SUMMARY
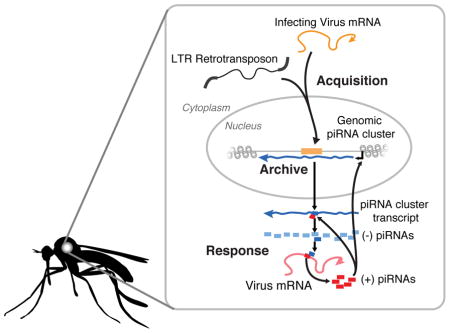
The Aedes aegypti mosquito transmits arboviruses including dengue, chikungunya and Zika virus. Understanding the mechanisms underlying mosquito immunity could provide new tools to control arbovirus spread. Insects exploit two different RNAi pathways to combat viral and transposon infection: short-interfering RNA (siRNAs) and PIWI-interacting RNAs (piRNAs) [1, 2]. Endogenous viral elements (EVEs) are sequences from non-retroviral RNA viruses that are inserted into the mosquito genome and act as templates for the production of piRNAs [3, 4]. EVEs therefore represent a record of past infections and a reservoir of potential immune memory [5]. The large-scale organization of EVEs has been difficult to resolve with short-read sequencing because they tend to integrate into repetitive regions of the genome. To define the diversity, organization and function of EVEs, we took advantage of the contiguity associated with long-read sequencing to generate a high-quality assembly of the Ae. aegypti-derived Aag2 cell line genome, an important and widely used model system. We show EVEs are acquired through recombination with specific classes of LTR retrotransposons and organize into large loci (>50kbp) characterized by high LTR density. These EVE-containing loci have increased density of piRNAs compared to similar regions without EVEs. Furthermore, we detected EVE-derived piRNAs consistent with a targeted processing of persistently infecting virus genomes. We propose that comparisons of EVEs across mosquito populations may explain differences in vector competence and further study of the structure and function of these elements in the genome of mosquitoes may lead to epidemiological interventions.
eTOC Blurb
Whitfield, et al. perform a genome-wide characterization of the endogenous viral elements (EVEs) present in the Aedes aegypti-derived Aag2 cell line. Using long-read sequencing suited to the highly repetitive mosquito genome, they explore the origin of these sequences and their potential role in mosquito immunity.
RESULTS
Identification of EVEs in the Aag2 genome
Due to the highly repetitive nature of the Ae. aegypti genome and EVEs’ tendency to cluster among transposable elements within such repetitive regions, many EVEs are likely to be missing from the current Ae. aegypti assemblies, which are based on relatively short read lengths[6–8]. To overcome the limitations of these earlier assemblies, we employed single-molecule, real-time (SMRT) sequencing technology (Pacific Biosystems) to generate a long-read assembly of the Aag2 cell line genome. Our draft assembly improves upon previous Aedes assemblies as measured by N50, L50, and by contig number (Table S1 and Figure S1a).
We used this assembly to better define the complete set of EVEs contained within the Aag2 genome, here referred to as the “EVEome”. We developed a BLASTx-based approach to identify acquired viral coding sequences with respect to each virus’ protein coding/(+)-sense strand (see STAR Methods). This approach identified 472 EVEs in our assembly, representing at least 8 annotated viral families. These were dominated by sequences derived from the Rhabdoviridae, Flaviviridae, and Chuviridae families. The identified EVEs covered 338,251 bp and ranged from 50 to 2,520 bp length with a median length of 620 bp (Figure 1a and b). Analysis of the viral coding regions revealed asymmetric incorporation of certain viral ORFs (Figure 1c). Flaviviridae-derived EVEs (Figure 1c, i) primarily mapped toward the 5′ end of the single Flaviviral ORF. EVEs derived from Rhabdoviridae primarily originate from the Nucleoprotein (N) and Glycoprotein (G) coding sequences, with only a few originating from the RNA-dependent RNA polymerase (L) (Figure 1c, ii). The lack of EVEs mapping to the polymerase may be the result of RNA expression levels, with L being the least expressed gene [9], suggesting that the template for EVE integration is viral mRNA.
Figure 1. Identification of Endogenous Viral Elements (EVEs) in the Aag2 assembly.
(A) Bar plots showing the number (top) and total length (bottom) of EVEs derived from each viral family. (B) Histogram showing the size distribution of EVEs in the Aag2 genome (top) and the total number of bases pairs derived from EVEs of a given size. The median EVE size (620bp) is indicated with a black bar.
(C) Coverage plots of EVEs derived from the viral families (i) Flaviviridae, (ii) Rhabdoviridae, and (iii) Chuviridae. Each bar represents a single EVE, while its length and position denotes the region of the indicated ORF from which its sequence is derived. (D) Phylogenetic relationship between 61 members of Flaviviridae. EVEs present in (i) Ae. aegytpi or (ii) Ae. albopictus which align to the indicated virus are marked with a colored circle. Size corresponds to abundance of EVEs derived from given species.
Comparison of EVEs in Ae. aegypti and Aedes albopictus genomes
EVEs serve as a record of viral infection over time. Ae. aegypti and Ae. albopictus species of mosquito occupy distinct (yet overlapping) regions around the globe [10] and have, therefore, faced different viral challenges throughout their evolution. The EVEome of these species of mosquito should reflect these differences [4, 11]. Indeed, in comparison to EVEs present in the Ae. aegypti-based genomes, which correspond well (see STAR methods), Ae. aegypti and Ae. albopictus do not share any specific EVEs. Closer examination of EVEs derived from the Flaviviridae family revealed that Ae. aegypti and Ae. Albopictus EVEs are derived from distinct but overlapping sets of viral species (Figure 1d).
Insights into the mechanism of EVE integration
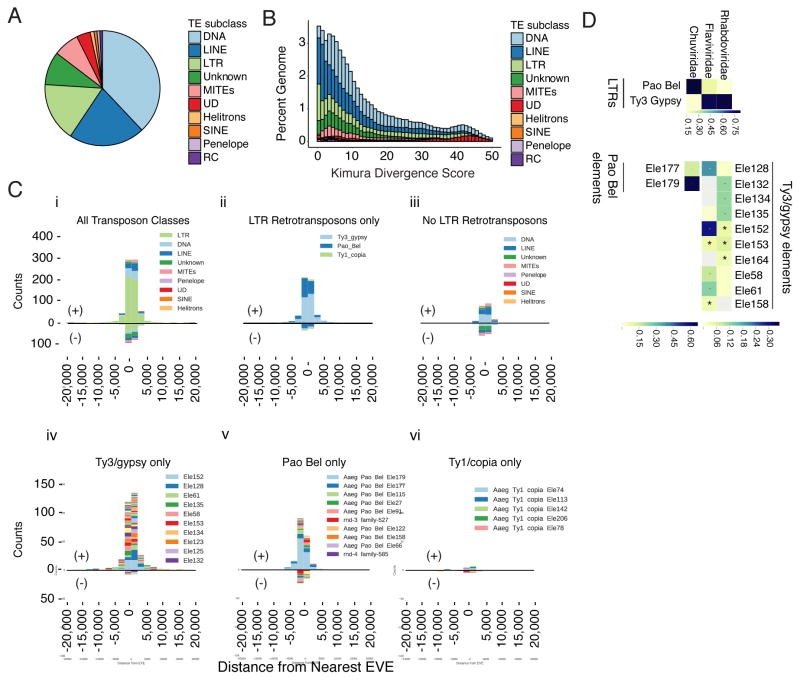
Transposable elements (TEs) provide an important source of genomic variation that drive evolution by modifying gene regulation and genome organization, and through the acquisition of new sequence information [12–14]. The acquisition, endogenization, and expansion of EVEs within the genome are also thought to be the result of transposable element activity [4, 5, 15–22]. TEs in the Aag2 genome are derived from several different families (Figure 2a). Kimura-distance based pair divergence analysis can be used to estimate the relative age of elements in the genome (Figure 2b) [23, 24]. Low Kimura scores indicate that sequences are closer to their shared consensus sequence, or ‘younger’, while higher scores indicate they are more divergent from each other, or ‘older’. The distributions of Kimura scores for TEs in our assembly indicate relatively recent expansions of LINE, LTR, and MITEs elements (Figure 2b). LTRs show a pronounced peak at the origin, indicating recent activity. This is consistent with reports that LTR-retrotransposon transcripts and proteins are readily detected in Aag2 cells [25].
