Abstract
The binding of nitric oxide (NO) to the heme cofactor of heme-nitric oxide/oxygen binding (H-NOX) proteins can lead to the dissociation of the heme-ligating histidine residue and yield a five-coordinate nitrosyl complex, which is an important step for NO-dependent signaling. In the five-coordinate nitrosyl complex, NO can reside either on the distal or proximal side of the heme, which could have a profound influence over the lifetime of the in vivo signal. To investigate this central molecular question, the Shewanella oneidensis H-NOX (So H-NOX)–NO complex was biophysically characterized under limiting and excess NO. The results show that So H-NOX preferably forms a distal NO species under both limiting and excess NO. Therefore, signal strength and complex lifetime in vivo will be dictated by the dissociation rate of NO from the distal complex and the return of the histidine ligand to the heme.
Introduction
Nitric oxide (NO) has emerged as a signaling molecule with selective control over important physiological functions in both prokaryotes and eukaryotes. Given the chemical reactivity and toxicity of NO, using this diatomic gas to bring about a selective biological response represents a formidable challenge. Nature’s answer to this challenge has been the evolution of highly selective and sensitive receptors for NO, such as the soluble isoform of guanylate cyclase (sGC)1. In response to NO, sGC catalyzes the conversion of guanosine 5′-triphosphate (GTP) to guanosine 3′,5′-cyclic monophosphate (cGMP); cGMP then acts as a second messenger that initiates a signaling cascade targeting downstream proteins including cGMP-dependent kinases and cGMP-gated ion channels1,2. In mammals, endothelial cell generation of NO by nitric oxide synthase leads to activation of sGC in smooth muscle cells and subsequent vasodilation3–6. sGC is a heterodimer composed of an α- and β-subunit. The N-terminus of the β-subunit contains a ferrous heme that serves as the NO binding site. CO also binds to the heme, albeit with relatively weak affinity. Remarkably, sGC shows no measurable affinity for O2 despite the presence of a ferrous heme with a histidine axial ligand and an open coordination site at the distal position7.
Sequence-related receptors with ligand binding properties similar to the heme-bound sGC N-terminal domain are also present in prokaryotes8. In addition, some of these bacterial proteins form stable, high-affinity complexes with O29,10. Given these ligand binding properties, these proteins are termed H-NOX (Heme-Nitric oxide/OXygen) domains in anticipation of physiological functions involving NO and O2. The bacterial H-NOXs are often found in an operon with a histidine kinase and could typically inhibit the autophosphorylation of the operon-partner kinase upon NO binding11. The effect of this inhibition on the respective signaling pathways has received some attention, most notably the outcome of biofilm formation12,13.
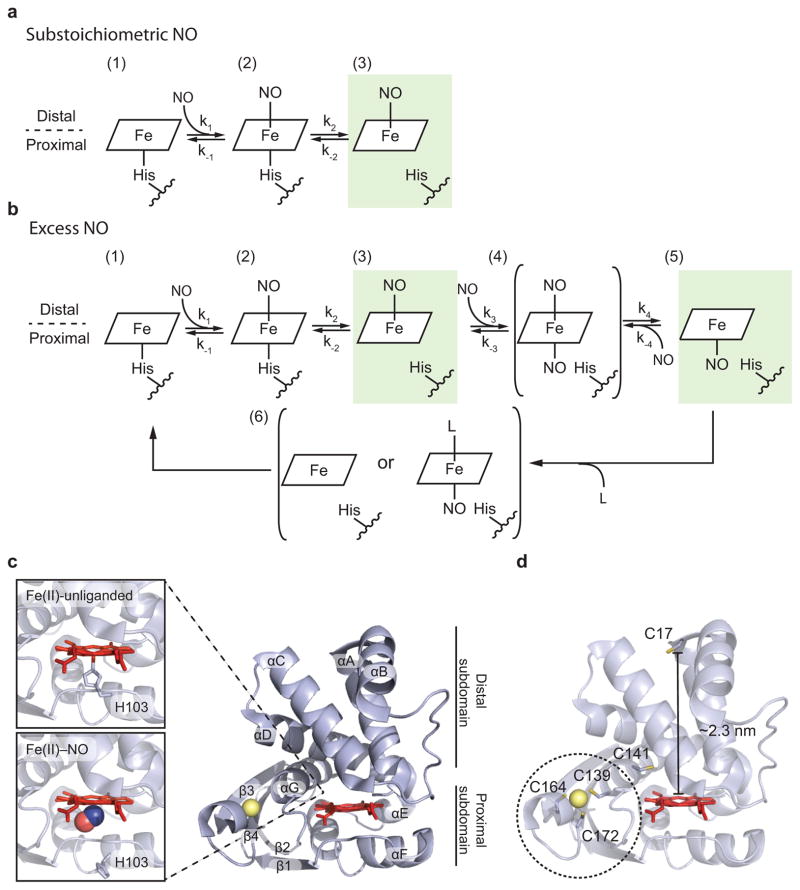
The kinetics of NO binding in full-length sGC, sGC truncations containing the H-NOX domain, and bacterial H-NOXs share common features. A transient six-coordinate complex is initially formed with NO bound in the distal heme site, axial to the heme-ligating histidine7,14–17. This six-coordinate complex then converts to a five-coordinate NO complex upon the breakage of the Fe(II)–His bond (Fig. 1a,b)7,8,18–21. The intermediacy of the six-coordinate complex is observed with the addition of a stoichiometric amount (relative to the heme) of NO or with excess NO (Fig. 1a,b). However, in the presence of excess NO, the formation of the final five-coordinate complex occurs at a faster rate7,18–20. This observation was initially explained by the formation of a di-nitrosyl intermediate that then has the potential to form a distal or proximal final five-coordinate complex depending on which NO is lost from the di-nitrosyl complex (Fig. 1b), and was supported by structures of NO-bound heme proteins. Structures of both cytochrome c′ from Alcaligenes xylosoxidans (Ax cyt c′), a protein with similar ligand properties to the non-O2 binding H-NOXs but with no sequence or structural similarity22–25, and a bacterial H-NOX from Shewanella oneidensis26, contained NO exclusively in the proximal positions when crystallized with excess NO (Fig. 1c). However, under stoichiometric conditions, NO is expected to bind in the distal pocket. Additionally, NO-bound Nostoc sp. H-NOX27 and NO-bound So H-NOX with manganese protoporphyrin IX in place of the natural iron porphyrin formed six-coordinate NO complexes in crystal structures, providing a good model of the unstable six-coordinate NO complex26.
Figure 1. NO-dependent activation of H-NOX protein.
(a,b) Proposed mechanism for NO-dependent activation of H-NOX protein. In the presence of a substoichiometric NO concentration (a), NO initially binds to the distal pocket of H-NOX and forms a six-coordinate complex (state 2). NO binding weakens the Fe(II)–histidine bond, which leads to the cleavage of this bond and results in a five-coordinate distal-bound Fe(II)–NO complex (state 3). If excess NO is present (b), another NO molecule can bind to the proximal pocket (state 4) and form a di-nitrosyl intermediate that then has the potential to form a distal or proximal final five-coordinate Fe(II)–NO complex, depending on which NO molecule dissociates from the di-nitrosyl complex (states 3 or 5, in green). Return of the distal NO complex to the five-coordinate histidyl resting state would be a microscopic reverse of the forward reaction (k−2, k−1). In the absence of excess NO, returning to the resting state of the proximal complex would be more complicated. The proximal-bound NO could dissociate with the assistance of a solution or protein nucleophile (L), or through a four-coordinate porphyrin to which the proximal histidine would then rebind (state 6). (c) Structure of the So H-NOX with its secondary structure designation and with detailed views of the heme binding pocket for the Fe(II)–unliganded state (PDB: 4U99) and the Fe(II)–NO state (PDB: 4U9B). The distal and proximal sub-domains are defined according to their locations relative to His103. (d) Cysteines in the So H-NOX Fe(II) structure. Cys17 was chosen as the MTSL labeling site.
