Abstract
N-methyl-D-aspartate (NMDA) receptors are the main calcium-permeable excitatory receptors in the mammalian central nervous system. The NMDA receptor gating is complex, exhibiting multiple closed, open, and desensitized states; however, the central questions regarding the conformations and energetics of the transmembrane domains as they relate to the gating states are still unanswered. Here, using single molecule Förster Resonance Energy Transfer (smFRET), we map the energy landscape of the first transmembrane segment of the Rattus norvegicus NMDA receptor under resting and various liganded conditions. These results show kinetically and structurally distinct changes associated with apo, agonist-bound, and inhibited receptors linked by a linear mechanism of gating at this site. Furthermore, the smFRET data suggest that allosteric inhibition by zinc occurs by an uncoupling of the agonist-induced changes at the extracellular domains from the gating motions leading to an apo-like state, while dizocilpine, a pore blocker, stabilizes multiple closely packed transmembrane states.
Introduction
For over 30 years, single ion channel recordings have suggested that channel proteins exist in multiple closed and open conformations. As electrophysiological techniques have advanced, the set of ion channel states and our ability to distinguish them have grown to encompass a multitude of long-lived and short-lived shut states1–3. Despite the advances in single channel recording approaches, the ability to distinguish transitions between shut states is limited by the fact that these shut states are all electrophysiologically silent. The multiplicity of closed states is not restricted only to single-channel data but has also become increasingly prominent with the rise of cryo-electron microscopy. States previously assumed homogenous increasingly reveal themselves to reflect a variety of underlying conformations4.
Single molecule FRET of surface-immobilized molecules is uniquely suited to probing the conformational heterogeneity associated with these predicted closed and open states. N-methyl-D-aspartate (NMDA) receptors are an ideal candidate for such smFRET studies as they have a rich history of single channel studies with several sound reaction mechanisms involving discrete shut states across multiple time scales5–7. Further, there exist a number of full-length structures of NMDA receptors in apo (unliganded), antagonist-bound, and allosteric modulator-bound states4,8,9, and NMDA receptor channel gating is relatively slow and thus approaches the temporal resolution of smFRET10,11. Importantly, previous smFRET studies of intact NMDA receptors exist12 that utilize simultaneous patch-clamp electrophysiology and FRET microscopy imaging. While these previous studies provide an excellent means of correlating functional activity with conformational change, the use of fluorophore-conjugated agonist and antibodies as donor and acceptor, respectively, does not allow for more detailed structure-function studies at specific domains of the receptor.
NMDA receptors are obligate heterotetramers with two glycine-binding GluN1 and two glutamate-binding GluN2 subunits9,13. Each subunit is modular, with extracellular amino-terminal and agonist-binding domains, a transmembrane domain, and an intracellular carboxyl-terminal tail9,14. The available structural information provides excellent insight into the structure-function relationships within the extracellular domains of the NMDA receptor; however, the transmembrane segments which comprise the ion channel pore itself are less well-resolved, especially in the open-channel configuration. Consequently, structure-function analysis of this region has been more challenging. Past functional studies probing the transmembrane segments have focused primarily on conformational changes between the apo and open states15,16, and implicate the disordered linker region connecting the agonist-binding domain and transmembrane segments as being crucial for coupling. Still unclear though, is whether desensitization or allosteric inhibition themselves induce any conformational rearrangements in the transmembrane segments, or how such conformational changes might be driven by the binding of extracellular agonists and allosteric ligands. Furthermore, most functional studies using macroscopic recordings have, out of necessity, treated the apo, desensitized, and inhibited states as discrete conformations, while single channel analysis reveals each of these classes to be a collection of interconverting states7,17,18.
To explore these issues, we have performed smFRET on full-length GluN1/GluN2A NMDA receptors at residue 554 of the GluN1 linker region proximal to the first transmembrane segment, examining this site in resting (apo), agonist-bound, and zinc-bound (allosterically inhibited) conditions. This method revealed conformational changes in the transmembrane domain that are associated with channel opening. In addition, we also observed that the receptor occupies multiple closed states that have different kinetic and structural properties under apo, agonist-bound, and inhibited conditions. These data provide the first experimental evidence for the range of transmembrane conformations that the receptor adopts and moreover shed light on the structural landscape associated with the functional data. The data also provide the first evidence for differences in the closed channel conformations adopted by apo, agonist-bound, and inhibited receptors.
Results
Functional characterization of the smFRET construct
To investigate the conformational changes of the R. norvegicus NMDA receptor transmembrane domain in various functional states using smFRET, we introduced a fluorophore-labeling site using the mutation F554C in GluN1. We chose residue 554, found within the linker region connecting the agonist-binding domain to the first transmembrane segment of the transmembrane domain, for its accessibility to labeling as well as for minimal expected perturbation of receptor function (Figure 1a). To minimize non-specific labelling by donor and acceptor fluorophores (Alexa 555 maleimide and Alexa 647 maleimide, respectively) we mutated the accessible cysteines Cys15 and Cys22 in GluN1 and Cys231, Cys399, and Cys460 in GluN2A to serines, with the resulting background constructs hereafter referred to as GluN1* and GluN2A*19–22. Electrophysiological characterization of labeled GluN1*F554C/GluN2A* receptors show that activation, desensitization, and inhibition (Figure 1b) are all preserved. Specifically, responses to a 1-ms pulse of 1 mM glutamate with constant glycine in outside-out patches deactivated with a weighted time constant of 43 ± 6 ms (n = 11, Figure 1b, left) as compared to wild-type deactivation of 55 ± 6 ms (n = 12). In response to a 5-second long 1 mM glutamate application, the smFRET construct showed rapid activation (10–90% rise-time, 7 ± 1 ms, n = 11) and desensitized to 20 ± 3% of the peak response with a weighted time constant of the 110 ± 20 ms (n = 11, Figure 1b, left). Wild-type receptors also activate rapidly (4.9 ± 0.6 ms, n = 9) but decay slower (1800 ± 200, n = 9) to 68 ± 2% of the peak response. The channel block by both dizocilpine, which is (5S,10R)-(+)-5-Methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine hydrogen maleate (MK-801), (1 μM, GluN1*F554C/GluN2A*: 93 ± 2% steady-state inhibition, n = 8, Figure 1b, middle; wild-type: 95 ± 1%, n =5) and inhibition by Zn2+ (10 μM, GluN1*F554C/GluN2A*: 83 ± 5% steady-state-inhibition, n = 5, Figure 1b, right; wild-type: 84 ± 5%, n =5) were intact in whole cell recordings of the smFRET construct.
Figure 1.
