Summary
Ciliary microtubules (MTs) are extensively decorated with post-translational modifications (PTMs), such as glutamylation of tubulin tails. PTMs and tubulin isotype diversity act as a “Tubulin Code” that regulates cytoskeletal stability and the activity of MT-associated proteins such as kinesins. We previously showed that, in C. elegans cilia, the deglutamylase CCPP-1 affects ciliary ultrastructure, localization of the TRP channel PKD-2 and the kinesin-3 KLP-6, and velocity of kinesin-2 OSM-3/KIF17, while a cell-specific α-tubulin isotype regulates ciliary ultrastructure, intraflagellar transport, and ciliary functions of extracellular vesicle (EV)-releasing neurons. Here, we examine the role of PTMs and the Tubulin Code in the cililary specialization of EV-releasing neurons using genetics, fluorescence microscopy, kymography, electron microscopy, and sensory behavioral assays. Although the C. elegans genome encodes five tubulin tyrosine ligase-like (TTLL) glutamylases, only ttll-11 specifically regulates PKD-2 localization in EV-releasing neurons. In EV-releasing cephalic male (CEM) cilia, TTLL-11 and the deglutamylase CCPP-1 regulate remodeling of 9+0 MT doublets into 18 singlet MTs. Balanced TTLL-11 and CCPP-1 activity fine-tunes glutamylation to control velocity of kinesin-2 OSM-3/KIF17 and kinesin-3 KLP-6 without affecting the IFT kinesin-II. TTLL-11 is transported by ciliary motors. TTLL-11 and CCPP-1 are also required for the ciliary function of releasing bioactive EVs, and TTLL-11 is itself a novel EV cargo. Therefore, MT glutamylation, as part of the tubulin code, controls ciliary specialization, ciliary motor-based transport, and ciliary EV release in a living animal. We suggest that cell-specific control of MT glutamylation may be a conserved mechanism to specialize the form and function of cilia.
eTOC Blurb
O’Hagan et al report that fine-tuning of microtubule glutamylation by the glutamylase TTLL-11 and the deglutamylase CCPP-1 regulates ciliary function by controlling ciliary receptor localization, the velocity of particular kinesin-2 and kinesin-3 motors, the release of extracellular vesicles, and sculpting a specialized axonemal ultrastructure.
Introduction
Cilia and flagella are antenna-like organelles that protrude from most eukaryotic cells and serve sensory and motility functions that are important for development, physiology, and behavior. Cilia have a conserved structural core called an axoneme, composed of microtubules (MTs) that typically form a ring of nine outer A–B doublet MTs surrounding two or zero inner singlets—the so-called “9 + 2” or “9 + 0” formations in motile or primary/sensory cilia, respectively [1]. Although virtually all cilia are built by a conserved intraflagellar transport (IFT) process and share a similar architecture, cilia and flagella adopt morphological specializations and serve diverse functions [1]. For example, the rods and cones of the retina are elaborately shaped cilia [2], while sperm have simple whip-like flagella that are variable in length and axoneme structure [3]. C. elegans amphid channel cilia, mammalian olfactory cilia, and mammalian renal primary cilia possess a proximal doublet region followed by a distal A-tubule singlet region [1, 2, 4, 5]. Another ciliary specialization is the ability to produce extracellular vesicles (EVs) called ectosomes [6–11]. The molecular underpinnings and functions of these specializations are only beginning to be appreciated.
Regulation of the function of conserved ciliogenesis proteins by post-translational modifications (PTMs) of MTs could provide a mechanism for generating structural and functional diversity of cilia. Ciliary MTs are marked by diverse PTMs that have been proposed act as a “Tubulin Code” to regulate particular motors, MT-binding proteins, and MAPs (microtubule associated proteins) [12–14]. The tubulin tyrosine ligase-like (TTLL) family of proteins includes glutamylases, which act as writers of the Tubulin Code by adding or elongating glutamate side-chains on MTs [15]. Carboxypeptidases of the M14D deglutamylase subfamily act as erasers of the Tubulin Code that remove or reduce the length of glutamate side-chains on tubulins [15, 16]. Hence, MT glutamylation is a reversible modification, and a balance of glutamylase and deglutamylase activity may fine- tune the extent or pattern of glutamylation in tubulin C-terminal tails [13]. Ciliary MTs are heavily glutamylated [15]. Defects in glutamylation are implicated in human ciliopathies. Joubert syndrome [17], blindness [18], and schizophrenia [19], are associated with defects in TTLL glutamylases. Defects in the Ccp1 deglutamylase cause neuronal degeneration in mice [20]. How dysregulated glutamylation might contribute to human disease is largely unknown.
C. elegans possess variant 9+0 cilia whose ultrastructures can be simultaneously analyzed using transmission electron microscopy (TEM) and electron tomography [4, 21]. Variant 9+0 cilia are not nematode-specific oddities. Variations from the “typical” 9+2 and 9+0 doublet structure may be more common than appreciated, largely due to technical difficulty of serial section TEM of mammalian cilia. Of the 302 neurons in the C. elegans hermaphrodite, 60 have dendritic endings that terminate in cilia [22]. In addition to the shared ciliated nervous system (common between hermaphrodites and males), the C. elegans male possesses 48 ciliated neurons of 385 total neurons [22]. Specialized male-specific cilia shed and release bioactive EVs that contain the polycystin receptor LOV-1 and TRP channel PKD-2 [7]. The diversity of C. elegans sensory cilia enables study of ciliary specialization and the role of the Tubulin Code in this process in a living animal.
CCPP-1, a homolog of the mammalian deglutamylase Ccp1, is required in C. elegans sensory neuronal cilia to regulate MT stability [23]. In nematodes lacking CCPP-1, EV-releasing cephalic male-specific CEM cilia contain fewer MTs [23]. In CEM cilia, CCPP-1 regulates localization of the ciliary TRP channel PKD-2 and the kinesin-3 KLP-6, and the velocity of homodimeric kinesin-2 OSM-3/KIF17 without affecting the anterograde heterotrimeric kinesin-II motor [23]. These pleiotropic defects are likely to result from MT hyperglutamylation.
CEM cilia display an ultrastructural specialization in which nine MT doublets splay into nine A-tubule and nine B-tubule singlets in middle regions of the axoneme, but remain joined in distal and proximal regions [24]. The α-tubulin isotype TBA-6 is essential for B-tubule singlet formation, hence the tubulin code is implicated in generating this specialized EV-releasing cilium [24].
The mammalian and C. elegans genomes encode nine and five ttll glutamylases, respectively [15, 25]. TTLL glutamylases are biochemically distinguished by their preferences for the C-terminal tails of α- or β-tubulin and whether they are initiases (adding the first E) or elongases (extending chains of poly-E) [15]. However, the true physiological function of each TTLL enzyme is not known. Here, we focus on how hypoglutamylation resulting from genetic ablation of the TTLL glutamylases affects the cilia of a set of C. elegans male-specific EV-releasing neurons (EVNs) that express PKD-2 [26]—specifically, the four CEM neurons in the head; the HOB (Hook B-type) neuron in the tail, and 16 RnB (Ray B-type, where n=1–9, excluding ray 6) neurons that innervate the copulatory fan structure of the male tail. We show that CCPP-1 deglutamylase and TTLL-11 glutamylase act in concert to sculpt the CEM axoneme and regulate ciliary kinesin-2 OSM-3/KIF17 and kinesin-3 KLP-6 motors. CEM cilia are functionally specialized to shed and release bioactive EVs. We find that CCPP-1 and TTLL-11 are required for environmental release of EVs from ciliated neurons in C. elegans males, and that TTLL-11 itself is a novel EV cargo. Our results suggest that CCPP-1 and TTLL-11 fine-tune glutamylation to regulate ciliary transport, EVs, and axonemal structure in cilia.
Results
The tubulin glutamylase TTLL-11B isoform regulates PKD-2∷GFP ciliary localization and is specifically expressed in EV releasing neurons
ccpp-1 is widely expressed in ciliated sensory neurons in hermaphrodites and males [23]. Here we focus on the role of glutamylation in ciliary specialization of the male-specific EVNs, where CCPP-1-mediated regulation of MT glutamylation is important for appropriate localization of the ciliary TRP channel PKD-2∷GFP. PKD-2∷GFP abnormally accumulates in ccpp-1 mutant cilia and distal dendrites (Figure 1A; [23]). To identify the TTLL glutamylase that opposes CCPP-1 in EVNs, we hypothesized that loss of a TTLL glutamylase might suppress the ccpp-1 PKD-2∷GFP ciliary (Cil) defective phenotype caused by hyperglutamylation. The C. elegans genome encodes five TTLL family members TTLL-4, TTLL-5, TTLL-9, TTLL-11, and TTLL-15 [27]. (We exclude TTLL-12 for two reasons: It is an ortholog to mammalian TTLL12, which lacks glutamylase and glycylase activity [15], and is not neuronally expressed [27]). Mutant alleles that delete portions of coding regions were available for each of the five ttll- genes: ttll-4(tm3310); ttll-5(tm3360); ttll- 9(tm3389); ttll-11(tm4059); ttll-11(gk482); ttll-15(tm3871). We examined ttll mutants for the ability to suppress the ccpp-1 PKD-2∷GFP Cil phenotype.
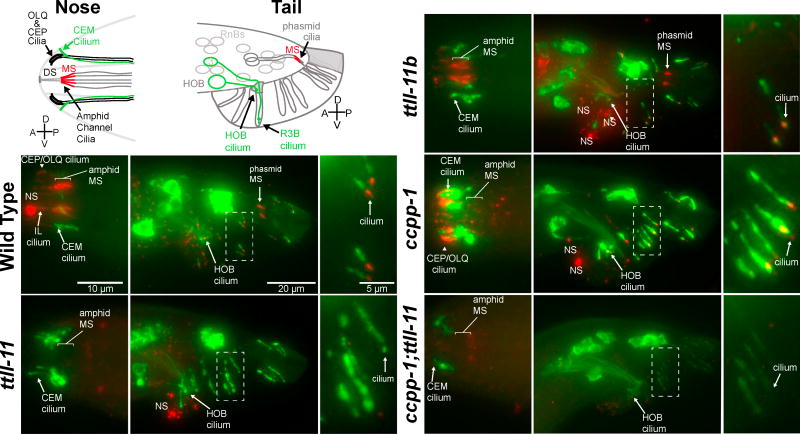
Figure 1. ttll-11 regulates PKD-2∷GFP ciliary localization and encodes two TTLL family glutamylase isoforms with non-overlapping expression patterns.