The “sidedness” of the NO complex (distal vs. proximal) could have profound biological consequences in terms of the signal half-life. Return of the distal NO complex to the five-coordinate histidyl resting state would be a simple microscopic reverse of the forward reaction (Fig. 1a,b). The return to the resting state of the proximal complex is more complicated. In the absence of excess NO, which would be the case under biological signaling, the proximal bound NO could dissociate, leaving a four-coordinate porphyrin to which the proximal histidine would then have access to rebind. While possible, the energetics of this pathway are unfavorable; a solution or protein nucleophile may bind distally to facilitate NO loss, but it is not obvious what nucleophile could serve in this role. Therefore, it is expected that the off-rate (signal time) of a distal NO complex would be faster than that of proximal NO. In fact, it may be that the biological signal lifetime is tailored to the concentration of NO generated in a particular signaling context, i.e. distal and short lifetime at low NO, proximal and long lifetime at high NO.
To address this question, off-rates were determined from complexes formed with substoichiometric NO and excess NO with the So H-NOX. Distal-bound NO would be expected under substoichiometric concentration with a faster off-rate and proximal-bound NO would be expected under excess NO concentration with a slower off-rate. As reported here, surprisingly, the off-rates from complexes formed under both conditions are identical. Further characterization by electron paramagnetic resonance (EPR) spectroscopic experiments unambiguously show that under both conditions, distal-bound NO is the dominant species. Only a minor portion of proximal-bound NO complex can be observed when excess NO is still present. The impact of these results on biological NO signaling is important both in molecular and cellular terms, as the lifetime of NO induced signal is independent of NO concentration.
Results
So H-NOX NO-dissociation rates
To examine the properties of complexes formed under substoichiometric and excess NO conditions, the dissociation rates of NO from So H-NOX were measured using two different NO traps. The first trap comprised of sodium dithionite saturated with carbon monoxide (COsat)8,14,16,28. In the presence of this trap, dissociation of NO from the heme of So H-NOX resulted in an increase in absorbance at 424 nm due to formation of a six-coordinate heme-CO complex (Supplementary Results, Supplementary Fig. 1). Two rates were observed for each sample: a faster rate constant k1 and a slower rate constant k2. This is consistent with the biphasic NO dissociation rates measured from sGC, representing “open” and “closed” conformations of five-coordinate So H-NOX Fe(II)–NO complex (Supplementary Table 1)8,14,29,30.
So H-NOX treated with excess NO (10–20 eq. of NO relative to the heme) yielded average rate constants and amplitudes of NO dissociation of 0.00035 s−1 (k1, 29%) and 0.00007 s−1 (k2, 71%) at 10 °C, which are very similar to the values measured from So H-NOX treated with substoichiometric NO [0.00109 s−1 (k1, 19% and 0.00010 s−1 (k2, 81%)] (Supplementary Table 1). There is slight temperature dependence of the observed rate constants and amplitudes when tested at 15 and 20 °C.
Ferro-di(N-(dithiocarboxy)sarcosine [Fe2+(DTCS)2] was used as an alternative NO trap to eliminate the potential influence of CO31,32. In the presence of Fe2+-(DTCS)2/dithionite, dissociation of NO from the heme of So H-NOX resulted in an increase in absorbance at 430 nm due to loss of NO and formation of the Fe(II)–unliganded complex (Supplementary Fig. 1). The dissociation rate constants and corresponding amplitudes measured here match those values obtained using the COsat/dithionite trap, indicating that the dissociation rates are not trap-dependent or influenced by the NO concentration in the sample.
DEER analysis of proximal vs. distal NO
As noted above, H-NOXs could possibly form either distal- or proximal-bound NO complexes19,26,33. However, the ability to definitively differentiate between these two species in solution has proven difficult, as both distal and proximal five-coordinate NO complexes are indistinguishable by electronic absorption spectroscopy. We therefore sought to employ EPR spectroscopic methods to distinguish distal- and proximal-NO.
The field-swept Q-band pulse EPR spectrum of So H-NOX treated with substoichiometric NO shows the expected pseudo-axial EPR signal with g = [2.11, 2.02, 2.01] and the characteristic 1:1:1 14N hyperfine splitting observed at the high-field edge of the spectrum (A = 51 MHz) (Supplementary Fig. 2)34. Samples prepared with excess NO followed by a buffer-exchange step to remove excess NO exhibited a nearly identical EPR spectrum (Supplementary Fig. 2). Both signals displayed some degree of spectral broadening, likely originating from some site-to-site inhomogeneity of the Fe(II)–NO unit, as has been observed previously30. In addition, there was a small signal corresponding to a six-coordinate Fe(II)–NO species. These minor species were present in all samples and their intensities did not vary with NO concentration; as such, we simulated them as a single species. Overall, samples treated with substoichiometric NO or excess NO followed by buffer-exchange have one major signal: a five-coordinate Fe(II)–NO species that we expect arises from NO binding to the distal face of the heme-Fe cofactor (vide infra). In contrast, the EPR spectra of samples treated with excess NO without a subsequent buffer-exchange step contained two major signals (Supplementary Fig. 2). One signal was readily identified as the major species in the substoichiometric NO sample with g = [2.11, 2.02, 2.01]. The other signal is more rhombic and characterized by g = [2.09, 2.01, 1.97], which is consistent with a six-coordinate Fe(II)–NO species (vide infra)34.
In order to directly identify the NO binding sites in So H-NOX, double electron-electron resonance (DEER) spectroscopy was used to measure the distance between the S = 1/2 Fe(II)–NO group and another paramagnetic center introduced by a covalently attached spin label, S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL), on the protein surface. We chose C17 as the labeling site because it is the most solvent-exposed cysteine and is not involved in coordinating a structurally important zinc ion26. The thiol moiety in C17 is also located ~2.3 nm from the Fe(II) center and almost perpendicular to the heme (Fig. 1c,d). The MTSL labeling reaction was optimized to achieve high labeling efficiency on C17 while minimizing the extent of labeling on the other cysteine residues. Mass spectrometry measurements of the intact and trypsin-digested protein–MTSL conjugates identified C17-MTSL as the main product, while the other four cysteines in the protein were largely unmodified (Supplementary Figs. 3 and 4).
