Abstract
Background
Plasmodium vivax is one of the leading causes of malaria worldwide. Infections with this parasite cause diverse clinical manifestations, and recent studies revealed that infections with P. vivax can result in severe and fatal disease. Despite these facts, biological traits of the host response and parasite metabolism during P. vivax malaria are still largely underexplored. Parasitemia is clearly related to progression and severity of malaria caused by P. falciparum, however the effects of parasitemia during infections with P. vivax are not well understood.
Results
We conducted an exploratory study using a high-resolution metabolomics platform that uncovered significant associations between parasitemia levels and plasma metabolites from 150 patients with P. vivax malaria. Most plasma metabolites were inversely associated with higher levels of parasitemia. Top predicted metabolites are implicated into pathways of heme and lipid metabolism, which include biliverdin, bilirubin, palmitoylcarnitine, stearoylcarnitine, phosphocholine, glycerophosphocholine, oleic acid and omega-carboxytrinor-leukotriene B4.
Conclusions
The abundance of several plasma metabolites varies according to the levels of parasitemia in patients with P. vivax malaria. Moreover, our data suggest that the host response and/or parasite survival might be affected by metabolites involved in the degradation of heme and metabolism of several lipids. Importantly, these data highlight metabolic pathways that may serve as targets for the development of new antimalarial compounds.
Keywords: Metabolomics, host-pathogen interactions, heme, glycerophospholipid
Background
Malaria is a life-threatening vector-borne disease caused by Plasmodium parasites that affects millions of individuals every year (WHO, 2016). Infections with P. vivax account for almost half of all cases of malaria outside Sub-Saharan Africa, leading to significant morbidity worldwide, besides causing severe and fatal disease (Lacerda et al., 2012; Mahgoub et al., 2012). While many efforts have been made to understand the burden and host response to infections with P. falciparum (Mueller et al., 2009), the biologic perturbations induced by infections with P. vivax are still largely unknown. P. falciparum’s blood-stages invade and replicate inside of red blood cells (RBCs) at any age, whereas P. vivax preferentially targets reticulocytes (immature erythrocytes that typically comprise about 1–2% of circulating RBCs) (Lim et al., 2016). A recent study demonstrated that reticulocytes exhibit a more complex metabolic phenotype than mature erythrocytes, suggesting that Plasmodium host cell tropism of distinct species may be caused by differences in the parasite’s intrinsic metabolism (Srivastava et al., 2015). The P. vivax preference for reticulocytes contributes to lower parasitemias, which likely accounts for slower progression of the disease and reduced lethality rates of P. vivax malaria when compared with P. falciparum (Anstey et al., 2012; Howes et al., 2016; Mueller et al., 2009).
Hyperparasitemia, defined by a peripheral blood slide showing ≥4% infected RBCs, has been long considered as a criterion of severe falciparum malaria (Severe Malaria, 2014), however, the association between parasitemia and disease severity during P. vivax malaria is not clear. A recent study demonstrated that parasitemia is a poor predictor of three or more severity criteria in P. vivax malaria patients (Siqueira et al., 2015). Nevertheless, high parasitemia was associated with fatal disease (Valecha et al., 2009), while another study demonstrated that peripheral P. vivax biomass has been underestimated and is associated with both systemic inflammation and disease severity (Barber et al., 2015). Indeed, increased parasitemia reflects the availability of an optimal nutritional environment together with the inability of the host to limit and control parasite growth. In this context, parasitemia correlates with levels of inflammatory and immunomodulatory mediators in the plasma of individuals with P. vivax malaria (Barber et al., 2015; Mendonça et al., 2013). Overall, these findings suggest that progression of P. vivax malaria can be affected by the density of parasites, and indicate that the metabolic response of the host and pathogen are also influenced by the parasite biomass during infections with P. vivax.
Metabolomics represents a powerful analytical approach to uncover the activity of physiological and pathological processes (Li et al., 2016) for the discovery of biomarkers of infectious diseases, including malaria (Park et al., 2015; Salinas et al., 2014). Nuclear magnetic resonance or gas/liquid chromatography coupled with mass spectrometry (GC/LC-MS) have been applied to understand the metabolic changes in models of host-Plasmodium interactions in vitro (Lakshmanan et al., 2012; MacRae et al., 2013; O’Hara et al., 2014; Olszewski et al., 2009; Park et al., 2015; Sana et al., 2013) and in vivo (Basant et al., 2010; Ghosh et al., 2012, 2013; Olszewski et al., 2009; Sengupta et al., 2013; Tritten et al., 2013). However, studies of human malaria are limited to evaluation of plasma from patients infected with P. falciparum (Lakshmanan et al., 2012; Surowiec et al., 2015; Sengupta et al., 2016) or urine from patients infected with P. vivax (Sengupta et al., 2011).
In view of the lack of understanding about host-pathogen interactions during infections with P. vivax, we used a high resolution metabolomics platform to investigate the associations between parasitemia and the plasma metabolome from patients with P. vivax malaria. The abundance of several metabolites varied according to the levels of parasitemia, whereby top predicted metabolites are involved in the metabolism of heme and lipids. This indicates that perturbations of these metabolic pathways are linked to the dynamics of parasite burden, and they may have an impact on the host response and parasite survival.
Methods
Study population and clinical evaluation
In this study, we used retrospective plasma samples obtained from patients with P. vivax malaria at the Fundação de Medicina Tropical Dr Heitor Vieira Dourado (FMT-HVD), as reported previously (Melo et al., 2014; Siqueira et al., 2015). Plasma samples were obtained under standard laboratorial procedures and stored at −80 ºC. Samples were shipped in dry ice to the Clinical Biomarkers Laboratory at Emory University, where they were also stored at −80 ºC. Briefly, all patients underwent an initial clinical characterization and physical examination followed by antimalarial treatment according to appropriate guidelines. At admission, laboratorial evaluation consisted of full blood count and biochemical analyses (hemoglobin, alanine aminotransferase, aspartate aminotransferase and creatinine), as reported (Melo et al., 2014; Siqueira et al., 2015). Diagnosis and quantification of parasitemia were performed with thick blood smear (TBS) microscopy. Each blood slide was analyzed independently at least two times, whereby the number of asexual and sexual parasites were quantified by counting either 500 leukocytes or 500 parasites. Parasitemia was determined by the mean of two readings of parasites and the white blood cell counts from total blood. Mono-infections with P. vivax were confirmed by PCR (Melo et al., 2014; Siqueira et al., 2015). Patients were classified into three categories of parasitemia: low (10–999 parasites/μL), moderate (1,000–9,999 parasites/μL) and high (> 10,000 parasites/μL) according to previous studies (Ketema and Bacha, 2013; Kotepui et al., 2015).