(A) Diagrams (adapted from [23]) of male-specific extracellular vesicle-releasing neurons (“EVNs”: CEMs in head, and HOB and RnBs, where n=1–9, except 6 in tail), which express PKD-2∷GFP. In the diagram of the nose, only the left side CEM neurons of the two bilateral pairs is shown. Boxed area indicates region shown in micrographs. Abbreviations: d, dendrite; cb, ciliary base; c, cilium. Cell bodies are further posterior, out of view. In the tail, each ray is innervated by a single RnB neuron; the dendrite and cilium of R3B are shown as an example. Panels show PKD-2∷GFP localization in nose and tail for indicated genotypes. See also Figure S1 and Table S1. (B) Genomic structure of ttll-11 locus, which encodes two isoforms. Bars below the diagram show the gk482 and tm4059 deletion alleles used in this study. Both TTLL-11B and TTLL-11A contain a TTL domain of 364 amino acids (black region). MYR indicates a predicted myristoylation site [29] in TTLL-11B. By analogy with other TTLL proteins, the C-terminal gray area is expected to bind microtubules. (C, D) A ttll-11b promoter drove GFP expression in the EVNs, marked by klp-6 promoter-driven tdTomato. Expression of both transgenes was somewhat mosaic, and therefore did not completely overlap; for example, one of the CEM ventral neurons in the head was not labeled by tdTomato. (E, F) GFP expression driven by the ttll-11a promoter was not visible in the EVNs, marked by expression of Pklp-6∷tdTomato. (G) Behavioral response to hermaphrodites in male mating was scored. ccpp-1 and ccpp-1;ttll-11 mutant males responded significantly less frequently than wild-type. Data represents mean ± sem; N = 4 or 5 trials, n = 35 – 56 males tested for each genotype. ***indicates p<0.0001 by ANOVA and Tukey post-hoc tests. (H) Vulva location behavior was scored for 10 – 25 males for all genotypes. ** p<0.001, *** p<0.0001 by Kruskal-Wallis and Dunn’s Multiple Comparison Test.
None of the tested TTLL glutamylase deletion mutations suppressed the ccpp-1 PKD-2∷GFP localization defect. However, ttll-11(tm4059) and ttll-11(gk482) deletion mutants displayed a PKD-2∷GFP Cil phenotype similar to ccpp-1 mutants, with PKD-2∷GFP abnormally accumulating in ciliary bases and distal dendrites (Figures 1A and S1). These results suggest that regulated MT glutamylation is essential for normal ciliary localization and abundance of PKD-2. We conclude that different TTLL enzymes may act in a cell-specific manner and may possess different enzymatic activities in vivo.
The C. elegans ttll-11 locus encodes two isoforms: the long TTLL-11B and short TTLL-11A proteins [25, 27]. A BLAST search of the Human TTLL11 long isoform (NCBI reference sequence NP_001132914) against C. elegans WS259 protein database identified both TTLL-11B and TTLL-11A as top hits [28], and alignments showed that amino acids 213 – 742 from the human TTLL11 long isoform are 35% identical and 58% similar to C. elegans TTLL-11B amino acids 119 – 642 and TTLL-11A amino acids 19 - 542. TTLL-11A lacks the first 100 amino acids of TTLL-11B, but is otherwise identical (Figure 1B; [25, 27]). TTLL-11B contains a putative myristoylation sequence at its N-terminus [29]. We previously showed that myristoylation is necessary for targeting and function of the EV regulator and EV cargo CIL-7 [30].
The ttll-11(tm4059) deletion allele is presumably a null allele for both ttll-11 isoforms, and is predicted to produce an early stop codon after 90 amino acids in TTLL-11A and 190 amino acids in TTLL-11B (Figure 1B; [27]), removing all except seven amino acids of the predicted ATP-grasp_4 domain [31]. The gk482 deletion allele affects the coding region of only the TTLL-11B long isoform (Figure 1B; [27]). Both the tm4059 and the gk482 alleles produced a PKD-2∷GFP Cil phenotype (Figures 1A and S1). Therefore, at least the TTLL-11B isoform is required for normal localization of PKD-2. Hereafter, unless specifically noted as the ttll-11b(gk482) allele, reference to mutation of ttll-11 indicates the tm4059 allele, which affects both isoforms.
To examine where the ttll isoforms function, we created transcriptional reporters. The ttll-11b reporter was exclusively expressed in ciliated EV-releasing neurons. Expression of GFP (green fluorescent protein; [32]) driven by the ttll-11b promoter was observed in the IL2 ciliated sensory neurons in both males and hermaphrodites, as well as the male-specific PKD-2-expressing neurons (CEMs in the head; HOB and Ray type B neurons, or “RnBs,” in the tail; Figure 1C, D). The IL2, CEM, HOB, and RnB neurons comprise the EVNs, which release bioactive EVs to the environment [7, 8]
The ttll-11a promoter drove GFP expression in a distinct and non-overlapping set of ciliated sensory neurons in the head, including the IL1s, OLQs, CEPs, and amphids, but not in the EVNs (Figure 1E). In the male tail, expression was seen in HOA, Ray type A neurons (RnAs), and phasmids, but not the HOB and RnB neurons (Figure 1F). Expression was also seen in phasmid neurons in the hermaphrodite tail (data not shown). These expression patterns suggest that TTLL-11B functions in the EVNs and is essential for normal localization of PKD-2, whereas TTLL-11A functions in other ciliated sensory neuronal types.
The polycystin PKD-2 and the male-specific EVNs mediate male mating behaviors [26]. Because ttll-11b expressed in these neurons and the ttll-11 mutant displayed abnormal accumulation of PKD-2∷GFP, we examined the mating behaviors of ttll-11 mutant males (Figure 1G, H). ttll-11 mutant males were “Lov” defective (i.e., abnormal location of vulva substep of mating behavior) but not “Rsp” defective (i.e. the response substep of mating behavior was normal). These results suggest that abnormal glutamylation impairs the function of these male-specific EVNs, and that hyperglutamylation caused greater impairment than hypoglutamylation. The RnBs play a role in response behavior, while the HOB functions in location of vulva behavior [26]. The fact that ttll-11 mutants are Lov defective but not Rsp defective suggests that requirements for MT glutamylation may not be identical even among the neurons that express ttll-11 and ccpp-1 and mediate mating behaviors. Alternatively, location of vulva behavior may be more sensitive to abnormal glutamylation because a single pair of neurons (HOB and HOA) senses the vulva, whereas multiple ray neurons redundantly sense the hermaphrodite for response behavior [26].
TTLL-11 is essential for MT glutamylation
ttll-11 encodes a glutamylase and ttll-11 mutations are predicted to reduce glutamylation. To determine if TTLL-11 regulates glutamate side-chain length, we analyzed glutamylation state by immunodetection with a polyclonal polyglutamylation (polyE) antibody (IN105) that recognizes chains of 3 or more glutamates [33]. Glutamylation in cephalic CEM and CEP cilia was undetectable by polyE staining in ttll-11(tm4059) mutant males, which lack both TTLL-11B and TTLL-11A. (Figure S2). However, in the absence of TTLL-11B only, some ciliary polyE staining remained in cephalic cilia.
To determine if TTLL-11 is required for glutamylation branch point initiation, we stained animals with the monoclonal antibody GT335, which detects the branch point of glutamylation side-chains on tubulin C-terminal tails [23, 34]. In ttll-11(tm4059) mutants, glutamylation was undetectable by GT335 staining in all cilia, including those in male-specific EVNs (Figure 2). In ttll-11b(gk482) mutant males, GT335 still stained cilia, consistent with TTLL-11A-mediated glutamylation of ciliary MTs in amphid, phasmid, CEP, and RnA cilia when only the ttll-11b isoform is mutated (Figure 2).
Figure 2. TTLL-11 was required for ciliary MT glutamylation.
Fixed young adult males expressing PKD-2∷GFP (green) stained with monoclonal antibody GT335 (red), which detects the branch point of glutamylation side-chains. Cartoon depicts cilia observed in the nose and tail. In the tail, each ray B-type neuron is equipped with a sensory cilium, but cartoon only shows cilium for the left R3B neuron. For each genotype, left panel shows ciliated endings in the tip of the nose; middle panel shows male tail fan, and right panel shows enlargements of the boxed areas in tails to show ray cilia. In wild type panels, GT335 staining of amphid ciliary MS (middle segments, which contain doublet MTs), CEP cilia, and IL cilia in the head, and phasmid cilia in the tail, are indicated. NS indicates puncta of non-specific staining, where antibody stuck to the cuticle or cellular debris on some animals. See also Figure S2 and Table S1.
Taken together, our immunofluorescence data (Figures 2 and S2) and isoform expression patterns (Figure 1C–F) suggest that TTLL-11B is an EVN-specific glutamylase and TTLL-11A functions in a distinct and non-overlapping set of neurons. Although a previous study showed that a mammalian TTLL11 prefers elongation over initiation of glutamate side-chains in vitro [33], our results suggest that initiation of glutamate side-chains is an important function of C. elegans TTLL-11 in vivo.
ccpp-1 and ttll-11 modulate OSM-3/KIF17 and kinesin-3 KLP-6 but not heterotrimeric kinesin-II
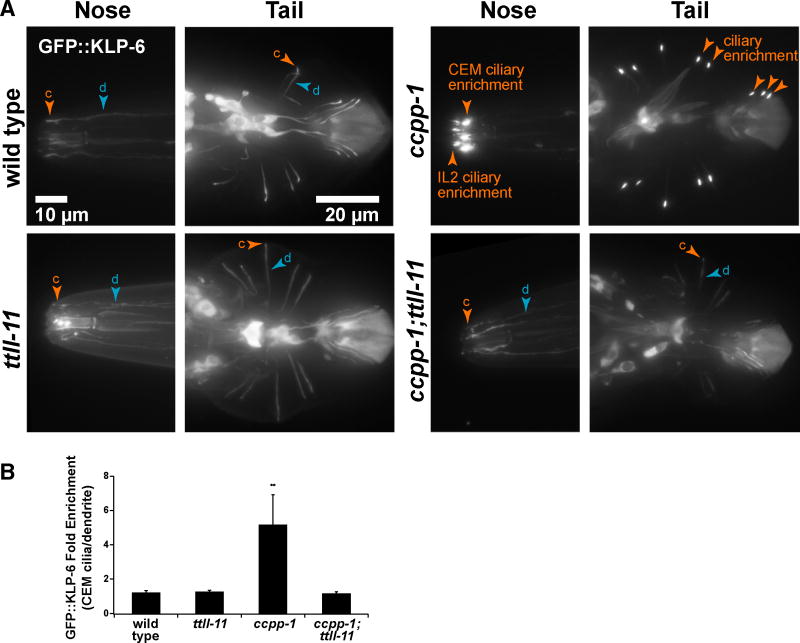
In CEM cilia, anterograde IFT is driven predominantly by kinesin-II with minimal modulation from accessory kinesin-2 OSM-3/KIF17 and kinesin-3 KLP-6 [35]. KLP-6 is similar to mammalian KIF13B [36] and KIF28 [35]. Hyperglutamylation of ciliary MTs in ccpp-1 mutants causes abnormal accumulation of the kinesin-3 motor KLP-6 in EVNs (Figure 3 A, B; [23]). Therefore, we tested if TTLL-11 was required for normal localization of GFP∷KLP-6 in EVNs. Although GFP∷KLP-6 localization was not affected in ttll-11 mutant males, loss of TTLL-11 did suppress the GFP∷KLP-6 accumulation defect in ccpp-1 mutants (Figure 3 A, B).
Figure 3. Loss of TTLL-11 function suppressed GFP∷KLP-6 kinesin-3 ciliary accumulation of ccpp-1 mutants.
