Abstract
Several Complement proteins exacerbate prion disease, including C3, C1q, and CD21/35. These proteins of the Complement cascade likely increase uptake, trafficking, and retention of prions in the lymphoreticular system, hallmark sites of early prion propagation. Complement regulatory protein Factor H (fH) binds modified host proteins and lipids to prevent C3b deposition and thus autoimmune cell lysis. Previous reports show fH binds various conformations of the cellular prion protein, leading us to question the role of fH in prion disease. Here we report transgenic mice lacking Cfh alleles exhibit delayed peripheral prion accumulation, replication and pathogenesis and onset of terminal disease in a gene-dose manner. We also report a biophysical interaction between purified fH and prion rods enriched from prion-diseased brain. Factor H also influences prion deposition in brains of infected mice. We conclude from these data and previous findings that the interplay between Complement and prions likely involves a complex balance of prion sequestration and destruction via local tissue macrophages, prion trafficking by B and dendritic cells within the lymphoreticular system, intranodal prion replication by B and Follicular Dendritic cells, and potential prion strain selection by CD21/35 and fH. These findings reveal a novel role for Complement regulatory proteins in prion disease.
Introduction
Prions encipher and transmit pathogen information via structural conversion of the soluble, cellular prion protein, PrPC, to a misfolded, aggregated and pathologic prion form, PrPSc. Examples of prion diseases include Creutzfeldt-Jakob disease in humans; Scrapie in sheep and goats; bovine spongiform encephalopathy (BSE) in cattle; chronic wasting disease (CWD) in deer, elk, and moose; and transmissible mink spongiform encephalopathy (TME) in mink. Hallmark clinical signs of prion diseases can include dementia, ataxia, and weight loss. Neuropathological manifestations of prion disease include astrogliosis, spongiform degeneration, and prion amyloid deposition. Many prion strains, including mouse-adapted scrapie strain RML5 and one TME strain, Hyper (HY), propagate in lymphoid tissues during a clinically silent carrier phase prior to neuroinvasion and clinical manifestations. Interestingly, BSE and another TME prion strain, Drowsy (DY) lack this phenotype, implicating alternate routes to the central nervous system independent of lymphoid propagation (1, 2).
The (in)ability of certain prions to breech species barriers, as well as distinct biochemical and pathological characteristics among prions derived from the same PrPC primary sequence (2, 3) suggest prions can exist as different strains or quasi-species. However, whether these prion strains arise from enhanced thermodynamic stability of certain conformations in the original inoculum, or host factors that preferentially promote certain prion conformations, remains inconclusive. Interestingly, prions derived from spleen display broader species tropism when compared to prions from brain (4). These data highlight the capacity of strain selection within a single host and suggest certain host factors likely select for certain prion strains during the transition between establishing infection in extraneural tissues to ultimate neuroinvasion. However, whether selection occurs at the protein conformational level, post-translational modification status, protein: protein interactions, or cell-type level, remains unknown.
Several elegant studies implicate the Complement system as a facilitator of early prion propagation and spread. CD21/35- and PrPC-expressing follicular dendritic cells (FDC) and B cells capture pathogens opsonized with C3 and C4 cleavage products, decreasing the threshold to activate B cells to induce antibody production. Both FDCs and B cells promote prion disease (5–11). Many studies suggest Complement promotes trafficking of infectious prions to cells expressing PrPC, a substrate for prion replication, in lymphoid follicles. Specifically, mice deficient in C3, C4, C1q, and Complement receptors CD21/35 accumulate less splenic prions early in disease and partially or completely resist terminal disease (7, 9–13).
Considering that Complement effector proteins can bind host cell surfaces, hosts require regulatory mechanisms to prevent self-recognition and subsequent autoimmunity. Complement regulatory protein Factor H (β1H globulin, CFH, or fH) prevents Complement activation on host tissues by binding sialic acids and polyanionic carbohydrates, such as glycosaminoglycan, which are largely absent on pathogen surfaces. Although C3b marks virtually all cell surfaces in circulation, fH bound to host polyanionic carbohydrates mediates C3b inactivation through co-factor activity with Factor I, as well as limits activity of the C3 convertase, C3bBb (14). Mutations or deficiency in fH lead to autoimmune diseases such as membranoproliferative glomerulonephritis, atypical hemolytic uremic syndrome and age-related macular degeneration (15–17).