Liquid chromatography and high-resolution mass spectrometry
Sample processing and high-resolution mass spectrometry analysis was performed as described previously (Park et al., 2012; Soltow et al., 2013; Go et al., 2015b; Jin et al., 2016; Walker et al., 2016; Cribbs et al., 2016; Li et al., 2017). Acetonitrile (2:1, v/v) was added to 65 μL of plasma containing 14 stable isotope internal standards ([13C6]-D-glucose, [15N]-indole, [2-15N]-L-lysine dihydrochloride, [13C5]-L-glutamic acid, [13C7]-benzoic acid, [3,4-13C2] cholesterol, [15N]-L-tyrosine, [trimethyl-13C3]-caffeine, [15N2]-uracil, [3,3-13C2]-cystine, [1,2-13C2]-palmitic acid, [15N, 13C5]-L-methionine, [15N]-choline chloride, and 2′-deoxyguanosine-15N2,13C10-5′-monophosphate). Proteins were removed by centrifugation (13,200 x rpm at 4 °C) for 10 min. The resulting supernatant was transferred to an autosampler vial for LC-MS analysis, using a LTQ Velos Orbitrap mass spectrometer (Thermo Fisher). Injection volume was 10 μl for each run. Reverse phase chromatography was accomplished using a 100x2.1mm, 5 μM C18 column (Higgins Analytical) and, an acetonitrile gradient (where FA = 2% formic acid, W = water, ACN = acetonitrile). During the first 2 minutes, the gradient consisted of 5% FA, 60% W and 35% ACN, followed by an 8 min gradient of 5% FA, 0% W and 95% ACN. During the first 6 min, the flow rate was 0.35 ml/min and then changed to 0.5 ml/min for the remaining 4 min. Before a new injection, the column was subjected to solution consisting of 2% formic acid in acetonitrile and equilibrated to initial conditions for 2 min. Mass spectral data was acquired with positive electrospray ionization and the full scan of mass-to-charge ratio (m/z) ranged from 85 to 2000 at a resolution of 60,000. Operating conditions were used as follow: spray voltage of 4.5 kV, sheath gas flow 45 (arbitrary units), auxiliary gas flow of 5 (arbitrary units), capillary temperature of 275°C, maximum injection time of 500 milliseconds and AGC target of 5 x 105. Xcalibur software (Thermo Fisher Scientific) was used to convert raw files to .cdf format and peak detection, noise filtering, m/z, and retention time alignment, and feature quantification were performed with apLCMS (Yu et al., 2009) and xMSanalyzer (Uppal et al., 2013). Quality control samples (pooled human plasma used to evaluate instrument stability, without clinical significance) was included in every batch of 20 samples, and each sample was run in triplicate. Each metabolite feature is defined by m/z and retention time, with intensity values associated with each replicate. Further quality control included the exclusion of technical replicates with overall Pearson correlation coefficient r <0.7. Data were averaged among analytical replicates, log2 transformed and normalized by the mean. Only features detected in more than 70% of all samples (3,670) were used in further analysis. Missing values were imputed using half mean of the feature across all samples.
Five different levels of metabolite identification are observed in the scientific literature (Schrimpe-Rutledge et al., 2016; Schymanski et al., 2014; Sumner et al., 2007). These include metabolite identification through comparison of 2 or more orthogonal properties between experimental data and authenticated chemical standards (level 1); putatively annotated compounds matching to both m/z and retention time of a previously characterized authenticated chemical standard (level 2); putatively annotated compounds matching to m/z from databases (level 3); putatively characterized compound classes (level 4), and unknown compounds (level 5). Therefore, putative annotation (level 3) of top significant metabolite features was determined by m/z matching to METLIN and KEEG databases (mass accuracy under 10 ppm, including multiple adducts). Over forty metabolites in this dataset matched to our in-house library constructed with LC-MS/MS of commercially available reference standards (annotation of level 2 confidence), including palmitoylcarnitine, stearoylcarnitine, which were significantly associated with P. vivax parasitemia. The mummichog software (version 1.0.7) was used for pathway analysis (mass accuracy under 10 ppm) (Li et al., 2013). The metabolomics data used in this study are available at the public repository Metabolomics Workbench, under the accession number ST000578.
Statistical analyses
Statistical analyses were carried out using the R Language and Environment for Statistical Computing (R) 3.2.0 (Ihaka and Gentleman, 1996). Only data from patients with P. vivax malaria were used in statistical analyses. ANOVA or Kruskal-Wallis followed by Dunn’s pairwise multiple comparisons procedure were used to identify significant differences between independent groups of parasitemia. Linear regression models adjusted by age and gender were used to determine the associations between plasma metabolites and parasitemia. False discovery rate (FDR) was computed using Benjamini-Hochberg method. Spearman’s rank correlations were used to evaluate associations between parasitemia and clinical features. Euclidian distance method and ward linkage algorithm were used for hierarchical clustering.
Results and Discussion
Study subjects and clinical characteristics
All individuals in this study, which included 50 females and 100 males with mean age of 35.9 years and standard deviation of 15.3, presented symptoms of malaria and had confirmed mono-infection with P. vivax. Evaluation of clinical data between categorical groups of parasitemia identified significant differences in levels of hemoglobin (P = 0.0005), RBC (P < 0.0001), white blood cell (WBC) (P = 0.0055), platelets (P = 0.0211), ALT (P = 0.033) and AST (P = 0.034). Consistent with previous reports, we also identified significant correlations between parasitemia and levels of clinical parameters that include hemoglobin, RBCs, WBCs and platelets (Ketema and Bacha, 2013; Kotepui et al., 2015; Demissie and Ketema, 2016; Rodrigues-da-Silva et al., 2014). Demographics and clinical characteristics of the study population are described in Table 1.
Table 1.
Demographics and clinical characteristics of the study population
| Parasitemia classification | Low (n = 50) (10–999 parasites/μL) | Moderate (n = 61) (1,000–9,999 parasites/μL) | High (n = 39) (> 10,000 parasites/μL) |
|---|---|---|---|
| Age, y | 37.7 (13.4) | 36.2 (16.0) | 33.1 (16.2) |
| Male, n(%) | 34 (68) | 44 (72) | 22 (56) |
| Female, n(%) | 16 (32) | 17 (28) | 17 (44) |
| Hemoglobin (g/dL) | 13.0 (2.2) | 13.0 (2.2) | 10.9 (3.0) |
| RBC (106/μL) | 4.7 (0.9) | 4.6 (0.8) | 3.9 (1.0) |
| WBC (103/μL) | 4.6 (1.8) | 5.7 (2.8) | 5.6 (2.0) |
| Platelets (103/μL) | 117.5 (96.8) | 94.7 (71.4) | 94.6 (114.3) |
| ALT (U/L) | 72.1 (58.3 | 62.6 (75.8) | 36.2 (31.2) |
| AST (U/L) | 105.9 (84.3) | 77.6 (78.8) | 68.5 (62.7) |
| Creatinine (mg/dL) | 0.8 (0.5) | 0.8 (0.4) | 0.8 (0.4) |
Mean values and standard deviation are shown. RBC – red blood cells; WBC – white blood cells; ALT - alanine aminotransferase; AST - aspartate aminotransferase.