Figure 2. The repeat landscape of the Aedes aegypti genome is predominantly made up of transposable elements.
(A) Pie chart representing relative numbers of repetitive elements in the Aag2 genome. Further detail can be found in Table S2.
(B) Stacked histogram of Kimura divergence for classes of TEs found in the Aag2 assembly, expressed as a function of percentage of the genome.
(C) Histograms showing counts of nearest, non-overlapping TEs closest to EVEs binned by distance, both upstream (negative x-axis values) and downstream (positive x-axis values). Positive y-axis counts refer to TE/EVE ‘pairs’ with the same strandedness, while negative y-axis counts are EVEs where the closest TE has the opposite strandedness.
(D) Heatmap showing categories of TEs nearest EVEs, categorized by the viral family from which the EVEs were derived. Scale indicates proportion of TEs of the indicated transposon family (see STAR methods for further discussion of Chuviridae).
The relationship between transposable elements and EVE integration [3, 26, 27] led us to explore the large-scale organization of TEs, specifically focusing on those proximal to EVE sequences. To identify mobile elements likely to be responsible for EVE genomic integration, we characterized the TEs surrounding EVE sequences (as called by RepeatMasker and BLASTX respectively, see STAR methods). LTR retrotransposons are greatly enriched among the nearest upstream and downstream, non-overlapping TEs nearest EVEs (Figure 2c(i)), most sharing the same polarity as their nearest EVE. This shared polarity (among EVEs derived from positive or negative stranded RNA viruses) suggests the main substrate for TE-virus recombination is viral mRNA, possibly due to the relative stoichiometry of positive- and negative-sense transcripts, or by a mechanism that selects coding strands.
Within the LTR retrotransposon family, both Ty3/gypsy and Pao Bel TEs are enriched surrounding EVEs (Figure 2civ,v). An association between LTR Ty3/gypsy elements and integrated viral sequence has also been observed previously in plants (and recently in adult mosquitoes) [17, 19, 20], suggesting a conserved mechanism for the acquisition and endogenization of viral sequences. We also observed an association between LTR and viral families. Rhabdoviridae and Flaviviridae associated with Ty3/Gypsy elements. However, Pao-Bel elements associated with Chuviridae EVEs are likely the result of recombination event that lead to acquisition of a Chuviridae-like glycoprotein by the Pao-Bel retrotransposon.(Figure 2d, see STAR methods for further discussion of Chuviridae) [28, 29].
EVEs associate with piRNA clusters
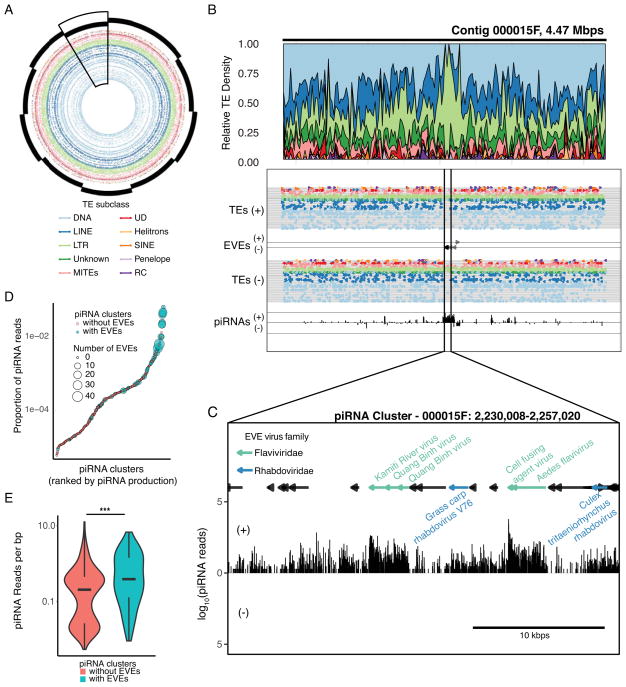
The strong enrichment for multiple LTRs around EVEs (Figure 2c, S3) led us to examine the genomic context of EVE-TE integration sites in the genome. The large contig sizes associated with our long-read sequencing approach allow us to assess the large-scale spatial distribution of TEs and EVEs in the genome. Strikingly, we identified numerous loci where many EVE sequences overlapped with large regions of increased LTR density, some larger than 50kbp in length (Figure 3a). In some cases, these large loci are so densely packed with a single LTR type they effectively “crowd out” any other repetitive elements (Figure 3b, S2). Within these loci, EVE sequences are interspersed with TE fragments in unidirectional orientations (Figure 3c). They contain large numbers of EVEs derived from a diverse set of viral families (Figure 3c) suggesting that these regions occasionally capture new TE-virus hybrids and are not solely the result of TE-EVE duplications in the genome.
Figure 3. EVEs are primarily associated with LTR transposable elements.
(A) Circle plot showing the arrangement and diversity of TE subclasses in the ten largest contigs in the Aag2 assembly. Individual contigs are denoted with staggered black bars. Specific TE elements are shown as dots, concentric rings represent individual families within each TE subclass. Large scale fluctuations in TE density can be seen in specific contigs (contig 000015F, boxed).
(B) Local density plots of a representative region of local LTR density in contig 000015F.
(C) The regions of local LTR density corresponds to the location of numerous EVEs (black arrows) and high piRNA density (bar chart, bottom track). Bioinformatically predicted piRNA cluster corresponding to a portion of the large LTR density in contig 000015F. LTRs (shown as black bars) are interspersed with EVE sequences (colored by virus family). piRNA production (black bars, below) shows highest density in regions corresponding to EVE sequences. See also Figure S2.
(D) Dotplot showing the relationship between piRNA cluster EVE content and piRNA production. piRNA clusters are ranked by piRNA production. (E) Violin plot comparing the distribution of piRNA density in piRNA clusters with or without EVEs. See also Figure S3.
The organization of these large, LTR-dense loci is similar to that of piRNA clusters [30]: piRNA-producing loci in the genome that result from the accumulation of TE fragments (due to non-random LTR integrase-directed integration)[31]. To assess the ability of these loci to produce piRNAs, we performed small RNA sequencing, employing a procedure to enrich for bona fide piRNAs (see STAR methods). Furthermore, small RNAs were extracted in the context of a Sindbis virus (SINV) infection, to better assess the small RNA response from EVEs in the context of an acute viral infection. Indeed, EVE loci produce a large number of piRNAs in a predominantly anti-sense orientation, consistent with the transcription of piRNA clusters [2, 30] (Figure S4).
