Summary
Maintenance of protein homeostasis, or proteostasis, is crucial for organismal health. Disruption of proteostasis can lead to the accumulation of protein aggregates, which are associated with aging and many human diseases such as Alzheimer’s disease [1–3]. Through analysis of the C. elegans host response to intracellular infection, we describe here a novel response pathway that enhances proteostasis capacity and appears to act in parallel to well-studied proteostasis pathways. These findings are based on analysis of the transcriptional response to infection by the intracellular pathogen Nematocida parisii [4]. The response to N. parisii is strikingly similar to the response to infection by the Orsay virus, another natural intracellular pathogen of C. elegans, and is distinct from responses to extracellular pathogen infection [4–6]. We have therefore named this common transcriptional response the Intracellular Pathogen Response (IPR), and it includes upregulation of several predicted ubiquitin ligase complex components such as the cullin cul-6. Through a forward genetic screen we found pals-22, a gene of previously unknown function, to be a repressor of the cul-6/Cullin gene and other IPR gene expression. Interestingly, pals-22 mutants have increased thermotolerance and reduced levels of stress-induced polyglutamine aggregates, likely due to upregulated IPR gene expression. We found the enhanced stress resistance of pals-22 mutants to be dependent on cul-6, suggesting that pals-22 mutants have increased activity of a CUL-6/Cullin-containing ubiquitin ligase complex. pals-22 mutant phenotypes appear independent of the well-studied heat shock and insulin signaling pathways, indicating that the IPR is a distinct pathway that protects animals from proteotoxic stress.
eTOC
Reddy et al report a pathway in C. elegans called the Intracellular Pathogen Response, or IPR, which appears independent of known stress response pathways. They identify PALS-22 as a negative regulator of the IPR, with pals-22 mutants showing increased resistance to proteotoxic stress dependent on the ubiquitin ligase component CUL-6.
Results
pals-22 is a negative regulator of IPR gene expression
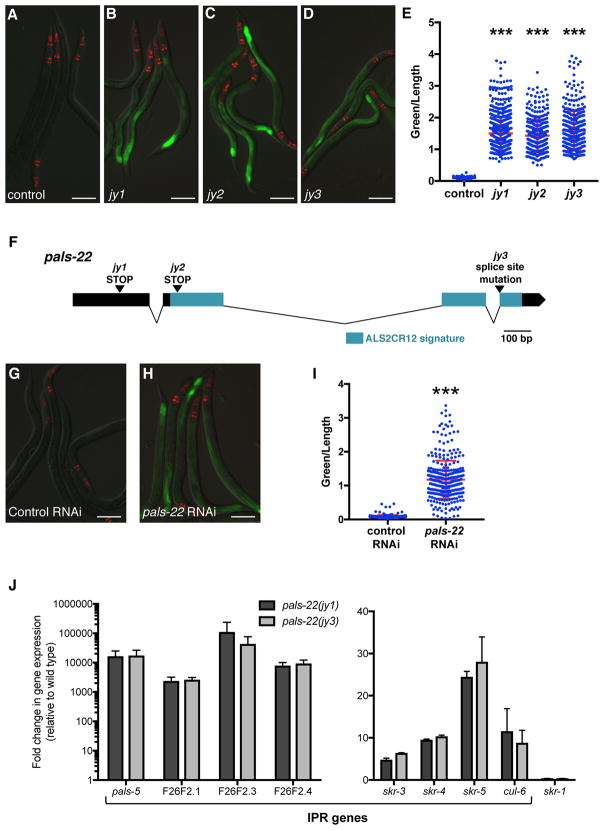
To investigate the transcriptional regulation of Intracellular Pathogen Response (IPR) genes, we performed a forward genetic screen for mutations that cause altered expression of a GFP reporter for pals-5, which is among the genes most highly induced by microsporidia infection [4]. pals-5 is a gene of unknown function that is a member of an expanded gene family in C. elegans (Figure S1A) called ‘pals’ for protein containing ALS2CR12 signature [7]. This signature is named for its identification in the human gene ALS2CR12, which is located in a region implicated in the human disease amyotrophic lateral sclerosis 2 (ALS2) [8] though is not believed to be the causative gene underlying the ALS2 disease [9]. pals-5, as well as many other pals genes, are highly induced by both microsporidia and viral infection (Figure S1A) [4–6]. In our screen, we examined F2 progeny of mutagenized pals-5p::GFP animals and identified three alleles (jy1, jy2, and jy3) that cause a strong increase in pals-5p::GFP expression in basal conditions (Figures 1A–E). As jy1, jy2 and jy3 all have essentially identical phenotypes, we used only the jy1 and jy3 alleles for further analyses. Of note, we observed that the jy1 and jy3 mutants develop more slowly than wild-type worms during normal well-fed conditions (Figures S1B–D).
Figure 1. pals-22 is a negative regulator of pals-5p::GFP expression.
(A–D) Mutants isolated from a screen show increased expression of the pals-5p::GFP reporter. Shown are (A) wild-type, (B) pals-22(jy1), (C) pals-22(jy2), and (D) pals-22(jy3) animals. (E) pals-5p::GFP expression quantified in pals-22 mutants using a COPAS Biosort machine to measure the mean GFP signal and length of individual animals, indicated by blue dots. Mean signal of the population is indicated by red bars, with error bars as SD. Graph is a compilation of three replicates, with at least 100 animals analyzed in each replicate. *** p < 0.001 with Student’s t-test. (F) pals-22 gene exon/intron structure. jy1 and jy2 encode stop mutations, and jy3 alters a splice site for the fourth exon. The ALS2CR12 signature is indicated in blue. (G–H) pals-5p::GFP animals treated with either (G) L4440 RNAi control or (H) pals-22 RNAi. (I) pals-5p::GFP expression in animals treated with L4440 RNAi control or pals-22 RNAi, quantified using a COPAS Biosort machine to measure the mean GFP signal and length of animals. Parameters the same as in (E). (J) qRT-PCR measurement of gene expression in pals-22(jy1) and pals-22(jy3) animals, shown as the fold change relative to wild-type control. Results shown are the average of two independent biological replicates, error bars are SD. In (A–D) and (G–H), green is pals-5p::GFP, red is myo-2p::mCherry expression in the pharynx as a marker for presence of the transgene. Images are overlays of green, red and Nomarski channels and were taken with the same camera exposure for all. Scale bar, 100 μm. Related to Figure S1.
We used whole-genome sequencing analysis and identified the gene defective in jy1, jy2, and jy3 mutants to be another member of the pals gene family, pals-22. Interestingly, the phylogenetic structure of the pals genes correlates with their regulation by infection, with pals-22 in a clade of pals genes that are not induced by intracellular infection, in contrast to pals-5, which is in a clade that is induced by intracellular infection (Figure S1A) [4–6]. The predicted PALS-22 protein contains 284 amino acids with no predicted function or identifiable domains except the ALS2CR12 signature. The jy1 and jy2 alleles cause early stop mutations in pals-22, and jy3 is a mutation predicted to affect splicing between exons 3 and 4 (Figure 1F). To confirm that pals-22 is the gene responsible for the altered pals-5 reporter expression in jy1, jy2, and jy3 mutants, we performed RNAi of pals-22 and found that it caused increased expression of the pals-5p::GFP reporter (Figures 1G–I), similar to the phenotypes of the jy1, jy2, and jy3 mutants. In addition, we found that a fosmid containing the wild-type pals-22 sequence tagged with GFP in the context of native cis-regulatory elements [10] could rescue the developmental delay of the jy3 mutant (Figure S1E).