Significant associations between plasma metabolites and P. vivax parasitemia
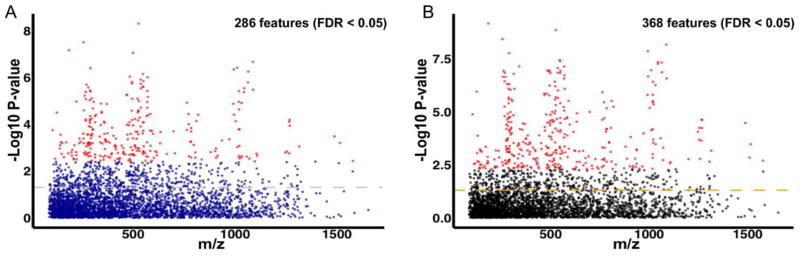
Our untargeted, high-resolution metabolomics platform measured over 20,000 metabolite features. After filtering by missing values, 3,670 features were used in the subsequent analysis. Unsupervised principal component analysis (PCA) of all samples revealed a clear pattern of two clusters among malaria patients and of replicates of quality control samples (Fig. S1), which demonstrates the robustness of the analytical method. Using the method of MWAS (Metabolome Wide Association Study (Bictash et al., 2010; Chadeau-Hyam et al., 2010; Nicholson et al., 2008), we investigated the metabolic phenotypes associated with distinct levels of parasitemia in P. vivax malaria. Categorical comparison of the plasma metabolome of patients with distinct levels of parasitemia using a univariate analysis of variance (ANOVA) identified the differential abundance of 286 metabolite features with FDR < 0.05 (Fig. 1A). One-way hierarchical clustering of highly significant metabolite features (50 features with FDR < 0.001) demonstrates that the intensity of plasma metabolites is mainly reduced with higher levels of parasitemia (Fig. 2A). To gather further insights into the associations between plasma metabolites with distinct levels of parasitemia, we fitted linear regression models adjusted by age and gender using parasitemia as a continuous variable. This identified 368 metabolite features associated with levels of parasitemia with FDR < 0.05 (Fig. 1B). Of note, linear regression models resulted in twice as many of highly significant metabolite features (103 features with FDR <0.001), whereby hierarchical clustering analysis retrieved two major clusters, one small cluster of metabolites with intensities increasing with elevated levels of parasitemia (Fig. 2B); and one composed of several sub-clusters, confirming that the majority of plasma metabolites are reduced with increasing levels of parasitemia (Fig. 2B). Over 230 metabolite features were selected by both statistical methods (Fig S2). However, corrections for age and gender in linear regression analyses retrieved a larger number of metabolite features (Fig. 1B and Fig. S2). Biological factors such as age or gender influence the plasma metabolome of humans (Jové et al., 2015; Psychogios et al., 2011) and could impact the results from univariate ANOVA. Therefore, further analyses were focused on metabolite features selected by linear regression models, while those selected by ANOVA were retained only for comparison purposes.
Figure 1.
Significant associations between plasma metabolites and P. vivax parasitemia. (A) m/z features selected with univariate analysis of variance (ANOVA). (B) m/z features selected with linear regression adjusted by age and gender. Significant features were identified with a FDR < 0.05 and colored in red. Dashed lines represent a P-value of 0.05.
Figure 2.
Dynamics of the abundance of metabolite features associated with P. vivax parasitemia. (A) One-way hierarchical clustering based on the intensity of highly significant metabolite features selected by ANOVA (FDR < 0.001, 50 m/z features). (B) One-way hierarchical clustering based on the intensity of highly significant metabolite features selected by liner regression models (FDR < 0.001, 103 m/z features). The yellow to red scale indicates lower to higher intensity levels based on a Z-score.
By m/z matching to METLIN database (Smith et al., 2005), putative annotations (level 3) of top significant metabolites shown in Fig. 1 and Fig. 2 include phospholipids such as phosphocholine, glycerophosphocholine and several lysophosphatidylcholines (lysoPC). Of interest, the second most significant peak selected by linear regression (m/z - 526.3279, P-value = 1.37E-09, FDR = 2.52E-06) matched to a cyanobacterium toxin denominated antillatoxin A. The possibility of compounds similar to a cyanobacterial metabolite (not necessarily antillatoxin) being produced by P. vivax parasites and, secreted into the bloodstream during the course of the infection is intriguing, as Plasmodium’s apicoplast enzymes share high similarity to that of cyanobacteria (Okada, 2009).
To gather another perspective about the nature of top significant metabolite features (Fig. 2), m/z matching was performed with the KEEG database. This returned 106 or 231 matches to features depicted in Fig 2A or Fig 2B, respectively (Supplementary Table 1). Several m/z peaks matched to more than one compound and, a large number of features did not match to any chemical entity in any database. Considering the challenges of metabolomics analyses, such as metabolite identification, a systematic and comprehensive methodological approach is required for appropriate interpretation and conclusions about these data (Li et al., 2013).
Pathways of heme and lipid metabolism are associated with P. vivax parasitemia
A recent development in the metabolomics field is the mummichog software, which uses pathway and network patterns to prioritize metabolite annotation (Li et al., 2013). We examined the biological context reflected by the metabolic profiles associated with P. vivax parasitemia using this computational tool. The most significant pathways overrepresented by metabolite features selected with both ANOVA and linear regression models include the porphyrin and heme metabolism, carnitine shuttle and glycerophospholipid metabolism (Table 2). Of note, metabolite features selected with univariate ANOVA identified an enrichment of the bile acid biosynthesis pathway that was not identified from the metabolites selected by linear regression. Accordingly, this result highlight that the synthesis of bile acids is indeed influenced by age (Einarsson et al., 1985), which indicates an advantage and higher accuracy of using linear regression models adjusted for confounding factors such as age and gender.
Table 2.