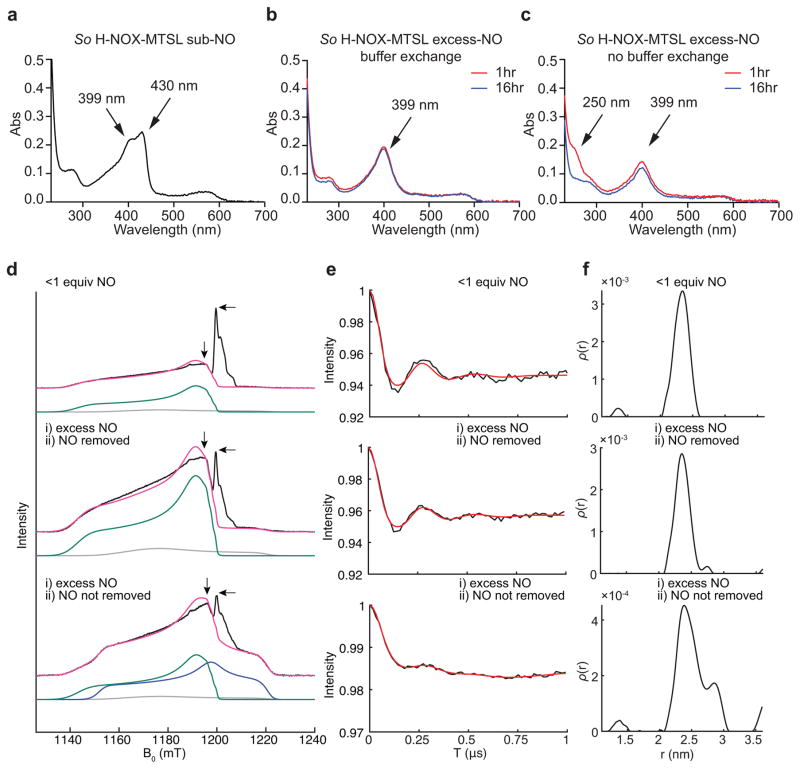
DEER samples were then prepared with So H-NOX-MTSL incubated with substoichiometric NO (0.75 eq. of NO) or excess NO (20 eq. of NO) concentrations (relative to heme). Both NO-treated samples had absorbance maxima at 399 nm, indicating five-coordinate NO complex formation (Fig. 2a–c). Additionally, the absorption spectrum from the substoichiometric NO treated sample contained a shoulder at 430 nm, showing that some protein, as expected, remains in the Fe(II)-unliganded state.
Figure 2. Distance measurement of Fe(II)–NO to So H-NOX C17-MTSL.
(a–c) Electronic absorption spectra of So H-NOX-MTSL treated with substoichiometric NO (a), excess NO with buffer exchange (b) and excess NO without buffer exchange (c). The shoulder at 430 nm indicated the partially bound five-coordinate So H-NOX-MTSL-NO complex. The absorbance at 399 nm indicated formation of five-coordinate NO complexes. The absorbance at ~250 nm indicated incomplete decomposition of DETA NONOate (t1/2 ~ 56 hours at 25 °C). (d) Q-band EPR spectra with data (black), total simulation (magenta), five-coordinate distal Fe(II)–NO simulation (green), six-coordinate proximal Fe(II)–NO simulation (blue) and minor species (gray). Simulation parameters are the same as in Supplementary Figure 2. The “pump” and “probe” positions are denoted by horizontal (at g = 2.01) and vertical arrows (at g = 2.0165), respectively. Note that the Fe(II)–NO signal in the substoichiometric NO spectrum is relatively weak because not all of the heme-Fe cofactor has NO bound. (e) DEER spectra corresponding to the samples in Figure 2d. Black, data; red, fit. The DEER spectrum of the substoichiometric NO sample shows a clean modulation in the time domain that corresponds to a distance of 2.3 nm. The DEER spectra of the non-buffer-exchanged excess NO sample displays a valley in the spin-echo intensity at 144 ns (arising from the frequency that corresponds to the 2.3 nm distance); however the peak at 272 ns is dampened due to the presence of an additional, lower-frequency contribution that corresponds to a longer distance. (f) Distance distributions corresponding to the DEER spectra in e.
The field-swept Q-band pulse EPR spectra of the MTSL-labeled samples contained the expected Fe(II)–NO signals as well as a narrow signal corresponding to the spin label (Fig. 2d). As was observed for the unlabeled samples, the MTSL-labeled samples that were treated with substoichiometric NO or excess NO followed by buffer exchange displayed EPR spectra that are essentially identical, with one major five-coordinate Fe(II)–NO species. Samples treated with excess NO without buffer exchange contained a mixture of five- and six-coordinate species.
As anticipated, the DEER spectrum of the substoichiometric NO sample indicated a Fe(II)–NO–nitroxide distance of 2.3 nm (Fig. 2e,f and Supplementary Fig. 5), matching the Fe(II)–thiol distance observed in the crystal structure and suggesting distal NO binding (Fig. 1d, Supplementary Fig. 6). Although we anticipated that the spin delocalization on the Fe(II)–NO moiety would somewhat shorten the effective Fe(II)–NO–nitroxide distance while the covalent attachment of the nitroxide spin label would lengthen it, these two effects appeared to largely cancel each other. MTSL conformer simulations supported a 2.3 nm average distance from the spin label to the distal-bound NO (Supplementary Figs. 5–9). It should be noted that there can be a strong orientation dependence to DEER measurements owing both to the g-anisotropy of the nitroxide EPR signal and the more substantial g-anisotropy of the Fe(II)–NO EPR signal; as such, the distances measured herein do not necessarily correspond to the precise interatomic Fe(II)–NO–MTSL distance distribution. However, changes in the effective distances measured for different samples can be used to track changes in the Fe(II)–NO binding mode (vide infra).
The DEER spectrum of the MTSL-labeled So H-NOX sample treated with excess NO followed by buffer exchange was nearly identical to that of the sample treated with substoichiometric NO (Fig. 2e). The corresponding distance distribution (Fig. 2f) showed a major feature centered at 2.3 nm and a very weak feature at a longer distance (too weak to be accurately determined). The DEER spectrum of the MTSL-labeled sample treated with excess NO without buffer exchange was markedly different (Fig. 2e). Analysis of the distance distribution reveals the expected 2.3 nm distance (corresponding to the distal, five-coordinate Fe(II)–NO complex) as well as a distance at 2.8 nm (Fig. 2f, Supplementary Fig. 6). We propose that this longer distance arises from a proximal, six-coordinate Fe(II)–NO complex, and emphasize that this longer distance is only observed in samples containing excess NO without buffer exchange.
To confirm that the measured DEER distance was truly generated by the coupling between Fe(II)–NO and MTSL and not from non-specifically bound NO radicals resulting from the high NO concentration, intact protein mass spectrometry was conducted to determine if there was any mass increase after the NO treatment. The sample with 16 h NO incubation only showed a mass increase < 5 Da, which was likely due to hydrogen/deuterium exchange with D2O buffer (Supplementary Fig. 10). This result demonstrated that the high NO concentration did not non-specifically modify the So H-NOX and interfere with the distances determined from the DEER experiments.