We next confirmed that these mutants have altered expression of endogenous pals-5 mRNA expression using qRT-PCR (Figure 1J). In addition, we measured levels of a subset of other genes induced as part of the IPR, including genes of unknown function F26F2.1, F26F2.3, and F26F2.4 as well as the SCF ubiquitin ligase components cul-6, skr-3, skr-4, and skr-5. Interestingly, we found that all of the IPR genes in this subset have increased expression in the jy1 and jy3 mutants as compared to wild type (Figure 1J). We also used qRT-PCR to measure expression of another SCF ubiquitin ligase component, skr-1 (which is not induced by intracellular infection or proteasomal inhibition) [4], and found that this gene did not have increased expression in the pals-22(jy1) and pals-22(jy3) mutants (Figure 1J). These results suggest that PALS-22 acts as a negative regulator specifically of IPR gene expression.
pals-22 mutants have increased thermotolerance, dependent on the cullin cul-6
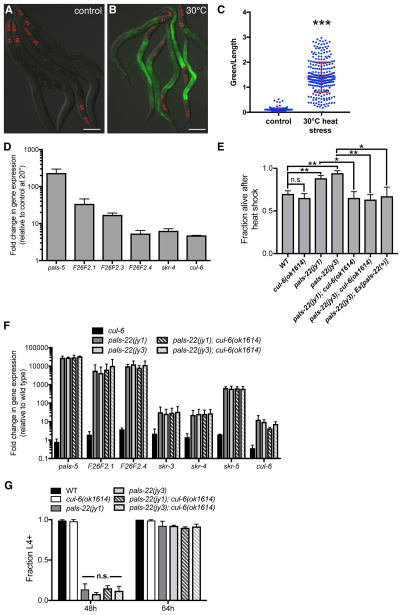
Genome-wide transcriptional profiling indicated there is a partial overlap between the response to intracellular infection and to a prolonged heat stress at 30°C, including genes in our IPR subset (pals-5, F26F2.3, F26F2.4, skr-4, and skr-5) [4, 11]. Here we show that the pals-5p::GFP reporter has increased expression after prolonged heat stress (Figures 2A–C). We also used qRT-PCR to show that genes in the IPR subset all have increased expression after prolonged heat stress treatment (Figure 2D). This result led us to test whether pals-22 mutants have increased survival after heat shock treatment. Indeed, we found that the pals-22(jy1) and pals-22(jy3) mutants have enhanced resistance to heat shock treatment compared to wild-type animals (Figure 2E), suggesting that constitutive IPR gene expression may be protective against the toxic effects of heat shock. We were able to rescue this phenotype using a transgene containing the wild-type pals-22 gene (Figure 2E).
Figure 2. IPR gene expression is increased by heat stress, and pals-22 mutants have increased survival after heat shock, dependent on the cullin cul-6.
(A,B) pals-5p::GFP animals grown at (A) 20°C or (B) shifted to 30°C for 24h. Green is pals-5p::GFP, red is myo-2p::mCherry expression in the pharynx as a marker for presence of the transgene. Images are overlays of green, red and Nomarski channels and were taken with the same camera exposure for all. Scale bar, 100 μm. (C) pals-5p::GFP expression quantified in pals-22 mutants using a COPAS Biosort to measure the mean GFP signal and length of individual animals, indicated by blue dots. Mean signal of the population is indicated by red bars, with error bars as SD. Graph is a compilation of three replicates, with at least 100 animals analyzed in each replicate. *** p < 0.001 with Student’s t-test. (D) qRT-PCR measurement of gene expression in wild-type animals shifted to 30°C for 24h, shown as the fold change relative to wild-type worms grown at 20°C. Results shown are the average of two independent biological replicates, error bars are SD. (E) pals-22 mutants have increased survival after heat shock, which is suppressed by a cul-6(ok1614) deletion. Animals were treated for 2h at 37°C followed by 24 hours at 20°C, and then assessed for survival. Strains were tested in triplicate, with at least 30 animals per plate. Mean fraction alive indicates the average survival among the triplicates, errors bars are SD. ** p < 0.01, * p < 0.05, n.s., not significant with Student’s t-test. Assay was repeated three independent times with similar results, and data from a representative experiment are shown. (F) qRT-PCR measurement of IPR gene expression, shown as the fold change relative to wild-type control. Results shown are the average of two independent biological replicates, error bars are SD. (G) pals-22 mutants have a developmental delay, which is not suppressed by cul-6(ok1614) deletion. Percentage of animals reaching the L4 larval stage at timepoints after eggs were laid is indicated. Results shown are the average of three independent biological replicates, with 100 animals assayed in each replicate. Error bars are SD. n.s., not significant with Student’s t-test. Related to Figures S2 and S3.
We next investigated whether the pals-22 thermotolerance phenotype was dependent on any of the IPR genes. Mutation of skr-3, skr-4, or skr-5 in a pals-22 mutant background did not alter survival after heat shock (Figures S2A–B). We also tested a large deletion encompassing 11 pals genes (see Experimental Procedures) and did not see effects on pals-22 thermotolerance (Figure S2C). In contrast, predicted loss-of-function alleles of the ubiquitin ligase component Cullin/cul-6 completely abrogated the increased thermotolerance of pals-22 mutants back to wild-type levels (Figures 2E and S2D). This result indicates that the increased thermotolerance of pals-22 mutants is dependent on CUL-6 function. We introduced a fosmid containing wild-type cul-6 [10] into the pals-22(jy1);cul-6(ok1614) double mutant to test for rescue but could not assay heat shock tolerance due to the decreased viability of this strain. Worms carrying the cul-6 mutations alone had no reduction of thermotolerance as compared to wild type in our assays (Figures 2E and S2D), indicating that this gene does not significantly affect heat shock resistance in a wild-type context, only in a pals-22 mutant background.
To address the possibility that the suppression of pals-22 thermotolerance by cul-6 was through its effects on gene expression, we used qRT-PCR to measure mRNA levels of a set of IPR genes in pals-22;cul-6 double mutants. As IPR genes are expressed at such a high level in pals-22 mutants, it is difficult to see a further increase in these genes upon heat stress and thus to test whether cul-6 affects a heat stress-induced change in IPR gene expression in pals-22 mutants. However, we did find that mutation of cul-6 did not alter the constitutive expression of any of the genes tested (Figure 2F). We also found that the cul-6 deletion does not suppress the developmental delay of pals-22 mutants (Figure 2G). Taken together, these results suggest that increased activity of CUL-6 in pals-22 mutants promotes thermotolerance.
pals-22 phenotypes are independent of known stress response pathways
We next investigated the interaction between pals-22 and previously characterized stress response pathways, focusing first on the conserved transcription factor Heat Shock Factor HSF-1. HSF-1 is a master regulator of the Heat Shock Response (HSR) and controls expression of heat shock proteins (HSPs), which are chaperones that facilitate protein refolding to protect cells against proteotoxic stress [12, 13]. Interestingly, we found that the double mutant hsf-1(sy441); pals-22(jy1) has a larval arrest phenotype. Given that pals-22(jy1) is a likely null mutation, this synthetic lethal phenotype supports the model that pals-22 and hsf-1 act in parallel pathways. We were not able to test hsf-1 RNAi in the heat shock assay (see STAR Methods), but using RNAi we found that hsf-1 is not required for the constitutive IPR gene expression phenotype of pals-22 mutants (Figures S3A–B). In addition, we found that pals-22 mutants do not have upregulation of HSP gene expression, which is controlled by hsf-1 (Figure S3C).