Metabolic pathway analysis
| Features selected with ANOVA | Features selected with linear regression | |||
|---|---|---|---|---|
| Pathways | Overlap size | P value | Overlap size | P value |
| Porphyrin and heme metabolism | 3 | 0.00427 | 7 | < 0.0001 |
| Carnitine shuttle | 4 | 0.0025 | 6 | < 0.0001 |
| Bile acid biosynthesis | 5 | 0.0022 | ||
| Glycerophospholipid metabolism | 4 | 0.0025 | 4 | 0.00056 |
| Glycosphingolipid metabolism | 3 | 0.00777 | 3 | 0.00181 |
| Sialic acid metabolism | 3 | 0.01356 | 3 | 0.00334 |
| Fatty acid activation | 3 | 0.00074 | ||
| De novo fatty acid biosynthesis | 3 | 0.00074 | ||
| Squalene and cholesterol biosynthesis | 3 | 0.00146 | ||
| Xenobiotics metabolism | 3 | 0.00484 | ||
| Tyrosine metabolism | 3 | 0.02098 | ||
Only pathways with three or more overlapping features are shown.
Some of the top predicted metabolites such as biliverdin, bilirubin, bilirubin-glucoronoside and bilirubin beta-diglucuronide are involved in porphyrin and heme metabolism (Table 2). The m/z peak matching to bilirubin was annotated with a confidence level 2, as this feature also matched to retention time and m/z of an authenticated chemical standard characterized previously in our laboratory (Go et al., 2015a). These metabolites exhibited higher abundance in the plasma of patients with increased levels of parasitemia (Fig. 3). Of interest, malaria has been characterized by hemolysis of both infected and uninfected erythrocytes (Fonseca et al., 2016; Joyner et al., 2016; Severe Malaria, 2014), leading to the release of cell-free hemoglobin and further heme prosthetic groups (Pamplona et al., 2007). The relatively low parasitemia observed in infections with P. vivax, and their preference to reticulocytes questions the relative contributions of parasite burden to hemolysis, especially of uninfected RBCs. However, several studies indicate that P. vivax exhibits a lower threshold of parasitemia associated with fever (Anstey et al., 2009; Karyana et al., 2008), and induces a greater inflammatory response when compared to P. falciparum (Karunaweera et al., 1992; Hemmer et al., 2006; Anstey et al., 2007). Moreover, the stability of RBCs are profoundly affected during active disease (Handayani et al., 2009; Lee et al., 2014). Noteworthy, patients with P. vivax malaria exhibit elevated levels of cell-free hemoglobin in plasma (Barber et al., 2016) and up-regulate levels of heme oxygenase-1 (HO-1) (Mendonça et al., 2013), which breaks down heme into biliverdin, carbon monoxide and iron. Levels of HO-1 and several other immunomodulatory and inflammatory mediators also correlate with P. vivax parasitemia during severe disease (Mendonça et al., 2013). Our data extend these findings, indicating that the abundance of metabolites generated by the metabolism of heme change according to levels of blood parasitemia. This association can be complex and affect the host in different ways. Heme oxygenase-1 is crucial for the survival of mouse models of malaria (Pamplona et al., 2007; Seixas et al., 2009), antagonizing the pathogenic effects of free heme, such as oxidative stress (Vinchi et al., 2013) and excessive inflammation (Dutra et al., 2014). However, another perspective is given by the direct immunomodulatory properties of biliverdin and bilirubin (Liu et al., 2008; Wegiel and Otterbein, 2012) and the fact that Plasmodium parasites also metabolize heme into bilirubin (Okada, 2009). This has the potential to impact the function of leukocytes recruited to limit replication and facilitate parasite evasion.
Figure 3.
Distribution of top predicted metabolites in patients with low, moderate and high levels of P. vivax parasitemia. Additional statistics were performed with Kruskal-Wallis followed by Dunn’s pairwise multiple comparisons procedure; mean values, standard deviation (SD) and significance levels are shown (*, P < 0.05 and **, P < 0.01).
Previous studies identified the differential abundance of serum lipids in malaria patients (Mesquita et al., 2016; Visser et al., 2013), however the mechanisms underlying this phenomenon are not clear. Our data provide insights into this process by demonstrating significant associations between parasitemia and pathways such as the carnitine shuttle and glycerophospholipid metabolism (Table 2). The abundance of metabolites putatively annotated as palmitoylcarnitine, stearoylcarnitine, heptadecanoyl carnitine and docosa-pentaenoyl carnitine decreased with higher levels of P. vivax parasitemia (Fig. 3). Both palmitoylcarnitine and stearoylcarnitine were also confirmed (annotation confidence level 2) by matched retention time and m/z to authenticated chemical standards characterized previously in our laboratory (Uppal et al., 2017). Of interest, those metabolites are involved in the carnitine shuttle pathway. The transfer of long-chain fatty acids across the inner mitochondrial membrane for β-oxidation is mediated by the carnitine shuttle pathway, whereas decreased abundance of related metabolites identified here might reflect increased uptake of host fatty acids by the parasite. Consistent with this hypothesis, we observed that levels of putatively annotated metabolites such as glycerophosphocholine and phosphocholine were also reduced with higher levels of parasitemia (Fig. 3). In contrast, we identified a significant increase in the abundance of the m/z peak 283.2627 (putatively annotated as oleic acid) with higher levels of parasitemia (Fig. 3), a fatty acid that is required for the intra-erythrocytic proliferation of P. falciparum (Mi-Ichi et al., 2007) and might also be essential for the growth of P. vivax. Indeed, malarial parasites scavenge, modify, and incorporate fatty acids and phospholipids from the host (Moll et al., 1988; Krishnegowda and Gowda, 2003), whereas elevated P. vivax parasitemia correlates with reduced levels of low and high-density lipoprotein in the serum of patients (Mesquita et al., 2016). Lipids are essential nutrients for parasite’s proliferation, and for the conversion of heme into hemozoin (Ambele and Egan, 2012; Pisciotta et al., 2007), which is a detoxification strategy used by the parasite to survive inside the RBC while digesting hemoglobin. Our data highlight the importance of lipid metabolism during malaria episodes, while supporting the development of antimalarial compounds targeting pathways related to the uptake and metabolism of fatty acids and phospholipids (Ben Mamoun et al., 2010).
The metabolite tentatively annotated as omega-carboxy-trinor-leukotriene B4 (OCTLB4) exhibited decreased abundance with higher levels of parasitemia (Fig. 3). This eicosanoid has been associated with peroxisome deficiency disorders and chronic kidney disease (Mayatepek et al., 1993; Zhang et al., 2016); more recently it was detected in resting human platelets, but not in thrombin activated platelets (Slatter et al., 2016). While leukotriene B4 (LTB4) is an inflammatory molecule and chemoattractant that mediates the migration of neutrophils induced by heme (Monteiro et al., 2011), omega-oxidation is the main pathway used by human neutrophils to catabolize LTB4, regulating the inflammatory profiles of these cells (Shak and Goldstein, 1984). Taken together, those results suggest that the inflammatory stimulus induced by higher levels of parasitemia could impact the metabolism and signaling of activated leukocytes or platelets, leading to reduced abundance of OCTLB4.