(A) Localization of the kinesin-3 motor GFP∷KLP-6 was diffuse in wild type and ttll-11 single mutants. In ccpp-1 mutants, GFP∷KLP-6 accumulated in cilia of EVNs in the head (CEMs and IL2s) and tail (HOB and RnBs). Mutation of ttll-11 suppressed the abnormal ciliary enrichment of GFP∷KLP-6 in ccpp-1 mutants. (Tail images were normalized so that brightness of autofluorescent posterior tip of acellular tail fan was similar across genotypes. c, cilium; d, dendrite.) (B) The enrichment of GFP∷KLP-6 was quantified across genotypes by calculating the ratio of the maximum pixel value in cilia over the maximum pixel value in the distalmost 10 μm of dendrites for the head neurons only. Data represents Mean ± sem for N = 3 animals for each genotype; ** indicates significantly different from wild type with p= 0.002 by ANOVA and post-hoc Tukey test. See also Table S1.
To determine if glutamylation state regulates the velocity of the kinesin-3 KLP-6, we analyzed the motility of GFP∷KLP-6 puncta in CEM cilia of young adult males (less than four hours after the L4 larval stage; see Methods). In wild type, GFP∷KLP-6 particles moved at 0.8μm/s (Table 1; Figure S3). In ccpp-1 GFP∷KLP-6 velocity increased to 0.88 (p<0.05 vs. wild type). By contrast, in ttll-11 cilia, GFP∷KLP-6 velocity decreased to 0.71 (p<0.05 vs wild type; Table 1; Figure S3). In the ccpp-1;ttll-11 double mutant, GFP∷KLP-6 velocity was similar to wild type. To date, perturbing glutamylation is the only genetic manipulation found to affect KLP-6 motility in vivo [24, 35].
Table 1.
CCPP-1 and TTLL-11 Regulate Velocities of GFP-tagged Kinesin-2 (OSM-3), Kinesin-3 (KLP-6), but not Heterotrimeric Kinesin-II, in CEM cilia
| Genetic Background | Anterograde IFT velocity (μms−1 + sd) | N worms/n particles | |
|---|---|---|---|
| Kinesin-3 | Wild type | 0.80 + 0.15 | 9/51 |
| GFP∷KLP-6 | ccpp-1(*vs wt) | 0.88 + 0.15 | 5/48 |
| ttll-11 (*vs wt;***vs ccpp-1) | 0.71 + 0.13 | 6/41 | |
| ccpp-1; ttll-11(***vs ccpp-1) | 0.77 + 0.14 | 8/43 | |
|
| |||
| Kinesin-2 | Wild type | 0.74 + 0.21 | 7/53 |
| OSM-3∷GFP | ccpp-1 (*** vs wt) | 0.92 + 0.24 | 6/46 |
| ttll-11 (*** vs ccpp-1) | 0.67 + 0.22 | 8/63 | |
| ccpp-1; ttll-11 (*** vs ccpp-1) | 0.65 + 0.21 | 6/60 | |
|
| |||
| Heterotrimeric | Wild type | 0.55 + 0.08 | 16/125 |
| Kinesin-II | ccpp-1 | 0.57 + 0.13 | 20/133 |
| KAP-1∷GFP | ttll-11 | 0.56 + 0.11 | 10/79 |
| ccpp-1; ttll-11 | 0.63 + 0.17 | 14/154 | |
In ccpp-1 CEM cilia, OSM-3∷GFP but not kinesin-II-driven IFT moves abnormally rapidly [23], suggesting that glutamylation specifically regulates OSM-3/KIF17. To test this, we examined OSM-3 and kinesin-II ciliary velocity in ttll-11 mutant CEM cilia. While ttll-11 mutants displayed normal OSM-3∷GFP velocity, ttll-11 suppressed the abnormally fast OSM-3∷GFP velocity of ccpp-1 mutants (Table 1; Figure S3).
C. elegans heterotrimeric kinesin-II comprises motor subunits KLP-11 and KLP-20 and accessory subunit KAP-1 [37]. In ccpp-1 single, ttll-11 single, and ccpp-1; ttll-11 double mutants, KAP-1∷GFP velocity was similar to wild type, indicating that kinesin-II is not regulated by hyper-glutamylation or hypo-glutamylation [23] (Table 1; Figure S3).
CCPP-1 and TTLL-11 regulate release of EVs
C. elegans EVNs produce bioactive EVs [7]. In the male head, the cephalic sensillum is comprised of two ciliated sensory neurons (CEM and CEP) surrounded by the glial socket and sheath cells. The CEM neuron secretes EVs from the ciliary base into the extracellular lumenal space formed by glial support cells (Figure 4A; [7]). We call this process EV “shedding.” In a process we call EV “release,” EVs exit through cuticular pores to the outside environment, where they can evoke a behavioral response in other males (Figure 4A; [7]). EVs shed and released by the CEMs and RnBs contain polycystins, and can be visualized by PKD-2∷GFP (Figure 4A, B; [7]). The kinesin-3 KLP-6 and α-tubulin TBA-6 regulate release of ciliary EVs, with mutants releasing fewer PKD-2∷GFP labeled EVs and also accumulating excessive amounts of EVs in the cephalic lumen surrounding the CEM cilium, as visualized by the PKD-2∷GFP reporter in living animals and transmission electron microscopy (TEM) in fixed animals [7, 24].
Figure 4. The MT glutamylase TTLL-11 and deglutamylase CCPP-1 were required for release of PKD-2∷GFP-labeled extracellular vesicles (EVs).
(A) Cartoon (modified from [7] shows CEM EV “shedding,” which produces the EVs surrounding the CEM ciliary base inside a lumen formed by the sheath and socket glial cells; and EV “release” from CEM ciliated neurons to the environment. (B) Diagram shows PKD-2∷GFP-labeled EVs released from CEMs float and accumulate at the cover slip. (C) Abundant PKD-2∷GFP-labeled EVs were released outside from CEM neurons in wild-type adult males (several EVs indicated by arrowheads). Few EVs were seen in ttll-11 mutants. (D) Quantification of EVs released to the local environment by sensory ciliated CEM neurons in adult male head. N animals scored in parentheses for each genotype. See Figure S4 for images of additional genotypes, as well as images of PKD-2∷GFP-labeled EVs released by neurons in the male tail in adults and L4 larvae. (E) EVs shed into the glial lumen were reconstructed and counted from serial TEM sections. Representative sections are shown in Fig. S5. See also Table S1. In parentheses, N = cilia scored. (Mean ± sem;* p=0.0153; ** p=0.0065; ***p<0.001 by ANOVA and post-hoc Tukey tests.)
To determine if the PKD-2∷GFP Cil phenotype of ccpp-1 and ttll-11 mutants is due to defects in EV shedding and/or release, we visualized and counted environmentally released PKD-2∷GFP-labeled EVs (Figure 4B – D; Figure S4). In wild-type males expressing PKD-2∷GFP, 102 ± 12 fluorescently marked EVs surrounded the nose, and 100 ± 11 surrounded the tails. In contrast, ccpp-1, ttll-11, and ttll-11b single mutants, and the ccpp-1;ttll-11 double mutant released few PKD-2∷GFP-labeled EVs from ciliated neurons in the head or tail of adult males. We also counted PKD-2∷GFP-labeled EVs trapped in the molting cuticles of L4 male tails. Wild-type L4 male tails contained 34 ± 4.7 EVs, compared with 9.6 ± 2.4 in ccpp-1; 3.4 ± 0.9 in ttll-11; 4.8 ± 2.1 in ccpp-1; ttll-11 double mutants; and 5.8 ± 1.0 in ttll-11b(gk482) mutants (Figure S4).
To examine EV shedding into the glial lumen, we counted EVs from TEM serial sections of ccpp-1 single, ttll-11 single, and ccpp-1; ttll-11 double mutants. The lumen surrounding CEM cilia in ccpp-1;ttll-11 double-mutant males contained on average the same number of EVs as wild type (Figures 4D and S5). In contrast, both ccpp-1 and ttll-11 single mutants contained an abnormally high number of EVs in the lumenal space surrounding CEM cilia– almost ten-fold more than wild type (Figures 4E and S5). We conclude that glutamylation enzymes regulate EV environmental release (Figures 4 and S4).
TTLL-11 moves in dendrites and cilia and is an EV cargo
To observe the subcellular localization of TTLL-11 in EVNs, we expressed TTLL-11∷GFP in the CEM, HOB, and RnB neurons under control of the pkd-2 promoter (Figure 5A–C). TTLL-11∷GFP localized in puncta throughout sensory neurons, including axons, cell bodies, and dendrites, and was enriched in cilia. TTLL-11∷GFP was packaged into EVs that were shed and released from CEMs and RnBs to the environment (Figure 5B, C). Therefore, TTLL-11 is a regulator of EV release as well as an EV cargo.
Figure 5. TTLL-11∷GFP was enriched in cilia and a cargo of environmentally released EVs.
(A) TTLL-11∷GFP expressed under the pkd-2 promoter was enriched in CEM cilia and ciliary bases. TTLL-11∷GFP appeared in puncta in dendrites and cell bodies of C. elegans males. CEM cilia and ciliary base are indicated by a bracket and arrow, respectively. (B, C) TTLL-11∷GFP was observed in EVs released from CEM neurons in the head (B) and RnB neurons in the tail (C). Yellow arrowheads point to TTLL-11∷GFP-labeled EVs. (D) Kymograph and histograms of TTLL-11∷GFP movement in CEM dendrites. Mean ± sem dendritic velocities shown for 15 animals, 153 anterograde particles; 98 retrograde particles. (E) Mean ± sem anterograde velocity of TTLL-11∷GFP in CEM cilia for genotypes shown, and representative kymographs created using Kymograph Direct [43]. See also Table S1.
Time-lapse fluorescence microscopy was used to see if TTLL-11∷GFP puncta move in dendrites or cilia of CEM neurons. In dendrites, TTLL-11∷GFP exhibited bidirectional and saltatory movement (Figure 5D, E). Depending on directionality, TTLL-11∷GFP puncta moved at different rates. Anterograde TTLL-11∷GFP movement (from cell body to ciliary base) was approximately 0.87 μm/sec and retrograde movement (from ciliary base to cell body) was 0.97 μm/sec (Fig 5D). TTLL-11 dendritic transport rates are similar to dendritic velocities of IFT components and different from the dendritic velocities of PKD-2 and the ODR-10 G-protein coupled receptor [38, 39]. We previously proposed that the dendritic transport of ciliary receptors and the IFT machinery involves different mechanisms [38]. Our new data support this hypothesis and suggest that TTLL-11 and the IFT machinery are transported along dendrites in a similar manner.
TTLL-11∷GFP also moved bidirectionally in CEM cilia. The average velocity of anterograde TTLL-11∷GFP transport was 0.87, which was faster that the velocity of KLP-6, OSM-3, or KAP-1 in wild-type CEM cilia, but similar to KLP-6 and OSM-3 rates in ccpp-1 hyper-glutamylated cilia (Figure 5E, Table 1). To identify the motors that transport TTLL-11∷GFP in cilia, we examined TTLL-11∷GFP transport in mutants lacking the OSM-3, KLP-6, or KLP-11 kinesin motor. In osm-3 and klp-6 mutants, TTLL-11∷GFP velocity was similar to wild type, but mutation of klp-11 significantly increased TTLL-11∷GFP velocity to approximately 1 μm/s (Figure 5E), which matches the faster 1 μm/s velocity observed previously for IFT-A and IFT-B polypeptides in klp-11 mutant CEM cilia [35]. Combined, these results indicate that TTLL-11 is moved by IFT and that TTLL-11 is a promiscuous cargo that does not rely on a single ciliary kinesin for its transport.