HSF-1 acts together with the conserved insulin/IGF-associated FOXO transcription factor DAF-16, to control HSP expression and promote thermotolerance. Previous work indicated that the genes upregulated by intracellular infection are not regulated by DAF-16 [4]. Importantly, we found that mutation of daf-16 did not affect the pals-22 increased thermotolerance phenotype (Figure S3D). In addition to the HSR and insulin/IGF pathways that promote protein refolding, there are proteostasis responses that promote protein degradation through upregulation of proteasome subunits [14]. We used qRT-PCR to examine expression of proteasome genes (Figure S3E) and found no increase in proteasome subunit mRNA levels in pals-22 mutants as compared to wild type.
Many manipulations in C. elegans that increase resistance to heat shock, such as increased activity of hsf-1 and daf-16, also confer a prolonged lifespan [15]. We therefore analyzed the lifespan of pals-22(jy1) and pals-22(jy3) mutant worms at 25°C. Surprisingly, we found that these strains have a decreased lifespan as compared to wild type (Figure S3F), and a similar finding was reported in another study [7]. Together these results suggest that the mechanisms underlying the increased stress resistance of pals-22 mutants are distinct from those of previously described stress response pathways.
pals-22 mutants have decreased polyglutamine aggregation, dependent on cul-6
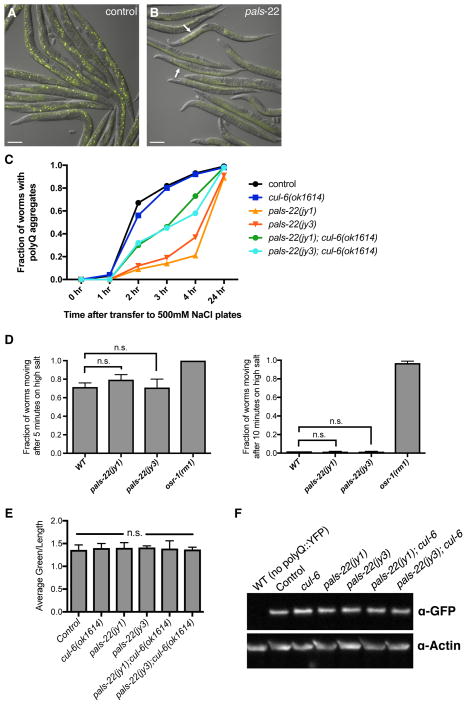
Next we examined aggregation of a polyglutamine (polyQ) repeat - YFP fusion protein, which is another phenotype commonly altered in stress-resistant animals [16] We found that pals-22 mutants have a greatly reduced number of osmotic-induced polyQ aggregates in the intestine compared to wild-type animals (Figures 3A–C). As a control, we tested whether the reduced polyQ aggregation phenotype of pals-22 mutants could simply be due to a general resistance to osmotic stress. Here we found that pals-22 mutants have osmotic stress resistance comparable to wild-type worms (Figure 3D), indicating the reduced aggregates are not simply due to increased osmotic stress resistance.
Figure 3. pals-22 mutants have decreased polyQ aggregation, dependent on cul-6.
(A,B) Images of (A) control and (B) pals-22(jy3) animals expressing polyQ(44)::YFP at 3 hours after transfer to high salt plates. Arrows indicate osmotic stress-induced aggregates. Images are overlays of yellow and Nomarski channels and were taken with the same camera exposure. Scale bar, 100 μm. (C) Fraction of animals with polyQ aggregates after transfer to high salt plates. At least 60 worms were scored for each strain. Assay was repeated two independent times with similar results, and data from a representative experiment shown. (D) Fraction of animals scored as moving after transfer to high salt plates at the indicated timepoints. At least 50 worms were scored for each strain. Results shown are the average of two independent replicates. Error bars are SD. n.s., not significant with Student’s t-test. (E) Quantification of fluorescence levels in the intestine of strains of the indicated genotypes, all expressing a polyQ(44)::YFP transgene. Fluorescence was measured with a COPAS Biosort machine. Results shown are the average of two independent replicates, with at least 500 animals measured for each sample. Error bars are SD. n.s., not significant with Student’s t-test. (F) Western blot analysis on total protein lysate from indicated strains. polyQ(44)::YFP protein is detected with anti-GFP, and anti-actin antibody is used as a loading control.
We next tested if this reduced aggregate formation phenotype of pals-22 mutants was dependent on cul-6, similar to the thermotolerance phenotype of pals-22 mutants. We found that pals-22;cul-6 double mutants have an intermediate aggregate formation phenotype (Figure 3C), suggesting that cul-6 functions to reduce polyQ aggregation in pals-22 mutants, although there are likely other factors contributing to this phenotype as well.
Finally, we quantified the amount of polyQ(44)::YFP protein expressed in the strains used in our analysis to ensure that the altered aggregate formation was not due to changes in polyQ protein levels. We measured YFP fluorescence and performed Western blot analysis, and in both cases found that mutation of pals-22 or cul-6 does not alter the amount of polyQ(44)::YFP protein (Figures 3E–F). Thus, pals-22 does not appear to affect the overall levels of polyQ(44)::YFP protein, just the propensity to form stress-induced aggregates.