We then used bioinformatic prediction to identify putative piRNA clusters in the genome based upon piRNA mapping density. This analysis identified 469 piRNA-encoding loci (piRNA clusters) using proTRAC [32, 33], accounting for 5,774,304 bp (0.335%) of the genome. To examine the functional significance of piRNA cluster-resident EVEs, we measured the relative piRNA output from piRNA clusters throughout the genome. piRNA clusters that contain EVEs tend to produce more piRNAs (Figure 3d,e, S3A), suggesting that EVEs tend to integrate into the most active piRNA clusters in the genome, and selection may drive these to maintain higher expression.
piRNA abundance reflects the cellular immune state
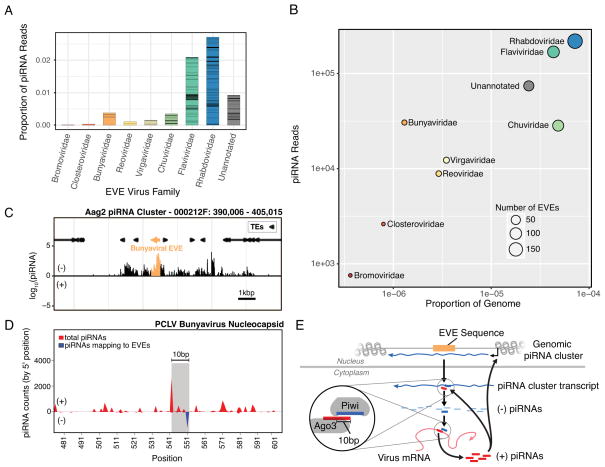
EVEs, in combination with their associated piRNAs, make up an immune archive of small of RNAs. In the Aag2 cell line, piRNA production from EVEs derived from a given viral family does not completely correlate with genomic footprint of those EVEs (Figure 4a,b). This suggests that the antiviral potential of the EVEome against a given viral family is a function of the amount of viral genetic information stored in the host genome and the transcriptional activity of individual piRNA clusters. Another important determinant of the antiviral potential of EVE sequences is their sequence identity to circulating viral challenges.
Figure 4. The antiviral potential of the cellular piRNA repertoire.
(A) Bar plot showing the proportion of piRNA mapping to EVE sequences from a given viral family. Bars are split to show the relative contribution of specific EVE sequences.
(B) Plot showing correlation the genomic footprint of EVEs from specific viral families in the Aag2 genome and their piRNA production. Note Bunyavirus and Chuvirus fall on opposite sides of the trend. See further discussion of Chuviridae in the STAR methods.
(C) The EVE responsible for producing the anti-sense piRNA in (D). A spike of piRNAs is produced from the EVE (highlighted in orange) within the overall piRNA cluster. See also Figure S4.
(D) Mapping of cellular piRNAs to the bunyavirus PCLV nucleocapsid coding sequence reveals the pattern of piRNA processing. The sense-piRNA peak is offset by 10-bp from the antisense-piRNA peak (which also maps to an EVE within the Aag2 genome; blue line), showing a distinct ping-pong like pattern (highlighted by the grey rectangle). Interestingly, although the sequence of the antisense piRNAs map perfectly to the Aag2 genome/EVE, the sense piRNAs map perfectly to the PCLV virus sequence. See also Figure S4. (E) Schematic showing the process of ping-pong piRNA amplification in the cell. An EVE sequence in a piRNA cluster is transcribed to yield an antigenomic transcript. This transcript is processed into piRNAs which bind the genome (or mRNA) of infecting viruses. Binding triggers processing of the viral target into piRNAs. These positive sense piRNAs bind antigenomic transcripts leading to further processing, but can also regulate piRNA cluster transcription epigenetically.
To examine the potential antiviral activity of cellular piRNAs that originate from genomic EVEs, we mapped the same piRNA libraries (allowing for up to 3 mismatches) to contemporary viral sequences from which EVEs were expected to have derived (Figure 4d). Aag2 cells are known to be persistently infected with cell-fusing agent virus (CFAV; flaviviridae), and were recently shown to also be persistently infected with Phasi Charoen-like virus (PCLV), a bunyavirus [25]. These viruses constitute available targets for recognition by EVE-derived piRNAs and subsequent processing. EVE-derived anti-sense piRNAs (Figure 4c) only mapped to a single site on the PCLV nucleocapsid. However, we identified numerous sense piRNAs derived from PCLV, including a prominent peak which is offset from the EVE-derived, anti-sense piRNA binding site by 10bps (Figure 4d,e). This pattern is consistent with canonical processing of EVE-derived piRNAs by the ping-pong amplification mechanism, which begins with their successful loading onto the Piwi proteins, methylation, and subsequent cleavage and processing of the PCLV viral mRNA. The subsequent feedback mechanism (Figure 4e) for amplification potentially explains the increased piRNA output of Bunyavirus-derived EVEs (Figure 4b). Consistent with this, the vast majority of these piRNAs bear the molecular signature of this amplification and are dependent on its machinery (Figure S4A, Bii/iii). Furthermore, levels of viral PCLV RNA increase around two-fold upon knockdown of Piwi4 (Figure S4C). These observations support the notion that an organism’s EVEome produces piRNAs capable of recognizing viruses and initiating an active response.
DISCUSSION
A solid foundation with which to study the genetic determinants of vector competence is of utmost importance as arboviruses become an increasing burden globally. With this in mind, we generated a long-read assembly of the Aedes aegypti cell line, Aag2 and used this highly contiguous assembly to identify a more complete set of endogenous viral elements and their surrounding genomic context in the Aag2 cell line at a genome-wide scale. The Aag2 cell line is an important model system for the characterization of arboviral replication in mosquito hosts. Considering the potential impact of EVE sequences and their associated piRNAs on viral infection, understanding the diversity of EVEs in commonly used cell lines is especially important.
Surveying the genome-wide collection of EVEs in the Aag2 genome provides not only a view of the historical interactions between host and virus, but also the repertoire of acquired sequences that define the piRNA-based immune system of this important model system. Our analysis refines our understanding of EVEs in the Ae. aegypti genome, their relationship to transposable elements, and the potential breadth of antiviral protection they provide. We propose that a mosquito’s EVEome, together with the piRNA system, represents a potentially long-lasting branch of its RNAi anti-viral defense system. Although all mosquito species share the same basic RNAi-based immune system, the differences in the EVEome of a given species, subpopulation, or individual, such as those observed between Ae. aegypti and Ae. albopictus (Figure 1a) [17], may represent a factor contributing to inherent differences in vector competence across many different scales. Indeed, the EVEome of wild mosquito populations appears to be in rapid flux [34].
The presence of piRNA producing EVEs in the Ae. aegypti genome is reminiscent of other nucleic acid-based antiviral defense systems [35]. These systems take advantage of the invading pathogen’s genetic material to create small RNAs capable of restricting an invading virus’ replication. Furthermore, both involve endogenization of viral sequences, potentially providing protection against infection across generations. The extent of conservation of this pathway across Eukarya is not yet clear, however recent publications have highlighted the endogenization of genetic material from non-retroviral RNA viruses into the genome of many different host species [5, 17, 36]. The antiviral activity of these sequences has not been established, however a subset of EVEs found in mammalian systems appear to be under purifying selection, suggesting some potential benefit to the host [22]. In contrast to the evolutionary repurposing of retroviral sequences [12–14], the direct integration, transcription and processing of EVE sequences into antiviral small RNAs constitutes a mechanism by which these acquired sequences can be rapidly repurposed for host immune purposes.