TTLL-11 sculpts CEM axonemal MT architecture
The structure of the wild-type CEM axoneme is distinctive (Figure 6A; [24]). Like most ciliary axonemes, the CEM ciliary transition zone contains a ring of nine doublet MTs, connected to the membrane by Y-link structures, followed by a proximal region with nine doublet MTs without Y-links [24]. In the middle region, the nine doublet MTs splay into 18 singlets [24]. Many of these A-tubule and B-tubule singlets remain adjacent to the ciliary membrane (Figure 6A). In the distal region, the MT A- and B-tubule singlets remain joined as doublets, and A-tubules extend to the distal most ciliary tip [24]. Therefore, ciliary doublets splay in the middle to create singlets that are fused at both the proximal and distal ends.
Figure 6. CCPP-1 and TTLL-11 regulators of MT glutamylation were required for normal CEM neuronal ciliary ultrastructure.
(A) Representative TEM sections through middle regions of CEM cilia in genotypes indicated, characterized by singlets in the wild type. Illustrations indicate position of CEM cilium and MTs, as well as the CEP cilium. Scale bar = 250nm. (B) A series of CEM middle sections from ccpp-1 mutants. Open “C-shaped” tubules in section 2 are indicated by arrows. Approximate locations of numbered sections indicated by red dotted lines in panel C. Scale bar = 50nm. (C) Cartoon model of a single outer doublet microtubule along the ciliary length for wild type (WT), ttll-11, ccpp-1, and ccpp-1; ttll-11 double mutant genotypes. See also Table S1.
Serial section TEM analysis of ttll-11 mutant male CEM cilia revealed that MT doublets extended along the entire length of the axoneme and failed to splay into singlets (Figure 6A). We previously reported that mutation of ccpp-1 causes loss of some of the singlets in CEM cilia, and that remaining MT singlets were more distant from the ciliary membrane [23]. We observed an additional ccpp-1 mutant phenotype using high-pressure freeze tannic acid staining of serial sections: open C-shaped singlets in middle segments that may represent B-tubules that separated from partner A-tubules and neither sealed to form B-tubule singlets nor remained joined as doublets in distal segments (Figure 6B). Remaining A-tubule and B-tubule singlets with 13 and 10 protofilaments, respectively, were visible (Figure 6B). In ccpp-1; ttll-11 double mutant males, CEM ciliary MTs resembled those in ttll-11 single mutants, with A–B doublets extending along the length of the axoneme and not splaying to A-tubule and B-tubule singlets in middle sections (Figure 6 A). Cilia were visible in the cuticular pore of all genotypes, and were therefore full-length (data not shown). These results indicate that loss of the TTLL-11 glutamylase is epistatic to loss of the deglutamylase CCPP-1, and that MT glutamylation by TTLL-11 promotes formation of A- and B-tubule singlets via splaying of MT doublets in cilia (Figure 6C).
Discussion
The Tubulin Code posits that tubulin isotypes and PTMs encode information on MTs needed for specific cytoskeletal functions [12, 13]. In many organisms, ciliary MTs are decorated with PTMs, including polyglutamylation and polyglycylation [13]. C. elegans lacks MT polyglycylation and polyglycylase homologs [15, 23, 27], and therefore may use a simplified Tubulin Code. We demonstrate here that writers and erasers of the tubulin code – the TTLL-11 positive and CCPP-1 negative regulators of glutamylation – specialize the form and function of EV-releasing neuronal sensory cilia in C. elegans.
An initially surprising result of this study was that loss of the tubulin glutamylase TTLL-11, which has hypoglutamylated MTs, results in a PKD-2∷GFP ciliary localization defect similar to ccpp-1, which has hyperglutamylated MTs. In fact, attempts to “balance” glutamylation by creating ccpp-1;ttll-11 double mutants demonstrated that these genes interact for some, but not all, functions in male-specific sensory cilia (Table S1). We suggest that exquisitely balanced glutamylation is essential for specialization of EVN cilia.
The influence of the Tubulin Code on ciliary motor traffic is not well understood [12]. The use of maleimide chemistry to attach glutamate side chains to tubulins at common polyglutamylation sites was shown to increase the processivity and velocity of kinesin-2 in vitro [14]. Our data indicate that tubulin isotypes and MT glutamylation regulate ciliary motors in specific manners in vivo. α-tubulin TBA-6 regulates the velocities and cargo of the IFT kinesin-2 motors kinesin-II and OSM-3/KIF17 without affecting kinesin-3 KLP-6 velocity [24]. Conversely, we show here that glutamylation regulates the velocity of the kinesin-3 KLP-6 and the kinesin-2 OSM-3/KIF17 without impacting heterotrimeric kinesin-II.
Microtubule doublets consist of complete A-tubules with 13-protofilaments and attached incomplete 10-protofilament B-tubules [40]. B-tubules are considered to be the predominant site of glutamylation in doublet microtubules [41] and navigated by heterotrimeric kinesin-II in Chlamydomonas flagella [42]. Intriguingly, MT glutamylation impacts OSM-3 in CEM cilia, which is capable of travel on A-tubule singlets in C. elegans chemosensory amphid and phasmid cilia [37, 43]. In C. elegans phasmid cilia, localization of heterotrimeric kinesin-II and kinesin-2 OSM-3 is attributed to motor turnaround frequencies rather than MT composition or structure [43]. Therefore, the preferences of particular motors for A- or B-tubules remain an open question. Moreover, glutamylation enzymes and/or ciliary motors may follow different rules for A–B microtubule doublets found in “canonical cilia” versus 13 protofilament A-tubule and 10 protofilament B-tubule singlets observed in CEM cilia.
Might glutamylation on B-tubules affect A-tubule-mediated transport indirectly? Although this hypothesis seems unlikely, polyglutamylation can seemingly exert structural effects on different doublets. For example, loss of the glutamylase TTLL9 in mouse sperm flagella causes reduction of glutamylation on doublet 5 but selective loss of doublet 7 [3]. Perhaps even minimal A-tubule glutamylation is sufficient to regulate A-tubule-mediated transport. Alternatively, A-tubule glutamylation may be more prevalent in sensory cilia than previously appreciated from studies in motile cilia. MT glutamylation may regulate motors both directly and indirectly, for example, by altering the charge of tubulin C-terminal tails and motor affinities, and by sculpting the ultrastructure of MTs.
A striking finding in this work is that TTLL-11, CCPP-1, and MT glutamylation regulate axonemal structure at a fundamental level. In wild-type CEM cilia, MT doublets splay into A-tubule and B-tubule singlets that remain joined as doublets at proximal and distal ends [24]. In ccpp-1 mutant CEM cilia, hyperglutamylation may cause abnormally aggressive splaying as demonstrated by loss of some singlets and C-shaped MTs that may be B-tubule singlets that fail to seal [23]. In contrast, ttll-11 and ccpp-1; ttll-11 mutants contain hypoglutamylated doublet MTs that fail to splay, indicating that mutation of ttll-11 acts epistatically to ccpp-1. We propose that MT glutamylation regulates splaying of A–B doublets to 13-protofilament A-tubule and 10-protofilament B-tubule singlets in CEM cilia. This implies the existence of MT-associated proteins that promote sealing of B-tubule singlets that have transiently separated from A-tubule partners.
The mechanisms by which TTLL enzymes are transported and located to discrete subcellular compartments remain relatively unexplored. However, a recent report suggested that pathological loss of dendritic spines and synapse activity may be caused by mislocalization of TTLL6 (and possibly of TTLL1, 5, or 11), leading to abnormal glutamylation patterns and excessive spastin-mediated severing of dendritic MTs [44].
In this first characterization of TTLL-11 transport and localization to cilia, we find that TTLL-11 moves bidirectionally in CEM cilia. TTLL-11 undergoes anterograde transport at a rate that is faster than IFT-A or B components (~0.5μm/s) in CEM cilia [35], but similar to both OSM-3∷GFP and GFP∷KLP-6 in ccpp-1 mutants. Because both loss of CCPP-1 and overexpression of TTLL-11 could lead to hyperglutamylated microtubules (Table 1), we therefore propose that TTLL-11∷GFP overexpression affects the PTM readers, namely the OSM-3 and KLP-6 accessory ciliary motors. In CEM cilia, loss of kinesin-II accelerates IFT-A and IFT-B components from 0.5 – 0.6 μm/s to approximately 0.9 – 1.06 μm/s [35] and TTLL-11∷GFP from 0.87 to 1.0 μm/s (Table 1). These data suggest that IFT-A, IFT-B, and the TTLL-11 glutamylase can associate with OSM-3 and KLP-6 motors. Our observations also suggest a mechanism whereby transport of TTLL-11 (a writer of the code) by ciliary kinesins (readers of the code) may provide feedback to regulate MTs upon which the kinesins move.
TTLL and CCPP enzymes may be evolutionarily conserved regulators of ciliary specialization. For example, splaying of axonemal doublets into singlets reminiscent of MTs in C. elegans CEM cilia has also been described in mammalian sperm [24, 45]. TTLLs and tubulin glutamylation are required for normal sperm MT structure and motility in mice [46]. TTLL9 [3] and TTLL5 [46] are required for normal mammalian sperm flagella ultrastructure, which supports this hypothesis. We speculate that mutations affecting glutamylation may impact human sperm motility and fertility.
What are the MT substrates of glutamylation? The most highly enriched tubulins in the EV-releasing ciliated neurons are α-tubulin TBA-6 and β-tubulin TBB-4 [8]. TBA-6 is essential for normal morphology and MT ultrastructure of the CEM cilia [24]. In tba-6 mutants, the CEM cilium has a middle segment with nine doublet microtubules followed by abrupt termination of the B-tubule and extension of nine A-tubule singlets distally. TBA-6 might be a non-glutamylatable tubulin subunit due to its unusual C-terminal tail that lacks glutamate residues [24]. Conversely, the C-terminal tail of TBB-4 may be a substrate for glutamylation. Possibly, TBA-6 insertion into the MT lattice in CEM cilia fine-tunes glutamylation to regulate the doublet-to-singlet transition. Particular tubulins have been found to be important for protofilament number and MT structure in C. elegans non-ciliated touch receptor neurons [47, 48]. Our results, together with previous findings, support the Tubulin Code hypothesis [12], in that tubulin function can be specialized by PTMs such as glutamylation. Therefore, both PTMs and particular tubulin isotypes may sculpt axonemes for diverse forms and functions. Further research is needed to determine which tubulins are true substrates for glutamylation in vivo.
In addition to TBA-6 and KLP-6, CCPP-1 and TTLL-11 regulate EV shedding and release, with mutants accumulating excessive amounts of lumenal EVs and failing to release EVs to the environment [7, 24]. Combined, these data suggest that writers, erasers, and readers of the Tubulin Code regulate EV-based signaling. EVs are sub-micron sized membrane-enclosed packages of proteins, lipids, and nucleic acids [7] used for internal communication between distant cells [49] and external communication between conspecific animals [7]. EV shedding and release is an evolutionarily conserved function of cilia and observed in Chlamydomonas, C. elegans, and mammals [6, 7, 9–11]. Mechanisms controlling formation, shedding and release of these ciliary EVs called ectosomes are poorly understood.