PALS-22 is broadly expressed and functions in the intestine to regulate IPR gene expression and thermotolerance
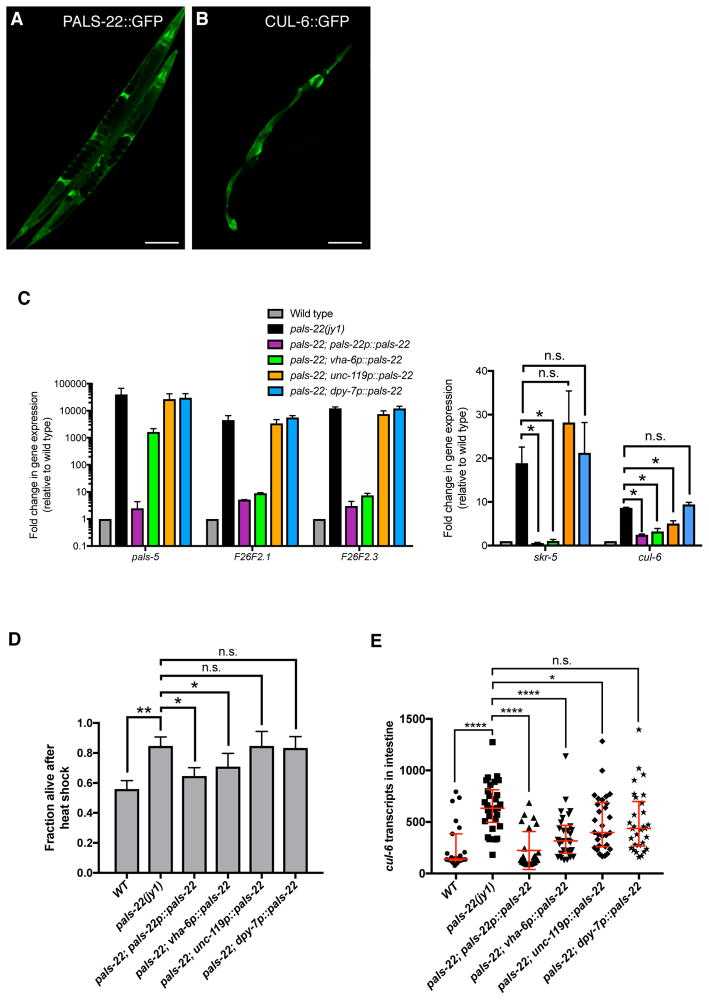
Using fosmids containing GFP-3xFLAG-tagged versions of pals-22 and cul-6 with native cis-regulatory elements [10] we observed PALS-22::GFP expression throughout the animal, including expression in the intestine, neurons, and hypodermis (Figure 4A), while CUL-6::GFP was seen only in the pharynx and intestine (Figure 4B). We next investigated in which tissues the PALS-22 protein acts to regulate gene expression and thermotolerance. Single-copy expression of GFP-tagged pals-22 cDNA expressed from its endogenous promoter showed similar tissue distribution as the analysis with multi-copy arrays of a fosmid described above [7]. Expression of the pals-22 cDNA from this endogenous promoter rescued the constitutive IPR gene expression phenotype, as measured by qRT-PCR (Figure 4C) as well as the increased thermotolerance phenotype (Figure 4D), indicating that this tagged cDNA is functional. We found that intestinal expression of pals-22 could strongly rescue the constitutive IPR gene expression phenotype of most genes tested, while pan-neuronal and hypodermal expression of pals-22 did not result in large effects on IPR gene expression (Figure 4C). Similarly, we saw a partial rescue of the pals-22 increased thermotolerance phenotype with intestinal rescue but no significant effect with neuronal or hypodermal rescue of pals-22 (Figure 4D).
Figure 4. PALS-22 is broadly expressed, and functions in the intestine to regulate IPR expression and thermotolerance.
(A,B) Confocal fluorescence images of adult animals with fosmid transgenes expressing (A) PALS-22::GFP and (B) CUL-6::GFP from endogenous promoters. (C) qRT-PCR measurement of IPR gene expression in animals with tissue-specific expression of a wild-type pals-22 cDNA driven by endogenous (pals-22), intestinal (vha-6), neuronal (unc-119), or hypodermal (dpy-7) promoters in a pals-22(jy1) mutant background. Results are shown as the fold change relative to wild-type control and are the average of two independent biological replicates. Error bars are SD. * p < 0.05, n.s., not significant with Student’s t-test. (D) Survival of animals with pals-22 tissue-specific rescue after 2h heat shock treatment at 37°C followed by 24 hours at 20°C. Strains were tested in triplicate. Mean fraction alive indicates the average survival among the triplicates, errors bars are SD. ** p < 0.01, * p < 0.05, n.s., not significant with Student’s t-test. Heat shock assay was repeated three independent times with similar results, and data from a representative experiment are shown. (E) Measurement of intestinal cul-6 mRNA transcripts using single molecule fluorescence in situ hybridization (smFISH). Graph is a compilation of three independent biological replicates. Each symbol represents the smFISH spot count from the four anterior-most intestinal cells of an individual worm. Statistical analysis was performed using one-way ANOVA. **** p < 0.0001, * p < 0.05, n.s., not significant. Related to Figure S4.
We used single molecule fluorescent in situ hybridization (smFISH) to quantify mRNA expression of cul-6, which is required for the pals-22 mutant enhanced proteostasis phenotypes. In pals-22(jy1) worms, we observed increased expression of cul-6 mRNA in the intestine as compared to wild-type worms, which was rescued by expression of pals-22 from the endogenous promoter (Figure 4E). Expression of pals-22 from either intestinal or neuronal promoters decreased the amount of cul-6 mRNA seen in the intestine, with the intestinal pals-22 expression having a larger effect. Taken together our results suggest that pals-22 acting in the intestine regulates cul-6 expression in this tissue, which may contribute to the increased thermotolerance of pals-22 mutants.
Interaction between pals-22 and small RNA-mediated gene silencing
Interestingly, a recent study found that pals-22 mutants have increased transgene silencing and an enhanced exogenous RNAi response, suggesting that the PALS-22 protein can regulate levels of small RNAs [7]. The transgene silencing and RNAi phenotypes are suppressed by a mutation in rde-4, which is required for small RNA-mediated gene silencing. We therefore investigated IPR gene expression and thermotolerance in pals-22 rde-4 double mutants. Here we found that an rde-4 mutation could partially suppress the increased IPR gene expression of pals-22 mutants (Figure S4A), indicating that small RNA-mediated silencing may be responsible for some of the effects of pals-22 on IPR gene expression. When we examined the pals-22 rde-4 double mutants for thermotolerance, we found that an rde-4 mutation led to a slight but not significant reduction of the increased thermotolerance of pals-22 mutants (Figure S4B). These results indicate that the increased proteostasis capacity of pals-22 mutants may also depend partially on small RNA pathways.
Discussion
In response to diverse kinds of natural intracellular infection, C. elegans induces a distinct transcriptional signature we are calling the Intracellular Pathogen Response (IPR). The IPR is characterized by transcriptional upregulation of genes of unknown function, such as genes in the large pals gene family (Figure S1A), as well as genes that are predicted to encode ubiquitin ligase components in the SCF family, such as cul-6. Through a forward genetic screen we identified another pals gene called pals-22, which acts as a repressor of IPR gene expression. Previous work showed that a subset of IPR genes can be induced by proteotoxic stressors such as proteasome inhibition and chronic heat stress [4, 11]. These findings suggest that the IPR is induced in response to proteotoxic stress, which led us to analyze pals-22 proteostasis phenotypes. Indeed, we found that pals-22 mutants have increased resistance in thermotolerance and polyglutamine aggregation stress assays, and these phenotypes are dependent on cul-6. Our genetic analyses suggest that the IPR is distinct from previously described stress response pathways, like the HSR. We propose that the IPR is a novel pathway that mediates protective responses to chronic stressors like intracellular infection and prolonged heat stress, to improve proteostasis capacity via PALS-22 and CUL-6 (Figure S4C).