Conclusions
In this study, we obtained high-resolution metabolomes of patients with P. vivax malaria, and successfully identified metabolites and pathways that are associated with parasitemia. Our data indicate that individuals with high parasitemia display increased activity heme degradation, and an overall reduction in the abundance of several lipids in the plasma. These results provide important insights into the host metabolic responses in P. vivax malaria. Despite of relatively low parasitemia, the inflammatory response induced by P. vivax might be responsible for the associations identified in this study. Future studies applying high-resolution metabolomics platforms will further enhance the understanding of the biology of infections with P. vivax and contribute to the identification of new drug targets and biomarkers.
Supplementary Material
Acknowledgments
Funding This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases; National Institutes of Health, Department of Health and Human Services [Contract No. HHSN272201200031C and NIH1S10OD18006-01], and the National Center for Research Resources [ORIP/OD P51OD011132]. J.L.S. and F.F.V. were supported by the Fogarty International Center Global Health Fellowship [R25TW009337]. M.V.G.L. and W.M.M. were supported by CAPES, CNPq and FAPEAM.
We thank Karan Uppal for assistance with LC-MS data processing and discussion, and Esmeralda VS Meyer for administrative assistance. We also thank the members of the Malaria Host Pathogen Interaction Center for assistance with data management.
Footnotes
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The study was approved by the Ethics Institutional Review Board (IRB) from the Fundação de Medicina Tropical Dr Heitor Vieira Dourado and CONEP, Manaus, Brazil (IRB approval #: CAAE: 12516713.8.0000.0005); and all protocols and documentation were reviewed and sample shipments approved by the Emory IRB. Written informed consent was obtained from all of the participants or their parents or legal guardians.
Authors' contributions L.G.G. performed the data analysis and wrote the manuscript. J.L.S., F.F.V., M.V.G.L., W.M.M., G.C.M. and A.M. S. performed the clinical study, including patient assessment and sample collection. V.T. performed the metabolomics sample analysis. R.J.C and S.L. supervised the data analysis. M.V.G.L., W.M.M., D.P.J., M.R.G. and S.L. designed and supervised the study. All authors participated in manuscript preparation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambele MA, Egan TJ. Neutral lipids associated with haemozoin mediate efficient and rapid β-haematin formation at physiological pH, temperature and ionic composition. Malar J. 2012;11:337. doi: 10.1186/1475-2875-11-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstey NM, Douglas NM, Poespoprodjo JR, Price RN. Plasmodium vivax: clinical spectrum, risk factors and pathogenesis. Adv Parasitol. 2012;80:151–201. doi: 10.1016/B978-0-12-397900-1.00003-7. [DOI] [PubMed] [Google Scholar]
- Anstey NM, Handojo T, Pain MCF, Kenangalem E, Tjitra E, Price RN, Maguire GP. Lung Injury in Vivax Malaria: Pathophysiological Evidence for Pulmonary Vascular Sequestration and Posttreatment Alveolar-Capillary Inflammation. J Infect Dis. 2007;195:589–596. doi: 10.1086/510756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstey NM, Russell B, Yeo TW, Price RN. The pathophysiology of vivax malaria. Trends Parasitol. 2009;25:220–227. doi: 10.1016/j.pt.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Barber BE, William T, Grigg MJ, Parameswaran U, Piera KA, Price RN, Yeo TW, Anstey NM. Parasite biomass-related inflammation, endothelial activation, microvascular dysfunction and disease severity in vivax malaria. PLoS Pathog. 2015;11:e1004558. doi: 10.1371/journal.ppat.1004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber BE, William T, Grigg MJ, Piera KA, Chen Y, Wang H, Weinberg JB, Yeo TW, Anstey NM. Nitric Oxide–Dependent Endothelial Dysfunction and Reduced Arginine Bioavailability in Plasmodium vivax Malaria but No Greater Increase in Intravascular Hemolysis in Severe Disease. J Infect Dis. 2016;214:1557–1564. doi: 10.1093/infdis/jiw427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basant A, Rege M, Sharma S, Sonawat HM. Alterations in urine, serum and brain metabolomic profiles exhibit sexual dimorphism during malaria disease progression. Malar J. 2010;9:110. doi: 10.1186/1475-2875-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Mamoun C, Prigge ST, Vial H. Targeting the Lipid Metabolic Pathways for the Treatment of Malaria. Drug Dev Res. 2010;71:44–55. doi: 10.1002/ddr.20347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bictash M, Ebbels TM, Chan Q, Loo RL, Yap IKS, Brown IJ, de Iorio M, Daviglus ML, Holmes E, Stamler J, Nicholson JK, Elliott P. Opening up the “Black Box”: metabolic phenotyping and metabolome-wide association studies in epidemiology. J Clin Epidemiol. 2010;63:970–979. doi: 10.1016/j.jclinepi.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadeau-Hyam M, Ebbels TMD, Brown IJ, Chan Q, Stamler J, Huang CC, Daviglus ML, Ueshima H, Zhao L, Holmes E, Nicholson JK, Elliott P, De Iorio M. Metabolic profiling and the metabolome-wide association study: significance level for biomarker identification. J Proteome Res. 2010;9:4620–4627. doi: 10.1021/pr1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs SK, Uppal K, Li S, Jones DP, Huang L, Tipton L, Fitch A, Greenblatt RM, Kingsley L, Guidot DM, Ghedin E, Morris A. Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome. 2016;4:3. doi: 10.1186/s40168-016-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demissie Y, Ketema T. Complicated malaria symptoms associated with Plasmodium vivax among patients visiting health facilities in Mendi town, Northwest Ethiopia. BMC Infect Dis. 2016;16:436. doi: 10.1186/s12879-016-1780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra FF, Alves LS, Rodrigues D, Fernandez PL, de Oliveira RB, Golenbock DT, Zamboni DS, Bozza MT. Hemolysis-induced lethality involves inflammasome activation by heme. Proc Natl Acad Sci U S A. 2014;111:E4110–4118. doi: 10.1073/pnas.1405023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsson K, Nilsell K, Leijd B, Angelin B. Influence of age on secretion of cholesterol and synthesis of bile acids by the liver. N Engl J Med. 1985;313:277–282. doi: 10.1056/NEJM198508013130501. [DOI] [PubMed] [Google Scholar]