Finally, we note that the identification of a proximal, six-coordinate Fe(II)–NO species formed under excess NO (without removal of excess NO)—rather than a proximal, five-coordinate Fe(II)–NO species—was somewhat surprising, given that a six-coordinate species was not observed by UV-Vis spectroscopy at room temperature. We suspect that binding of the sixth ligand in the empty distal pocket is entropically driven and occurs during the freezing step in sample preparation. To probe if amines derived from NONOate decomposition could function as the sixth ligand, samples were prepared with varying NO(g) concentrations. The Q-band EPR spectra of the NO(g)-treated samples were not notably different from the NONOate-treated samples (Supplementary Fig. 11): the distal, five-coordinate Fe(II)–NO species was observed at low NO concentration, and the proximal, six-coordinate Fe(II)–NO species grew in only at higher NO(g) concentration. These results reassured us that the conclusions drawn from the samples prepared using NONOates are identical to those prepared using NO(g) and that NONOate-derived byproducts do not function as the sixth ligand for the proximal, six-coordinate Fe(II)–NO species.
Inhibition of kinase activity by NO-bound So H-NOX
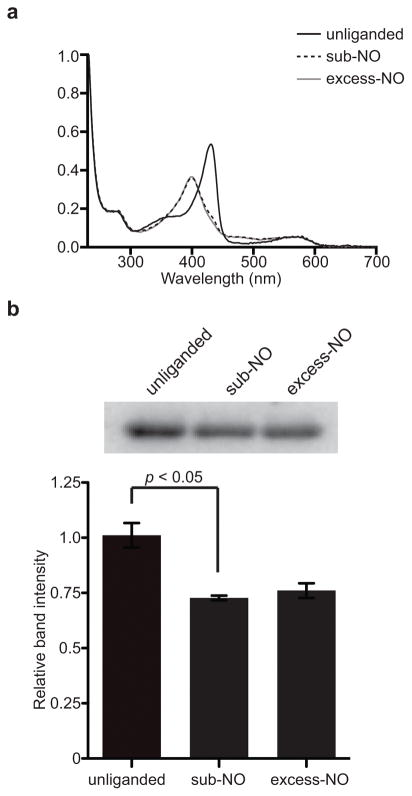
So H-NOX serves as a sensor protein of an associated histidine kinase (So HK) in vivo to form a multi-component signaling network11,12. The autophosphorylation of So HK is inhibited by So H-NOX upon NO binding. As shown above, the distal-NO complex is the major species in solution regardless of the NO concentration. Therefore, it would be expected that the in vivo biological readout, namely cognate kinase inhibition, would be the same at substoichiometric or excess NO concentrations if the same amount of NO-bound H-NOX is formed. To test this hypothesis, a kinase autophosphorylation activity assay using [γ-32P] ATP as the substrate was carried out. UV-Vis spectra of substoichiometric and excess NO-treated samples showed identical absorbance at 399 nm, indicating that in both conditions, the H-NOX formed a five-coordinate complex (Fig. 3a). As predicted, both substoichiometric and excess NO-treated So H-NOX were able to inhibit So HK autophosphorylation activity to ~70%, compared to the So H-NOX Fe(II)-unliganded sample (p<0.05) (Fig. 3b, Supplementary Fig. 12). Therefore, substoichiometric and excess NO concentrations do not affect the signaling strength of So H-NOX, since the distal-NO complex is preferably formed under both conditions.
Figure 3. So H-NOX signaling assay with different NO concentrations.
(a) Electronic absorption spectra of So H-NOX used in the assay: So H-NOX Fe(II) treated without NO (black line), substoichiometric NO (dashed line) or excess NO (gray line). (b) Measurement of So HK autophosphorylation activity regulated by So H-NOX treated with different NO concentrations. P-values of 0.0075, 0.0182 and 0.3975 were calculated to compare relative kinase autophosphorylation with unliganded/sub-NO, unliganded/excess-NO and sub-NO/excess-NO treated So H-NOX, respectively. The experiment was done in triplicate and data represent mean ± SEM.
Discussion
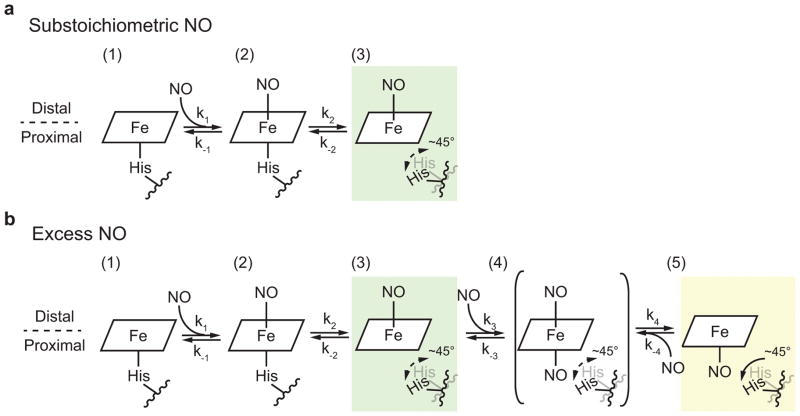
Based on these results, we can now propose a physiological model of NO-dependent activation and deactivation of H-NOX proteins (Fig. 4). At substoichiometric concentrations, NO binds to the open coordination site in the H-NOX distal pocket, forming a six-coordinate complex. The Fe(II)–histidine bond is weakened by NO binding, which leads to the cleavage of this bond and results in a five-coordinate distal-bound Fe(II)–NO complex. When NO is in excess, a di-nitrosyl heme intermediate could form by interacting with another NO molecule in the proximal pocket. This di-nitrosyl intermediate could form either a distal or proximal final five-coordinate Fe(II)–NO complex, depending on which NO molecule dissociates. However, based on our observation that the distal Fe(II)–NO complex is the dominant species in the presence of excess NO and the only five-coordinate Fe(II)–NO complex when excess NO is depleted, we conclude that this di-nitrosyl intermediate and the proximal Fe(II)–NO complex are kinetically less stable in solution, suggesting that the reverse rate constants k–3 and k–4 are dominant (k3 < k−3, k4 < k−4). Bacteria typically experience NO concentrations ranging from picomolar to sub-micromolar13,35. Thus, the proximal Fe(II)–NO complex is not likely to be physiologically relevant, since environmental concentrations of NO will not be sufficiently high. Deactivation of the signal could be accomplished through a simple histidine rebinding followed by NO dissociation from the distal Fe(II)–NO complex. In conclusion, the lifetime of the signal will be determined by the off-rate from the distal complex at physiological concentrations of NO.
Figure 4. Proposed mechanism for NO-dependent activation and deactivation of H-NOX protein.
(a) Under substoichiometric NO concentrations, the NO molecule initially binds to the distal pocket of H-NOX and forms a six-coordinate complex (state 2), which is followed by the cleavage of the Fe(II)–histidine bond, resulting in a five-coordinate distal-bound Fe(II)–NO complex as the only product (state 3). (b) Under excess NO concentrations, another NO molecule can bind to the proximal pocket to form a di-nitrosyl intermediate (state 4) which could potentially form either a distal (state 3) or proximal final five-coordinate Fe(II)–NO complex (state 5). This intermediate and the proximal Fe(II)–NO complex (highlighted in yellow) are likely to be kinetically unstable due to the faster reverse rate constants (k3 < k−3, k4 < k−4), making the distal Fe(II)–NO complex (state 3, highlighted in green) the only species under physiological concentration of NO. Dissociation of the distal NO would be accomplished through proximal histidine rebinding. The position of the proximal histidine in the proximal NO complex is known from the crystal structure and noted on in state 5, but is unknown in the distal or proposed di-NO complexes and so is shown as being in motion.