Both PrPC and PrPSc contain sialic acid residues, and splenic PrPSc is more sialylated than brain PrPSc (18). Furthermore, fH binds various conformations of recombinant PrPC, suggesting fH potentially impacts prion disease outcomes (19–20). Collectively, these previous findings led us to investigate the role of fH in a mouse model of Scrapie.
Inactivation of the Cfh gene renders mice deficient in plasma C3 (15, and confirmed in serum of mice reported here; data not shown). Since C3 promotes prion disease (9,12,13) its absence in mice also lacking fH confounds the ability to assess the role of fH in prion pathogenesis. To ascertain the specific effect of fH deficiency, we peripherally inoculated mice with a high dose of RML5 mouse-adapted Scrapie prions previously shown to affect C3 deficient mice nearly identically to wild type mice (9). Transgenic mice expressing zero, one, or two allelic copies of Cfh exhibited a gene-dose effect of early prion accumulation and onset of terminal disease after challenge with a high dose of RML5 prions. Knockout and hemizygous mice accumulated fewer splenic and brain prions at early time points and resisted the onset of terminal disease in comparison to their fH sufficient cohorts. We confirmed a direct interaction between fH and recombinant PrP using surface plasmon resonance (SPR), and for the first time demonstrate fH interacts with prion amyloid enriched from prion-infected brain. Overall, these surprising data demonstrate fH may play a direct role in prion disease by promoting certain prion strains’ lymphotropism and subsequent neuroinvasion.
Materials and Methods
Mice
All mice were bred and maintained at Lab Animal Resources, accredited by the Association for Assessment and Accreditation of Lab Animal Care International and approved on January 14, 2016 by the Institutional Animal Care and Use Committee at Colorado State University. Factor H knockout mice on the C57/Bl6 background were kindly provided by Dr. Joshua Thurman (University of Colorado – Denver). We crossed Cfh−/− mice to hemizygosity with C57/Bl6 wild type mice from Jackson Laboratories. Hemizygous breeders generated pups of various Cfh genotypes.
Genotyping transgenic mice
We determined Cfh genotype from DNA extracted from tail clips (Qiagen 69504) using the following primers: 5′ CTACAAATGCCGCCCTGGAT 3′ (mzBAP3), 5′ CTGCCAGCCTAAAGGACCC 3′ (mzBAP4r), 5′ GTAAAGGTCCTCCTCCAAGAG 3′ (fHReg200), and 5′ GGGGATCGGCAATAAAAAGAC 3′ (NEO). mzBAP3 and mzBAP4r primers were designed using NCBI’s primer design website and blasted against the available C57BL/6 genome to ensure specificity. PCR conditions were as follows for 35 cycles: denaturation at 95°C, annealing at 55°C, and elongation at 72°C. PCR products were resolved using agarose gel electrophoresis. Amplicons of 400 bp (fHReg200 and NEO) indicated the presence of a knockout allele, and amplicons of 451 bp (BAP3/4) indicated the presence of a wild type allele.
Mouse inoculations
Age- and sex- matched mice (n ≥ 6 per genotype) ranging from 6 weeks to 1 year intraperitoneally received 100 μl of approximately 106 lethal dose50 (LD50) units of mouse-adapted scrapie strain RML5 prions or uninfected normal brain homogenate (NBH) as mock infection controls.
Clinical scoring
Mice were monitored daily and sacrificed at the onset of terminal disease or specified time points. We employed a composite scoring system to assess the severity of disease, including: tail rigidity (0–2), akinesia (0–4), ataxia (0–4), tremors (0–4), and weight loss (0–2). Mice scored above 10 or 4 in any category were euthanized via CO2 inhalation replacing 20% of air per minute to effect.