- Fonseca LL, Alezi HS, Moreno A, Barnwell JW, Galinski MR, Voit EO. Quantifying the removal of red blood cells in Macaca mulatta during a Plasmodium coatneyi infection. Malar J. 2016;15:410. doi: 10.1186/s12936-016-1465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sengupta A, Sharma S, Sonawat HM. Metabolic Perturbations of Kidney and Spleen in Murine Cerebral Malaria: 1H NMR-Based Metabolomic Study. PLOS ONE. 2013;8:e73113. doi: 10.1371/journal.pone.0073113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sengupta A, Sharma S, Sonawat HM. Metabolic fingerprints of serum, brain, and liver are distinct for mice with cerebral and noncerebral malaria: a 1H NMR spectroscopy-based metabonomic study. J Proteome Res. 2012;11:4992–5004. doi: 10.1021/pr300562m. [DOI] [PubMed] [Google Scholar]
- Go YM, Liang Y, Uppal K, Soltow QA, Promislow DEL, Wachtman LM, Jones DP. Metabolic Characterization of the Common Marmoset (Callithrix jacchus) PLOS ONE. 2015a;10:e0142916. doi: 10.1371/journal.pone.0142916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y-M, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V, Strobel F, Quyyumi AA, Ziegler TR, Pennell KD, Miller GW, Jones DP. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol Sci Off J Soc Toxicol. 2015b;148:531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handayani S, Chiu DT, Tjitra E, Kuo JS, Lampah D, Kenangalem E, Renia L, Snounou G, Price RN, Anstey NM, Russell B. High Deformability of Plasmodium vivax Infected Red Blood Cells under Microfluidic Conditions. J Infect Dis. 2009;199:445–450. doi: 10.1086/596048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmer CJ, Holst FGE, Kern P, Chiwakata CB, Dietrich M, Reisinger EC. Stronger host response per parasitized erythrocyte in Plasmodium vivax or ovale than in Plasmodium falciparum malaria. Trop. Med. Int. Health TM IH. 2006;11:817–823. doi: 10.1111/j.1365-3156.2006.01635.x. [DOI] [PubMed] [Google Scholar]
- Howes RE, Battle KE, Mendis KN, Smith DL, Cibulskis RE, Baird JK, Hay SI. Global Epidemiology of Plasmodium vivax. Am J Trop Med Hyg. 2016:16–0141. doi: 10.4269/ajtmh.16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. J Comput Graph Stat. 1996;5:299–314. doi: 10.1080/10618600.1996.10474713. [DOI] [Google Scholar]
- Jin R, Banton S, Tran VT, Konomi JV, Li S, Jones DP, Vos MB. Amino Acid Metabolism is Altered in Adolescents with Nonalcoholic Fatty Liver Disease-An Untargeted, High Resolution Metabolomics Study. J Pediatr. 2016;172:14–19.e5. doi: 10.1016/j.jpeds.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jové M, Maté I, Naudí A, Mota-Martorell N, Portero-Otín M, Fuente M, Dla Pamplona R. Human Aging Is a Metabolome-related Matter of Gender. J Gerontol A Biol Sci Med Sci. 2015:glv074. doi: 10.1093/gerona/glv074. [DOI] [PubMed] [Google Scholar]
- Joyner C, Moreno A, Meyer EVS, Cabrera-Mora M, Kissinger JC, Barnwell JW, Galinski MR. Plasmodium cynomolgi infections in rhesus macaques display clinical and parasitological features pertinent to modelling vivax malaria pathology and relapse infections. Malar J. 2016;15:451. doi: 10.1186/s12936-016-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunaweera ND, Grau GE, Gamage P, Carter R, Mendis KN. Dynamics of fever and serum levels of tumor necrosis factor are closely associated during clinical paroxysms in Plasmodium vivax malaria. Proc Natl Acad Sci U S A. 1992;89:3200–3203. doi: 10.1073/pnas.89.8.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyana M, Burdarm L, Yeung S, Kenangalem E, Wariker N, Maristela R, Umana KG, Vemuri R, Okoseray MJ, Penttinen PM, Ebsworth P, Sugiarto P, Anstey NM, Tjitra E, Price RN. Malaria morbidity in Papua Indonesia, an area with multidrug resistant Plasmodium vivax and Plasmodium falciparum. Malar J. 2008;7:148. doi: 10.1186/1475-2875-7-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketema T, Bacha K MaHPIC Consortium. Plasmodium vivax associated severe malaria complications among children in some malaria endemic areas of Ethiopia. BMC Public Health. 2013;13:637. doi: 10.1186/1471-2458-13-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotepui M, Piwkham D, PhunPhuech B, Phiwklam N, Chupeerach C, Duangmano S. Effects of malaria parasite density on blood cell parameters. PloS One. 2015;10:e0121057. doi: 10.1371/journal.pone.0121057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnegowda G, Gowda DC. Intraerythrocytic Plasmodium falciparum incorporates extraneous fatty acids to its lipids without any structural modification. Mol Biochem Parasitol. 2003;132:55–58. doi: 10.1016/j.molbiopara.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Lacerda MVG, Fragoso SCP, Alecrim MGC, Alexandre MAA, Magalhães BML, Siqueira AM, Ferreira LCL, Araújo JR, Mourão MPG, Ferrer M, Castillo P, Martin-Jaular L, Fernandez-Becerra C, del Portillo H, Ordi J, Alonso PL, Bassat Q. Postmortem characterization of patients with clinical diagnosis of Plasmodium vivax malaria: to what extent does this parasite kill? Clin Infect Dis Off Publ Infect Dis Soc Am. 2012;55:e67–74. doi: 10.1093/cid/cis615. [DOI] [PubMed] [Google Scholar]