How can the NO concentration effect on the rate of five-coordinate complex formation be explained? Under substoichiometric NO concentrations, the rate of the distal complex formation will be dependent on k2 and k−2 since the NO on-rate is very fast. The conformational changes induced by distal NO binding are likely very similar to the proximal-NO structure reported previously where a ~45° rotation of αF helix moved the iron-bound histidine well outside of the proximal pocket (Fig. 4), since the iron–His bond is severed in both complexes26. The locations of the proximal histidine in the distal NO complex and the proposed di-NO complex are unknown, but this residue is likely to be in motion before reaching the position observed in the crystal structure. The relative flexibility of the distal complex may prevent crystal formation. The increase in the apparent formation rate of the five-coordinate complex (Fig. 4) observed with excess NO may be due to the ability of NO to effectively compete with histidine rebinding (i.e. k3 > k−2). Previous studies attempted to answer the central question addressed here with sGC using a mixture of stable NO isotopes19. Their conclusions, which differ from those reported here, are based on a model in which active sGC has a proximally bound NO, while the distal NO complex is unstable and forms an inactive “desensitized” six-coordinate complex with a histidine residue, different from that in the resting state, in the proximal position. However, direct evidence for this scheme is lacking.
While the H-NOX domain in sGC shares the essential features of heme ligand-binding properties and kinetics with bacterial H-NOX proteins, activation of sGC to full activity is more complicated. NO occupancy of the heme activates sGC to ~15 % maximal activity2,7. Evidence here suggests that this NO molecule would be in the distal site. Dissociation from the heme is slow, such that it is considered irreversible in a physiological context. Full activation of sGC requires additional NO that is transiently bound in a rapidly reversible complex36. Thus, sGC activation requires NO bound at the heme in steps like we have described for bacterial H-NOX proteins and toggles in vivo between the resulting low activity form (15% maximal activity) and full activation.
Timing is fundamental to any signaling pathway. A major component of signal strength is the length of time that the signal is on; therefore, the focus on the molecular details of NO capture by the heme in H-NOX receptor proteins is critical for understanding the molecular mechanism of NO signaling. The results presented above show that the distal NO complex is the predominate five-coordinate Fe(II)–NO species in solution, whereas the proximal NO complex was only observed in the presence of a large excess of NO. Moreover, the signaling strength of So H-NOX only depends on the concentration of five-coordinate So H-NOX Fe(II)–NO, but not on the overall NO concentration in the environment. Hence, these results bear directly on in vivo signaling.
With regards to the broader context of this work, our direct observation of NO sidedness in So H-NOX has provided a new insight into how NO interacts with H-NOX-containing proteins. This discovery leads to an in vivo model for NO activation and deactivation, which has immediate implications on NO-dependent signal transduction in bacterial H-NOXs and sGC-involved pathways at both the molecular and cellular levels13.
Online methods
Protein expression and purification
For bacterial expression, So H-NOX (1–181) was cloned into pET20b vector with a TEV-cleavable His6 tag at the C-terminus. Protein was expressed from RP523(DE3) cells, grown in Terrific broth (Research Products International) supplemented with trace metals (1 mM CaCl2, 10 mM MgSO4, 20 μM FeCl3 and 150 μM ZnCl2) at 37 °C and induced with 1 mM IPTG at OD600 of 0.7. After overnight growth at 18 °C, cells were harvested by spinning at 4°C, 4700 ×g for 25 min. The pellet was re-suspended in buffer (50 mM phosphate, pH 8.0, 300 mM NaCl, 5% glycerol) and stored at −80°C. Purification was conducted using affinity chromatography (His60 Ni superflow resin, Clontech), followed by size-exclusion chromatography (Superdex 75, GE Healthcare) in 50 mM HEPES pH 8.0, 150 mM NaCl, 5% glycerol and 2 mM TCEP. Following gel filtration, protein was transferred to a glove bag (Coy Laboratory Products) under an argon: hydrogen (95:5%) atmosphere, at ~25 °C. So H-NOX was fully oxidized (λmax 403 nm) using 1–2 equivalents of potassium ferricyanide (K3Fe(CN)6), followed by buffer exchange through a PD10 desalting column (GE healthcare) to remove excess K3Fe(CN)6. Protein was then fully reduced (λmax 430 nm) [Fe(II)] using 1 mM sodium dithionite (Na2S2O4; final concentration). Residual dithionite was removed by exchanging the protein into fresh buffer (50 mM TEA (pH 8.0), 50 mM NaCl, 5% (v/v) glycerol for NO dissociation experiments or 50 mM phosphate, pH 8.0, 150 mM NaCl, 20% (v/v) glycerol for EPR studies).
So HK was expressed and purified as previously described37. So HK (1–311) was cloned into a pET-GSTx with an N-terminal glutathione-S-transferase (GST) tag and expressed in E. coli BL21(DE3) cells in Terrific broth (Research Products International) in the presence of 100 μg/mL ampicillin. Protein was induced at OD600 ~0.6 with 400 μM IPTG and harvested after overnight growth at 18 °C. The pellet was re-suspended in buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10 mM MgCl2, 25 mM L-glutamate, 5 mM β-mercaptoethanol, 10% v/v glycerol) and stored at −80°C. Purification was conducted using affinity chromatography (Pierce Glutathione Agarose, Thermo Scientific), followed by size-exclusion chromatography (Superdex 200, GE Healthcare) in 50 mM Tris pH 8.0, 150 mM NaCl, 10 mM MgCl2, 25 mM L-glutamate, 5 mM DTT, and 10% v/v glycerol.
NO dissociation - carbon monoxide/dithionite Trap
Protein-NO complexes were generated by mixing 5 μM So H-NOX Fe(II) (heme concentration) with DEA-NONOate (Cayman Chemical) (in 10 mM NaOH) at a concentration to obtain the desired NO:protein ratio (0.6–0.8 eq. for substoichiometric and 10–20 eq. for excess) (assuming 1.5 moles NO per moles parent compound; t1/2 ~16 min at room temperature, pH 7.4) in a septum-sealed anaerobic cuvette (Starna Cells). Protein was allowed to react with the NO donor for ~30 minutes to ensure nearly complete decomposition of the NO donor. Conversion to the five-coordinate NO species (λmax 399 nm) was monitored by UV-Vis absorbance spectroscopy, following the shift in Soret maximum from 430 to 399 nm. Protein samples were used immediately for subsequent experiments.