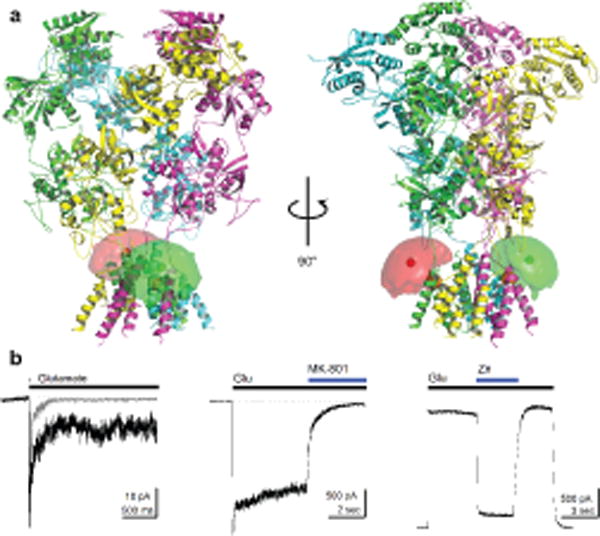
smFRET constructs and characterization. (a) GluN1*F554C/GluN2A* NMDA receptors were labeled with donor and acceptor fluorophores at site 554 of GluN1, proximal to the first transmembrane segment of GluN1 (mean fluorophore positions shown as green or red hard spheres surrounded by a fluorophore cloud, and Cα of F554 on GluN1 shown as an orange sphere). (b) Representative electrophysiological responses from the smFRET construct showing deactivation (gray) and desensitization (black) (left) with 1 mM glutamate and constant 100 μM glycine recorded with outside-out patches at −60 mV, inhibition by 1 μM MK-801 recorded in whole cell mode at −60 mV (middle), and inhibition by 10 μM Zn2+ recorded in whole cell mode at +50 mV (right). Dotted lines indicate baseline current.
smFRET identified distinct and stable states
For smFRET experiments, we expressed GluN1*F554C/GluN2A* receptors in a derivative of human embryonic kidney cells (HEK293T cells), labelled them with donor and acceptor fluorophores, and immunopurified the labeled receptors in situ on prepared coverslips using an antibody toward the C-terminus (abcam ab64572). To ensure that conjugation of the antibody to the C2 cassette of GluN1 did not adversely affect data acquisition, we generated an additional construct, GluN1*F554C-TS, to allow for attachment to the prepared coverslip via a Twin-Strep-tag (see Online Methods), generating comparable results (Supplementary Results, Supplementary Figure 1).
As with prior experiments using soluble iGluR domains23–26, sample scanning confocal microscopy showed clearly resolved single spots on these coverslips, which were not present when unmutated GluN1*/GluN2A* was used (Supplementary Figure 2). We collected single molecule FRET trajectories from these full-length labelled GluN1*F554C/GluN2A* receptors under various liganded conditions, and show the resulting ensemble-averaged denoised FRET efficiency histograms plotting the normalized occurrence at each apparent FRET efficiency (EA) in Figure 2. We denoised the raw trajectories through wavelet decomposition27,28, and identified specific states using the previously described Step Transition and State Identification (STaSI) analysis23,28–34 (Table 1, Figure 2 insets). In brief, the STaSI analysis analyzes piecewise constant signals based on a function described as the minimum description length (MDL). The MDL function then analyzes the data to optimize the goodness of the fit and complexity of the fitting model simultaneously. Thus, the MDL reaches a minimum when the fitting model ensures sparsest approximation and minimal fitting error. In other words, the minimum of the MDL function provides the optimum number of states to be assigned to the system and ensures a balance between the simplicity and accuracy of the fitting model29. STaSI analysis proceeded on each ligand dataset independently, and the MDL functions obtained from each dataset guided the optimum assignments of states (Supplementary Figure 3). For comparison, states with similar FRET efficiencies are grouped under same letter designations (States A–G from low to high FRET efficiency, Table 1).
Figure 2.
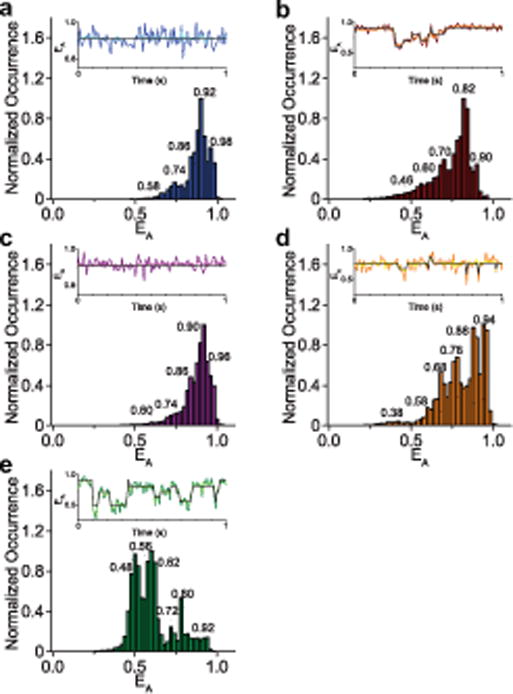
Denoised smFRET histograms of the NMDA receptor. smFRET data of the NMDA receptor were obtained under varying ligand conditions, denoised via wavelet decomposition27,28, and used to generate smFRET efficiency histograms plotting the normalized occurrence of the receptor exhibiting a given apparent FRET efficiency (EA). STaSI analysis was performed on the individual trajectories to reveal the underlying conformational states. Peaks corresponding to each STaSI state are labeled with the state efficiency. Shown in an inset above each histogram is a representative observed efficiency trace, the denoised trace (lighter shade), and the STaSI fit (black). The different conditions studied were the apo, unliganded receptor (a, 70 molecules), the agonist-bound (Glu-Gly) receptor (b, 36 molecules), and agonist-bound receptor inhibited by the allosteric inhibitor zinc (c, 26 molecules), and the agonist-bound receptor in the presence of the open-channel blocker MK-801 at 50 μM (d, 32 molecules), and 1 μM (e, 20 molecules).
Table 1.