Our analysis revealed a strong relationship between LTRs and EVE sequences. Template switching during reverse transcription has previously been proposed to play a role in transposon-virus hybrid generation and subsequent integration [37, 38]. However, the apparent specificity of Ty3/gypsy TEs for EVE integration may suggest a deeper relationship (Figure 2d). This could have occurred by chance (recent LTR replication and virus infection happened to coincide), or may hint at a specific mechanism directing capture of viral sequences by LTRs. In some cases these TEs and viruses may share increased sequence homology leading to more frequent template switching [39], or replicate in a similar subcellular location. Given the difference in piRNA production among piRNA clusters with EVEs and without (Figure 3e, S3a), selection may act primarily at the level of piRNA production, suggesting that specific piRNA-producing LTR/EVE pairs were maintained by selection after EVE integration.
piRNA abundance reflects the underlying transcriptional activity of individual piRNA clusters. Whether EVEs integrate into transcriptionally active piRNA clusters, or whether selection drives EVE-containing piRNA clusters to become more active is unknown. However, LTRs (and specifically Ty3 elements) located in piRNA clusters without EVEs produce, on average, more piRNAs per bp than other classes of TEs (Figure S3B,C), likely reflecting the increased LTR activity in Aag2 cells [25]. The regulation of the transcriptional activity of piRNA clusters is mediated by epigenetic changes that respond, in part, to active ping-pong processing of target transcripts in the cytoplasm [40]. Therefore, the presence of active LTRs in the cell line will likely facilitate EVE insertion into activated piRNA clusters targeting these elements, potentially explaining the overwhelming prevalence of EVEs among LTR/Ty3_gypsy elements.
Uncovering the genomic context of EVEs highlights the potential for the piRNA system to shape the mosquito immune system. It also provides a foundation for future investigations into EVE function. Comparative genomic approaches that incorporate long-read sequencing to understand the diversity of the EVEome across populations will allow us to better understand the forces that underlie the epidemiology and population dynamics of arboviruses. Moreover, the potential to manipulate this heritable, anti-viral immune system could present opportunities for epidemiological interventions in natural settings, or as a genetic system to understand the insect immune system in the laboratory.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Raul Andino (raul.andino@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Aedes aegypti Aag2 [41, 42] cells were cultured at 28 °C without CO2 in Schneider’s Drosophila medium (GIBCO-Invitrogen), supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1X non-essential amino acids (NEAA, UCSF Cell Culture Facility, 100X stock is 0.1 μM filtered, 10 mM each of Glycine, L-Alanine, L-Asparagine, L-Aspartic acid, L-Glutamic Acid, L-Proline, and L-Serine in de-ionized water), and 1X Penicillin-Streptomycin-Glutamine (Pen/Strep/G, 100X = 10,000 units of penicillin, 10,000 μg of streptomycin, and 29.2 mg/ml of L-glutamine, Gibco). Cells were authenticated by genomic sequencing (as presented in this paper).
METHOD DETAILS
DNA sequencing
Aag2 cells were grown in T-150 flasks until ~80% confluent. Cells were then washed with dPBS twice and scrapped off in dPBS + 10 μg/ml RNase A (ThermoFisher). Genomic DNA (gDNA) was extracted from ~10^8 Aag2 cells using the QIAamp DNA Mini Kit according to the manufacturer’s instructions with the optional RNase A treatment. Aag2 gDNA was re-suspended in 10mM Tris pH8, and the quality and quantity of the sample was assessed using the Agilent DNA12000 kit and 2100 Bioanalyzer system (Agilent Technologies), as well as the Qubit dsDNA Broad Range assay kit and Qubit Fluorometer (Thermo Fisher) and visualized by gel electrophoresis (1% TBE gel). After purification and quality control, a total of 130 ug of DNA was available for library preparation and sequencing.
SMRTbell libraries were prepared using the PacBio SMRTbell Template Prep Kit 1.0 and a slightly modified version of the PacBio protocol, “Procedure & Checklist - 20-kb Template Preparation Using BluePippin Size-Selection System (15-kb Size Cutoff)”. Specifically, 52.5ug of gDNA were hydrodynamically sheared to target sizes of 30kb (26 μg) and 35 kb (26 μg) using the Megaruptor® (Diogenode) with long hydropores according to the manufacturer’s protocols. Size distributions of the final sheared gDNA were verified by pulse field electrophoresis of a 100ng sub-aliquot through 0.75% agarose using the Pippin Pulse (Sage Science), run according to the manufacturer’s “10–48 kb protocol” for 16 hrs. The two sheared samples were then pooled, for a total of 37ug sheared DNA to be used as input into SMRTbell preparation. Sheared DNA was subjected to DNA damage repair and ligated to SMRTbell adapters. Following ligation, extraneous DNA was digested with exo-nucleases and the resulting SMRTbell library was cleaned and concentrated with AMPure PB beads (Pacific Biosciences). A total of 20.5ug of library was available for size selection.
Approximately half (10ug) of the SMRTbell pooled SMRTbell library was size-selected using the BluePippin System (Sage Science) using a 15 kb cutoff and 0.75% agarose cassettes. To obtain longer read lengths, an additional 5ug of the library was selected using a 17kb cutoff.
Library quality and quantity were assessed using the Agilent 12000 DNA Kit and 2100 Bioanalyzer System (Agilent Technologies), as well as the Qubit dsDNA Broad Range Assay kit and Qubit Fluorometer (Thermo Fisher). An additional DNA Damage Repair step and AMPure bead cleanup were included after size-selection of the libraries.
Annealed libraries were then bound to DNA polymerases using 3nM of the SMRTbell library and 3X excess DNA polymerase at a concentration of 9nM using PacBio DNA/Polymerase Binding Kit P6 v2.
Bound libraries were sequenced on the PacBio RSII using P6/C4 chemistry (PacBio DNA Sequencing Reagent Kit 4.0 v2), magnetic bead loading (PacBio MagBead kit v2) and 6 hour collection times. 84 SMRTcells (v3) of the > 15 kb library were loaded at concentrations of 75–100 pM on-plate. 32 SMRTcells of the > 17 kb library was prepared separately and loaded at on-plate concentrations of 40 pM and 60 pM. These 116 SMRTcells generated 92.7 GB of sequencing data, which resulted in approximately 76X coverage of the Aag2 genome. Average polymerase read lengths, and average subread lengths were 15.5KB and 13.2 kb, respectively.. Assembly was performed using FALCON [43] and polished with Quiver [44].
Genome assembly statistics
Basic statistics (e.g. Size, Gaps, N50, L50, # contigs) for each genome analyzed was produced using Quast [45].
As a complementary approach Benchmarking sets of Universal Single-Copy Orthologs (BUSCO) [46] was also run using the Arthropod dataset in order to assess the completeness of genome assembly. Of the 2675 BUSCO groups searched only 81 were missing from the Aag2 assembly, indicating good assembly completeness. Of the 2315 BUSCOs found only 279 of them were annotated as fragmented, emphasizing the continuity of the assembly.