We found that PALS-22 functions in the intestine to repress intestinal expression of cul-6 mRNA, and to regulate thermotolerance. The simplest model for how PALS-22 and CUL-6 work together is that PALS-22 is a negative regulator of cul-6 transcription. However, as neither our work nor another study [7] found PALS-22 protein enriched in the nucleus, PALS-22 is likely not a transcription factor but rather may be an indirect regulator of cul-6 mRNA expression. Indeed, we found that PALS-22 may regulate expression of cul-6 and other IPR genes in part through small RNA-mediated pathways. It is possible that the PALS-22 protein also functions biochemically with the CUL-6 protein as a more direct inhibitor of a process mediated by a CUL-6-containing ubiquitin ligase that targets proteins in the intestine for ubiquitylation. Future studies will focus on the mechanism used by PALS-22 to repress expression of IPR genes like cul-6, as well as the biochemical mechanism by which PALS-22 and CUL-6 increase proteostasis capacity to promote resistance to diverse stressors.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact Emily Troemel (etroemel@ucsd.edu).
Experimental model and subject details
The nematode Caenorhabditis elegans was used as the experimental model for this study. All experiments were performed with hermaphrodite animals; males were used only for crosses. Unless otherwise indicated, all experiments were carried out with L4 stage animals. Strains were maintained at 20°C on Nematode Growth Media (NGM) plates seeded with Streptomycin-resistant E. coli OP50-1 bacteria according to standard methods [17]. Mutant or transgenic strains were backcrossed at least three times.
Method details
EMS screen and cloning of alleles
Worms carrying the jyIs8[pals-5p::GFP, myo-2::mCherry] transgene were mutagenized with ethyl methane sulphonate (EMS) (Sigma) using standard procedures as described [18]. L4 stage P0 worms were incubated in 47 mM EMS for 4 hours at 20°C. Worms were screened in the F2 generation for increased expression of GFP under a fluorescence dissecting microscope (Zeiss Discovery V8). Complementation tests were carried out by generating worms heterozygous for two mutant alleles and scoring pals-5p::GFP fluorescence. Linkage group mapping with visible markers was done using the strains ERT507, ERT508, and ERT509 and scoring pals-5p::GFP fluorescence. For whole-genome sequencing analysis of mutants, genomic DNA was prepared using a Puregene Core kit (Qiagen) and 20X sequencing coverage was obtained using a 90 bp paired-end Illumina HiSeq 2000 at Beijing Genomics Institute. We identified only one gene (pals-22) on LGIII containing variants predicted to alter function in all three mutants sequenced (jy1, jy2, and jy3). The jy1 mutation is an early stop at amino acid 51, jy2 is an early stop at amino acid 105, and jy3 is a G to A mutation of the splice acceptor before the 4th exon.
RNA interference
RNA interference was performed using the feeding method. Overnight cultures of RNAi clones in the HT115 bacterial strain were seeded onto NGM plates supplemented with 5mM IPTG and 1mM carbenicillin, and incubated at 25°C for 1 day. Synchronized L1 stage animals were fed RNAi until the L4 stage at 20°C. All RNAi experiments had an unc-22 positive control RNAi clone, which resulted in twitching animals in all experiments. RNAi clones were verified by sequencing. For experiments in Figures 1G and 1H, at least 100 animals were analyzed for each condition in each replicate, and experiments were repeated three times.
Generation of transgenic strains
Transgenic strains expressing TransgeneOme fosmids [10] as extrachromosomal arrays (strains ERT365 and ERT422) were generated by microinjecting fosmids at 100 ng/ul into strain EG6699 and selecting non-unc animals. Mos1-mediated Single Copy Insertion (MosSCI) of transgenes was performed as previously described [19]. Briefly, plasmids pCFJ601 (50 ng/ul), pMA122 (10 ng/ul), pGH8 (10 ng/ul), pCFJ90 (2.5 ng/ul), and pCFJ104 (5 ng/ul) were mixed with a plasmid containing the transgene to be inserted (25 ng/ul) (see Key Resources Table) and microinjected into strain EG6699. After injection, worms were incubated on NGM plates at 25°C for 1 week and then subjected to a 2 hour heat shock at 34°C. Non-unc animals that were not killed by the heat shock were selected.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-actin, clone JLA20 | Millipore | RRID: AB_11203498 |
| Goat anti-mouse IgM HRP conjugate | Millipore | RRID: AB_92481 |
| Goat anti rabbit IgG HRP conjugate | Millipore | RRID: AB_10682749 |
| Rabbit anti-GFP | Gift from the Karen Oegema lab | N/A |
| Bacterial and Virus Strains | ||
| E. coli: OP50-1 | Caenorhabditis Genetics Center | N/A |
| E. coli: HT115 | Caenorhabditis Genetics Center | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRI Reagent | Molecular Research Center | Cat#TR118 |
| Critical Commercial Assays | ||
| RETROscript Reverse Transcription Kit | Ambion | Cat#AM1710 |
| qScript cDNA SuperMix | Quanta Biosciences | Cat#95048 |
| iQ SYBR Green Supermix | Bio-Rad | Cat#1708880 |
| ECL Prime Western Blotting Detection Reagent | Amersham | Cat#89238-012 |
| Experimental Models: Organisms/Strains | ||
| C.elegans: Strain wild-type N2 | Caenorhabditis Genetics Center | N2 |
| C.elegans: Strain ERT54: jyIs8[pals-5p::GFP, myo-2::mCherry] X | [4] | ERT54 |
| C.elegans: Strain ERT286: pals-22(jy1) III; jyIs8[pals-5p::GFP, myo-2::mCherry] X | This paper | ERT286 |
| C.elegans: Strain ERT287: pals-22(jy2) III; jyIs8[pals-5p::GFP, myo-2::mCherry] X | This paper | ERT287 |
| C.elegans: Strain ERT288: pals-22(jy3) III; jyIs8[pals-5p::GFP, myo-2::mCherry] X | This paper | ERT288 |
| C.elegans: Strain ERT507: rol-6(e187) II; jyIs8 X | This paper | ERT507 |
| C.elegans: Strain ERT508: dpy-5(e61) I; unc-32(e189) III; jyIs8 X | This paper | ERT508 |
| C.elegans: Strain ERT509: unc-5(e53) IV; dpy-11(e224) V; jyIs8 X | This paper | ERT509 |
| C.elegans: Strain ERT356: pals-22(jy1) III | This paper | ERT356 |
| C.elegans: Strain ERT415: pals-22(jy3) III | This paper | ERT415 |
| C.elegans: Strain ERT365: unc-119(ed3) III; jyEx193[pals-22::EGFP::3xFLAG, unc-119(+)] | This paper | ERT365 |
| C.elegans: Strain ERT422: unc-119(ed3) III; jyEx224[cul-6::EGFP::3xFLAG, unc-119(+)] | This paper | ERT422 |
| C.elegans: Strain ERT442: pals-22(jy3) unc-119(ed3) III; jyEx193[pals-22::EGFP::3xFLAG, unc-119(+)] | This paper | ERT442 |