Conformational states sampled by the NMDA receptor under various liganded conditions. Conformational states were identified using Step Transition and State Identification analysis23,29. Distances for the states were calculated with an R0 of 51 Å.
| State Designa- tion |
Apo | Glutamate-Glycine | Glutamate-Glycine/Zn2+ | Glutamate-Glycine/ MK-801, 50 μM |
Glutamate-Glycine/ MK-801, 1 μM |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FRET Efficien- cy |
Percent Occur- rence |
FRET distance (Å) |
FRET Efficien- cy |
Percent Occur- rence |
FRET distance (Å) |
FRET Efficien- cy |
Percent Occur- rence |
FRET distance (Å) |
FRET Efficien- cy |
Percent Occur- rence |
FRET distance (Å) |
FRET Efficien- cy |
Percent Occur- rence |
FRET distance (Å) |
|
| A | 0.46 ± 0.02 | 7.1 | 52 ± 1 | 0.38 ± 0.02 | 3.4 | 55 ± 1 | 0.48 ± 0.02 | 23.6 | 52 ± 1 | ||||||
| B | 0.56 ± 0.02 | 24.8 | 49 ± 1 | ||||||||||||
| C | 0.58 ± 0.02 | 1.6 | 48 ± 1 | 0.60 ± 0.02 | 9.2 | 48 ± 1 | 0.60 ± 0.02 | 4.3 | 48 ± 1 | 0.58 ± 0.02 | 6.6 | 48 ± 1 | 0.62 ± 0.02 | 26.0 | 47 ± 1 |
| D | 0.74 ± 0.02 | 14.6 | 43 ± 1 | 0.70 ± 0.02 | 18.4 | 44 ± 1 | 0.74 ± 0.02 | 5.6 | 43 ± 1 | 0.68 ± 0.02 | 14.4 | 45 ± 1 | 0.72 ± 0.02 | 8.0 | 44 ± 1 |
| E | 0.86 ± 0.02 | 17.4 | 38 ± 1 | 0.82 ± 0.02 | 53.4 | 40 ± 1 | 0.82 ± 0.02 | 20.5 | 40 ± 1 | 0.78 ± 0.02 | 28.2 | 41 ± 1 | 0.80 ± 0.02 | 11.3 | 40 ± 1 |
| F | 0.92 ± 0.02 | 50.3 | 34 ± 2 | 0.90 ± 0.02 | 11.8 | 35 ± 1 | 0.90 ± 0.02 | 45.9 | 35 ± 1 | 0.88 ± 0.02 | 21.7 | 37 ± 2 | 0.92 ± 0.02 | 6.4 | 34 ± 2 |
| G | 0.98 ± 0.02 | 16.0 | 27 ± 5 | 0.96 ± 0.02 | 23.7 | 30 ± 3 | 0.94 ± 0.02 | 25.8 | 32 ± 2 | ||||||
The agonist-bound conformational landscape
The FRET efficiency histogram of the apo NMDA receptor (Figure 2a) shows five discrete states, with the predominant state (State F, 50.3% occupancy) at a FRET efficiency of 0.92 ± 0.02 (Table 1). The glutamate-glycine bound histogram (Figure 2b) also shows five states; however, the receptor occupancy distribution is shifted toward lower efficiency states (Supplementary Figure 4a and 4b). This may be due to channel opening, though the higher-efficiency states are still prominent, consistent with the high level of desensitization by the GluN1*F554C construct. These higher-efficiency states explored by the glutamate-glycine-bound receptor (States E and F) have distances of 40 ± 1 Å and 35 ± 1 Å, respectively, between the fluorophores. These distances are similar to the X-ray and electron microscopy (EM)-based structures of the closed-channel receptor, which show a Cα–Cα distance of 30 Å. Additionally, the distance change of 17 ± 1 Å between the highest glutamate-glycine bound FRET efficiency state F (0.90 ± 0.02 efficiency, 35 ± 1 Å) and its lowest FRET efficiency state A (0.46 ± 0.02 efficiency, 52 ± 1 Å) is similar not only to the 22 Å change at the M3 helix between the closed-channel EM structure and the low-resolution EM structure thought to represent the open receptor4, but also to the changes seen at an identical site in high resolution crystal structures of the NaK at similar sites35,36. Combined, these further suggest that the lower FRET efficiency states could correlate to open channel forms of the receptor. Consistent with the above observation, States A and C represent 16.3% of the total occupancy for the glutamate-glycine-bound receptor (State B was not identified as a highly populated state for the glutamate-glycine-bound receptor). This percent correlates well with the percent of 20 ± 3% residual steady-state current after desensitization relative to peak response in our electrophysiological measurements of the smFRET construct (Figure 1b). Furthermore, the glutamate-glycine states A and C are rarely observed with the apo receptor (1.6% occupancy for apo State C with no observed population corresponding to State A), as would be expected for the apo receptor, which does not spontaneously open.
The apo receptor explores an additional high-efficiency State G at 0.98 ± 0.02 (27 ± 5 Å) (Figure 2a, Table 1). This closed state has no corresponding equivalent in the glutamate-glycine bound receptor. This is not surprising given the fact that the agonist-binding domain of the apo state of the receptor is able to probe more open-cleft states relative to the agonist bound form the receptor24,37,38, and prior indirect functional studies have shown tight coupling between the agonist-binding domain and the transmembrane segments15.
Comparison of agonist and inhibitor binding
To obtain a fuller understanding of the gating motions of the NMDA receptor, we also examined the accessible conformational states of the transmembrane domain in the presence of the divalent cation zinc, which acts as an allosteric inhibitor18–20,39. Like other allosteric inhibitors, zinc binds to the amino-terminal domain of GluN2 subunits, inducing cleft closure and a reduction in channel activity18–20,40, though the precise nature of the conformational changes that occur at the transmembrane segments upon zinc binding are still unknown. Figure 2c shows the ensemble histogram of the denoised FRET data of the glutamate/glycine/Zn2+-bound receptors, which are primarily shifted toward more closed states as compared to the agonist-only-bound receptor (Supplementary Figure 4 c and d). However, in addition to stabilizing the agonist-bound closed states, another high-efficiency, more closed state is observed at 0.96 ± 0.02 (Figure 2c, Table 1), which is structurally distinct from the closed states probed with glutamate and glycine. The efficiency of this more closed state in the presence of zinc is similar to the 0.98 ± 0.02 efficiency State G observed in the apo form of the receptor (Figure 2a, Table 1), and in fact their histograms are remarkably similar (Supplementary Figure 4 e and f). Thus, rather than simply shifting the conformational equilibrium of the glutamate-glycine-bound receptor toward its more closed states, the binding of zinc results in the more compact “apo-like” state of the receptor’s transmembrane segments. These data are consistent with the structural data showing that the allosteric inhibition allows for the decoupling of the extracellular domains from the transmembrane segments, but additionally show that the transmembrane segments are able to occupy apo-like closed conformations.