The dissociation of NO from the heme of So H-NOX was performed using a carbon monoxide-saturated (COsat)/dithionite solution as previously described8,14,16,28. The trapping solution was prepared anaerobically as follows: a solution of 60 mM sodium dithionite in 50 mM TEA, pH 8.0, 50 mM NaCl, 5% (v/v) glycerol buffer was prepared in a Teflon-sealed Reacti-Vial (Pierce). The solution was removed from the anaerobic chamber and then saturated with CO (99.99%; Praxair, Inc.) by bubbling the gas through the solution for 10 min prior to immediate use. Cuvette and trap solutions were equilibrated at assay temperature for 5 min prior to initiation. The reaction was initiated by equal addition of the COsat/dithionite solution to the anaerobic cuvette using an anaerobic buffer-purged Hamilton gas-tight syringe followed by thorough mixing. The final dithionite concentration in the reaction mixture was 30 mM. Final five-coordinate NO protein concentrations were 2.5 μM and 1.5–2 μM for excess and substoichiometric NO-treated So H-NOX, respectively. Final reaction volumes were 500 μL. The reaction was monitored by electronic absorption spectroscopy using a Cary 100 spectrophotometer equipped with a temperature controller (Agilent Technologies) set to varying temperatures (10–20 °C). Data were acquired by scanning periodically over a range of 350–700 nm at 600 nm/min with a 1 nm interval. Spectra were corrected for baseline drift by normalizing the 690–700 nm range. The dissociation of NO from the heme was monitored by the formation of the Fe(II)–CO complex (Soret maximum at 424 nm). Difference spectra were calculated by subtracting the first spectrum from each subsequent spectrum (normalized). The values for ΔAbs424 extracted from the difference spectra were plotted versus time and fit to single or two parallel exponentials [of the form f(x) = A×(1−e−kx)] to obtain rates of NO dissociation. Measurements at 10 °C were repeated 11 times for sub-NO sample and 7 times for excess-NO sample. Other conditions were repeated 5 times. Protein used in the experiments was obtained from two different purifications. It should be noted that the relative populations of these two exponentials do not fluctuate substantially between different So H-NOX preparations.
Synthesis of N-(dithiocarboxy)-sarcosine
Synthesis of N-(dithiocarboxy)-sarcosine (DTCS) was performed as described previously38,39. Sarcosine (Sigma Aldrich, 2.5 g, 28.05 mmol) was added to a solution of NaOH (1.25 g, 31.25 mmol in 20 mL of water) cooled to 10°C with constant stirring. To this mixture, a solution of carbon disulfide (2.8 mL, 46.5 mmol) dissolved in 7.5 mL absolute ethanol was slowly added while carefully maintaining the temperature at 10 °C. The reaction was allowed to stir for an additional 30 min followed by chloroform extraction (3 × 25 mL). The remaining aqueous solution was rotary-evaporated to dryness yielding a caramel-like oil. Absolute ethanol was added, mixed to homogeneity, and again rotary-evaporated to dryness. This step was repeated until a solid material was obtained. The material was further dried overnight using a high vacuum pump. This entire procedure was repeated with the isolated material in place of sarcosine. NMR analysis was used to monitor reaction completeness and verify that the final compound was N-(dithiocarboxy)-sarcosine38,39.
NO dissociation - dithiocarbamate/dithionite Trap
The trapping solution was prepared immediately prior to use as follows: a solution of 10 mM DTCS, 2 mM Fe(II) (FeCl2) in buffer B supplemented with 60 mM dithionite was prepared in a Teflon-sealed Reacti-Vial anaerobically. Cuvette and trap solutions were equilibrated at the assay temperature for 5 min prior to initiation. The reaction was initiated by equal addition of the ferro-di(N-(dithiocarboxy) sarcosine) [Fe2+(DTCS)2] solution to the anaerobic cuvette using an anaerobic buffer-purged Hamilton gas-tight syringe followed by thorough mixing. The dissociation of NO from the heme was monitored as formation of the Fe(II)–unliganded complex (Soret maximum at 430 nm). Data acquisition and processing were performed as described for the COsat/dithionite trap. Special care was taken to eliminate oxygen contamination to prevent oxidation of the (DTCS)2-chelated iron and formation of insoluble ferric chelates.
MTSL labeling on So H-NOX C17
The MTSL (Toronto Research Chemicals) labeling reaction was optimized to avoid labeling cysteines other than C17. All labeling reactions were carried out in EPR buffer (50 mM phosphate, pH 8.0, 150 mM NaCl, 20% (v/v) glycerol) anaerobically at room temperature. Optimization of MTSL concentration was done by incubating 10 μM reduced So H-NOX with MTSL at protein : MTSL molar ratios = 1:5, 1:10, 1:25 and 1:500 for 15 min. Reactions were quenched by desalting the reaction mixture through a PD10 column to remove excess MTSL and products were analyzed by intact protein mass spectrometry. Protein : MTSL molar ratio = 1:10 (100 μM MTSL) resulted in the highest yield of So H-NOX +1 MTSL product (+184 Da) (Supplementary Fig. 3). Reaction time was then optimized by time course experiments of incubating 10 μM So H-NOX with 100 μM MTSL for 30 min, 1 h, 2 h, 3 h, 4 h and 5 h. Mass spectrometry measurements showed that the 30 min reaction produced the highest level of +1 MTSL product and a relatively low level of multi-MTSL products (Supplementary Fig. 3).
Trypsin digestion
So H-NOX-MTSL was first denatured with 8 M urea, and then diluted with EPR buffer until the urea concentration was less than 1 M with a final protein concentration ~1 mg/mL. 1 mg/mL trypsin stock was made by dissolving 2 mg trypsin powder (Sigma Aldrich, trypsin from bovine pancreas) in 2 mL of 50 mM acetic acid. Four trypsin digestion reactions were carried out using different So H-NOX-MTSL and trypsin ratios as follows: 1) 30 μL So H-NOX-MTSL + 0.5 μL trypsin; 2) 30 μL So H-NOX-MTSL + 1 μL trypsin; 3) 15 μL So H-NOX-MTSL + 15 μL EPR buffer + 0.5 μL trypsin; 4) 15 μL So H-NOX-MTSL + 15 μL EPR buffer + 1 μL trypsin. Reactions were incubated at 37 °C in a water bath overnight and flash frozen in liquid nitrogen.
Liquid chromatography-mass spectrometry analysis of intact proteins
Intact proteins were analyzed using a Synapt G2-Si mass spectrometer that was equipped with an electrospray ionization (ESI) source and C4 protein ionKey (150-μm inner diameter, 50-mm length, 300-Å pore size, 1.7-μm particle size), and connected in line with an Acquity M-class liquid chromatograph (Waters, Milford, MA). Acetonitrile, formic acid (Fisher Optima grade, 99.9%), and water purified to a resistivity of 18.2 MΩ·cm (at 25 °C) using a Milli-Q Gradient ultrapure water purification system (Millipore, Billerica, MA) were used to prepare mobile phase solvents. Solvent A was 99.9% water/0.1% formic acid and solvent B was 99.9% acetonitrile/0.1% formic acid (v/v). The elution program consisted of a linear gradient from 1% to 85% (v/v) B over 12 min, isocratic flow at 85% B for 1 min, a linear gradient from 85% to 1% B over 2 min, and isocratic flow at 1% B for 15 min, at a flow rate of 2 μL/min. Mass spectra were acquired in the positive ion mode over the range m/z = 400 to 5000. Mass spectral deconvolution was performed using ProMass software (version 2.5 SR-1, Novatia, Monmouth Junction, NJ).
Liquid chromatography-tandem mass spectrometry analysis of proteolytically digested proteins
Method details have been published elsewhere40. Briefly, trypsin-digested protein samples were analyzed using a Thermo-Dionex UltiMate3000 RSLCnano liquid chromatograph that was connected in line with an LTQ-Orbitrap-XL mass spectrometer equipped with a nanoelectrospray ionization source (Thermo Fisher Scientific, Waltham, MA). Data acquisition was controlled using Xcalibur software (version 2.0.7, Thermo) and data analysis was performed using Proteome Discoverer software (version 1.3, Thermo).