| C.elegans: Strain ERT441: pals-22(jy1) III; cul-6(ok1614) IV | This paper | ERT441 |
| C.elegans: Strain ERT439: pals-22(jy3) III; cul-6(ok1614) IV | This paper | ERT439 |
| C.elegans: Strain OG412: drIs20[vha-6p::Q44::YFP, rol-6(su1006)] | [16] | WB strain: OG412 |
| C.elegans: Strain ERT467: pals-22(jy1) III; drIs20 | This paper | ERT467 |
| C.elegans: Strain ERT448: pals-22(jy3) III; drIs20 | This paper | ERT448 |
| C.elegans: Strain ERT506: cul-6(ok1614) IV; drIs20 | This paper | ERT506 |
| C.elegans: Strain ERT474: pals-22(jy3) III; cul-6(ok1614) IV; drIs20 | This paper | ERT474 |
| C.elegans: Strain ERT500: pals-22(jy1) III; cul-6(ok1614) IV; drIs20 | This paper | ERT500 |
| C.elegans: Strain ERT501: C17H1 deletion I | This paper | ERT501 |
| C.elegans: Strain ERT502: C17H1 deletion I; pals-22(jy1) III | This paper | ERT502 |
| C.elegans: Strain VC40679 skr-4(gk759439) V | Caenorhabditis Genetics Center | WB strain: VC40679 |
| C.elegans: Strain ERT479: pals-22(jy3) III; skr-4(gk759439) V | This paper | ERT479 |
| C.elegans: Strain ERT537: daf-16(mgDf47) I; pals-22(jy1) III | This paper | ERT537 |
| C.elegans: Strain ERT526: jySi37[pals-22p::pals-22::GFP, unc-119 (+)] II; pals-22( jy1) unc-119(ed3) III | This paper | ERT526 |
| C.elegans: Strain ERT527: jySi38[vha-6p::pals-22::GFP, unc-119 (+)] II; pals-22( jy1) unc-119(ed3) III | This paper | ERT527 |
| C.elegans: Strain ERT534: jySi39[unc-119p::pals-22::GFP, unc-119 (+)] II; pals-22( jy1) unc-119(ed3) III | This paper | ERT534 |
| C.elegans: Strain ERT539: jySi41[dpy-7p::pals-22::GFP, unc-119(+)] II; pals-22(jy1) unc-119(ed3) III | This paper | ERT539 |
| C.elegans: Strain RB640: skr-3(ok365) V | Caenorhabditis Genetics Center | WB strain: RB640 |
| C.elegans: Strain RB2266: skr-5(ok3068) V | Caenorhabditis Genetics Center | WB strain: RB2266 |
| C.elegans: Strain VC40063: cul-6(gk214864) IV | Caenorhabditis Genetics Center | WB strain: VC40063 |
| C.elegans: Strain VC40034: cul-6(gk214867) IV | Caenorhabditis Genetics Center | WB strain: VC40034 |
| C.elegans: Strain ERT554: pals-22(jy1) III; skr-3(ok3068) V | This paper | ERT554 |
| C.elegans: Strain ERT555: pals-22(jy1) III; skr-5(ok3068) V | This paper | ERT555 |
| C.elegans: Strain ERT556: pals-22(jy1) III; cul-6(gk214864) IV | This paper | ERT556 |
| C.elegans: Strain ERT557: pals-22(jy1) III; cul-6(gk214867) IV | This paper | ERT557 |
| C.elegans: Strain AM1: osr-1(rm1) I | Caenorhabditis Genetics Center | WB strain: AM1 |
| C.elegans: Strain CL2070: dvIs70[hsp-16.2p::GFP + rol-6(su1006)] | Caenorhabditis Genetics Center | WB strain: CL2070 |
| C.elegans: Strain EG6699: ttTi5605 II; unc-119(ed3) III | Caenorhabditis Genetics Center | WB strain: EG6699 |
| C.elegans: Strain OH11062: otIs381[ric-19p::NLS::gfp, elt-2p::DsRed] V | [7] | OH11062 |
| C.elegans: Strain OH15179: pals-22(ot811) III; otIs381 V | [7] | OH15179 |
| C.elegans: Strain OH15180: rde-4(ne301) III; otIs381 V | [7] | OH15180 |
| C.elegans: Strain OH15181: pals-22(ot811) rde-4(ne301) III; otIs381 V | [7] | OH15181 |
| Oligonucleotides | ||
| See Table S1 for oligonucleotide sequences used in this study | ||
| Recombinant DNA | ||
| Plasmid: pCFJ601 eft-3p::Mos1 transposase | [19] | Addgene Plasmid #34874 |
| Plasmid: pMA122 peel-1 negative selection | [19] | Addgene Plasmid #34873 |
| Plasmid: pGH8 rab-3p::mCherry | [19] | Addgene Plasmid #19359 |
| Plasmid: pCFJ90 myo-2p::mCherry | [19] | Addgene Plasmid #19327 |
| Plasmid: pCFJ104 myo-3p::mCherry | [19] | Addgene Plasmid #19328 |
| Plasmid: pCFJ151-pals-22p::pals-22::gfp | This paper | pET631 |
| Plasmid: pCFJ151-vha-6p::pals-22::gfp | This paper | pET636 |
| Plasmid: pCFJ151-unc-119p::pals-22::gfp | This paper | pET637 |
| Plasmid: pCFJ151-dpy-7p::pals-22::gfp | This paper | pET640 |
| Plasmid: pL4440 RNAi control | Ahringer RNAi library | N/A |
| Plasmid: pL4440-hsf-1 | Ahringer RNAi library | N/A |
| Plasmid: pL4440-pals-22 | Ahringer RNAi library | N/A |
| Plasmid: pL4440-unc-22 | Ahringer RNAi library | N/A |
| Plasmid: pDD162 eft-3p::Cas9 | Gift from Goldstein lab | Addgene Plasmid #47549 |
| Plasmid: pU6::klp-12_sgRNA | Gift from Nonet lab | N/A |
| Plasmid: C17H1.3 guide RNA | This paper | pC17H1.3_sg2 |
| Plasmid: C17H1.7 guide RNA | This paper | pC17H1.7_sg2 |
| Fosmid: pals-22::EGFP::3xFLAG | [10] | 9830596596427236 B12 |
| Fosmid: cul-6::gfp::EGFP::3xFLAG | [10] | 873721959883807 G09 |
| Software and Algorithms | ||
| ImageJ | NIH Image | RRID:SCR_003070 |
| StarSearch | Arjun Raj lab | http://rajlab.seas.upenn.edu/StarSearch/launch.html |
| GraphPad Prism 7 | GraphPad Software, Inc. | RRID:SCR_002798 |
| MUSCLE version 3.8.31 | [25] | N/A |
| trimA1 version 1.2 | [26] | N/A |
| BEAST version 1.8.3 | [27] | N/A |
CRISPR deletion of 11 pals genes
To construct a 34kb C17H1 region deletion mutant, we used the co-conversion strategy as described [20] with dpy-10 sgRNA as the selection marker. We first constructed the short guide RNA (sgRNA) plasmids (pC17H1.3_sg2 and pC17H1.7_sg2) by designing the sgRNA (5′-gaacagagtgaagcaggaag-3′) to target C17H1.3/pals-2 and sgRNA (5′-acgggcagatatacagagac-3′) to target C17H1.7/pals-6. The short oligonucleotides were synthesized (IDT), annealed and ligated into a modified version of DR274 (Addgene Plasmid #42250) where the sgRNA site was flanked by BsaI and C. elegans U6 promoter and terminator from pU6::klp-12_sgRNA (Gift from Michael Nonet). The constructed sgRNA expression plasmids (20 ng/uL each) were co-injected into N2 worms with C17H1 region single stranded donor DNA (5′-attttgctcttatcacatttatagaaatgacaaaagtcaccgagccctcggtttttctttgcgatagttcagagcttctcaaatctctca-3′) (500nM), pDD162 (Addgene Plasmid #47549) that expresses Cas9 with empty sgRNA (50ng/uL), dpy-10(cn64) single stranded donor DNA (500nM), and dpy-10(cn64) sgRNA (20 ng/uL). Dumpy or roller F1s were selected to ensure that CRISPR/Cas9 was expressed and dpy-10 was successfully modified. Worms were subsequently genotyped for the deletion with single worm 3-primer PCR where a primer pair flanking the whole deletion region and a third primer in the proposed deleted region (GW503 (5′-gttagaaatgcgctgtgacgt-3′), GW504, (5′-agctcgctcagcattgttg-3′), GW515 (5′-ggaatggtactaccagtgctg-3′)). The dpy-10 animal was crossed with N2 to obtain the final ~34kb deletion mutant strain without the dumpy phenotype. pals genes in the deleted region are pals-2, pals-3, pals-4, pals-5, pals-6, pals-7, pals-8, pals-9, pals-10, pals-11, and pals-12.
