been attributed to allosteric binding of glutathione to the transcriptional regular PrfA (43). Soon after entry into the host cell cytoplasm, actin surrounds L. monocytogenes forming a “cloud” (44), and actin tails form only after polar expression of ActA is initiated (45,46). We found little evidence of ActA-mediated localization of actin into polar tails in cDC infected ex vivo; however, many of the intracellular bacteria were surrounded by actin (data not shown). Thus, it is possible ActA expression is not induced in the cytosol of cDC. If this were true, it would suggest that the cytosol of cDC may have a lower glutathione concentration than normally found in cells that are permissive for intracellular growth, such as macrophages.
Although it has long been understood that bone marrow-derived GM-CSF cultured cells contained a heterogeneous mixture of cells, the abundant yield of cells and the reproducibility of results when used for in vitro assays has led to widespread acceptance of these cells as prototypic dendritic cells. The results presented here further confirm the assertion made by Helft et al. that most GM-CSF cultures contain a large percentage of cells with attributes more closely resembling macrophages, which they referred to as GM-Macs (32). Indeed, L. monocytogenes replicate exponentially in the cytosol of bone marrow-derived CD64hiF4/80+ macrophages, but not in monocytes (19), or as demonstrated here, in cDC analyzed directly ex vivo. Prolonged maintenance of cultured cells in vitro may alter the properties of DC-like cells due to extended feedback regulation from cytokines secreted into the media (47). Alternatively, the function of the cultured cells may be altered over time due to the absence of regulatory factors normally expressed by other immune cells or stromal cells in vivo. This study further highlights the need to define cells within the myeloid lineage based on functional attributes such as susceptibility to infection, rather than by surface marker phenotype, and to use primary cells analyzed directly ex vivo whenever possible.
Supplementary Material
Acknowledgments
We thank Greg Baumann and Jennifer Strange for assistance with flow cytometry and Michelle Pitts for critical analysis of the manuscript.
This work was supported by National Institutes of Health grant AI101373. Grant Jones was supported by an AAI Careers in Immunology Fellowship.
Abbreviations
- BALB
BALB/c/By/J
- cDC
conventional dendritic cell
- gentR
resistant to 10 μg/ml gentamicin
- hpi
hours post-infection
- MLN
mesenteric lymph nodes
References
- 1.Neuenhahn M, Kerksiek KM, Nauerth M, Suhre MH, Schiemann M, Gebhardt FE, Stemberger C, Panthel K, Schroder S, Chakraborty T, Jung S, Hochrein H, Russmann H, Brocker T, Busch DH. CD8α+ dendritic cells are required for efficient entry of Listeria monocytogenes into the spleen. Immunity. 2006;25:619–30. doi: 10.1016/j.immuni.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell LM, Brzoza-Lewis KL, Henry CJ, Grayson JM, Westcott MM, Hiltbold EM. Distinct responses of splenic dendritic cell subsets to infection with Listeria monocytogenes: maturation phenotype, level of infection, and T cell priming capacity ex vivo. Cell Immunol. 2011;268:79–86. doi: 10.1016/j.cellimm.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid CD, Stackpoole A, Meager A, Tikerpae J. Interactions of tumor necrosis factor with granulocyte-macrophage colony-stimulating factor and other cytokines in the regulation of dendritic cell growth in vitro from early bipotent CD34+ progenitors in human bone marrow. J Immunol. 1992;149:2681–8. [PubMed] [Google Scholar]
- 5.Scheicher C, Mehlig M, Zecher R, Reske K. Dendritic cells from mouse bone marrow: in vitro differentiation using low doses of recombinant granulocyte-macrophage colony-stimulating factor. J Immunol Methods. 1992;154:253–64. doi: 10.1016/0022-1759(92)90199-4. [DOI] [PubMed] [Google Scholar]