States occupied by open channel blocker MK-801
As an additional exploration of the conformational landscape accessible by the NMDA receptor, we employed MK-801, an open-channel blocker that binds uncompetitively to the activated receptors leading to channel block41,42. Structural data of NMDA receptors with MK-801 bound show a closed-pore conformation43. This is consistent with previous data that demonstrate that NMDA receptors can close and trap MK-801 within the transmembrane pore44. Consistent with this prior data, our smFRET histogram with saturating concentrations of MK-801 (50 μM) show occupancies shifted toward higher efficiencies, suggesting a more compact transmembrane arrangement (Figure 2d). Saturation with MK-801 was established by investigating the fractional occurrence of the high efficiency FRET state (Supplementary Figure 5). The histogram for the MK-801 bound state of the receptor was not as compact as those observed in zinc-bound states of the receptor indicating differences at the transmembrane segments for the two kinds of inhibition. Additionally, when using a sub-saturating concentration of 1 μM MK-801, we see a notable shift in the smFRET histogram toward lower efficiencies compared to the histograms with higher concentrations of MK-801 (Figure 2e). Two possible mechanisms could account for this observation. These low efficiency conformational populations may be a reflection of binding and unbinding of MK-801, due to the slow kinetics of MK-801 block and recovery44. This would be consistent with the prior studies showing that MK-801 binding required the receptor to occupy an open channel state44. Alternatively, this could represent a second binding site, similar to what has been previously see with other uncompetitive pore blockers45–47, giving rise to a more loose transmembrane packing, with fully trapped conformations being more prevalent at higher MK-801 concentrations.
Dynamics of the NMDA receptor
Beyond providing the conformational landscape that the NMDA receptor probes, our smFRET data also provide insight into the dynamics of the receptor. The smFRET traces under apo conditions were relatively static, with fewer observed state transitions (Figure 2a, inset), whereas the traces of the liganded receptor showed more transitions (Figure 2b, inset). To quantify the differences in transition frequency, we first examined the average number of transitions occurring in time-windows of varying length for each condition (Figure 3a), which confirm that the apo condition indeed exhibited a much lower average number of transitions than receptors in other conditions. Interestingly, the allosterically inhibited zinc-bound form of the receptor exhibits a comparable number of transitions to the other liganded conditions, despite being electrically less active and with an overall histogram reminiscent of the apo condition. These transitions of the inhibited receptor, however, are primarily between the highest-efficiency closed states, as discussed later. Such behavior is reminiscent of single-channel recordings, which display long-lived shut states interspersed with clusters of electrical activity48. As a second measure of conformational fluctuation, the cumulative probability plot of the coefficient of variation of each single molecule trajectory is shown in Figure 3b. Static, stable traces exhibit low coefficient of variation, whereas dynamic traces exhibit a higher coefficient of variation. The coefficient of variation of each smFRET efficiency trajectory versus that trajectory’s length was studied to ensure that the length of the smFRET trajectories did not affect the CV-ligand condition relationship (Supplementary Figure 6). As with the average transitions over time, apo receptors show much less variation and the cumulative probability rises quickly at a low coefficient of variation value. In contrast, a much larger portion of the agonist-bound traces show higher variation and so the rise is slower. Zinc-inhibited receptors, though they undergo as many transitions as does the uninhibited agonist-bound receptor receptors (Figure 3a), show a steeper rise, intermediate between the glutamate-glycine and the apo receptors. As described below, this behavior is consistent with the zinc-bound receptors exhibiting transitions between a smaller range of states, fluctuating mainly among the closed states, and rarely visiting open states.
Figure 3.
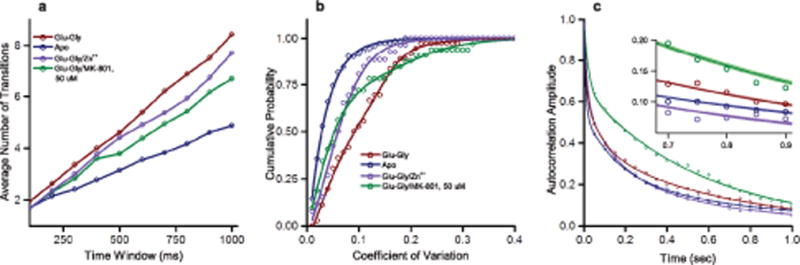
Dynamics of the NMDA receptor show differences in transitional behavior under different ligand conditions. The dynamics of the NMDA receptor in the apo, glu-gly, glu-gly with zinc, and glu-gly with 50 μM MK-801 bound states, as probed by (a) the average number of transitions seen per time window, (b) the cumulative probability plot of the coefficient of variation and (c) autocorrelation of the single molecule data fit to three-term exponential decay. The thickness of the lines represents the fitting errors (zoomed in for visibility in inset). Data for each molecule was obtained over a total acquisition time of, in seconds, 506.14, 330.36, 255.48, and 216.08, respectively.
The coefficient of variation data for the MK-801 bound state exhibits a biphasic behavior suggesting two populations—one which exhibits a high level of transitions and a second which is more stable. This is evident in the individual traces, where high FRET receptors show very few transitions, while lower FRET receptors exhibit a large number of transitions (Supplementary Figure 7). These data are consistent with the idea that there could be more than one MK-801 binding modes: the high FRET mode representing trapped MK-801 in a closed channel like state exhibiting low transitions, and a second “pre-trapped” mode exhibiting a large number of transitions.
As a final measure to examine the dynamics of the transmembrane domain, we calculated the autocorrelation of each of the smFRET trajectories to generate an average autocorrelation curve for each set of ligand conditions. We then fit these average autocorrelation curves to exponential decay, with the exponential fits yielding three different fluctuation timescales for the apo, glu-gly, and glu-gly/Zn2+ condition: short (τ1), intermediate (τ2), and long (τ3) (Figure 3c and Table 2). Two exponentials, short (τ1) and long (τ3), best fit the glu-gly/50 μM MK-801 data. Short autocorrelation decay times indicate a more rigid nature of the molecules, whereas longer decay times indicate the opposite26. The process associated with the longest timescale (τ3) arise from conformational transitions or protein fluctuation events49. As expected, apo and Zn2+-inhibited receptors have smaller amplitudes for this timescale, whereas agonist-bound and MK-801-bound NMDA receptors have larger amplitudes for this timescale. Furthermore, the trend in overall weighted average fluctuation timescales indicates that the agonist-bound and MK-801-bound receptors exhibit a longer overall decay compared to the apo and zinc-inhibited receptors, again reflecting the greater stability of electrically inactive receptors under these conditions. MK-801 bound receptors specifically show the highest amplitude, 70 percent, in the longest timescale (Table 2), and the highest weighted time scale of 380 milliseconds, which most likely reflect the large number of transitions observed in the low FRET states.
Table 2.