Quantitative RT-PCR
To measure endogenous mRNA expression changes, synchronized L1 worms were grown at 20°C to the L4 stage on OP50 or RNAi bacteria. RNA was extracted using TriReagent (Molecular Research Center, Inc.) and reverse transcribed using either the Retroscript kit (Ambion) or qScript cDNA SuperMix (Quanta Biosciences). Quantitative PCR was performed with iQ SYBR Green Supermix (Bio-Rad) using a CFX Connect Real-Time PCR Detection System (Bio-Rad). Primer sequences for each gene are provided in Table S1. At least two independent biological replicates were measured for each condition, and each biological replicate was measured in duplicate and normalized to the snb-1 control gene, which did not change upon conditions tested. The Pffafl method was used for quantifying data [21].
Heat stress assays
Worms were grown on standard NGM plates until the L4 stage at 20°C. To assess thermotolerance, worms were shifted to 37°C for two hours. Following heat shock, plates were laid in a single layer on the bench top for 30 minutes to recover, and then moved to 20°C incubator overnight. Worms were scored in a blinded manner for survival 24 hours after heat shock; animals not pumping or responding to touch were scored as dead. Three plates were assayed for each strain in each replicate, with at least 30 worms per plate, and three independent assays were performed for each condition. To assess response to prolonged heat stress, worms were shifted to 30°C for 24 hours and then imaged or harvested for RNA extraction. Attempts to use feeding RNAi to assay gene function in heat shock assays were not successful, as we found that pals-22 mutants do not have have increased thermotolerance when feeding on RNAi bacteria (E. coli strain HT115), likely due to effects of diet on physiology.
Osmotic stress assays
Assays were performed as described previously [16, 22]. Worms were raised under standard conditions until 24 hours post-L4 stage, then transferred to NGM plates containing 500 mM NaCl and incubated at 20°C. For polyQ aggregation assays, worms were imaged on a fluorescence dissecting microscope (Zeiss Discovery V8) and scored for the presence of aggregates at the indicated timepoints. For osmotic stress resistance assays, worms were scored for motility over a period of 10 minutes. Two plates were assayed for each strain in each replicate, with at least 30 worms per plate. Each experiment was repeated at least three times.
Fluorescence measurement
Synchronized L1 stage animals were placed on NGM plates seeded with OP50-1 bacteria and incubated at 20°C to the L4 stage. The COPAS Biosort machine (Union Biometrica) was used to measure the time of flight (length) and fluorescence of individual worms. At least 100 worms were measured for each strain, and all experiments were repeated three times.
Generation of pals-22 rescue constructs
Using standard PCR fusion [23] and Gibson cloning [24], constructs were generated in which either the pals-22, vha-6, unc-119, or dpy-7 promoter drives expression of a pals-22 cDNA fused to GFP and the unc-54 3′UTR, in the vector pCFJ151 to enable Mos1-mediated single copy insertion [19]. Sequences of primers used for promoter amplification are in Table S1.
Bioinformatic Analysis
Amino acid sequences of 36 genes were aligned using MUSCLE (version 3.8.31) [25] and trimmed with trimAl (version 1.2) [26]. Bayesian Markov chain Monte Carlo inference (LG + I + G + F) was performed using BEAST (version 1.8.3) [27]. Analyses were performed with a chain length of 10 million states (sampled every 1,000 iterations) applying an uncorrelated relaxed clock (lognormal distribution) and a Yule model tree prior. Convergence and mixing was assessed with Tracer (version 1.5) and maximum clade credibility tree generated after a 25% burn-in. Posterior probability values greater than 0.5 are marked on branch labels.
Lifespan
L4 stage worms were transferred to 6 cm NGM plates seeded with OP50-1 bacteria and incubated at 25°C. Three plates were assayed for each strain in each replicate, with at least 30 worms per plate, and experiments were repeated two independent times with similar results. Worms were scored every day, and animals that did not respond to touch were scored as dead. Animals that died from internal hatching or crawled off the plate were censored. Worms were transferred to new plates every day throughout the reproductive period.
Single molecule fluorescent in situ hybridization (smFISH)
For each strain, 1000 synchronized L1 larvae were placed on each of three standard 10 cm NGM plates seeded with OP50-1. Worms were grown at 20°C for 2 days, washed off plates using M9 + 0.1% Triton X-100, and fixed in 1mL 4% paraformaldehyde in PBST (1X PBS + 0.1% Tween-20) for 30min at room temperature. After washing three times in PBST, worms were resuspended in 1mL of 70% ethanol, incubated on a rotator at 4°C overnight, and then stored at 4°C in ethanol until staining. The fixed sample was washed with 500 μl wash buffer (10% formamide, 2X SSC) and then incubated in 1mL wash buffer at room temperature for 3min. 50μL of hybridization solution (10% formamide, 2X SSC, 10% dextran sulfate, 2mM vanadyl ribonucleoside complex, 0.02% RNAse free BSA, 50μg E. coli tRNA) containing 1μM cul-6 CalFluor610 smFISH probes was added to worms and incubated at 30°C in the dark overnight. Worms were washed with 500μL wash buffer and then incubated in 1mL wash buffer for 30min in the dark. After washing with 500μL 2X SSC, 2μL Vectashield + DAPI was added to each sample. 2μL of stained worms were added to the center of a glass microscope slide, and covered with a 22x22mm square glass coverslip. Imaging was performed using a Zeiss AxioImager M1 upright fluorescent microscope with a 63X oil immersion objective. Z-stacks of worm heads and anterior intestines were acquired with a z-spacing of 1 μm, collecting signal for GFP (GFP-tagged PALS-22 protein), CAL Fluor Red 610 (cul-6 smFISH probe), DAPI (DNA staining), and DIC. Image processing was performed using ImageJ (https://imagej.nih.gov/ij/). smFISH spot quantification was performed by blinded scorers using StarSearch (http://rajlab.seas.upenn.edu/StarSearch/launch.html). smFISH probes targeting the cul-6 transcript were designed using the Stellaris RNA FISH Probe Designer from Biosearch (https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer). The resulting set of 48 probes tagged with Calfluor610 were obtained from Biosearch. Experiments were repeated three times.