- 6.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179:1109–18. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity. 1999;11:753–61. doi: 10.1016/s1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 8.Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, Carotta S, O’Keeffe M, Bahlo M, Papenfuss A, Kwak JY, Wu L, Shortman K. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–26. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 9.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–16. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 10.Maraskovsky E, Brasel K, Teepe M, Roux ER, Lyman SD, Shortman K, McKenna HJ. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J Exp Med. 1996;184:1953–62. doi: 10.1084/jem.184.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerovic V, Houston SA, Scott CL, Aumeunier A, Yrlid U, Mowat AM, Milling SW. Intestinal CD103(−) dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 2013;6:104–13. doi: 10.1038/mi.2012.53. [DOI] [PubMed] [Google Scholar]
- 12.Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, Fuchsberger M, Lahoud MH, O’Keeffe M, Shao QX, Chen WF, Villadangos JA, Shortman K, Wu L. Cutting edge: generation of splenic CD8+ and CD8− dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005;174:6592–7. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- 13.Brasel K, De Smedt T, Smith JL, Maliszewski CR. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood. 2000;96:3029–39. [PubMed] [Google Scholar]
- 14.Rescigno M. Intestinal dendritic cells. Adv Immunol. 2010;107:109–38. doi: 10.1016/B978-0-12-381300-8.00004-6. [DOI] [PubMed] [Google Scholar]
- 15.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, Yona S. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–8. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGFβ and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Forster R, Agace WW. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202:1063–73. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones GS, Bussell KM, Myers-Morales T, Fieldhouse AM, Ghanem EN Bou, D’Orazio SE. Intracellular Listeria monocytogenes comprises a minimal but vital fraction of the intestinal burden following foodborne infection. Infect Immun. 2015;83:3146–56. doi: 10.1128/IAI.00503-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones GS, D’Orazio SE. Monocytes are the predominant cell type associated with Listeria monocytogenes in the gut, but they do not serve as an intracellular growth niche. J Immunol. 2017;198:2796–804. doi: 10.4049/jimmunol.1602076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bou Ghanem EN, Myers-Morales T, D’Orazio SE. A mouse model of foodborne Listeria monocytogenes infection. Curr Protoc Microbiol. 2013;31:9B 3 1–9B 3 16. doi: 10.1002/9780471729259.mc09b03s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bou Ghanem EN, Myers-Morales T, Jones GS, D’Orazio SEF. Oral transmission of Listeria monocytogenes in mice via ingestion of contaminated food. J Vis Exp. 2013;75 doi: 10.3791/50381. doi:3791/50381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Resendiz-Albor AA, Esquivel R, Lopez-Revilla R, Verdin L, Moreno-Fierros L. Striking phenotypic and functional differences in lamina propria lymphocytes from the large and small intestine of mice. Life Sci. 2005;76:2783–803. doi: 10.1016/j.lfs.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 23.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. Journal of Immunological Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 24.Naik SH, O’Keeffe M, Proietto A, Shortman HH, Wu L. CD8+, CD8−, and plasmacytoid dendritic cell generation in vitro using flt3 ligand. Methods Mol Biol. 2010;595:167–76. doi: 10.1007/978-1-60761-421-0_10. [DOI] [PubMed] [Google Scholar]
- 25.Mayer CT, Ghorbani P, Nandan A, Dudek M, Arnold-Schrauf C, Hesse C, Berod L, Stuve P, Puttur F, Merad M, Sparwasser T. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood. 2014;124:3081–91. doi: 10.1182/blood-2013-12-545772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui KR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 2010;32:557–67. doi: 10.1016/j.immuni.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lecuit M, Ohayon H, Braun L, Mengaud J, Cossart P. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect Immun. 1997;65:5309–19. doi: 10.1128/iai.65.12.5309-5319.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–14. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts AJ, Williams SK, Wiedmann M, Nightingale KK. Some Listeria monocytogenes outbreak strains demonstrate significantly reduced invasion, inlA transcript levels, and swarming motility in vitro. Appl Environ Microbiol. 2009;75:5647–58. doi: 10.1128/AEM.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westcott MM, Henry CJ, Cook AS, Grant KW, Hiltbold EM. Differential susceptibility of bone marrow-derived dendritic cells and macrophages to productive infection with Listeria monocytogenes. Cell Microbiol. 2007;9:1397–411. doi: 10.1111/j.1462-5822.2006.00880.x. [DOI] [PubMed] [Google Scholar]