Autocorrelation data under the various ligated conditions. The average autocorrelation decay of the single molecule FRET data was fit to a three-term exponential decay, resulting in short, intermediate and long timescale decays for each condition.
| Ligand | Short timescale τ1 (ms) | Amplitude (%) | Intermediate timescale τ2 (ms) | Amplitude (%) | Long timescale τ3 (ms) | Amplitude (%) | Weighted average lifetime τ (ms) |
|---|---|---|---|---|---|---|---|
| Apo | 8 ± 1 | 46.4 ± 0.7 | 170 ± 10 | 30 ± 1 | 860 ± 60 | 23 ± 2 | 250 ± 20 |
| Glu-Gly | 9 ± 3 | 37 ± 1 | 82 ± 6 | 24.3 ± 0.9 | 650 ± 10 | 38.9 ± 0.7 | 275 ± 6 |
| Gly-Gly/Zn2+ | 13 ± 1 | 39 ± 1 | 150 ± 10 | 34 ± 2 | 640 ± 40 | 26 ± 2 | 220 ± 20 |
| Glu-Gly/MK-801, 50 μM | 20 ± 1 | 32 ± 1 | — | — | 560 ± 6 | 66.4 ± 0.6 | 378 ± 8 |
Transitions among the conformational states
The smFRET trajectories further allow us to determine the transitions between and among the different conformational states. Figure 4a–c shows the transition maps representing the relative number of transitions from one state to the other. The data show that transitions primarily occur between states of nearest FRET efficiency, whereas transitions between non-adjacent states are less common. Thus, a linear mechanism connecting the different states best describes the observed transition maps. Such a mechanism has a striking similarity to the mechanism proposed based on single channel analysis which show several closed states interconverting, but a single closed state transitioning to an open state18. Thus our data show multiple structural states that could possibly be correlated to the multiple functional states identified in single channel recordings, allowing us to place these states along a plausible structural reaction coordinate corresponding to the functionally relevant distinction between open and closed.
Figure 4.
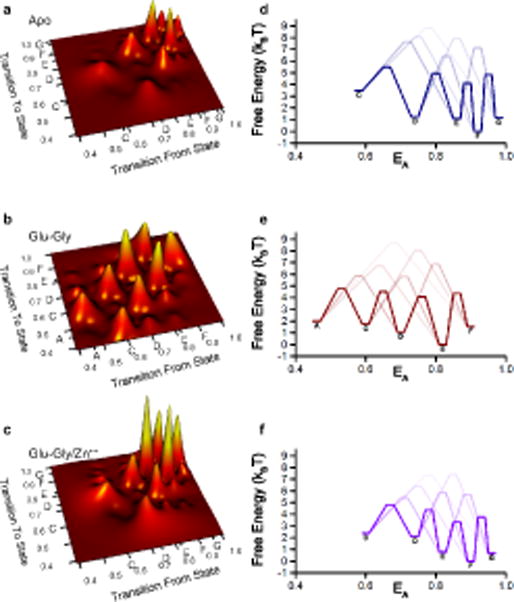
Transition maps and free energy diagrams of the NMDA receptor smFRET data. Examination of the individual single molecule traces show primarily transitions between adjacent states, portrayed here in terms of the normalized number of transitions for (a) apo, (b) glutamate-glycine bound, and (c) glutamate-glycine-bound zinc-inhibited NMDA receptors. Data from these transition maps were used to generate free energy diagrams (d–f). State designations as noted in Table 1 are shown on the axes in a–c and next to the energy well for the corresponding state in d–f.
Energetics of the NMDA receptor
Based on the distribution of states (Figure 2a–e, Table 1) and the transition maps (Figure 4a–c), we then calculated the relative free energies of the different states along with the relative free energy barriers for transitions between pairs of states50 (Figure 4d–f). These smFRET-derived data are again strikingly similar to the free energy profiles proposed based on functional single-channel recordings18 with one difference being that our data show a higher free energy for the low efficiency FRET states than seen in the single-channel recording model. These differences may result from the fact that the smFRET construct has a slightly higher degree of desensitization than the wild type receptor, or from the acquisition of smFRET measurements in the presence of detergents rather than the cellular environment used in single channel recordings. The energy profiles of the apo state also support our conclusion that the resting apo receptor remains primarily in high FRET closed conformational states with a high activation energy between states and thus fewer transitions. The zinc-inhibited receptor also occupies primarily closed states, but the high transition frequency noted in the transition maps (Figure 4) indicate a relatively lower activation energy between the states that allows the zinc-inhibited receptor to exhibit more transitions than apo. The ability of uninhibited agonist-bound receptors to activate the ion channel follows from the lower activation energies between the pre-open closed state and the open state, which facilitates transitions between them.
Discussion
Here we have used single molecule FRET to examine the conformational landscape of full-length intact NMDA receptors under various liganded conditions. In contrast to previous smFRET examinations of full-length NMDA receptors12, the use of small fluorescent dyes directly coupled to protein residues allowed for a more detailed investigation specifically at the linker preceding the first transmembrane segment. Our analysis revealed a multiplicity of FRET states in each dataset. The smFRET data show similar efficiencies between different liganded conditions but with different occupancies, consistent both with functional studies using single channel recordings and with the model of conformational selection at the level of the transmembrane domain. Agonist binding lowers the energy barrier preventing the transitions of the apo receptor, allowing the glutamate-glycine bound receptor to explore the low-FRET efficiency conformations that could correlate to channel opening, whilst destabilizing the tightest high-FRET conformations, which could correspond to the closed channel arrangement. Prior structural and FRET studies show that the agonist-binding domain explores multiple closed-cleft conformations, and one question has been whether and how the propagation of the multiplicity of states occurs beyond the linker connecting the agonist-binding domain and the transmembrane region. Here, we are able to show the transmembrane segments do exhibit multiple stable states which, when propagated to the channel segments, may contribute to the multiple states observed in the single channel functional recordings.
Our studies also show that the binding of the allosteric modulator zinc inhibits the NMDA receptor by lowering the energy barrier of the glutamate-glycine bound receptor leading into an apo-like tightest closed-state arrangement, essentially decoupling the closed-cleft tension of the agonist-binding domains from the transmembrane segments. The pore blocker MK-801 stabilizes a less compact transmembrane segment relative to the zinc bound form of the receptor and the low FRET efficiency state of MK-801 bound receptor exhibits more transitions indicating that it is more dynamic.
Online Methods
Generation of smFRET constructs
Wildtype R. norvegicus GluN1-1a and GluN2A plasmids in pcDNA3.1 were kindly provided by Shigetada Nakanishi (Osaka Bioscience Institute, Osaka, Japan). All mutations were introduced using standard PCR-based mutagenesis methods. To create the background constructs, non-disulfide-bonded cysteines at sites 15 and 22 in GluN1 and 231, 399, and 460 in GluN2A were mutated to serines, resulting in GluN1* and GluN2A*. In order to label these receptors, a reactive cysteine was mutated to replace the native phenylalanine at site 554 in GluN1*. To provide an alternative epitope to pull down NMDA receptors, an additional construct, GluN1*F554C-TS, was generated by affixing a thrombin cleavage site followed by a Twin-Strep-tag to the C-terminus of GluN1*F554C.