Western blot
For each strain, 1500 synchronized L1 worms were placed on NGM plates seeded with OP50-1 bacteria and incubated at 20°C. After 72 hours, worms were washed off plates with M9 buffer and washed 2 times with 15 ml of M9 buffer. The worm pellet was then mixed with 20ul of 6X SDS-PAGE sample buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 0.01% Bromophenol blue, 10% glycerol, 10 mM dithiothreitol), incubated 10 minutes at 95°C, and then centrifuged 15 minutes at 16,000g. Protein extracts were electrophoresed on a 4–20% gradient gel (Bio-Rad) at 180 V for approximately 40 minutes, and then transferred onto PVDF membrane (BioRad) using a semi-dry transfer system (Transblot SD BioRad) for 20 minutes at 15V. Non-specific binding was blocked using 5% nonfat dry milk in PBS-Tween (0.2%) for 1 hour at room temperature. The membranes were incubated with primary antibodies overnight at 4°C (rabbit anti-GFP diluted 1:5,000 and mouse anti-actin diluted 1:1,000). Following three 15 minute washes in PBS-Tween, membranes were blotted in horseradish peroxidase-conjugated secondary antibodies at room temperature for 2 hours (Goat anti-mouse IgM diluted 1:2,000 and Goat anti-rabbit IgG diluted 1:10,000). Membranes were then washed and treated with ECL reagent (Amersham), and imaged using a Bio-Rad Chemidoc XRS+ with Image Lab software.
Microscopy
Worms were anesthetized with 10 uM levamisole in M9 buffer and mounted on 2% agarose pads for imaging. Images in Figure 4 were captured with a Zeiss LSM700 confocal microscope. All other C. elegans images were captured with a Zeiss AxioImager M1.
Quantification and Statistical Analysis
All statistical details (statistical test used, number of samples, and p values) for each experiment can be found in the Figure labels. Graphic representation and statistical analyses were performed using GraphPad Prism 7 software. Data shown in graphs indicate mean, and error bars represent standard deviation unless otherwise indicated. Data were considered statistically significant when p < 0.05 by statistical test performed as indicated in the Figure labels. Two-tailed unpaired Student’s t-test was used to compare two different groups of samples when indicated. The log-rank test was used to analyze survival curves in Figure S3F, and one-way ANOVA was used to compare samples in Figure 4E. Asterisks indicate corresponding statistical significance: * p< 0.05; ** p< .01; *** p<0.001. **** p<0.0001.
Supplementary Material
Highlights.
The Intracellular Pathogen Response (IPR) defines a distinct proteostasis program
PALS-22 is a negative regulator of IPR gene expression, including the cullin cul-6
pals-22 mutants have increased tolerance of proteoxic stress, dependent on CUL-6
PALS-22 acts in the intestine to regulate CUL-6 expression and thermotolerance
Acknowledgments
We thank Lakshmi Somasundaram and Lianne Cohen for technical assistance, and Oliver Hobert for communicating unpublished results and providing strains. Some C. elegans strains were provided by the Caenorhabditis Genetics Center (CGC), funded by the NIH Office of Research Infrastructure Programs P40 OD010440. We thank Oliver Hobert, Aaron Reinke, Eric Bennett, and Robert Luallen for providing helpful comments on the manuscript. J.N.S. is an IRACDA fellow and was supported by NIGMS/NIH award K12GM068524. This work was supported by NIH R21 AI115442 to D.W. and by GM114139, AG052622, and a Burroughs Wellcome Fund fellowship to E.R.T.
Footnotes
Author Contributions
Conceptualization: K.C.R. and E.R.T.; Investigation: K.C.R., T.D., J.N.S., J.P., K.C., and E.S.L.; Writing: K.C.R. and E.R.T.; Supervision: E.R.T. and D.W.; Funding Acquisition: E.R.T. and D.W.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and-aggregation diseases. Trends in cell biology. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & development. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harbor perspectives in biology. 2011;3:a004440. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TL, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10:e1004200. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen K, Franz CJ, Jiang H, Jiang Y, Wang D. An evolutionarily conserved transcriptional response to viral infection in Caenorhabditis nematodes. BMC genomics. 2017;18:303. doi: 10.1186/s12864-017-3689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkies P, Ashe A, Le Pen J, McKie MA, Miska EA. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome research. 2013;23:1258–1270. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leyva-Díaz E, Stefanakis N, Carrera I, Glenwinkel L, Wang G, Driscoll M, Hobert O. Silencing of Repetitive DNA Is Controlled by a Member of an Unusual Caenorhabditis elegans Gene Family. Genetics. 2017 doi: 10.1534/genetics.117.300134. 117.300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hentati A, Bejaoui K, Pericak-Vance MA, Hentati F, Speer MC, Hung WY, Figlewicz DA, Haines J, Rimmler J, Hamida CB. Linkage of recessive familial amyotrophic lateral sclerosis to chromosome 2q33–q35. Nature genetics. 1994;7:425–428. doi: 10.1038/ng0794-425. [DOI] [PubMed] [Google Scholar]
- 9.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nature genetics. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 10.Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, Angermann K, Hasse S, Rupprecht M, Vinis E, Tinney M. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell. 2012;150:855–866. doi: 10.1016/j.cell.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mongkoldhumrongkul N, Swain SC, Jayasinghe SN, Sturzenbaum S. Bio-electrospraying the nematode Caenorhabditis elegans: studying whole-genome transcriptional responses and key life cycle parameters. J R Soc Interface. 2010;7:595–601. doi: 10.1098/rsif.2009.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 13.Vihervaara A, Sistonen L. HSF1 at a glance. The Company of Biologists Ltd; 2014. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munoz MJ. Longevity and heat stress regulation in Caenorhabditis elegans. Mechanisms of ageing and development. 2003;124:43–48. doi: 10.1016/s0047-6374(02)00168-9. [DOI] [PubMed] [Google Scholar]
- 16.Mazzeo LEM, Dersh D, Boccitto M, Kalb RG, Lamitina T. Stress and aging induce distinct polyQ protein aggregation states. Proceedings of the National Academy of Sciences. 2012;109:10587–10592. doi: 10.1073/pnas.1108766109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutscher LM, Shaham S. WormBook: the online review of C. elegans biology. 2014. Forward and reverse mutagenesis in C. elegans; p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frøkjær-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature genetics. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solomon A, Bandhakavi S, Jabbar S, Shah R, Beitel GJ, Morimoto RI. Caenorhabditis elegans OSR-1 regulates behavioral and physiological responses to hyperosmotic environments. Genetics. 2004;167:161–170. doi: 10.1534/genetics.167.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 24.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.