Tissue collection and analysis
After euthanasia, the following samples were collected and frozen or fixed in 4% formaldehyde in 1X PBS: serum, spleen (half fixed, half frozen), kidneys (one fixed, one frozen), tail clip, and brain (half fixed, half frozen). We assessed the presence of protease-resistant prions (PrPSc) in 10% (w/v) homogenate after proteinase K (Roche) digestion (10 μg/mL for spleen and 50 μg/mL for brain) and western blotting using anti-PrP monoclonal antibody BAR224 (Cayman Chemical) conjugated to horseradish peroxidase (HRP). Blots were developed using chemiluminescent substrates hydrogen peroxide and luminol for 5 minutes at room temperature and visualized using a GE digital imager and ImageQuant software. Tissues negative for PrPSc on western blots were subjected to serial protein misfolding cyclic amplification (sPMCA, see Saborio and Soto, 2001). Briefly, PMCA uses 10% normal brain homogenate in PMCA buffer (1X PBS, 1% Triton X-100, 4 mMEDTA and 150 mM NaCL) from PrPC over-expressing transgenic mice, strain TgA20, as substrate for amplification of previously undetectable prions. We mixed 25 μl of normal brain homogenate (NBH) with 25 μl 10% sample homogenate, and sonicated samples for 40 seconds ~150 watts, followed by a 30-minute incubation, and repeated this cycle for 24 hours, constituting one round of PMCA. Serial rounds were set up similarly, transferring 25 μl of the previous round’s sample to 25 μl of fresh NBH. Each biological sample was run in at least technical duplicates, and round-to-positivity was determined by PK digestion and western blotting. Relative PMCA units (rpu) were assigned as previously described (21). Non-amplified samples containing detectable PrPSc via western blot were assigned a conservative amplification factor of 10 (equating to 1000 rpu) because we estimate one round of PMCA amplifies PrPSc at least 10-fold (22).
Histology
Paraffin-embedded, 5 μm sagittal brain sections were incubated in 53°C for 30 minutes prior to immersion in xylene for 10 minutes and repeated once. Sections were then rehydrated through an ethanol gradient consisted of 100%, 95% and 70% concentrations for 5 minutes each and then immersed in 88% formic acid for 10 minutes. Slides were rinsed in running water for 10 minutes, then autoclaved at 121 °C for 2h in citrate buffer, pH 7.4. Cooled slides were rinsed twice in PBS containing 0.1% Triton X for 5 minutes on a rocker. To extinguish exogenous peroxidase activity of the tissues, sections were immersed in a 3% hydrogen peroxide in methanol for 30 minutes before undergoing another rinse cycle. Tissues were encircled in a DAP pen and incubated with Superblock (ThermoFisher Scientific) for 30 minutes. Excess Superblock was removed and slides allowed to incubate overnight at 4°C with 1:1000 D18 antibody. Slides were rinsed in PBS and incubated with a biotinylated anti-human Ig (1:1000) for 1 hour at room temperature. They were then rinsed and incubated with an avidin-biotin complex for 30 minutes at room temperature. After 3 rinse cycles, slides were incubated with Diaminobenzidine reagent for 2 min, rinsed 2×2 min with PBS, counterstained with hematoxylin for 5 minutes, and then immersed in water for 10 minutes. Slides were dehydrated through the ethanol gradient and xylene before being mounted with a coverslip. Sections were visualized and digitally photographed using an Olympus BX60 microscope equipped with 10× and 40× objectives and a cool charge-coupled diode camera.
Prion rod preparation and surface plasmon resonance
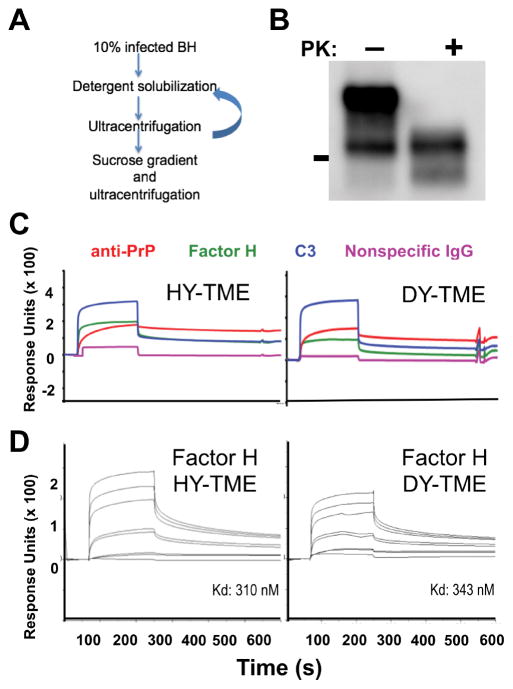
Prion rods were enriched from infected brain as previously described (23–25). Briefly, brains from hamsters (5) infected with hyper- or drowsy- transmissible spongiform encephalopathies (HY- or DY-TME) were homogenized in 1X PBS to 10% (w/v) concentration. Sucrose (1.2M) was added to 10 mL of clarified tissue to a final concentration of 165.5 mM, and samples were ultracentrifuged (100,000 × g) for one hour at 4°C. Pellets were resuspended to a final protein concentration of 5.0 mg/ml in 1X TBS containing 2.0% Triton X (TBST) and incubated on ice for 30 minutes. Samples were subjected to another round of ultracentrifugation for 20 minutes at 0°C, washed twice in 1X TBST, and twice with 1X TBS. The pellets were then resuspended in 1X PBS containing 1% Sarcosyl and protease inhibitor cocktail (Roche). Vortexed samples were incubated on a heated shaker at 37°C for two hours at 800 rpm. Samples were then gently overlaid on a 0.32 M sucrose in PMCA buffer 1 (PMCA buffer without Triton-X 100) cushion, ultracentrifuged for one hour at 4°C, and supernatants removed. Pellets were resuspended in 2.3M NaCl, 5% Sarcosyl in 1X PBS, centrifuges at 13,000 × g, washed three times in 50 mM Tris 150 mM NaCl, and were either stored dry or suspended in PBS at −80°C. Presence of PK resistant prion rods were confirmed by western blot.