EPR sample preparation
All samples were prepared anaerobically inside a glove bag. Unlabeled and MTSL-labeled So H-NOX was concentrated to ~400 μM and ~200 μM, respectively, using a 5 kDa spin concentrator (Sartorius) in D2O-based buffer. For substoichiometric NO samples, protein was treated with 0.75 eq. of NO from DEA NONOate stock solution (made in 10 mM NaOH) and incubated for 30 min or longer to form So H-NOX-MTSL Fe(II)–NO complexes. For excess NO samples, protein was treated with 20 eq. of NO from DEA NONOate stock plus 500 μM DETA NONOate stock solution and incubated overnight. Buffer exchange was performed using a spin-6 desalting column (Bio-Rad) following manufacturer instructions. Samples treated with NO(g) were prepared by diluting the protein with NO-saturated EPR buffer (2 mM NO concentration), which was obtained by purging NO(g) through a sealed reaction vial containing 3.5 mL EPR buffer for 30 min. Samples were transferred to Q-band EPR tubes and frozen in liquid nitrogen for data collection.
EPR spectroscopic methods
EPR spectra were recorded using a Bruker ELEXSYS E580 spectrometer, a CF935 cryostat (Oxford Instruments), and an ITC-5 temperature controller (Oxford Instruments). Spectra were acquired at 33.8 GHz and 15 K with a Bruker EN5107D2 resonator. Field-swept spectra were acquired using the pulse sequence π/2–τ–π–τ–echo with excitation and inversion pulse lengths of 16 ns and 32 ns, respectively. DEER spectra were acquired using the dead-time-free four-pulse sequence π1/2–τ1–π1–(τ1+T)–π2–(τ2–T)–π1–τ2–echo where π1 corresponds to the probe pulses and π2 corresponds to the pump pulse. The excitation and inversion probe pulse lengths were 16 and 32 ns, respectively, the pump pulse was 64 ns, and τ1 was 400 ns. T was stepped in 12 ns increments. The pump and probe pulses were positioned at g = 2.01 and g = 2.0165, respectively, corresponding to a frequency difference of 109 MHz. The field, B, at which the experiments were recorded was B = Bprobe + 1/3(Bpump − Bprobe). A shot repetition time of 1200 ns was employed. Spectral simulations of the field-swept EPR spectra were performed with MATLAB Release 2015a using the EasySpin 4.5.5 toolbox41. DEER data were processed using DeerAnalysis201542. Background subtraction was performed followed by least-squares fitting using Tikhonov regularization with L-curve selection. Rotamers of MTSL attached to Cys 17 of So H-NOX were analyzed using the MMM software package43 version 2015 with Matlab Release 2015a. Protein side chain conformations were treated dynamically as implemented in the SCWRL4 software package44. See the SI for further discussion of the DEER spectroscopy analysis and interpretation.
Histidine kinase activity assay
GST-So HK (4 μM ) was incubated with 40 μM of So H-NOX treated with 0, 0.8 eq. or 7.5 eq. NO (relative to the heme) in the final volume of 20 μL for 30 min inside a glove bag (Coy Laboratory Products) under an argon: hydrogen (95:5%) atmosphere, at ~25 °C. Assays were initiated by adding 2.5 μCi of [γ-32P] ATP (PerkinElmer) with 0.5 mM ATP and quenched by SDS loading dye with 20 mM EDTA (final concentration) after 30 min. Reactions were analyzed by SDS-PAGE as described previously45. Dried gels were exposed for 2 hours on a phosphor imager plate (GE Healthcare) and imaged using a Typhoon Trio (GE Healthcare) set to storage phosphor mode and imaging at 200 μm resolution. Phosphorylated So HK bands were quantified using ImageQuant software. Band intensities were normalized to the So H-NOX Fe(II)-unliganded control. Experiments were performed in three independent replicates and averaged. Error was reported in SEM and p-values were calculated using unpaired Student’s t-test.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of General Medical Sciences of the National Institutes of Health (F32GM111025 to D.L.M.S., R01GM104543 to R.D.B., and 1S10OD020062-01 to A.T.I.). We would like to thank current and former members of the laboratory of Michael A. Marletta for frequent helpful discussions and their invaluable insight (particularly Dr. Charles W. Hespen, Benjamin G. Horst, Dr. John A. Hangasky, Dr. Christopher M. Lemon and Dr. Joel J. Bruegger) and laboratory of John Kuriyan (particularly Dr. Tiago Barros and Dr. Jonathan Winger). We would also like to think Dr. Brian Michel and Dr. Alexander Nierth for help with synthesis and characterization of N-(dithiocarboxy)sarcosine. Finally, we thank Dr. Raquel Bromberg for proof reading the manuscript.
Footnotes
Author Contributions
Y.G., D.L.M.S., M.A.H., R.D.B., and M.A.M. designed research; M.A.H measured and analyzed the NO off-rates; Y.G. carried out the protein expression/purification, MTSL labeling of So H-NOX, EPR sample preparation and kinase activity assays; Y.G. and A.T.I. were responsible for the mass spectrometry and data analysis. D.L.M.S. and R.D.B carried out the EPR/DEER measurements and data analysis; Y.G., D.L.M.S., M.A.H., A.T.I., R.D.B. and M.A.M. wrote the paper.
Notes
The authors declare no competing financial interests.