Electrophysiology
HEK293T cells were plated into 30 mm dishes and transfected (jetPrime, PolyPlus) with GluN1*F554C and GluN2A* constructs at a mass ratio of 1.5:4.5 μg per 20 ml of media. 300 μM DL-AP5 (abcam) and 30 μM DCKA (abcam) were present in the media and recordings were performed 24 to 48 hours post-transfection. Prior to recording, cells were incubated in 150 nM Alexa 555 maleimide (ThermoFisher) and 600 nM Alexa 647 maleimide (ThermoFisher) for at least 1 hour to mimic smFRET labelling conditions. Outside-out patches were excised and piezo-driven solution exchange was performed as outlined elsewhere51. The external solution was (in mM) 150 NaCl, 20 HEPES, 10 Tricine, 1 CaCl2, and 0.1 glycine, pH 7.4 (NaOH). The pipette solution was 135 CsF, 33 CsOH, 11 EGTA, 10 HEPES, 2 MgCl2 and 1 CaCl2, pH 7.4. Lifted whole cell recordings were performed as described elsewhere52 using the same solutions as above with the addition of 2.5 mM KCl to the external solution.
Single molecule FRET sample preparation
HEK293T cells were transiently transfected with GluN1*F554C and GluN2A* DNA at a mass ratio of 2.5:7.5 μg per 10 cm dish. An equivalent mass of GluN1*F554C-TS was used in place of GluN1*F554C for experiments involving it. 300 μM DL-AP5 (abcam) and 30 μM DCKA (abcam) were present during transfection to limit excitotoxicity. One day post-transfection, cells from two 10-cm dishes were harvested and labeled for 1 hour at room temperature with 150 nM of donor fluorophore Alexa 555 maleimide (ThermoFisher) and 600 nM of acceptor fluorophore Alexa 647 maleimide (ThermoFisher) in 3 mL extracellular buffer. After washing, labeled cells were then solubilized for 1 hour at 4°C in buffer containing phosphate-buffered saline, 1% lauryl maltose neopentyl glycol (Anatrace), 2 mM cholesteryl hydrogen succinate (MP Biomedicals), and protease inhibitor (Pierce). Unsolubilized debris were then spun down for 1 hour at 100,000 × g at 4°C, and the supernatant used as the smFRET sample.
Flow Chamber Preparation
Plasma cleaned glass coverslips (22 × 22 mm No. 1) were aminosilanized through Vectabond treatment (Vectabond in acetone 2% vol/vol; Vector Laboratories). Silicone templates (Grace bio-Labs) were used to treat a small section of the coverslips with a PEG solution containing 5kDa biotin-terminated PEG (2.5% w/w in molecular biology grade (MB) water, NOF Corp.), and 5kDa mPEG succinimidyl carbonate (25% w/w in MB water, Laysan Bio Inc.) in 0.1M sodium bicarbonate (Sigma-Aldrich). The coverslips were then left to incubate in a dark and moist environment overnight. On the day of the experiment, the coverslips were treated with another round of PEGylation with a short chain 333 Da NHS-ester PEG (Thermo Scientific) and incubated for 2–3 hrs. After washing off excess PEG, the coverslips were dried with a mild flow of nitrogen. Custom hybriwell chambers (Grace bio-Labs) with dual silicon press-fit tubing connectors (Grace bio-Labs) were placed atop the coverslips to construct a flow chamber.
Protein preparation and attachment to coverslips
Streptavidin in buffer solution containing phosphate-buffered saline (PBS), 1mM DDM (n-dodecyl-β-D-maltoside) and 0.2 mM CHS (cholesteryl hemisuccinate), were introduced through the flow chamber and incubated for 10 minutes. 10nM of biotinylated goat Anti-Mouse IgG (H+L) secondary antibody (Jackson Immuno Research Laboratories, Inc., cat. no. 115-065-003) was then flowed into the chamber and incubated for 20–30 minutes. Next, 10nM of anti-NMDAR mouse monoclonal primary antibody (abcam, cat. no. ab64572) was flowed in. After each antibody addition, the chamber was flushed with buffer to get rid of the unbound antibodies. All dilutions were made in PBS buffer with 1 mg/mL bovine serum albumin (Sigma-Aldrich). No antibodies were added when studying GluN1*F554C-TS, with those proteins attaching directly to streptavidin via the Twin-Strep-tag.
Intact, solubilized NMDA receptors as prepared above were then attached to a glass slide for FRET data acquisition using in situ immunopreciption (SiMPull53) by passing the solubilized protein through the chamber in three 60 μL shots and incubating for 20–30 minutes before flushing the chamber with buffer containing Phosphate Buffered Saline (PBS), 1mM DDM (n-dodecyl-β-D-maltoside) and 0.2 mM CHS (cholesteryl hemisuccinate).
Control slides using unmutated GluN1*/GluN2A* receptors showed minimal background labeling, while slides prepared with GluN1*F554C/GluN2A* showed isolatable single molecules exhibiting fluorescence and energy transfer (Supplementary Figure 1).
smFRET data acquisition
For smFRET data acquisition the sample is excited at both 532nm and 637nm to colocalize the proteins having an acceptor tag and exhibiting FRET. All single molecule FRET measurements were acquired using a custom-built confocal microscope described previously26. The sample was excited either with a continuous wave 532nm laser (Compass 315M-100SL; Coherent) or a 637nm laser (OBIS-FP 637 LX; Coherent) and focused through an oil immersion objective (100 × 1.3 NA; Carl Zeiss) on the sample. The power density at the sample was kept at 50W/cm2. The sample position was controlled through a scanning x-y-z piezo stage (P-517.3CL; Physik Instrumente). Then the emission signal from the sample was collected back through the same objective and separated via a 640nm high-pass dichroic mirror (640 DCXR; Chroma Technology) to collect the donor and acceptor emission signals separately at two avalanche photodiodes (SPCM-AQR-15; PerkinElmer). The emission signal was tuned to 570 and 670 nm with band-pass filters (NHPF-532.0; Kaiser Optical Systems), placed before the detectors. All acquisitions were performed under a constant flow of a photostabilizer and oxygen scavenging solution buffer system (ROXS) consisting of 1mM methyl viologen, 1mM ascorbic acid, 0.01% w/w glucose oxidase, 0.001% w/v catalase, 3.3% w/w glucose (all from Sigma-Aldrich), 1mM DDM (Chem-Impex), and 0.2 mM CHS (MP Biomedicals, LLC) in PBS, pH 7.4. Ligands—1mM glutamate, 1mM glycine, MK-801, and/or 10 μM Zn2+—were added as appropriate to the ROXS to achieve the liganded conditions necessary for each experiment. Additionally, 10 mM tricine was included whenever Zn2+ was not added in order to chelate any unbound zinc ions. Donor and sensitized acceptor photon counts were acquired for individual molecules at 1 millisecond resolution, binned to 10 milliseconds, denoised with wavelet decomposition, and the calculated denoised efficiencies were then plotted as separate histograms24–26,54. Step Transition and State Identification (STaSI) analysis23,29 was run for each sample to obtain an unbiased determination of number and the identification of discrete conformational states within each sample23–26,29,54.