Loss of either the TTLL-11 glutamylase or the CCPP-1 deglutamylase virtually abolishes release of PKD-2∷GFP-labeled EVs with concomitant accumulation of EVs shed into the cephalic lumen. The kinesin-3 klp-6 mutant displays a similar EV phenotype [7]. In ccpp-1 mutants, GFP∷KLP-6 localization is defective, suggesting that KLP-6 may not be fully functional in hyperglutamylated cilia [23]. Furthermore, GFP∷KLP-6 velocity is affected by either ccpp-1 or ttll-11 single mutants. Therefore, the effect of ccpp-1 and ttll-11 single mutants on EV shedding and release may depend on KLP-6 function. The ccpp-1;ttll-11 double mutant fails to release PKD-2∷GFP-labeled EVs to the environment, but lumenal EVs do not accumulate. Therefore, EV shedding into the lumen might also be perturbed when both positive and negative regulation of MT glutamylation are lost.
TTLL-11∷GFP is an EV cargo. Glutamylases and deglutamylases have both tubulin and non-tubulin substrates [16, 50, 51]. Hence, TTLL-11 could act locally in EVs in addition to its role modifying ciliary MTs. N-myristoylation can anchor proteins to membranes and target proteins to cilia or EVs [30, 52, 53]. In C. elegans EVNs, the EV regulator CIL-7 requires an N-terminal myristoylation sequence for its localization and function in PKD-2∷GFP-labelled EV release [30]. The TTLL-11B isoform is expressed in the EVNs and possesses a predicted myristoylation site, which may target and package this isoform into EVs.
Both positive and negative regulators of tubulin glutamylation are implicated in human ciliopathies. Examples of hyperglutamylation leading to disease include mutations in the deglutamylase AGBL5, associated with retinal degeneration in humans [54], and loss of the deglutamylase Ccp1, which causes progressive neurodegeneration and sperm immotility in mice [20]. A growing list of reports links hypoglutamylation to disease. A chromosomal abnormality leading to loss of TTLL11 may contribute to schizophrenia [19]. A cause of Joubert syndrome is proposed to be hypoglutamylation of ciliary MTs, indirectly caused by mutation of the CEP41, which is needed for ciliary trafficking of the TTLL6 glutamylase [17]. Hypoglutamylation in mice also causes defects in synaptic function [55]. Recent work also identified TTLL5 mutation as a cause of recessive retinal dystrophy in humans [18] and sperm immotility in mice [46]. Ciliary specialization and diversity, mediated at least in part by Tubulin Code-based mechanisms, may play important roles as modifiers of ciliopathic genetic diseases. Our work, elucidating functions of the writers, erasers, and readers of the Tubulin Code at the cellular and molecular level, will have important impacts on our understanding of their effects on human health.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Robert O’Hagan (ohagan@dls.rutgers.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Culture of C. elegans nematodes
Nematodes were cultured on Nematode Growth Media (NGM) agar plates containing a lawn of E. coli (strain: OP50). All animals were incubated at room temperature or 20°C. In all experiments in which males were tested, we used animals in either the him-5(e1490) or myIs1 0pkd-2(sy606);him-5(e1490) background to generate a supply of males. These backgrounds were considered wild type. Males with the him-5(e1490) mutation exhibit normal mating behaviors and are commonly used as wild-type controls for mating assays. We also used males that were heterozygous for him-5(e1490) and made no distinction from homozygous him-5(e1490) males.
METHOD DETAILS
Molecular Biology and Transgenes
pRO125 was constructed using Gateway cloning (www.thermofisher.com) using genomic sequence of the ttll-11b promoter amplified using primers RO-P218F/RO-P220R (see Key Resources Table). pRO132 was constructed by Gibson assembly [56] fusing a PCR product generated with primers RO-P307F/RO-P305R with a plasmid backbone including the pkd-2 promoter, green fluorescent protein, and the unc-54 3′UTR, which was amplified using primers RO-P308F/RO-P306R. The Pttll-11a∷gfp PCR product was amplified using primers RO-P341F/RO-P344R.
We created transgenic strains by injection of plasmids and/or PCR products dissolved in M9 (22 mM KH2PO4, 42 mM Na2HPO4, 85 mM NaCl) into the germline in young adult hermaphrodites. We created PT3161 by injecting the Pttll-11a∷gfp PCR product, comprising 1122 bp upstream of the ttll-11a start codon fused to GFP coding sequence and unc-54 3′ UTR. PT3166 was created by germline injection of pRO125, which encodes a ttll-11b promoter of 1027 bp upstream of the start codon fused to GFP coding sequence and unc-54 3′ non-coding sequence. We created PT3159 by germline injection of pRO132, which encodes a pkd-2 promoter plus genomic ttll-11b sequence fused in-frame with gfp and the unc-54 3′ UTR. We used three transformation markers, individually or in combination, for each transgenic strain: 1) pBX, a plasmid containing the rescuing pha-1 transformation marker; 2) Punc-122∷gfp, which produces green fluorescence in coelomocytes visible in both males and hermaphrodites; and 3) Punc-122∷rfp, which produces red fluorescence in coelomocytes visible in both males and hermaphrodites. All other transgenes (noted in Key Resources Table) were introduced into ccpp-1 and ttll-11 mutant backgrounds by crossing.
Epifluorescence Microscopy
Nematodes were anaesthetized with 10 mM levamisole and mounted on agarose pads for imaging at room temperature. Epifluorescence images were acquired with Metamorph software (www.moleculardevices.com) using either a Zeiss Axio Imager.D1M (Zeiss, Oberkochen, Germany) or a Zeiss Axioplan2 microscope with 10×, 63× (NA 1.4), and 100× (NA 1.4) oil-immersion objectives, equipped with either a Retiga-SRV Fast 1394 digital camera (Q-Imaging, Surrey, BC, Canada), a Photometrics Cascade 512B CCD camera, or a Hamamatsu C11440-42U ORCA-Flash4.0 LT Digital CMOS camera (www.hamamatsu.com). We imported images into FIJI/ImageJ 2.0 (imagej.net/Fiji/) using the Bioformats 5.5.3 plugin (www-legacy.openmicroscopy.org/) to quantify fluorescence in some images, and to create optical Z-stack projections, add scale bars, rotate, and adjust contrast. Images were then exported as jpegs to Adobe Photoshop CS4 for cropping and saving as layered PSD files for assembly into figures in Adobe Illustrator CS5.
For analyzing expression patterns, subcellular localization of fluorescently tagged proteins, and quantification of GFP-labeled EV release, we isolated L4 (fourth larval stage) males from hermaphrodites 20–24 hours before observation by transferring them to plates seeded with OP50 bacteria. At the time of observation, all animals were young adult males. Still images were captured as Z-stacks. No blind analysis was performed. No sample size estimations were performed. Sample sizes are indicated in figures or legends where appropriate.
For PKD-2∷GFP localization, we used PT443, PT2168, PT2169, PT2170, PT2171, PT2172, PT2433, PT2488, PT2497, PT2498, PT2988, PT2499, PT2500, PT2666, PT2667, PT2668, PT2670, PT2920, PT2921, PT2958, and PT2988. The PKD-2∷GFP ciliary localization defective (Cil) phenotype was characterized by visible PKD-2∷GFP in dendrites and ciliary bases, especially in rays in the tail. Images of PKD-2∷GFP localization were captured as Z-stacks using the Zeiss Axio Imager.D1M with 100X objective and Retiga SRV camera with 200ms exposure. In wild-type CEMs, ciliary PKD-2∷GFP is very dim, but in the mutants, the cilia and ciliary bases are more clearly seen. We increased the brightness and contrast equally for all PKD-2∷GFP images, but to avoid oversaturation in mutant images, we increased contrast only to the point that wild-type cilia are dimly visible. We used the Bioformats plugin to import TIFF stacks into FIJI/ImageJ to quantify this phenotype from Z-projections of epifluorescence images by tracing along visible patches of PKD-2∷GFP from the tips of neuronal cilia into dendrites, and comparing the average length of the PKD-2∷GFP accumulation patches across genotypes.
For GFP∷KLP-6 localization, we used strains: PT2102, PT2149, PT2713, PT2715. Images were captured as Z-stacks using the 100X objective and 200ms exposure on the Zeiss Axio Imager.D1M with 100X objective and Retiga SRV camera with 200ms exposure. We used FIJI to quantify GFP∷KLP-6 localization defects from Z-projections of TIFF stacks of epifluorescence images by capturing the maximum pixel value from regions of interest (ROIs) containing the distalmost 10μm of dendrites in head (CEM and IL2), and from ROIs containing CEM and IL2 cilia. The ratio of ciliary maximum/dendrite maximum pixel value represents fold enrichment in cilia compared to dendrites.
For counting numbers of PKD-2∷GFP-labeled environmentally released EVs in synchronized adult males, we used strains: PT443, PT2168, PT2497, PT2498, PT2988. We acquired Z-stacks using the 100X objective on the Zeiss Axio Imager.D1M with the Retiga digital camera. We also counted EVs trapped in the molting cuticle of late L4 males as a means to unambiguously identify EVs that were released from individual animals. We picked late L4 males immediately before mounting for imaging, and imaged only those with molting cuticles visible on their tails. Although cuticles were in various stages of molting, as males gradually emerged from the old cuticle, we measured the area enclosed by the old cuticle of the Z-projected stacks to ensure that we compared EV counts of similarly aged animals. The enclosed areas (in square pixels) were indistinguishable by one-way ANOVA performed in IGOR 6 software (Wavemetrics, Inc.) across genotypes. For both young adult males and molting late L4 males, EVs were counted in FIJI/ImageJ using the ROI manager tool.
To determine the expression pattern of TTLL-11A and TTLL-11B, we crossed adult PT1722 males expressing Pklp-6∷tdTomato to mark EVNs (extracellular vesicle-releasing neurons) with PT3161 (Pttll-11b∷gfp) or PT3166 (Pttll-11a∷gfp) hermaphrodites and picked F1 males. We observed overlap of GFP and tdTomato cellular expression patterns (to identify Pttll-11b∷gfp expression in EVNs). Where patterns did not overlap, we used tdTomato-illuminated EVNs as landmarks to identify cells expressing Pttll-11a∷gfp by relative position. Images were acquired as Z-stacks using the 63X objective on the Axio Imager.D1M with the Orca-Flash 4.0 camera.
In vivo Motor Velocity Analysis
For all motor velocity experiments, except as noted for GFP∷KLP-6, we isolated L4 (fourth larval stage) males from hermaphrodites 20–24 hours before observation by transferring them to fresh plates seeded with OP50 bacteria. Animals were mounted on slides for imaging as young adult males. Velocities of motile particles were scored by a researcher blinded as to genotype. Blinding was done by another researcher, who renamed data files with numbers. For all motor velocity experiments, number of animals and motile particles analyzed are presented with the data. For all motor velocity experiments, CEM cilia that did not lie entirely within a focal plane were excluded from analysis. No sample size estimations were performed. Sample sizes are indicated where appropriate.
Motor velocity experiments for OSM-3∷GFP were conducted using time-lapse microscopy for 100–200 frames, each with 100ms exposure, Gain 1, on the Axio Imager.D1M/ Retiga-SRV camera, using the 100X objective. We used Metamorph software to convert the streaming video to kymographs and calculate the velocity of moving particles. For OSM-3∷GFP velocity experiments, strains used were PT2065, PT2098, PT2669, PT2678.