Viral stock and preparation
Original Sindbis stock [47] was propagated in BHK cells to generate the P1 stock at 24 hours post infection. Virus was titrated by plaque assay by infecting confluent monolayers of BHK cells with serial dilutions of virus. Cells were incubated under an agarose layer for 2 to 3 days at 37°C before being fixed in 2% formaldehyde and stained with crystal violet solution (0.2% crystal violet and 20% ethanol).
Repeat Identification and Kimura Divergence
In order to de novo identify and classify novel repetitive elements from the Aag2 genome, RepeatModeler[48] was run on the assembled genome using standard parameters. Outputs from RepeatModeler were cross-referenced with annotated entries for Aedes aegypti from TEfam. All entries from RepeatModeler that were >80% identical to TEfam entries were discarded as redundant. This combined annotated and de novo identified list of repeat elements was used to identify the genome wide occurrences of repeats using RepeatMasker[49] using standard parameters.
Kimura scores and corresponding alignment information were extracted from the “.align” file as output by RepeatMasker. Visualization of TE Kimura scores was done using R (version 3.30) [50] and the ggplot2[51] package (see Data and Software Availability section).
EVE identification
Identification of EVEs was achieved using standalone Blast+ [52]. Blast Searches were run using the Blastx command specifying the genome as the query and a refseq library composed of the ssRNA and dsRNA viral protein-coding sequences from the NCBI genomes as the database. The E-value threshold was set at 10−6 (see Data and Software Availability section).
The EVE with the lower E-value was chosen for further analysis to predict EVEs that overlapped. Several Blast hits to viral protein genes were identified as artifacts because of their homology to eukaryotic genes (e.g. closteroviruses encode an Hsp70 homologue). These artifacts were filtered by hand.
Identification of LTR enrichment near EVEs
Separate BED files containing all TEs in the Aag2 assembly and all EVEs in the Aag2 assembly were used as input to Bedtools [53] (bedtools closest command using the –io flag, and –id or -iu) to find the single closest non-overlapping TE to each EVE (both upstream and downstream).
An in-house script compiled these two output files together and filtered them for the TE content of interest. TE categories (subclass, family, element) were assigned by RepeatMasker. TEs with taxonomy assignments of LTR/Gypsy, LTR/Copia, LTR/Pao were renamed LTR/Ty3_gypsy, LTR/Ty1_copia, and LTR/Pao_Bel respectively for consistency with other TE assignments in the assembly. Enrichment was compared to the prevalence of the TE element genome wide based on a one-sided binomial test. The legend lists (up to) the 10 most prevalent TE elements of TE/EVE pairs in the same orientation. Plots were produced using Python (version 2.7.6) [54] with the pandas [55] and matplotlib [56] plugins.
Of 614 LTRs neighboring EVEs, 543 shared the same polarity as their nearest-neighbor EVE (i.e. both elements are located on the same genomic DNA strand). These 543 LTRs made up the vast majority of all TEs with the same polarity as their nearest EVE (543/746; Figure 2c(i); p-value = 1.09x10−252 by one-sided binomial test).
Enrichment for Ty3/gypsy and Pao Bel elements near EVE loci is strongest when the EVE and TE are in the same orientation (p-value = 6.90x10−29 and 1.71x10−3 respectively).
Total counts represented in each histogram: All classes (n=942); LTR only (n=614); No LTRs (n=328); Ty3/gypsy only (n=358); Pao Bel only (n=226); Ty1/copia only (n=30).
TEs nearest Chuviridae-derived sequences exhibit a distinct pattern from those surrounding other viral families (i.e. Flaviviridae and Rhabdoviridae). Chuviridae-derived EVEs (primarily originating from GP proteins) are chiefly associated with one Pao-Bel element (Pao-Bel element 179). Moreover, this element exhibits a high degree of spatial overlap in the genome with its respective EVEs (as called by RepeatMasker and BLASTx respectively). It is known that only a subset of Pao-Bel elements contain an envelope protein, and that LTR-retrotransposons typically acquire env sequences from viral glycoproteins [29]. The similarity and overlap between these Chuviridae EVEs and Pao-Bel element 179 raises the interesting possibility that many Chuviridae EVEs are in fact part of Pao Bel element 179.
Classification of nearest TE to EVEs by virus taxonomy
Taxonomy categories for viruses from which each EVEs derived were assigned using an in-house script (see Data and Software Availability section). Assignments were made based on NCBI’s taxonomy database (ftp://ftp.ncbi.nih.gov/pub/taxonomy/), with the following additional annotations by hand. Viruses assigned to Chuviridae family: Wuhan Mosquito Virus 8, Wuchang Cockraoch Virus 3, Lishi Spider Virus 1, Shayang Fly Virus 1, Wenzhou Crab Virus 2. Viruses assigned to Rhabdoviridae family: Bole Tick Virus 2, Shayang Fly Virus 2, Wuhan Ant Virus, Wuhan Fly Virus 2, Wuhan House Fly Virus 1, Wuhan Mosquito Virus 9, Yongjia Tick Virus 2. Viruses assigned to Virgaviridae family: Cilv-C, Citrus leprosis virus C, Blueberry necrotic ring blotch virus. Viruses assigned to Bunyaviridae: Wutai Mosquito Virus.
Heat maps were produced using the Seaborn [57] plugin for python. Only TEs with >=10% proportion in at least one sample (Flaviviridae, Chuviridae, or Rhabdoviridae) are shown. Color was assigned based on proportion of TE element/family in each viral category. Grey indicates the element was not found to be the closest TE to any EVEs derived from the indicated viral family. “Pao Bel elements” refers to Chuviridae, while “Ty3/gypsy elements” corresponds to Flaviviridae and Rhabdoviridae. Enrichment was scored as above using a one-sided binomial test against the background prevalence of a given TE category in the genome (e.g. among all LTRs nearest Chuviridae-derived EVEs, Pao Bel elements are specifically enriched compared to the genome-wide counts of Pao Bel among all LTRs). Only TEs with the same strandedness as its nearest EVE were used.
Total sample size of all TEs analyzed for each dataset: LTRs- Rhabdoviridae (n=130), Flaviviridae (n=181), Chuviridae (n=107); Pao Bel- Chuviridae (n=84); Ty3/gypsy- Rhabdoviridae (n=100), Flaviviridae (n=136).
dsRNA preparation
PCR primers including the T7 RNA polymerase promoter were used to amplify in vitro templates for RNA synthesis using Phusion polymerase (NEB). Manufacturer’s recommendations were used for the concentrations of all reagents in the PCR. Primers were synthesized by Integrated DNA Technologies, Inc. (IDT). The thermocycling protocols were as follows: 98°C 2:00, (98°C 0:15, 65°C 0:15, 72°C 0:45, these three cycles were repeated 10X with a lowering of the annealing temperature by 1°C per cycle); (98°C 0:15, 60°C 0:15, 72°C 0:45, these three steps were repeated 30 X), 72°C 2:00.