Free Energy Calculations
For each liganded condition, the free energy of the most populated STaSI-identified state was set to 0 kBT. The STaSI determined occupancies were then used to calculate the equilibrium constant Keq between each pair of states, and the free energy of every state relative to the most populated state was determined via the equation:
The transition probabilities between each pair of states, given our 10 ms bin time, was used to determine the reaction rate for each transition, and the heights of the energy of activation barriers were calculated assuming a first-order reaction rate and the Arrhenius equation:
Concentration of the starting state was taken as the STaSI-derived fractional occupancy of that state, and the value of the pre-exponential was chosen to be (10ms)−1. Forward and reverse energies of activation were averaged in the final figure.
Statistics
Data were analyzed using Origin (OriginLab Corp.), MATLAB (MathWorks), and Excel (MicrosoftCorp.). Desensitization and deactivation kinetics were fit with single-exponential functions in Clampfit 10.6 (Molecular Devices) as judged by eye. For outside-out patches, n was considered as a single patch. For whole cell experiments, n was considered as the response from the whole cell. Error in measurements is given as SEM. For smFRET experiments after filtering the molecules for those that passed cross- and auto-correlation checks, the number of particles for apo, glutamate-glycine, and glutamate-glycine/Zn2+ was, respectively, n = 70, 36, and 26. These led to a total acquisition time of, in seconds, 506.14, 330.36, and 255.48, respectively. For glutamate-glycine/MK-801 histograms and the dose-response curve, experiments were performed with MK-801 concentrations of 1μM, 10 μM, 20 μM, and 50 μM, with n = 20, 50, 42, and 32, for total acquisition times of, in seconds, 146.65, 193.27, 306.15, and 216.08, respectively.
Code Availability
Code for denoising and STaSI can be downloaded from the Landes Research Group website at http://www.lrg.rice.edu/Content.aspx?id=96.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
Methods and additional data can be found in the supplementary materials. This project was supported by NIH grant R35GM122528 to V.J., K99NS094761to D.M.M., the American Heart Association Fellowship 16POST30030007 to S.A.S., the Schissler Foundation Fellowship to D.M.D., and the Welch Foundation Grant C-1787 to C.F.L.
Footnotes
Author Contributions
D.M.D. performed the mutations and prepared the labeled proteins, analyzed the smFRET data, and contributed to designing the research, interpreting the results, and writing the manuscript. S.C. performed the smFRET measurements, analyzed the smFRET data, and contributed to designing the research, interpreting the results, and writing the manuscript. D.M.M. and S.A.S. performed the electrophysiology. C.F. and L.D.C.B. analyzed the smFRET data. C.F.L., and V.J. analyzed the smFRET data and contributed to designing the research, interpreting the results, and writing the manuscript.
Competing Financial Interests Statements
The authors declare no competing interests.
References
- 1.Zhang W, Howe JR, Popescu GK. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nature neuroscience. 2008;11:1373–1375. doi: 10.1038/nn.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lape R, Colquhoun D, Sivilotti LG. On the nature of partial agonism in the nicotinic receptor superfamily. Nature. 2008;454:722–727. doi: 10.1038/nature07139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Devi SP, Tomita S, Howe JR. Auxiliary proteins promote modal gating of AMPA- and kainate-type glutamate receptors. The European journal of neuroscience. 2014;39:1138–1147. doi: 10.1111/ejn.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tajima N, et al. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016;534:63–68. doi: 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popescu G, Robert A, Howe JR, Auerbach A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430:790–793. doi: 10.1038/nature02775. [DOI] [PubMed] [Google Scholar]
- 6.Banke TG, Traynelis SF. Activation of NR1/NR2B NMDA receptors. Nature neuroscience. 2003;6:144–152. doi: 10.1038/nn1000. [DOI] [PubMed] [Google Scholar]
- 7.Kussius CL, Popescu GK. Kinetic basis of partial agonism at NMDA receptors. Nature neuroscience. 2009;12:1114–1120. doi: 10.1038/nn.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu S, et al. Mechanism of NMDA Receptor Inhibition and Activation. Cell. 2016;165:704–714. doi: 10.1016/j.cell.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science (New York, NY) 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JY, Kim C, Lee NK. Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering. Nature communications. 2015;6:6992. doi: 10.1038/ncomms7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Meth. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasmal DK, Lu HP. Single-Molecule Patch-Clamp FRET Microscopy Studies of NMDA Receptor Ion Channel Dynamics in Living Cells: Revealing the Multiple Conformational States Associated with a Channel at Its Electrical Off State. Journal of the American Chemical Society. 2014;136:12998–13005. doi: 10.1021/ja506231j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monyer H, et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science (New York, NY) 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 14.Traynelis SF, et al. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacological Reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nature neuroscience. 2014;17:914–922. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobolevsky AI, Beck C, Wollmuth LP. Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron. 2002;33:75–85. doi: 10.1016/s0896-6273(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 17.Borschel WF, Cummings KA, Tindell LK, Popescu GK. Kinetic Contributions to Gating by Interactions Unique to N-methyl-d-aspartate (NMDA) Receptors. Journal of Biological Chemistry. 2015;290:26846–26855. doi: 10.1074/jbc.M115.678656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amico-Ruvio SA, Murthy SE, Smith TP, Popescu GK. Zinc effects on NMDA receptor gating kinetics. Biophysical journal. 2011;100:1910–1918. doi: 10.1016/j.bpj.2011.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirrieh RE, MacLean DM, Jayaraman V. A conserved structural mechanism of NMDA receptor inhibition: A comparison of ifenprodil and zinc. The Journal of General Physiology. 2015;146:173–181. doi: 10.1085/jgp.201511422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirrieh RE, MacLean DM, Jayaraman V. Amino-terminal domain tetramer organization and structural effects of zinc binding in the N-methyl-D-aspartate (NMDA) receptor. The Journal of biological chemistry. 2013;288:22555–22564. doi: 10.1074/jbc.M113.482356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirrieh RE, MacLean DM, Jayaraman V. Subtype-dependent N-Methyl-d-aspartate Receptor Amino-terminal Domain Conformations and Modulation by Spermine. The Journal of biological chemistry. 2015;290:12812–12820. doi: 10.1074/jbc.M115.649723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rambhadran A, Gonzalez J, Jayaraman V. Conformational changes at the agonist binding domain of the N-methyl-D-aspartic acid receptor. The Journal of biological chemistry. 