GFP∷KLP-6 motor velocity experiments were conducted using time-lapse microscopy for 100–200 frames, each with 100ms exposure, Gain 1, on the Axio Imager.D1M/ Retiga-SRV camera, using the 100X objective. Strains used: PT2102, PT2149, PT2713, PT2715 for GFP∷KLP-6 motility. Because of intense ciliary accumulation of GFP∷KLP-6 in ccpp-1 mutant CEM neurons, it was not possible to analyze how MT glutamylation affected kinesin-3 velocity using males isolated 20 – 24 hours prior to the experiment. Therefore, we tested motility of GFP∷KLP-6 in CEM cilia at an earlier time point by isolating L4 males and analyzing only those males that had molted into mature adults four hours later. GFP∷KLP-6 accumulation was less severe in these very young adult males, making it possible to visualize moving GFP∷KLP-6 particles in cilia. We used Metamorph software to convert the streaming video to kymographs and calculate the velocity of moving particles.
KAP-1∷GFP velocity experiments were conducted using time-lapse microscopy to acquire 200 frames with 200 ms exposure, Gain 20, on the Axio Imager.D1M/ Retiga-SRV camera, using the 100X objective. Strains used: PT2108, PT3179, PT3180, PT3181. For KAP-1∷GFP, we used FIJI/ImageJ with the Kymograph Clear macro toolset [57] to generate kymographs from streaming videos of CEM cilia and to manually trace lines on moving particles. Kymographs and traced lines were analyzed using Kymograph direct software [57] to calculate the velocity of moving particles.
To analyze TTLL-11∷GFP localization and motility, we used the following strains: PT3159, PT3256, PT3257, PT3258. Images were captured using the Axio Imager.D1M/Orca camera using the 100X objective. For localization of TTLL-11∷GFP in neurons and in EVs, Z-stacks were processed using FIJI/ImageJ and Photoshop. To analyze ciliary and dendritic motility, we acquired time-lapse 100 – 200 frame video streams (200ms exposure, Axio Imager.D1M/Orca-Flash 4.0 camera) using the 100X objective. Dendritic kymographs were created and analyzed using Metamorph software. For our blinded analysis of ciliary movement of TTLL-11∷GFP, we used FIJI/ImageJ with the Kymograph Clear macro toolset [57] to generate kymographs from streaming videos of CEM cilia and to manually trace lines on moving particles. Kymographs and traced lines were analyzed using Kymograph direct software [57] to calculate the velocity of moving particles. Scoring of motile TTLL-11∷GFP puncta was challenging because of a low signal-to-noise ration. Therefore, in our blinded analysis of TTLL-11∷GFP motility in wild-type, osm-3, klp-6, and kap-1 backgrounds, we also included a strain that expresses soluble GFP in CEM and other neurons. Our analysis revealed that the frequency at which we detected moving TTLL-11∷GFP particles was on average 0.15–0.2 particles per second of video in wild-type and mutant genotypes, but only approximately 0.002 per second for soluble GFP. Therefore, although scoring motile TTLL-11∷GFP puncta was challenging, we detected their movement 100X more frequently than spurious movement of soluble GFP.
Immunofluorescence Imaging
For immunofluorescence imaging, we synchronized animals (Strains PT443, PT2168, PT2497, PT2498, PT2988) by bleaching and fixed as 1-day-old adults. Fixation was accomplished by washing animals from 3 NGM plates using M9 buffer, then washing animals in a 15 ml conical tube 3 more times with M9 over one hour. Worms were chilled on ice before washing in ice-cold Ruvkun buffer (80 mM KCl, 20 mM NaCl, 10 mM EGTA, 5 mM spermidine-HCl, 15 mM Pipes, pH 7.4 and 25% methanol) plus 2% formaldehyde in 1.6ml centrifuge tubes. The tubes were immersed in liquid nitrogen, and melted under tap water to crack the worms’ cuticles. Worms were then washed with Tris-Triton buffer (100 mM Tris-HCl, pH 7.4, 1% Triton X-100 and 1 mM EDTA), suspended in Tris-Triton buffer+1% β-mercaptoethanol, and incubated overnight at 37°C. The next day, worms were washed with 1X BO3 buffer (50 mM H3BO3, 25 mM NaOH) + 0.01% Triton, and suspended in 1X BO3 + 0.01% Triton buffer + 10 mM DTT for 15 minutes with gentle agitation at room temperature. Worms were then washed with 1X BO3 buffer (50 mM H3BO3, 25 mM NaOH) + 0.01% Triton, and suspended in 1X BO3 + 0.01% Triton buffer + 0.3% H2O2 for 15 minutes with gentle agitation at room temperature. After washing once with 1X BO3 + 0.01% Triton buffer, worms were washed for 15 minutes in Antibody buffer B (1X PBS, 0.1% BSA, 0.5% Triton X-100, 0.05% sodium azide, 1mM EDTA) with gentle agitation at room temperature. Fixed worms were stored in Antibody buffer A (1X PBS, 1% BSA, 0.5% Triton X-100, 0.05% sodium azide, 1mM EDTA) at 4°C for up to one month before antibody staining.
Animals were stained overnight at room temperature with a 1:600 dilution (in Antibody Buffer A) of GT335, a monoclonal antibody which binds the branch point of both monoglutamylated and polyglutamylated substrates [34] or with a polyclonal polyglutamylation (polyE) antibody IN105 (both obtained from www.adipogen.com), which recognizes chains of 3 or more glutamates [33]. Stained worms were washed with several changes of Antibody B Buffer with gentle agitation at room temperature over several hours. After rinsing with Antibody Buffer A, either Alexa-fluor 568-conjugated donkey anti-mouse (for GT335) or Alexa-Fluor 568-conjugated anti-rabbit (for polyE) secondary antibodies (Invitrogen) were added at a dilution of 1:2000 and incubated for 2 hours at room temperature with gentle agitation. Worms were then washed with several changes of Antibody Buffer B over several hours before mounting on 2% agarose pads for imaging.
Male Mating Behavior
We conducted mating assays using strains CB1490, CB169, PT2281, PT3032, and PT3149. L4 males were picked to a fresh plate ~24 hours before behavior experiments to be assayed as young adults. CB169 hermaphrodites were also picked as L4 larvae ~24 hours before experiments. Male mating assays were conducted on a fresh NGM agar plate with a spot of E. coli (OP50) containing 25 young adult CB169 uncoordinated mutant hermaphrodites. One, two, or three males were placed in the center of the OP50 and observed for 4 minutes. Mating behavior requires a series of substeps [26] including “response,” in which the male senses the presence of a hermaphrodite mate and moves backwards, pressing his tail against her body, and “location of vulva,” in which males stop at the hermaphrodite vulva and prod with their spicules. A “response” was scored only if a male began scanning a hermaphrodite with his tail rays and maintained contact for at least 10 seconds. “Location of vulva” was scored if a male stopped at the hermaphrodite vulva and began prodding with his spicules, but insertion of spicules was not a requirement. For both response and vulva location assays, males were scored in random order with respect to genotype. Genotypes of males assayed were cycled over time on assay days so that individual genotypes were not scored in a single block of time. No blind analysis was performed. No sample size estimations were performed. No blind analysis was performed. Number of trials and sample size are indicated in Figure 1 legend.
Serial Section Transmission Electron Microscopy
We analyzed ciliary ultrastructure using the following strains: PT443, PT2168, PT2497, PT2498. Young adult animals were subjected to high-pressure freeze fixation and freeze substituted, in 0.5% glutaraldehyde, 0.25% tannic acid and 2% water in acetone at −90°C for 104 hours. Samples were then gradually warmed to −25°C over 13 hours, and held at that temperature for 16 hours. Samples were then warmed gradually to 0°C, over 5 hours and held for 2 hours. After this final warming, specimens were rinsed in cold acetone. Samples were re-stained in filtered 1% uranyl acetate in acetone in the cold, then rinsed 3X in pure acetone, stained again in filtered Reynold’s lead mixture in acetone, rinsed in acetone, and transferred to microporous type C holders for processing into plastic resin [58]. Samples were infiltrated into Embed812 plastic resin and embedded in an 8-well chamber slide for curing at 60°C for 24 hrs. Serial sections (70 nm thickness) of fixed animals were collected on copper slot grids and stained with 4% uranyl acetate in 70% methanol, followed by washing and incubating with aqueous lead citrate. Images were captured on a Philips CM10 transmission electron microscope at 80kV with a Morada 11 megapixel TEM CCD camera driven by iTEM software (Olympus Soft Imaging Solutions).