RNA was synthesized in a 100 μl in vitro transcription (IVT) reaction containing 30 μl of PCR product, 20 μl 5X IVT buffer ( 400 mM HEPES, 120 mM MgCl2, 10 mM Spermidine, 200 mM DTT), 16 μl 25 mM rNTPs, and 1 unit of T7 RNA polymerase. The IVT reaction was incubated @ 37°C f or 3–6 hours and then 1 μl of DNase-I (NEB) was added and the reaction was further incubated at 37°C for 30 min. The RNA was purified by phenol-chloroform-isoamyl alcohol followed by isopropanol precipitation. RNA was quantified using a Nanodrop (Thermo Scientific) and analyzed by agarose gel electrophoresis to ensure integrity and correct size.
dsRNA soaking
Prior to dsRNA soaking, Aag2 cells were washed once with phosphate buffered saline w/o calcium or magnesium (dPBS, 0.1 μM filtered, 0.2g/L KH2PO4, 2.16g/L Na2HPO4, 0.2g/L KCl, 8.0g/L NaCl). Cells were soaked in 5 μg /ml dsRNA in minimal medium (Schneider’s Drosophila medium, 0.5% FBS, 1% NEAA, and 1% Pen/Strep/G) for the time indicated by the experiment. All incubations were performed at 28 °C without CO2.
Sindbis virus infection of Aag2 cells treated with dsRNA
Three days after dsRNA soaking, dsRNA-treated Aag2 cells were infected with Sindbis virus (MOI = 0.1). Infection was done in culture media with 1% FBS for 1 hour at room temperature. Cells were then rinsed with PBS and incubated in complete culture media (10% FBS) for 4 days at 28°C before being harvested and processed for RNA extraction with Trizol (Ambion).
Beta-elimination and deep sequencing of small RNAs
7 x 106 Aag2 cells were seeded in each T-75 flask in complete medium and allowed to attach overnight. Cells were washed with dPBS three times, scraped off the dish in dPBS, and centrifuged at 2000 rcf for 5 min at 4°C. RNAs were isolated using the miRvana kit (Life technologies). The large RNA fraction was used for RT-qPCR. The small RNA fraction was precipitated by adding 1/10th volume 3M NaOaC pH 3.0, 1 μl gylcoblue (Life technologies), and 2.5 volumes 100% EtOH and incubated at −80°C at least 4 hours and then centrifuged at 12000 rcf for 10 min at 4°C. The pel let was washed with 80% EtOH and then resuspended in Gel Loading Buffer II (Life Technologies) and run on a 20% polyacrylamide gel containing 8M urea. Small RNAs (17–30 nt) were cut out from the gel and eluted overnight at 4°C an d precipitated by adding 1/10th volume 3M NaOaC pH 3.0, 1 μl gylcoblue (Life technologies), and 2.5 volumes 100% EtOH and incubated at −80°C at least 4 hours a nd then centrifuged at 12000 rcf for 10 min at 4°C. After gel size extract ion, 10 pmoles of small RNAs (17–30nt) in 6.75 μL RNAse-free water were mixed with 2 μl 5× borate buffer (0.12 M, pH 8.6) and 1.25 μL sodium periodate (42mg/mL made fresh). The reaction was incubated in the dark at room temperature for 10 min. 2μl 50% glycerol was added to the reaction and incubation was continued in the dark for 10 min at room temperature. The excess of liquid was then evaporated in a Speedvac at top speed for 1 hour (no heat) and then resuspended in 50 μl 1× borax buffer (0.06 M, pH 9.5), incubated for 90 min at 45°, and then precipitated. Small RNAs were cloned using microRNA Cloning Linker1 (IDT) for the 3′ ligation and the modified 5′ adapter with randomized 3′end (CCTTGrGrCrArCrCrCrGrArGrArArTrTrCrCrArNrNrNrNrN), and run on a HiSeq 2500 using the Rapid run protocol.
Small RNA bioinformatics and mapping conditions
Adaptors were trimmed using FASTX toolkit (fastx clipper) [58].
Small RNA reads as they mapped to the entire Aag2 assembly (using bowtie [59] with - v1 flag) were used to calculate normalized piRNA cluster piRNA output (Figure 3).
To further control for the repetitiveness of the transposon and EVE sequences prevalent throughout the Aag2 assembly, piRNAs were also mapped to a FASTA file consisting of non-TE, non-EVE sequences within piRNA clusters (using bowtie, -v 0 –m 1) (Figure S3a). The FASTA file containing only non-TE, non-EVE sequences was generated using bedtools’ subtract function, removing coordinates corresponding to TEs and EVEs from a .bed file initially containing coordinates of piRNA clusters (as determined by proTRAC, see below). piRNA cluster production was calculated on a per piRNA cluster basis, adding together counts of all uniquely mapping piRNAs to non-TE, non-EVE sequence of a given cluster, and dividing by the total length of a given piRNA cluster’s non-TE, non-EVE sequence. As the y-axis is logarithmic, only non-zero values are shown. Significance between the datasets was performed using a one-sided Mann Whitney U test using only non-zero values (because non-TE, non-EVE sequences tended to be significantly shorter in piRNA clusters without EVEs).
Determination of piRNA output from a given TE class or family was performed by mapping piRNAs to a FASTA file of all TE sequence contained within piRNA clusters (using bowtie; - m 1 v 0). Uniquely mapping piRNA counts were used to calculate piRNA production of TEs of a given family or class in piRNA clusters without EVEs. TE production was calculated on a per TE sequence basis, adding together counts of all uniquely mapping piRNAs to a given TE sequence fragment, and dividing by the total length of that TE sequence (yielding piRNA counts per TE per base pair). These values were then grouped by the class/family from which the TE derived for visualization (zero values are not shown on the plot). Order of x-axis is based on the mean values of each group, including zeros.
Small RNA size distributions were generated using the ‘.map’ output file of bowtie (and fastx_collapser [58]) and visualized with an in-house R script using the ggplot package. Sequence logo analysis of piRNAs mapping to EVEs and PCLV was done using the R package seqLogo [60] (see smallRNApopulationCharacterization.R). Visualization of ‘ping-pong piRNAs mapping to PCLV was done using an in-house R script (see piRNAmappingToViralGenomes_pingPongOneGenome.R). Mapping of piRNAs to PCLV nucleocapsid CDS allowed up to three mismatches (bowtie – v 3)
n-values for figures S3a,b,c
Figure S3a: piRNA clusters without EVEs: n = 169 (202 additional clusters had a value of 0); piRNA clusters with EVEs: n = 54 (8 additional clusters had a value of 0).
Figure S3b: DNA (n = 894; 759 of which are 0); Helitrons (n = 51; 37 of which are 0); LINE (n = 1158; 963 of which are 0); LTR (n = 1558; 908 of which are 0); MITEs (n = 209; 172 of which are 0); Penelope (n = 22; 14 of which are 0); RC (n = 69; 67 of which are 0); SINE (n = 15; 10 of which are 0); UD (n = 77; 63 of which are 0); Unknown (n = 573; 507 of which are 0);
Figure S3c: Pao Bel (n = 317; of which 260 are 0); Ty1/copia (n = 136; of which 92 are 0); Ty3/gypsy (n = 1105; of which 556 are 0)
piRNA cluster Analysis
piRNA clusters were identified using proTRAC [32] based on mapping with positions for beta-eliminated small RNAs libraries from Aag2 cells from sRNAmapper.pl. Based on these predictions, visualizations of clusters were produced using EasyFig [61] for visualization of TEs and R for comparison of TEs, piRNA abundance and EVE positions.
Depending on the mapping algorithm used, between 63% (bowtie) and 77% (sRNAmapper.pl, see Methods) of beta-eliminated small RNAs from Aag2 cells mapped to these loci. Of the identified piRNA clusters, 65 (14.1%) have EVE sequences associated with them and 64 of these piRNA cluster-resident EVE sequences act as the template for piRNAs. Of the 472 EVEs identified, 256 (66.7%) or 280,475 bp of the 411,239 EVE bp mapped to piRNA clusters (68.2%, Fisher’s test p<2.2e-16, OR=203.42).