- Lakshmanan V, Rhee KY, Wang W, Yu Y, Khafizov K, Fiser A, Wu P, Ndir O, Mboup S, Ndiaye D, Daily JP. Metabolomic analysis of patient plasma yields evidence of plant-like α-linolenic acid metabolism in Plasmodium falciparum. J Infect Dis. 2012;206:238–248. doi: 10.1093/infdis/jis339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Malleret B, Lau YL, Mauduit M, Fong MY, Cho JS, Suwanarusk R, Zhang R, Albrecht L, Costa FTM, Preiser P, McGready R, Renia L, Nosten F, Russell B. Glycophorin C (CD236R) mediates vivax malaria parasite rosetting to normocytes. Blood. 2014;123:e100–109. doi: 10.1182/blood-2013-12-541698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting Network Activity from High Throughput Metabolomics. PLOS Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sullivan NL, Rouphael N, Yu T, Banton S, Maddur MS, McCausland M, Chiu C, Canniff J, Dubey S, Liu K, Tran V, Hagan T, Duraisingham S, Wieland A, Mehta AK, Whitaker JA, Subramaniam S, Jones DP, Sette A, Vora K, Weinberg A, Mulligan MJ, Nakaya HI, Levin M, Ahmed R, Pulendran B. Metabolic Phenotypes of Response to Vaccination in Humans. Cell. 2017;169:862–877. e17. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Todor A, Luo R. Blood transcriptomics and metabolomics for personalized medicine. Comput Struct Biotechnol J. 2016;14:1–7. doi: 10.1016/j.csbj.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Pereira L, Saliba KS, Mascarenhas A, Maki JN, Chery L, Gomes E, Rathod PK, Duraisingh MT. Reticulocyte Preference and Stage Development of Plasmodium vivax Isolates. J Infect Dis. 2016;214:1081–1084. doi: 10.1093/infdis/jiw303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li P, Lu J, Xiong W, Oger J, Tetzlaff W, Cynader M. Bilirubin Possesses Powerful Immunomodulatory Activity and Suppresses Experimental Autoimmune Encephalomyelitis. J Immunol. 2008;181:1887–1897. doi: 10.4049/jimmunol.181.3.1887. [DOI] [PubMed] [Google Scholar]
- MacRae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, Kenny S, Bottova I, Tilley L, McConville MJ. Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol. 2013;11:67. doi: 10.1186/1741-7007-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahgoub H, Gasim GI, Musa IR, Adam I. Severe Plasmodium vivax malaria among sudanese children at New Halfa Hospital, Eastern Sudan. Parasit. Vectors. 2012;5:154. doi: 10.1186/1756-3305-5-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayatepek E, Lehmann WD, Fauler J, Tsikas D, Frölich JC, Schutgens RB, Wanders RJ, Keppler D. Impaired degradation of leukotrienes in patients with peroxisome deficiency disorders. J Clin Invest. 1993;91:881–888. doi: 10.1172/JCI116309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo GC, Monteiro WM, Siqueira AM, Silva SR, Magalhães BML, Alencar ACC, Kuehn A, del Portillo HA, Fernandez-Becerra C, Lacerda MVG. Expression Levels of pvcrt-o and pvmdr-1 Are Associated with Chloroquine Resistance and Severe Plasmodium vivax Malaria in Patients of the Brazilian Amazon. PLOS ONE. 2014;9:e105922. doi: 10.1371/journal.pone.0105922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça VR, Queiroz AT, Lopes FM, Andrade BB, Barral-Netto M. Networking the host immune response in Plasmodium vivax malaria. Malar J. 2013;12:69. doi: 10.1186/1475-2875-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita TC, Martin TGO, Alves ER, Mello MBC, Nery AF, Gomes LT, Fontes CJF. Changes in serum lipid profile in the acute and convalescent Plasmodium vivax malaria: A cohort study. Acta Trop. 2016;163:1–6. doi: 10.1016/j.actatropica.2016.07.010. [DOI] [PubMed] [Google Scholar]
- Mi-Ichi F, Kano S, Mitamura T. Oleic acid is indispensable for intraerythrocytic proliferation of Plasmodium falciparum. Parasitology. 2007;134:1671–1677. doi: 10.1017/S0031182007003137. [DOI] [PubMed] [Google Scholar]
- Moll GN, Vial HJ, Ancelin ML, Op den Kamp JA, Roelofsen B, van Deenen LL. Phospholipid uptake by Plasmodium knowlesi infected erythrocytes. FEBS Lett. 1988;232:341–346. doi: 10.1016/0014-5793(88)80765-8. [DOI] [PubMed] [Google Scholar]
- Monteiro APT, Pinheiro CS, Luna-Gomes T, Alves LR, Maya-Monteiro CM, Porto BN, Barja-Fidalgo C, Benjamim CF, Peters-Golden M, Bandeira-Melo C, Bozza MT, Canetti C. Leukotriene B4 mediates neutrophil migration induced by heme. J Immunol Baltim Md. 2011;1950–186:6562–6567. doi: 10.4049/jimmunol.1002400. [DOI] [PubMed] [Google Scholar]
- Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009;9:555–566. doi: 10.1016/S1473-3099(09)70177-X. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Elliott P. The metabolome-wide association study: a new look at human disease risk factors. J Proteome Res. 2008;7:3637–3638. doi: 10.1021/pr8005099. [DOI] [PubMed] [Google Scholar]
- O’Hara JK, Kerwin LJ, Cobbold SA, Tai J, Bedell TA, Reider PJ, Llinas M. Targeting NAD+ Metabolism in the Human Malaria Parasite Plasmodium falciparum. PLOS ONE. 2014;9:e94061. doi: 10.1371/journal.pone.0094061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K. The novel heme oxygenase-like protein from Plasmodium falciparum converts heme to bilirubin IXalpha in the apicoplast. FEBS Lett. 2009;583:313–319. doi: 10.1016/j.febslet.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Olszewski KL, Morrisey JM, Wilinski D, Burns JM, Vaidya AB, Rabinowitz JD, Llinás M. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamplona A, Ferreira A, Balla J, Jeney V, Balla G, Epiphanio S, Chora Â, Rodrigues CD, Gregoire IP, Cunha-Rodrigues M, Portugal S, Soares MP, Mota MM. Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria. Nat Med. 2007;13:703–710. doi: 10.1038/nm1586. [DOI] [PubMed] [Google Scholar]
- Park YH, Lee K, Soltow QA, Strobel FH, Brigham KL, Parker RE, Wilson ME, Sutliff RL, Mansfield KG, Wachtman LM, Ziegler TR, Jones DP. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295:47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YH, Shi YP, Liang B, Medriano CAD, Jeon YH, Torres E, Uppal K, Slutsker L, Jones DP. High-resolution metabolomics to discover potential parasite-specific biomarkers in a Plasmodium falciparum erythrocytic stage culture system. Malar J. 2015;14:122. doi: 10.1186/s12936-015-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisciotta JM, Coppens I, Tripathi AK, Scholl PF, Shuman J, Bajad S, Shulaev V, Sullivan DJ. The role of neutral lipid nanospheres in Plasmodium falciparum haem crystallization. Biochem J. 2007;402:197–204. doi: 10.1042/BJ20060986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. The Human Serum Metabolome. PLOS ONE. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-da-Silva RN, da Lima JC, Junior, de Fonseca BPFe, Antas PRZ, Baldez A, Storer FL, Santos F, Banic DM, de Oliveira-Ferreira J. Alterations in cytokines and haematological parameters during the acute and convalescent phases of Plasmodium falciparum and Plasmodium vivax infections. Mem. Inst. Oswaldo Cruz. 2014;109:154–162. doi: 10.1590/0074-0276140275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas JL, Kissinger JC, Jones DP, Galinski MR. Metabolomics in the fight against malaria. Mem Inst Oswaldo Cruz. 2014;109:589–597. doi: 10.1590/0074-0276140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana TR, Gordon DB, Fischer SM, Tichy SE, Kitagawa N, Lai C, Gosnell WL, Chang SP. Global Mass Spectrometry Based Metabolomics Profiling of Erythrocytes Infected with Plasmodium falciparum. PLOS ONE. 2013;8:e60840. doi: 10.1371/journal.pone.0060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J Am Soc Mass Spectrom. 2016;27:1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, Hollender J. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol. 2014;48:2097–2098. doi: 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Seixas E, Gozzelino R, Chora A, Ferreira A, Silva G, Larsen R, Rebelo S, Penido C, Smith NR, Coutinho A, Soares MP. Heme oxygenase-1 affords protection against noncerebral forms of severe malaria. Proc Natl Acad Sci U S A. 2009;106:15837–15842. doi: 10.1073/pnas.0903419106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Ghosh S, Basant A, Malusare S, Johri P, Pathak S, Sharma S, Sonawat HM. Global host metabolic response to Plasmodium vivax infection: a 1H NMR based urinary metabonomic study. Malar J. 2011;10:384. doi: 10.1186/1475-2875-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Ghosh S, Das BK, Panda A, Tripathy R, Pied S, Ravindran B, Pathak S, Sharma S, Sonawat HM. Host metabolic responses to Plasmodium falciparum infections evaluated by (1)H NMR metabolomics. Mol Biosyst. 2016;12:3324–3332. doi: 10.1039/c6mb00362a. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Ghosh S, Sharma S, Sonawat HM. 1H NMR Metabonomics Indicates Continued Metabolic Changes and Sexual Dimorphism Post-Parasite Clearance in Self-Limiting Murine Malaria Model. PLOS ONE. 2013;8:e66954. doi: 10.1371/journal.pone.0066954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severe Malaria. Trop Med Int Health. 2014;19:7–131. doi: 10.1111/tmi.12313_2. [DOI] [PubMed] [Google Scholar]
- Shak S, Goldstein IM. Omega-oxidation is the major pathway for the catabolism of leukotriene B4 in human polymorphonuclear leukocytes. J Biol Chem. 1984;259:10181–10187. [PubMed] [Google Scholar]
- Siqueira AM, Lacerda MV, Magalhães BML, Mourão MP, Melo GC, Alexandre MA, Alecrim MG, Kochar D, Kochar S, Kochar A, Nayak K, del Portillo H, Guinovart C, Alonso P, Bassat Q. Characterization of Plasmodium vivax-associated admissions to reference hospitals in Brazil and India. BMC Med. 2015;13:57. doi: 10.1186/s12916-015-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatter DA, Aldrovandi M, O’Connor A, Allen SM, Brasher CJ, Murphy RC, Mecklemann S, Ravi S, Darley-Usmar V, O’Donnell VB. Mapping the Human Platelet Lipidome Reveals Cytosolic Phospholipase A2 as a Regulator of Mitochondrial Bioenergetics during Activation. Cell Metab. 2016;23:930–944. doi: 10.1016/j.cmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics Off J Metabolomic Soc. 2013;9:S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Creek DJ, Evans KJ, Souza DD, Schofield L, Müller S, Barrett MP, McConville MJ, Waters AP. Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites. PLOS Pathog. 2015;11:e1004882. doi: 10.1371/journal.ppat.1004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics Off J Metabolomic Soc. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowiec I, Orikiiriza J, Karlsson E, Nelson M, Bonde M, Kyamanwa P, Karenzi B, Bergström S, Trygg J, Normark J. Metabolic Signature Profiling as a Diagnostic and Prognostic Tool in Pediatric Plasmodium falciparum Malaria. Open Forum Infect Dis. 2015;2:ofv062. doi: 10.1093/ofid/ofv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritten L, Keiser J, Godejohann M, Utzinger J, Vargas M, Beckonert O, Holmes E, Saric J. Metabolic Profiling Framework for Discovery of Candidate Diagnostic Markers of Malaria. Sci Rep. 2013;3:2769. doi: 10.1038/srep02769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, Jones DP. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14:15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K, Walker DI, Jones DP. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal Chem. 2017;89:1063–1067. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valecha N, Pinto RGW, Turner GDH, Kumar A, Rodrigues S, Dubhashi NG, Rodrigues E, Banaulikar SS, Singh R, Dash AP, Baird JK. Histopathology of fatal respiratory distress caused by Plasmodium vivax malaria. Am J Trop Med Hyg. 2009;81:758–762. doi: 10.4269/ajtmh.2009.09-0348. [DOI] [PubMed] [Google Scholar]
- Vinchi F, De Franceschi L, Ghigo A, Townes T, Cimino J, Silengo L, Hirsch E, Altruda F, Tolosano E. Hemopexin therapy improves cardiovascular function by preventing heme-induced endothelial toxicity in mouse models of hemolytic diseases. Circulation. 2013;127:1317–1329. doi: 10.1161/CIRCULATIONAHA.112.130179. [DOI] [PubMed] [Google Scholar]
- Visser BJ, Wieten RW, Nagel IM, Grobusch MP. Serum lipids and lipoproteins in malaria - a systematic review and meta-analysis. Malar J. 2013;12:442. doi: 10.1186/1475-2875-12-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DI, Uppal K, Zhang L, Vermeulen R, Smith M, Hu W, Purdue MP, Tang X, Reiss B, Kim S, Li L, Huang H, Pennell KD, Jones DP, Rothman N, Lan Q. High-resolution metabolomics of occupational exposure to trichloroethylene. Int J Epidemiol. 2016;45:1517–1527. doi: 10.1093/ije/dyw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel B, Otterbein LE. Go Green: The Anti-Inflammatory Effects of Biliverdin Reductase. Front Pharmacol. 2012;3 doi: 10.3389/fphar.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Malaria Report 2016 [WWW Document] WHO; n.d. [accessed 12.15.16]. URL http://www.who.int/malaria/publications/world-malaria-report-2016/report/en/ [Google Scholar]
- Yu T, Park Y, Johnson JM, Jones DP. apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-H, Chen H, Vaziri ND, Mao J-R, Zhang L, Bai X, Zhao Y-Y. Metabolomic Signatures of Chronic Kidney Disease of Diverse Etiologies in the Rats and Humans. J Proteome Res. 2016;15:3802–3812. doi: 10.1021/acs.jproteome.6b00583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.