The trakEM2 suite in FIJI [59] was used to quantify EVs shed into a lumen formed by glial cells. Serial TEM sections images were stacked, aligned, and annotated before spheres were manually fitted to each EV in cephalic sensilla to count EV abundance through all ciliary sections. No blind analysis was performed. No sample size estimations were performed. Serial TEM sections were included only when an entire sensillum could be reconstructed from a single animal per genotype. Number of cilia examined is indicated in Figure 4 legend.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data values are expressed as mean ± standard error unless indicated. To determine the statistical significance of differences in experimental results, we conducted statistical tests (as indicated) in IGOR 6 (Wavemetrics, Inc.) or Prism (Graphpad Software). Statistical tests used and P values are indicated for each comparison. P ≤ 0.05 was considered to indicate significant difference.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Polyglutamylation Modification, mAb (GT335) | www.adipogen.com | AG-20B-0020-C100 |
| anti-Polyglutamate chain (polyE), pAb (IN105) | www.adipogen.com | AG-25B-0030 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | www.thermofisher.com | Cat # A10037 |
| Donkey-anti-Rabbit-IgG-H-L-Highly-Cross-Adsorbed-Secondary Antibody, Alexa Fluor 568 | www.thermofisher.com | Cat # A10042 |
| Bacterial and Virus Strains | ||
| E. coli, OP50 strain | cgc.umn.edu | OP50 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Levamisole | www.acros.com | Cat # 187870100 |
| Bovine Serum Albumin (BSA) Fraction V; Protease Free | www.sigmaaldrich.com | Cat # 3117332001 |
| PIPES | www.sigmaaldrich.com | Cat # P6757-100G |
| Triton X-100 | www.sigmaaldrich.com | Cat # X100-500ML |
| Formaldehyde Solution 37% | www.sigmaaldrich.com | Cat # F1635-500ML |
| Embed812 plastic resin | www.emsdiasum.com | Cat # 14900 |
| Tannic Acid | www.sigmaaldrich.com | Cat # 16201-500G |
| Glutaraldehyde Solution 25% | www.emsdiasum.com | Cat # 16200 |
| Uranyl Acetate | www.emsdiasum.com | Cat # 22400 |
| Lead Citrate | www.emsdiasum.com | Cat # 17800 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| C. elegans Strain CB169: unc-31(e169) IV | CGC | RRID:WB-STRAIN:CB169 |
| C. elegans Strain CB1490: him-5(e1490) V | CGC | RRID:WB-STRAIN:CB1490 |
| C. elegans Strain PT443: myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;him-5(e1490) V | [34] | RRID:WB-STRAIN:PT443 |
| C. elegans Strain PT1722: pha-1(e2123) III; him-5(e1490) V; myEx632[Pklp-6∷tdTomato + pha-1(+)] | This paper | RRID:WB-STRAIN:PT1722 |
| C. elegans Strain PT2065: ccpp-1(ok1821) I;pha-1(e2123) III;him-5(e1490) V;myEx685[Pklp-6∷osm-3∷gfp + pha-1(+)] | This paper | RRID:WB-STRAIN:PT2065 |
| C. elegans Strain PT2098: pha-1(e2123) III;him-5(e1490) V;myEx685[Pklp-6∷osm-3∷gfp + pha-1(+)] | [48] | RRID:WB-STRAIN:PT2098 |
| C. elegans Strain PT2102: pha-1(e2123) III;him-5(e1490) V;myEx686[gfp∷klp-6+ pha-1(+)] | [48] | RRID:WB-STRAIN:PT2102 |
| C. elegans Strain PT2108: pha-1(e2123) III;him-5(e1490) V;myEx687[Ppkd-2∷kap-1∷gfp + pha-1(+)] | [48] | RRID:WB-STRAIN:PT2108 |
| C. elegans Strain PT2149: ccpp-1(ok1821) I;pha-1(e2123) III;him-5(e1490) V;myEx686[gfp∷klp-6+ pha-1(+)] | This paper | RRID:WB-STRAIN:PT2149 |
| C. elegans Strain PT2168: ccpp-1(ok1821) I; myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2168 |
| C. elegans Strain PT2169: myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;him-5(e1490) ttll-9(tm3889) V | [34] | RRID:WB-STRAIN:PT2169 |
| C. elegans Strain PT2170: ccpp-1(ok1821) I;myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;him-5(e1490) ttll-9(tm3889) V | [34] | RRID:WB-STRAIN:PT2170 |
| C. elegans Strain PT2171: ttll-4(tm3310) III;myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;him-5(e1490) V | [34] | RRID:WB-STRAIN:PT2171 |
| C. elegans Strain PT2172: ccpp-1(ok1821) I;ttll-4(tm3310) III;myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;him-5(e1490) V | [34] | RRID:WB-STRAIN:PT2172 |
| C. elegans Strain PT2281: ccpp-1(ok1821) I;him-5(e1490) V | [34] | RRID:WB-STRAIN:PT2281 |
| C. elegans Strain PT2433: myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV; ttll-5(tm3360) him-5(e1490) V | [34] | RRID:WB-STRAIN:PT2433 |
| C. elegans Strain PT2488: ccpp-1(ok1821) I; myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV; ttll-5(tm3360) him-5(e1490) V | [34] | RRID:WB-STRAIN:PT2488 |
| C. elegans Strain PT2497: ccpp-1(ok1821) I; myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11(tm4059) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2497 |
| C. elegans Strain PT2498: myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11(tm4059) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2498 |
| C. elegans Strain PT2499: ccpp-1(ok1821) I; myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;ttll-15(tm3871) him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2499 |
| C. elegans Strain PT2500: myIs1[pkd-2∷gfp + Punc-122∷gfp] pkd-2(sy606) IV;ttll-15(tm3871) him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2500 |
| C. elegans Strain PT2613: pha-1(e2123) III;ttll-11(tm4059) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2613 |
| C. elegans Strain PT2666: myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11(tm4059) IV; ttll-5(tm3360) him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2666 |
| C. elegans Strain PT2667: myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11(tm4059) IV;him-5(e1490) ttll-9(tm3889) V | This paper | RRID:WB-STRAIN:PT2667 |
| C. elegans Strain PT2668: myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11(tm4059) IV;ttll-15(tm3871) him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2668 |
| C. elegans Strain PT2669: ccpp-1(ok1821) I;pha-1(e2123) III;ttll-11(tm4059) IV;him-5(e1490) V;myEx685[Pklp-6∷osm-3∷gfp + pha-1(+)] | This paper | RRID:WB-STRAIN:PT2669 |
| C. elegans Strain PT2670: ttll-4 (tm3310) III; myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11 (tm4059) IV; him-5 (e1490) V | This paper | RRID:WB-STRAIN:PT2670 |
| C. elegans Strain PT2678: pha-1(e2123) III;ttll-11(tm4059) IV;him-5(e1490) V;myEx685[Pklp-6∷osm-3∷gfp + pha-1(+)] | This paper | RRID:WB-STRAIN:PT2678 |
| C. elegans Strain PT2713: pha-1(e2123) III;ttll-11(tm4059) IV;him-5(e1490) V;myEx686[gfp∷klp-6+ pha-1(+)] | This paper | RRID:WB-STRAIN:PT2713 |
| C. elegans Strain PT2715: ccpp-1(ok1821) I;pha-1(e2123) III;ttll-11(tm4059) IV;him-5(e1490) V;myEx686[gfp∷klp-6+ pha-1(+)] | This paper | RRID:WB-STRAIN:PT2715 |
| C. elegans Strain PT2920: ttll-4(tm3310) III; myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11(tm4059) IV; ttll-5(tm3360) him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2920 |
| C. elegans Strain PT2921: ccpp-1(ok1821) I; ttll-4(tm3310) III; myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11(tm4059) IV; ttll-5(tm3360) him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2921 |
| C. elegans Strain PT2958: ccpp-1(ok1821) I;myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11b(gk482) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2958 |
| C. elegans Strain PT2988: myIs1[pkd-2∷gfp + Punc-122∷gfp] ttll-11b(gk482) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT2988 |
| C. elegans Strain PT3032: ttll-11(tm4059) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT3032 |
| C. elegans Strain PT3149: ccpp-1(ok1821) I;ttll-11(tm4059) IV;him-5(e1490) V | This paper | RRID:WB-STRAIN:PT3149 |
| C. elegans Strain PT3159: pha-1(e2123) III;him-5(e1490) V;myEx899[Ppkd-2∷ttll-11b∷gfp + pha-1(+) + Punc-122∷rfp] | This paper | RRID:WB-STRAIN:PT3159 |
| C. elegans Strain PT3161: pha-1(e2123) III;him-5(e1490) V;myEx900[Pttll-11a∷gfp pcr product + pha-1(+) + Punc-122∷rfp] | This paper | RRID:WB-STRAIN:PT3161 |
| C. elegans Strain PT3166: pha-1(e2123) III;him-5(e1490) V;myEx902[Pttll-11b∷gfp + pha-1(+)] | This paper | RRID:WB-STRAIN:PT3166 |
| C. elegans Strain PT3179: ccpp-1(ok1821) I;pha-1(e2123 ) III;him-5(e1490) V;myEx687[Ppkd-2∷kap-1∷gfp + pha-1(+)] | This paper | RRID:WB-STRAIN:PT3179 |
| C. elegans Strain PT3180: pha-1(e2123) III;ttll-11(tm4059) IV;him-5(e1490) V;myEx687[Ppkd-2∷kap-1∷gfp + pha-1(+)] | This paper | RRID:WB-STRAIN:PT3180 |
| C. elegans Strain PT3181: ccpp-1(ok1821) I pha-1(e2123) III;ttll-11(tm4059) IV;him-5(e1490) V;myEx687[Ppkd-2∷kap-1∷gfp + pha-1(+)] | This paper | RRID:WB-STRAIN:PT3181 |
| C. elegans Strain PT3256: pha-1(e2123) III;osm-3(p802) IV; him-5(e1490) V; myEx899[Ppkd-2∷ttll-11b∷gfp + pha-1(+) + Punc-122∷rfp] | This paper | RRID:WB-STRAIN:PT3256 |
| C. elegans Strain PT3257: pha-1(e2123) III; klp-11(tm324) IV; him-5(e1490) V; myEx899[Ppkd-2∷ttll-11b∷gfp + pha-1(+) + Punc-122∷rfp] | This paper | RRID:WB-STRAIN:PT3257 |
| C. elegans Strain PT3258: pha-1(e2123) III; klp-6(my8) IV; him-5(e1490) V; myEx899[Ppkd-2∷ttll-11b∷gfp + pha-1(+) + Punc-122∷rfp] | This paper | RRID:WB-STRAIN:PT3258 |
| Oligonucleotides | ||
| GGGGACAACTTTGTACAAAAAAGTTG cgtgtgctgtgaaggagaag | This paper | RO-P218F |
| GGGGACAACTTTGTACAAGAAAGTTG taatcatattatcaccggttttaaatgaaatatc | This paper | RO-P220F |
| ggtcctcctgaaaatgttctatgttatg TGCAAGTCGCTCGTTGATTTTTG | This paper | RO-P305R |
| TTCTGTTGATATCTTGCAGCCCAT ggatatgttgtgttttacagtattatgtagtc | This paper | RO-P306R |
| gactacataatactgtaaaacacaacatatcc ATGGGCTGCAAGATATCAACAGAA | This paper | RO-P307F |
| CAAAAATCAACGAGCGACTTGCA cataacatagaacattttcaggaggacc | This paper | RO-P308F |
| cgaaatcacttggcattaacaca | This paper | RO-P341F |
| ACGGCCGACTAGTAGGAAAC | This paper | RO-P344R |
| Recombinant DNA | ||
| plasmid [Pttll-11∷gfp] | This paper | pRO125 |
| plasmid [Ppkd-2∷ttll-11∷gfp] | This paper | pRO132 |
| pcr product [Pttll-11a∷gfp] | This paper | N/A |
| Software and Algorithms | ||
| Metamorph | www.moleculardevices.com | N/A |
| FIJI/ImageJ 2.0 | www.imagej.net/Fiji/ | N/A |
| Kymograph Clear | www.nat.vu.nl/~erwinp/downloads.html | N/A |
| Kymograph Direct | www.nat.vu.nl/~erwinp/downloads.html | N/A |
| Prism 6 | www.graphpad.com | N/A |
| IGOR 6 | www.wavemetrics.com | N/A |
| iTEM software | OLYMPUS SOFT IMAGING SOLUTIONS GmbH Johann-Krane-Weg 39 48149 Münster, Germany |
N/A |
| Other | ||
| microporous type C holders | www.emsdiasum.com | Cat # 70187-20 |
| copper slot grids | www.emsdiasum.com | Cat # G2010-Cu |
Supplementary Material
Highlights.
TTLL-11 and CCPP-1 fine-tune ciliary microtubule glutamylation
Velocity of OSM-3/KIF17 and KLP-6/KIF28 motors sensitive to glutamylation defects
TTLL-11 and CCPP-1 required for ciliary extracellular vesicle release
TTLL-11 and CCPP-1 and microtubule glutamylation specialize ciliary ultrastructure
Acknowledgments
We thank members of the Barr lab, the Rutgers C. elegans community, Joel Rosenbaum, and three anonymous reviewers for helpful suggestions; Gloria Androwski for technical assistance; Leslie Gunther and Geoff Perumal for help in HPF-FS performed at Einstein; and WormBase (U41 HG002223) and WormAtlas (R24 OD010943) for online resources. This work was funded by NJCSCR postdoctoral fellowship 10-2951-SCR-E-0 and NJCSCR Grant CSCR15IRG014 to R.O.; NIH OD 10943 to D.H.H.; NIH DK059418 and DK074746 to M.M.B., and Waksman Institute Charles and Johanna Busch Fellowship to M.S. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), or by the Mitani Lab through the National Bio-Resource Project of the MEXT, Japan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
RO designed and performed experiments, analyzed data, and wrote the paper. MS designed and performed ultrastructure experiments and analyzed data. KCQN performed ultrastructure experiments. DHH designed ultrastructure experiments and wrote the paper. WZ performed some kymographic experiments. SB created strains and genetic constructs, and scored EVs from TEM sections. YHR analyzed antibody staining and fluorescence intensity data. MMB designed experiments and wrote the paper.