Phylogenetic analysis of Flaviviridae polyprotein sequences
For phylogenetic analysis of Flaviviridae, polyprotein sequences from 61 members of the Flaviviridae family were aligned with MUSCLE [62] and a maximum likelihood tree was generated with FastTree [63] using the generalized time reversible substitution model (“-gtr”). Trees were visualized and annotated with ggtree [64].
EVE coverage
Base R (version 3.3.0) was used to show regions individual EVEs span on the indicated viral family (and protein). EVE length is expressed as a percentage of the total ORF to normalize for varying ORF lengths among different members of a given viral family. The genome organization of CFAV is presented for reference in Figure 1C(i). In (ii) and (iii), a generic genome is presented to illustrate from where EVEs are derived within the genome and within each specific ORF.
QUANTIFICATION AND STATISTICAL ANALYSIS
Enrichment for types of TE elements near EVEs was determined with a one-sided binomial test (alternative hypothesis ‘greater’), comparing the TE type of interest near EVEs to its prevalence in the Aag2 assembly (e.g. among all LTRs nearest EVEs, Ty3/gypsy elements are specifically enriched compared to the genome-wide counts of Ty3/gypsy among all LTRs). Discussion of these statistical tests and n-values are in the STAR methods (‘Identification of LTR enrichment near EVEs’ section).
Enrichment for TE elements near EVEs derived from a particular viral family was determined in the same way (significance required a p-value < 0.0001). A “*” indicates significant enrichment by one-sided binomial test against the background prevalence of a given TE category in the genome (eg among all LTRs nearest Flaviviridae-derived EVEs, Ty3/gypsy elements are specifically enriched compared to the genome-wide counts of Ty3/gypsy among all LTRs). Color indicates proportion of a given TE category nearest EVEs derived from the indicated viral family. Grey indicates the element was not found to be the closest TE to any EVEs derived from the indicated viral family. Only TE elements which made up at least 10% of the dataset for a given viral family are shown. Discussion of these statistical tests is in the results section, while n-values are in the STAR methods (‘Classification of nearest TE to EVEs by virus taxonomy’ section).
Enrichment of EVEs in piRNA clusters was determined by Fisher’s Test. P- and n-values are discussed in the STAR methods.
The enrichment for piRNA production of EVE-containing piRNA clusters vs. those without EVEs (Figure 3) was performed using a Mann-Whitney U test.
Significance of increased piRNAs from non-TE, non-EVE sequence in piRNA clusters with EVEs vs. without EVEs was done with a one-sided Mann-Whitney U test. This test did not include any values of 0 (Figure S3a).
Significance of increased piRNAs from LTRs and Ty3/gypsy was done with a one-sided Mann-Whitney U test. This test did include values of 0 (Figure S3b,c).
DATA AND SOFTWARE AVAILABILITY
Any datasets or custom scripts not provided here are available upon request.
Datasets generated during this paper
The Aag2 genome (v 1.00) is available through VectorBase (https://www.vectorbase.org/organisms/aedes-aegypti/aag2/aag2).
The following datasets in Mendeley data (doi: doi:10.17632/d6zf6fvzwn.1) :
Aag_Contigs_EVEs_NEW.bed – A .bed file giving the coordinates of EVEs identified in our Aag2 assembly.
TEsClosestToEVEs_nearestOnly_withEVEtaxonomy.txt – For every EVE, gives nearest upstream and downstream TE. Overlapping TEs are ignored. The last column gives distance between EVE and TE.
TEsClosestToEVEs_overlapOnly_withEVEtaxonomy.txt – List of TEs which were found to overlap EVEs sequences (ie RepeatMasker designated TE coordinates overlapped with our EVE identification pipeline coordinates). The last column gives distance between EVE and TE. A distance of 0 indicates the two sequences overlap by at least one base pair.
TEsClosestToEVEs_overlapOrNearest_withEVEtaxonomy.txt - List of TEs which were found to overlap EVEs sequences. If an EVE does not overlap with an EVE, its nearest TE is listed (both upstream and downstream). The last column gives distance between EVE and TE. A distance of 0 indicates the two sequences overlap by at least one base pair.
Aag2_piClusters.bed- A .bed file of all piRNA clusters in our Aag2 assembly as called by protract
Aag2_TEs.bed- A .bed file of all TEs in our Aag2 assembly as called by repeat masker
small RNA sequencing reads (dsFluc + SINV, dsAgo3 + SINV, and dsPiwi4 + SINV) have been deposited in the NCBI SRA database under accession codes SAMN07513338, SAMN07514688, and SAMN07514689 respectively.
EVE_TE_pi_Analysis_byClust.csv – tabulation of EVEs and piRNAs mapping to individual piRNA clusters.
EVE_TE_pi_Analysis_byEVE.csv- tabulation of piRNAs mapping to individual EVEs.
fc_run.cfg – Settings used for genome assembly with FALCON.
Custom scripts used in this paper
NAMESdmp_cleanup.sh- Filters names.dmp to only include scientific names. A good idea to run this before ‘EVEgrouping.sh’.
EVEgrouping.sh – Uses the below scripts to take in a file where one column contains virus names (column headers must be manually specified), and then uses names.dmp and nodes.dmp from taxdump.tar.gz at ftp://ftp.ncbi.nih.gov/pub/taxonomy to return the taxonomy (order, family, genus, species, etc…) of that virus, each designation being its own column. These designations are then merged to the original file.
extractVirusNamesForEVEgrouping.sh
IdentifyRankForEVEgrouping.sh
ClassifyEVEtaxonomy.py
piRNAmappingToViralGenomes_pingPongOneGenome.R- Takes the .map file generated by bowtie (using a fastx collapsed small RNA fasta file) after mapping to a single (viral) genome, and quantifies piRNA mapping positions by their 5′ ends. Produces a graph showing sense and antisense peaks. Identifies sense and antisense piRNAs which are offset by 10bp. Also takes a .map file of small RNAs mapped to EVEs to check whether a piRNA also maps to an EVE.
smallRNApopulationCharacterization.R – Uses a .map file from bowtie (fastx collapsed) and creates a seqLogo, and a histogram of size distribution.
ExtractKimuraScores.sh – Uses ‘.align’ file from repeat masker to extract each repeat assignment and its corresponding Kimura score (when available) in two different files.
KimuraClustering.py – Merges the two files from ‘ExtractKimuraScores.sh’ for visualization.
KimuraVisualization.R – Plots Kimura score distributions TE classes vs. percent of genome.
EVE_finder.sh – coordinates the below scripts to identify EVEs in a genome assembly.
BLAST_NR_filter.py
Blast_to_Bed3.py
Index_Genome_Bedtools.sh
RNA_and_ssDNA_viral_TAXIDs
TAX_check4.py
Top_score_BED2.py
Supplementary Material
Highlights.
Long-read draft assembly of the repeat-rich Aag2 cell line genome.
Genome-wide identification of EVEs using the improved assembly.
Description of the large-scale organization of the genomic loci harboring EVEs.
Evidence supporting antiviral function of piRNAs produced from EVEs.
Acknowledgments
We thank members of Judith Frydman’s lab and the Andino lab for helpful discussions. PTD acknowledges support from F32 GM113483-01. ZJW was supported in part by MPHD T32 grant 5T32AI060537-14.