2011;286:16953–16957. doi: 10.1074/jbc.M111.224576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper DR, et al. Conformational Transitions in the Glycine-Bound GluN1 NMDA Receptor LBD via Single-Molecule FRET. Biophysical journal. 2015;109:66–75. doi: 10.1016/j.bpj.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolino DM, et al. Structural Dynamics of the Glycine-binding Domain of the N-Methyl-d-Aspartate Receptor. Journal of Biological Chemistry. 2015;290:797–804. doi: 10.1074/jbc.M114.605436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramaswamy S, et al. Role of conformational dynamics in alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor partial agonism. The Journal of biological chemistry. 2012;287:43557–43564. doi: 10.1074/jbc.M112.371815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landes CF, Rambhadran A, Taylor JN, Salatan F, Jayaraman V. Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nature chemical biology. 2011;7:168–173. doi: 10.1038/nchembio.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor JN, Makarov DE, Landes CF. Denoising single-molecule FRET trajectories with wavelets and Bayesian inference. Biophysical journal. 2010;98:164–173. doi: 10.1016/j.bpj.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor JN, Landes CF. Improved resolution of complex single-molecule FRET systems via wavelet shrinkage. The journal of physical chemistry. 2011;B 115:1105–1114. doi: 10.1021/jp1050707. [DOI] [PubMed] [Google Scholar]
- 29.Shuang B, et al. Fast Step Transition and State Identification (STaSI) for Discrete Single-Molecule Data Analysis. The journal of physical chemistry letters. 2014;5:3157–3161. doi: 10.1021/jz501435p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chizhik AM, et al. Super-Resolution Optical Fluctuation Bio-Imaging with Dual-Color Carbon Nanodots. Nano Letters. 2016;16:237–242. doi: 10.1021/acs.nanolett.5b03609. [DOI] [PubMed] [Google Scholar]
- 31.Hu Z, et al. Excitonic Energy Migration in Conjugated Polymers: The Critical Role of Interchain Morphology. Journal of the American Chemical Society. 2014;136:16023–16031. doi: 10.1021/ja508112k. [DOI] [PubMed] [Google Scholar]
- 32.Wedeking T, et al. Single Cell GFP-Trap Reveals Stoichiometry and Dynamics of Cytosolic Protein Complexes. Nano Letters. 2015;15:3610–3615. doi: 10.1021/acs.nanolett.5b01153. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Park H, Kaufman LJ. Highly Anisotropic Conjugated Polymer Aggregates: Preparation and Quantification of Physical and Optical Anisotropy. The Journal of Physical Chemistry C. 2017;121:13854–13862. [Google Scholar]
- 34.Zhang W, Caldarola M, Pradhan B, Orrit M. Gold Nanorod Enhanced Fluorescence Enables Single-Molecule Electrochemistry of Methylene Blue. Angewandte Chemie. 2017;129:3620–3623. doi: 10.1002/anie.201612389. [DOI] [PubMed] [Google Scholar]
- 35.Alam A, Jiang Y. High-resolution structure of the open NaK channel. Nature structural & molecular biology. 2009;16:30–34. doi: 10.1038/nsmb.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi N, Ye S, Alam A, Chen L, Jiang Y. Atomic structure of a Na+- and K+-conducting channel. Nature. 2006;440:570–574. doi: 10.1038/nature04508. [DOI] [PubMed] [Google Scholar]
- 37.Dolino DM, Rezaei Adariani S, Shaikh SA, Jayaraman V, Sanabria H. Conformational Selection and Submillisecond Dynamics of the Ligand-binding Domain of the N-Methyl-d-aspartate Receptor. The Journal of biological chemistry. 2016;291:16175–16185. doi: 10.1074/jbc.M116.721274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Y, Belcher J, Berger AJ, Mayer ML, Lau AY. Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure (London, England: 1993) 2013;21:1788–1799. doi: 10.1016/j.str.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters S, Koh J, Choi DW. Zinc selectively blocks the action of N-methyl-D-aspartate on cortical neurons. Science (New York, NY) 1987;236:589–593. doi: 10.1126/science.2883728. [DOI] [PubMed] [Google Scholar]
- 40.Low CM, Zheng F, Lyuboslavsky P, Traynelis SF. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-D-aspartate NR1/NR2A receptors. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11062–11067. doi: 10.1073/pnas.180307497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong EH, et al. The anticonvulsant MK-801 is a potent N-methyl-D-aspartate antagonist. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:7104–7108. doi: 10.1073/pnas.83.18.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foster AC, Wong EH. The novel anticonvulsant MK-801 binds to the activated state of the N-methyl-D-aspartate receptor in rat brain. British journal of pharmacology. 1987;91:403–409. doi: 10.1111/j.1476-5381.1987.tb10295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lü W, Du J, Goehring A, Gouaux E. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science (New York, NY) 2017;355 doi: 10.1126/science.aal3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huettner JE, Bean BP. Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: selective binding to open channels. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:1307–1311. doi: 10.1073/pnas.85.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobolevsky AI. Two-component blocking kinetics of open NMDA channels by organic cations. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1999;1416:69–91. doi: 10.1016/s0005-2736(98)00211-9. [DOI] [PubMed] [Google Scholar]
- 46.Sobolevsky A, Koshelev S. Two Blocking Sites of Amino-Adamantane Derivatives in Open N-Methyl-d-Aspartate Channels. Biophysical journal. 1998;74:1305–1319. doi: 10.1016/S0006-3495(98)77844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanpied TA, Boeckman FA, Aizenman E, Johnson JW. Trapping Channel Block of NMDA-Activated Responses By Amantadine and Memantine. Journal of Neurophysiology. 1997;77:309–323. doi: 10.1152/jn.1997.77.1.309. [DOI] [PubMed] [Google Scholar]
- 48.Mortensen M, Smart TG. Single-channel recording of ligand-gated ion channels. Nat Protocols. 2007;2:2826–2841. doi: 10.1038/nprot.2007.403. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Wolynes P. Instantons and the Fluctuating Path Description of Reactions in Complex Environments. The Journal of Physical Chemistry. 1996;100:1129–1136. [Google Scholar]
- 50.Pirchi M, et al. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nature communications. 2011;2:493. doi: 10.1038/ncomms1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacLean DM, Ramaswamy SS, Du M, Howe JR, Jayaraman V. Stargazin promotes closure of the AMPA receptor ligand-binding domain. J Gen Physiol. 2014;144:503–512. doi: 10.1085/jgp.201411287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaikh SA, et al. Stargazin Modulation of AMPA Receptors. Cell reports. 2016;17:328–335. doi: 10.1016/j.celrep.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nick Taylor J, Darugar Q, Kourentzi K, Willson RC, Landes CF. Dynamics of an anti-VEGF DNA aptamer: a single-molecule study. Biochemical and biophysical research communications. 2008;373:213–218. doi: 10.1016/j.bbrc.2008.05.191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