Highly enriched prion rods were coupled to CM-5 sensor chips outside of the instrument after generating the reference flow cells within the instrument by activating with M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride/N-hydroxysuccinimide (EDC/NHS) and deactivating with ethanolamine three to five times. The chip was then removed from the instrument and the gold chip disassembled from the cassette. The entire surface was activated with 100 μl of EDC/NHS for 12 minutes. A pellet of rods was resuspended in 100 μl of 10 mM sodium acetate pH 5.5, sonicated at 37°C for 40 seconds, and incubated on the gold chip at room temperature for one hour. The chip was then briefly rinsed with 1X PBS, and remaining active groups were deactivated with ethanolamine for 7 minutes. Prior to use in interaction analyses, four startup cycles of 50 mM sodium hydroxide served to remove any nonspecifically bound prion rods from the surface.) Purified C3 and fH was buffer exchanged into 1X running buffer (50 mM Tris base, 150 mM NaCl, pH 7.42), and protein concentration was determined by protein A280. 5 nM, 10 nM, 100 nM, 100 nM, 500 nM, 1000 nM, and 2000 nM fH and 5 μM C3 was passed over HY- or DY- coated CM5 Series S sensor chips for 180 seconds at 30 μl/min, followed by a 360 second dissocation phase. Affinity analyses were performed using Biacore T200 evaluation software. Baseline = 0 relative response units. Binding levels with monoclonal antibody BAR224 (200 nM, Cayman Chemical) confirmed equal amounts of prion amyloid were coated onto each chip.
All SPR experiments involved recombinant PrP (rPrP) or prion rods enriched from infected brain as the ligand coated to a CM5 Series S sensor chip, and commercially-available fH (CompTech) as the analyte. Factor H was buffer exchanged in Amicon filter devices into 1X Running Buffer (50 mM Tris HCl 150 mM NaCl pH 7.42). The rPrP-coated chip was kindly provided by Dr. Hae-Eun Kang in the Telling lab.
Statistical analyses
All statistical analyses were performed using GraphPad Prism software. One- or two- way ANOVAs were run to compare genotype, sex, or the interaction between these two variables. P-values < 0.05 were considered significantly different. In the terminal disease study, we observed one male outlier and subsequently removed this data point. All graphs represent mean ± SEM.
Results
fH promotes early splenic prion propagation
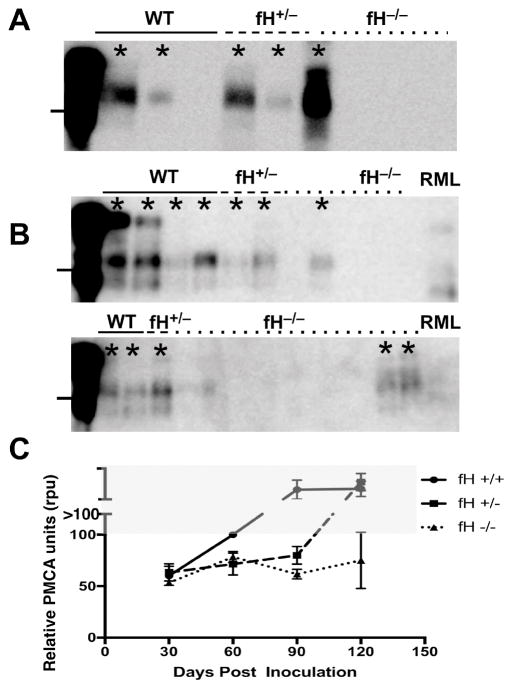
To assess the role of fH in early Scrapie trafficking and lymphoid propagation, we intraperitoneally inoculated littermates of various Cfh genotypes with 106 LD50 units of RML5 and assessed prion loads in spleens via PMCA. Relative PMCA units (rpu) were assigned as previously described (21). Samples are assigned rpu scores based on the first positive of PMCA after which we detect amplified prions by western blot. Samples requiring fewer PMCA rounds to reach the detection threshold receive higher rpu scores. We found increased splenic loads corresponded with increased Cfh gene dosage after 30 days post infection (dpi, Figure 1). Wild type, fH sufficient mice contained increasing splenic prion loads over time until reaching a plateau load by 90 dpi. Hemizygous mice displayed a lag phase, containing significantly less splenic prions than wild type at 60 and 90 dpi, then harboring prion loads similar to wild type by 120 dpi. Completely fH deficient mice exhibited significantly more protracted prion accumulation at these time points. We detected similar prion loads in mice of all genotypes by 150 dpi (data not shown).