References
- 1.Derbyshire ER, Marletta MA. Structure and regulation of soluble guanylate cyclase. Annu Rev Biochem. 2012;81:533–59. doi: 10.1146/annurev-biochem-050410-100030. [DOI] [PubMed] [Google Scholar]
- 2.Russwurm M, Koesling D. NO activation of guanylyl cyclase. EMBO J. 2004;23:4443–50. doi: 10.1038/sj.emboj.7600422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marletta MA. Nitric oxide synthase: function and mechanism. Adv Exp Med Biol. 1993;338:281–4. [PubMed] [Google Scholar]
- 4.Marletta MA, Hurshman AR, Rusche KM. Catalysis by nitric oxide synthase. Curr Opin Chem Biol. 1998;2:656–63. doi: 10.1016/s1367-5931(98)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33:829–37. 837a–837d. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munzel T, et al. Physiology and pathophysiology of vascular signaling controlled by guanosine 3′,5′-cyclic monophosphate-dependent protein kinase [corrected] Circulation. 2003;108:2172–83. doi: 10.1161/01.CIR.0000094403.78467.C3. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, Brandish PE, Ballou DP, Marletta MA. A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc Natl Acad Sci U S A. 1999;96:14753–8. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boon EM, et al. Nitric oxide binding to prokaryotic homologs of the soluble guanylate cyclase beta1 H-NOX domain. J Biol Chem. 2006;281:21892–902. doi: 10.1074/jbc.M600557200. [DOI] [PubMed] [Google Scholar]
- 9.Karow DS, et al. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochemistry. 2004;43:10203–11. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 10.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc Natl Acad Sci U S A. 2004;101:12854–9. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46:13677–83. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plate L, Marletta MA. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell. 2012;46:449–60. doi: 10.1016/j.molcel.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plate L, Marletta MA. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem Sci. 2013;38:566–75. doi: 10.1016/j.tibs.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winger JA, Derbyshire ER, Marletta MA. Dissociation of nitric oxide from soluble guanylate cyclase and heme-nitric oxide/oxygen binding domain constructs. J Biol Chem. 2007;282:897–907. doi: 10.1074/jbc.M606327200. [DOI] [PubMed] [Google Scholar]
- 15.Brandish PE, Buechler W, Marletta MA. Regeneration of the ferrous heme of soluble guanylate cyclase from the nitric oxide complex: acceleration by thiols and oxyhemoglobin. Biochemistry. 1998;37:16898–907. doi: 10.1021/bi9814989. [DOI] [PubMed] [Google Scholar]
- 16.Cary SP, Winger JA, Marletta MA. Tonic and acute nitric oxide signaling through soluble guanylate cyclase is mediated by nonheme nitric oxide, ATP, and GTP. Proc Natl Acad Sci U S A. 2005;102:13064–9. doi: 10.1073/pnas.0506289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharitonov VG, Sharma VS, Magde D, Koesling D. Kinetics of nitric oxide dissociation from five- and six-coordinate nitrosyl hemes and heme proteins, including soluble guanylate cyclase. Biochemistry. 1997;36:6814–8. doi: 10.1021/bi970201o. [DOI] [PubMed] [Google Scholar]
- 18.Stone JR, Marletta MA. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry. 1996;35:1093–9. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- 19.Martin E, Berka V, Sharina I, Tsai AL. Mechanism of binding of NO to soluble guanylyl cyclase: implication for the second NO binding to the heme proximal site. Biochemistry. 2012;51:2737–46. doi: 10.1021/bi300105s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, Liu W, Berka V, Tsai AL. The selectivity of Vibrio cholerae H-NOX for gaseous ligands follows the “sliding scale rule” hypothesis. Ligand interactions with both ferrous and ferric Vc H-NOX. Biochemistry. 2013;52:9432–46. doi: 10.1021/bi401408x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt AP, Lehnert N. Heme-nitrosyls: electronic structure implications for function in biology. Acc Chem Res. 2015;48:2117–25. doi: 10.1021/acs.accounts.5b00167. [DOI] [PubMed] [Google Scholar]
- 22.Lawson DM, Stevenson CE, Andrew CR, Eady RR. Unprecedented proximal binding of nitric oxide to heme: implications for guanylate cyclase. EMBO J. 2000;19:5661–71. doi: 10.1093/emboj/19.21.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson DM, Stevenson CE, Andrew CR, George SJ, Eady RR. A two-faced molecule offers NO explanation: the proximal binding of nitric oxide to haem. Biochem Soc Trans. 2003;31:553–7. doi: 10.1042/bst0310553. [DOI] [PubMed] [Google Scholar]
- 24.George SJ, Andrew CR, Lawson DM, Thorneley RN, Eady RR. Stopped-flow infrared spectroscopy reveals a six-coordinate intermediate in the formation of the proximally bound five-coordinate NO adduct of cytochrome c′. J Am Chem Soc. 2001;123:9683–4. doi: 10.1021/ja0158307. [DOI] [PubMed] [Google Scholar]
- 25.Yoo BK, Lamarre I, Martin JL, Andrew CR, Negrerie M. Picosecond binding of the His ligand to four-coordinate heme in cytochrome c′: a one-way gate for releasing proximal NO. J Am Chem Soc. 2013;135:3248–54. doi: 10.1021/ja312140f. [DOI] [PubMed] [Google Scholar]
- 26.Herzik MA, Jr, Jonnalagadda R, Kuriyan J, Marletta MA. Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins. Proc Natl Acad Sci U S A. 2014;111:E4156–64. doi: 10.1073/pnas.1416936111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, Sayed N, Beuve A, van den Akker F. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. EMBO J. 2007;26:578–88. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore EG, Gibson QH. Cooperativity in the dissociation of nitric oxide from hemoglobin. J Biol Chem. 1976;251:2788–94. [PubMed] [Google Scholar]
- 29.Derbyshire ER, et al. Characterization of two different five-coordinate soluble guanylate cyclase ferrous-nitrosyl complexes. Biochemistry. 2008;47:3892–9. doi: 10.1021/bi7022943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunn A, Derbyshire ER, Marletta MA, Britt RD. Conformationally distinct five-coordinate heme-NO complexes of soluble guanylate cyclase elucidated by multifrequency electron paramagnetic resonance (EPR) Biochemistry. 2012;51:8384–90. doi: 10.1021/bi300831m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azizi F, et al. Rates of nitric oxide dissociation from hemoglobin. Free Radic Biol Med. 2005;39:145–51. doi: 10.1016/j.freeradbiomed.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimura T, Kotake Y. Spin trapping of nitric oxide with the iron-dithiocarbamate complex: chemistry and biology. Antioxid Redox Signal. 2004;6:639–47. doi: 10.1089/152308604773934404. [DOI] [PubMed] [Google Scholar]
- 33.Tsai AL, Berka V, Sharina I, Martin E. Dynamic ligand exchange in soluble guanylyl cyclase (sGC): implications for sGC regulation and desensitization. J Biol Chem. 2011;286:43182–92. doi: 10.1074/jbc.M111.290304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehnert N, Scheidt WR, Wolf MW. Structure and Bonding in Heme–Nitrosyl Complexes and Implications for Biology. In: Mingos DMP, editor. Nitrosyl Complexes in Inorganic Chemistry, Biochemistry and Medicine II. Vol. 154. Springer; Berlin Heidelberg: 2013. pp. 155–223. [Google Scholar]
- 35.Hall CN, Garthwaite J. What is the real physiological NO concentration in vivo? Nitric Oxide. 2009;21:92–103. doi: 10.1016/j.niox.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernhoff NB, Derbyshire ER, Marletta MA. A nitric oxide/cysteine interaction mediates the activation of soluble guanylate cyclase. Proc Natl Acad Sci U S A. 2009;106:21602–7. doi: 10.1073/pnas.0911083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao M, Herzik MA, Jr, Iavarone AT, Marletta MA. Nitric Oxide-Induced Conformational Changes Govern H-NOX and Histidine Kinase Interaction and Regulation in Shewanella oneidensis. Biochemistry. 2017;56:1274–1284. doi: 10.1021/acs.biochem.6b01133. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura T, et al. In vivo EPR detection and imaging of endogenous nitric oxide in lipopolysaccharide-treated mice. Nat Biotechnol. 1996;14:992–4. doi: 10.1038/nbt0896-992. [DOI] [PubMed] [Google Scholar]
- 39.Pou S, et al. Spin trapping of nitric oxide by ferro-chelates: kinetic and in vivo pharmacokinetic studies. Biochim Biophys Acta. 1999;1427:216–26. doi: 10.1016/s0304-4165(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 40.Hershey DM, et al. Magnetite Biomineralization in Magnetospirillum magneticum Is Regulated by a Switch-like Behavior in the HtrA Protease MamE. J Biol Chem. 2016;291:17941–52. doi: 10.1074/jbc.M116.731000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Jeschke G, et al. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Applied Magnetic Resonance. 2006;30:473–498. [Google Scholar]
- 43.Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Physical Chemistry Chemical Physics. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 44.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins: Structure, Function, and Bioinformatics. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hespen CW, Bruegger JJ, Phillips-Piro CM, Marletta MA. Structural and Functional Evidence Indicates Selective Oxygen Signaling in Caldanaerobacter subterraneus H-NOX. ACS Chem Biol. 2016;11:2337–46. doi: 10.1021/acschembio.6b00431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files.