Footnotes
Author Contributions
ZJW, PTD, MK, MT, and RA designed the experiments; ZJW, PTD, MK, and MT conducted the experiments; ZJW, PTD, and RA wrote the paper; MK, MGS, SO, CH, and EP performed the genome sequencing/assembly.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mongelli V, Saleh MC. Bugs Are Not to Be Silenced: Small RNA Pathways and Antiviral Responses in Insects. Annu Rev Virol. 2016;3:573–589. doi: 10.1146/annurev-virology-110615-042447. [DOI] [PubMed] [Google Scholar]
- 2.Czech B, Hannon GJ. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci. 2016;41:324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miesen P, Joosten J, van Rij RP. PIWIs Go Viral: Arbovirus-Derived piRNAs in Vector Mosquitoes. PLoS Pathog. 2016;12:e1006017. doi: 10.1371/journal.ppat.1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parrish NF, Fujino K, Shiromoto Y, Iwasaki YW, Ha H, Xing J, Makino A, Kuramochi-Miyagawa S, Nakano T, Siomi H, et al. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA. 2015;21:1691–1703. doi: 10.1261/rna.052092.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017 doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicoso B, Bachtrog D. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 2015;13:e1002078. doi: 10.1371/journal.pbio.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conzelmann KK. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genet. 1998;32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 10.Kraemer MU, Sinka ME, Duda KA, Mylne AQ, Shearer FM, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife. 2015;4:e08347. doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes EC. The evolution of endogenous viral elements. Cell Host Microbe. 2011;10:368–377. doi: 10.1016/j.chom.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013;23:218–226. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson PJ, Macfarlan TS, Lorincz MC. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Mol Cell. 2016;62:766–776. doi: 10.1016/j.molcel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goic B, Stapleford KA, Frangeul L, Doucet AJ, Gausson V, Blanc H, Schemmel-Jofre N, Cristofari G, Lambrechts L, Vignuzzi M, et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat Commun. 2016;7:12410. doi: 10.1038/ncomms12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh MC. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol. 2013;14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 17.Palatini U, Miesen P, Carballar-Lejarazu R, Ometto L, Rizzo E, Tu Z, van Rij RP, Bonizzoni M. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics. 2017;18:512. doi: 10.1186/s12864-017-3903-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen XG, Jiang X, Gu J, Xu M, Wu Y, Deng Y, Zhang C, Bonizzoni M, Dermauw W, Vontas J, et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc Natl Acad Sci U S A. 2015;112:E5907–5915. doi: 10.1073/pnas.1516410112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee A, Nolan A, Watson J, Tristem M. Identification of an ancient endogenous retrovirus, predating the divergence of the placental mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120503. doi: 10.1098/rstb.2012.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staginnus C, Gregor W, Mette MF, Teo CH, Borroto-Fernandez EG, Machado ML, Matzke M, Schwarzacher T. Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species. BMC Plant Biol. 2007;7:24. doi: 10.1186/1471-2229-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert C, Feschotte C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 24.Chalopin D, Naville M, Plard F, Galiana D, Volff JN. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome biology and evolution. 2015;7:567–580. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maringer K, Yousuf A, Heesom KJ, Fan J, Lee D, Fernandez-Sesma A, Bessant C, Matthews DA, Davidson AD. Proteomics informed by transcriptomics for characterising active transposable elements and genome annotation in Aedes aegypti. BMC Genomics. 2017;18:101. doi: 10.1186/s12864-016-3432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda T, Tomonaga K. Endogenous non-retroviral RNA virus elements evidence a novel type of antiviral immunity. Mob Genet Elements. 2016;6:e1165785. doi: 10.1080/2159256X.2016.1165785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 28.Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, Qin XC, Xu J, Holmes EC, Zhang YZ. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4 doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik HS, Henikoff S, Eickbush TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka S, Siomi MC, Siomi H. piRNA clusters and open chromatin structure. Mob DNA. 2014;5:22. doi: 10.1186/1759-8753-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA Integration. Chem Rev. 2016;116:12730–12757. doi: 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenkranz D, Zischler H. proTRAC--a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformatics. 2012;13:5. doi: 10.1186/1471-2105-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenkranz D, Rudloff S, Bastuck K, Ketting RF, Zischler H. Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals. RNA. 2015;21:911–922. doi: 10.1261/rna.048603.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varjak M, Maringer K, Watson M, Sreenu VB, Fredericks AC, Pondeville E, Donald CL, Sterk J, Kean J, Vazeille M, et al. Aedes aegypti Piwi4 Is a Noncanonical PIWI Protein Involved in Antiviral Responses. mSphere. 2017;2 doi: 10.1128/mSphere.00144-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki Y, Frangeul L, Dickson LB, Blanc H, Verdier Y, Vinh J, Lambrechts L, Saleh MC. Uncovering the repertoire of endogenous flaviviral elements in Aedes mosquito genomes. J Virol. 2017 doi: 10.1128/JVI.00571-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM, Hangartner L. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science. 2009;323:393–396. doi: 10.1126/science.1167375. [DOI] [PubMed] [Google Scholar]
- 38.Cotton JA, Steinbiss S, Yokoi T, Tsai IJ, Kikuchi T. An expressed, endogenous Nodavirus-like element captured by a retrotransposon in the genome of the plant parasitic nematode Bursaphelenchus xylophilus. Sci Rep. 2016;6:39749. doi: 10.1038/srep39749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delviks-Frankenberry K, Galli A, Nikolaitchik O, Mens H, Pathak VK, Hu WS. Mechanisms and factors that influence high frequency retroviral recombination. Viruses. 2011;3:1650–1680. doi: 10.3390/v3091650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 41.Peleg J. Growth of arboviruses in monolayers from subcultured mosquito embryo cells. Virology. 1968;35:617–619. doi: 10.1016/0042-6822(68)90293-6. [DOI] [PubMed] [Google Scholar]
- 42.Lan Q, Fallon AM. Small heat shock proteins distinguish between two mosquito species and confirm identity of their cell lines. Am J Trop Med Hyg. 1990;43:669–676. doi: 10.4269/ajtmh.1990.43.669. [DOI] [PubMed] [Google Scholar]
- 43.Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 45.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 47.Strauss EG, Rice CM, Strauss JH. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 48.Smit A, Hubley R. RepeatModeler 2008–2015 [Google Scholar]
- 49.Smit A, Hubley R. RepeatMasker 2013–2015 [Google Scholar]
- 50.R core team. A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- 51.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2009. [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Python Software Foundation Python Language Reference, version 2.7.
- 55.McKinney W. Data Structures for Statistical Computing in Python. Proceedings of the 9th Python in Science Conference; 2010. pp. 51–56. [Google Scholar]
- 56.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–95. [Google Scholar]
- 57.Waskom M. seaborn: statistical data visualization. 2012–2017. [Google Scholar]
- 58.Hannon Lab FASTX Toolkit- FASTQ/A short-reads pre-processing tools
- 59.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bembom O. seqLogo: Sequence logos for DNA sequence alignments. 1.42.0 Edition. 2017. [Google Scholar]
- 61.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu GSD, Zhu H, Guan Y, Lam TT. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 2017;8:22–36. [Google Scholar]
- 65.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stéfan van der Walt SCCaGV. The NumPy Array: A Structure for Efficient Numerical Computation. Computing in Science & Engineering. 2011;13:22–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.