References
- 1.Fisch C, Dupuis-Williams P. Ultrastructure of cilia and flagella - back to the future! Biol Cell. 2011;103:249–270. doi: 10.1042/BC20100139. [DOI] [PubMed] [Google Scholar]
- 2.Falk N, Losl M, Schroder N, Giessl A. Specialized Cilia in Mammalian Sensory Systems. Cells. 2015;4:500–519. doi: 10.3390/cells4030500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konno A, Ikegami K, Konishi Y, Yang HJ, Abe M, Yamazaki M, Sakimura K, Yao I, Shiba K, Inaba K, et al. Ttll9−/− mice sperm flagella show shortening of doublet 7, reduction of doublet 5 polyglutamylation and a stall in beating. J Cell Sci. 2016;129:2757–2766. doi: 10.1242/jcs.185983. [DOI] [PubMed] [Google Scholar]
- 4.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 5.Tsuji T, Matsuo K, Nakahari T, Marunaka Y, Yokoyama T. Structural basis of the Inv compartment and ciliary abnormalities in Inv/nphp2 mutant mice. Cytoskeleton (Hoboken) 2016;73:45–56. doi: 10.1002/cm.21264. [DOI] [PubMed] [Google Scholar]
- 6.Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KC, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Kaletsky R, Silva M, Williams A, Haas LA, Androwski RJ, Landis JN, Patrick C, Rashid A, Santiago-Martinez D, et al. Cell-Specific Transcriptional Profiling of Ciliated Sensory Neurons Reveals Regulators of Behavior and Extracellular Vesicle Biogenesis. Curr Biol. 2015;25:3232–3238. doi: 10.1016/j.cub.2015.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long H, Zhang F, Xu N, Liu G, Diener DR, Rosenbaum JL, Huang K. Comparative Analysis of Ciliary Membranes and Ectosomes. Curr Biol. 2016;26:3327–3335. doi: 10.1016/j.cub.2016.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife. 2015;4:e05242. doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salinas RY, Pearring JN, Ding JD, Spencer WJ, Hao Y, Arshavsky VY. Photoreceptor discs form through peripherin-dependent suppression of ciliary ectosome release. J Cell Biol. 2017;216:1489–1499. doi: 10.1083/jcb.201608081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 13.Janke C. The tubulin code: molecular components, readout mechanisms, and functions. The J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu I, Garnham CP, Roll-Mecak A. Writing and Reading the Tubulin Code. J Biol Chem. 2015;290:17163–17172. doi: 10.1074/jbc.R115.637447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, Peris L, Gold ND, Lacroix B, Grau MB, et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143:564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, Olvera J, Brancati F, Iannicelli M, Ikegami K, et al. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet. 2012;44:193–199. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sergouniotis PI, Chakarova C, Murphy C, Becker M, Lenassi E, Arno G, Lek M, MacArthur DG, Consortium, U.C.-E. Bhattacharya SS, et al. Biallelic variants in TTLL5, encoding a tubulin glutamylase, cause retinal dystrophy. Am J Hum Genet. 2014;94:760–769. doi: 10.1016/j.ajhg.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fullston T, Gabb B, Callen D, Ullmann R, Woollatt E, Bain S, Ropers HH, Cooper M, Chandler D, Carter K, et al. Inherited balanced translocation t(9;17)(q33.2;q25.3) concomitant with a 16p13.1 duplication in a patient with schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2011;156:204–214. doi: 10.1002/ajmg.b.31157. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez de la Vega Otazo M, Lorenzo J, Tort O, Aviles FX, Bautista JM. Functional segregation and emerging role of cilia-related cytosolic carboxypeptidases (CCPs) FASEB J. 2013;27:424–431. doi: 10.1096/fj.12-209080. [DOI] [PubMed] [Google Scholar]
- 21.Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife. 2014;3:e01948. doi: 10.7554/eLife.01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inglis PN, Ou G, Leroux MR, Scholey JM. The sensory cilia of Caenorhabditis elegans. WormBook. 2007:1–22. doi: 10.1895/wormbook.1.126.2. www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 23.O’Hagan R, Piasecki BP, Silva M, Phirke P, Nguyen KC, Hall DH, Swoboda P, Barr MM. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr Biol. 2011;21:1685–1694. doi: 10.1016/j.cub.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva M, Morsci NS, Nguyen KC, Rizvi A, Rongo C, Hall DH, Barr MM. Cell-Specific α-Tubulin Isotype Regulates Ciliary Microtubule Ultrastructure, Intraflagellar Transport, and Extracellular Vesicle Biology. Current Biology. 2017;27:968–980. doi: 10.1016/j.cub.2017.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura Y, Kurabe N, Ikegami K, Tsutsumi K, Konishi Y, Kaplan OI, Kunitomo H, Iino Y, Blacque OE, Setou M. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs) J Biol Chem. 2010;285:22936–22941. doi: 10.1074/jbc.C110.128280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 27.www.wormbase.org
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Eisenhaber F, Eisenhaber B, Kubina W, Maurer-Stroh S, Neuberger G, Schneider G, Wildpaner M. Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Pi, NMT and PTS1. Nucleic Acids Res. 2003;31:3631–3634. doi: 10.1093/nar/gkg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maguire JE, Silva M, Nguyen KC, Hellen E, Kern AD, Hall DH, Barr MM. Myristoylated CIL-7 regulates ciliary extracellular vesicle biogenesis. Mol Biol Cell. 2015;26:2823–2832. doi: 10.1091/mbc.E15-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 33.van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Molecular cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Wolff A, de Nechaud B, Chillet D, Mazarguil H, Desbruyeres E, Audebert S, Edde B, Gros F, Denoulet P. Distribution of glutamylated α and β-tubulin in mouse tissues using a specific monoclonal antibody, GT335. European journal of cell biology. 1992;59:425–432. [PubMed] [Google Scholar]
- 35.Morsci NS, Barr MM. Kinesin-3 KLP-6 regulates intraflagellar transport in male-specific cilia of Caenorhabditis elegans. Curr Biol. 2011;21:1239–1244. doi: 10.1016/j.cub.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schou KB, Mogensen JB, Morthorst SK, Nielsen BS, Aleliunaite A, Serra-Marques A, Furstenberg N, Saunier S, Bizet AA, Veland IR, et al. KIF13B establishes a CAV1-enriched microdomain at the ciliary transition zone to promote Sonic hedgehog signalling. Nat Commun. 2017;8:14177. doi: 10.1038/ncomms14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nature cell biology. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 38.Bae YK, Qin H, Knobel KM, Hu J, Rosenbaum JL, Barr MM. General and cell-type specific mechanisms target TRPP2/PKD-2 to cilia. Development. 2006;133:3859–3870. doi: 10.1242/dev.02555. [DOI] [PubMed] [Google Scholar]
- 39.Dwyer ND, Adler CE, Crump JG, L’Etoile ND, Bargmann CI. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron. 2001;31:277–287. doi: 10.1016/s0896-6273(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 40.Linck R, Fu X, Lin J, Ouch C, Schefter A, Steffen W, Warren P, Nicastro D. Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2+-binding proteins, and stable protofilaments. J Biol Chem. 2014;289:17427–17444. doi: 10.1074/jbc.M114.568949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lechtreck KF, Geimer S. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil Cytoskeleton. 2000;47:219–235. doi: 10.1002/1097-0169(200011)47:3<219::AID-CM5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 42.Stepanek L, Pigino G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 2016;352:721–724. doi: 10.1126/science.aaf4594. [DOI] [PubMed] [Google Scholar]
- 43.Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJ. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol. 2015;17:1536–1545. doi: 10.1038/ncb3263. [DOI] [PubMed] [Google Scholar]
- 44.Zempel H, Mandelkow EM. Tau missorting and spastin-induced microtubule disruption in neurodegeneration: Alzheimer Disease and Hereditary Spastic Paraplegia. Mol Neurodegener. 2015;10:68. doi: 10.1186/s13024-015-0064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afzelius BA, Dallai R, Lanzavecchia S, Bellon PL. Flagellar structure in normal human spermatozoa and in spermatozoa that lack dynein arms. Tissue Cell. 1995;27:241–247. doi: 10.1016/s0040-8166(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 46.Lee GS, He Y, Dougherty EJ, Jimenez-Movilla M, Avella M, Grullon S, Sharlin DS, Guo C, Blackford JA, Jr, Awasthi S, et al. Disruption of Ttll5/stamp gene (tubulin tyrosine ligase-like protein 5/SRC-1 and TIF2-associated modulatory protein gene) in male mice causes sperm malformation and infertility. J Biol Chem. 2013;288:15167–15180. doi: 10.1074/jbc.M113.453936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng C, Diaz-Cuadros M, Nguyen KCQ, Hall DH, Chalfie M. Distinct effects of tubulin isotype mutations on neurite growth in Caenorhabditis elegans. Mol Biol Cell. 2017 doi: 10.1091/mbc.E17-06-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lockhead D, Schwarz EM, O’Hagan R, Bellotti S, Krieg M, Barr MM, Dunn AR, Sternberg PW, Goodman MB. The tubulin repertoire of C. elegans sensory neurons and its context-dependent role in process outgrowth. Mol Biol Cell. 2016 doi: 10.1091/mbc.E16-06-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lener T, Gimona M, Aigner L, Borger V, Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo HA, et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun X, Park JH, Gumerson J, Wu Z, Swaroop A, Qian H, Roll-Mecak A, Li T. Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc Natl Acad Sci U S A. 2016;113:E2925–2934. doi: 10.1073/pnas.1523201113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao KN, Anand M, Khanna H. The carboxyl terminal mutational hotspot of the ciliary disease protein RPGRORF15 (retinitis pigmentosa GTPase regulator) is glutamylated in vivo. Biol Open. 2016;5:424–428. doi: 10.1242/bio.016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen B, Wu N, Yang JM, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem. 2011;286:14383–14395. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Astuti GD, Arno G, Hull S, Pierrache L, Venselaar H, Carss K, Raymond FL, Collin RW, Faradz SM, van den Born LI, et al. Mutations in AGBL5, Encoding alpha-Tubulin Deglutamylase, Are Associated With Autosomal Recessive Retinitis Pigmentosa. Invest Ophthalmol Vis Sci. 2016;57:6180–6187. doi: 10.1167/iovs.16-20148. [DOI] [PubMed] [Google Scholar]
- 55.Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Taira S, Hatanaka K, Morone N, Yao I, et al. Loss of α-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci U S A. 2007;104:3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 57.Mangeol P, Prevo B, Peterman EJ. KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol Biol Cell. 2016;27:1948–1957. doi: 10.1091/mbc.E15-06-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hall DH, Hartwieg E, Nguyen KC. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 2012;107:93–149. doi: 10.1016/B978-0-12-394620-1.00004-7. [DOI] [PubMed] [Google Scholar]
- 59.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.