- 31.Na YR, Jung D, Gu GJ, Seok SH. GM-CSF Grown Bone Marrow Derived Cells Are Composed of Phenotypically Different Dendritic Cells and Macrophages. Mol Cells. 2016;39:734–41. doi: 10.14348/molcells.2016.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, Goubau D, Reis e Sousa C. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 2015;42:1197–211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Pron B, Boumaila C, Jaubert F, Berche P, Milon G, Geissmann F, Gaillard JL. Dendritic cells are early cellular targets of Listeria monocytogenes after intestinal delivery and are involved in bacterial spread in the host. Cell Microbiol. 2001;3:331–40. doi: 10.1046/j.1462-5822.2001.00120.x. [DOI] [PubMed] [Google Scholar]
- 34.del Rio ML, Bernhardt G, Rodriguez-Barbosa JI, Forster R. Development and functional specialization of CD103+ dendritic cells. Immunol Rev. 2010;234:268–81. doi: 10.1111/j.0105-2896.2009.00874.x. [DOI] [PubMed] [Google Scholar]
- 35.Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, Hashimoto D, Chow A, Price J, Greter M, Bogunovic M, Bellemare-Pelletier A, Frenette PS, Randolph GJ, Turley SJ, Merad M, C. Immunological Genome Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–99. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, Kc W, Belizaire R, Aoshi T, Schreiber RD, Miller MJ, Murphy TL, Unanue ER, Murphy KM. CD8α(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity. 2011;35:236–48. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muraille E, Giannino R, Guirnalda P, Leiner I, Jung S, Pamer EG, Lauvau G. Distinct in vivo dendritic cell activation by live versus killed Listeria monocytogenes. Eur J Immunol. 2005;35:1463–71. doi: 10.1002/eji.200526024. [DOI] [PubMed] [Google Scholar]
- 38.Verschoor A, Neuenhahn M, Navarini AA, Graef P, Plaumann A, Seidlmeier A, Nieswandt B, Massberg S, Zinkernagel RM, Hengartner H, Busch DH. A platelet-mediated system for shuttling blood-borne bacteria to CD8α+ dendritic cells depends on glycoprotein GPIb and complement C3. Nat Immunol. 2011;12:1194–201. doi: 10.1038/ni.2140. [DOI] [PubMed] [Google Scholar]
- 39.Aoshi T, Carrero JA, Konjufca V, Koide Y, Unanue ER, Miller MJ. The cellular niche of Listeria monocytogenes infection changes rapidly in the spleen. Eur J Immunol. 2009;39:417–25. doi: 10.1002/eji.200838718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumura F, Yamakita Y, Starovoytov V, Yamashiro S. Fascin confers resistance to Listeria infection in dendritic cells. J Immunol. 2013;191:6156–64. doi: 10.4049/jimmunol.1300498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 42.Shetron-Rama LM, Marquis H, Bouwer HG, Freitag NE. Intracellular induction of Listeria monocytogenes actA expression. Infect Immun. 2002;70:1087–96. doi: 10.1128/IAI.70.3.1087-1096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. Glutathione activates virulence gene expression of an intracellular pathogen. Nature. 2015;517:170–3. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilney LG, DeRosier DJ, Tilney MS. How Listeria exploits host cell actin to form its own cytoskeleton. I. Formation of a tail and how that tail might be involved in movement. J Cell Biol. 1992;118:71–81. doi: 10.1083/jcb.118.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–31. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 46.Smith GA, Portnoy DA, Theriot JA. Asymmetric distribution of the Listeria monocytogenes ActA protein is required and sufficient to direct actin-based motility. Mol Microbiol. 1995;17:945–51. doi: 10.1111/j.1365-2958.1995.mmi_17050945.x. [DOI] [PubMed] [Google Scholar]
- 47.Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, Chang MW, Beckman SK, Cook AD, Hamilton JA. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188:5752–65. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