Figure 1. Factor H promotes early prion propagation in spleen at early timepoints.
Mice (n ≥ 6 mice/genotype of both sexes) were sacrificed at 30, 60, 90 dpi, and 120 dpi, and their spleens (10% w/v homogenates) were first treated with proteinase K (PK, 10 μg/mL, panel A). Lines to the left of each blot mark 25 kD molecular weight. RML infected animals negative for PrPSc after straight western blotting (A) were subjected to serial rounds of PMCA (B,C). Asterisks indicate samples scored as positive for PrPSc. Each biological replicate was run in technical duplicates, and each lane represents one mouse. Western blot analysis of PMCA material (B) shows a higher proportion of RML-infected wild type mice (solid line) positive by round 1 of PMCA, (top blot), whereas most knockout mice (bottom blot, dotted line) required 2 or more rounds of PMCA to visualize PrPSc. RML (far right) represents 0.05% RML5 amplified prions and serves as a positive control for each PMCA round. ANOVA analysis revealed no significant differences at 30 dpi, but significant differences between the genotypes at 60, 90, and 120 dpi ((p=0.0236, 0.0021, and 0.0032, respectively). Samples in first lane of each blot were not PK digested. Numbers > 100 rpu in the grey area in graph (C) represent scores from samples detected without amplification, and therefore fall outside the dynamic range of PMCA.
fH promotes prion neuroinvasion
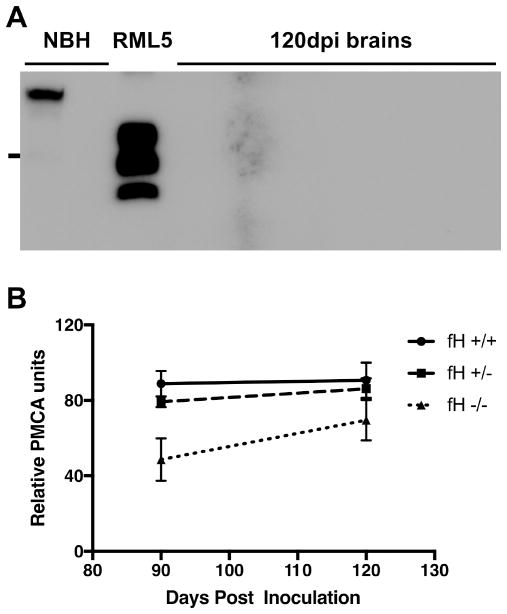
To determine the effects of fH on neuroinvasion, we assessed prion loads in brain at 90 and 120 dpi using sPMCA. Similar to the trend observed in spleen after i.p. challenge, prion loads in the brain positively correlated with Cfh expression. Factor H deficient mice contained significantly lower prion loads in brain at 90 and120 dpi (Figure 2). Interestingly, however, knockout mice reached hemizygous and wild type brain prion levels by 150 dpi as well.
Figure 2. Factor H promotes early prion propagation in brain at early time points.
(A) Mice (n ≥ 6 mice/genotype of both sexes) were sacrificed at 60, 90, and 120 dpi, and 10% w/v homogenates of their brains probed for PrPSc. We detected no PrPSc by direct western blotting and thus performed sPMCA on all samples. The blot shows representative samples from mice sacrificed at 120 dpi. All samples were digested with PK except lane 1. (B) Each biological replicate was run in technical duplicates. ANOVA analysis revealed a significant difference at 90 dpi (p=0.0024). While ANOVA analysis revealed no significant difference at 120 dpi, unpaired T-test between wild type and knockout PMCA scores revealed significant differences (p=0.0269).
Absence of fH delays clinical manifestations of terminal prion disease and affects prion deposition patterns
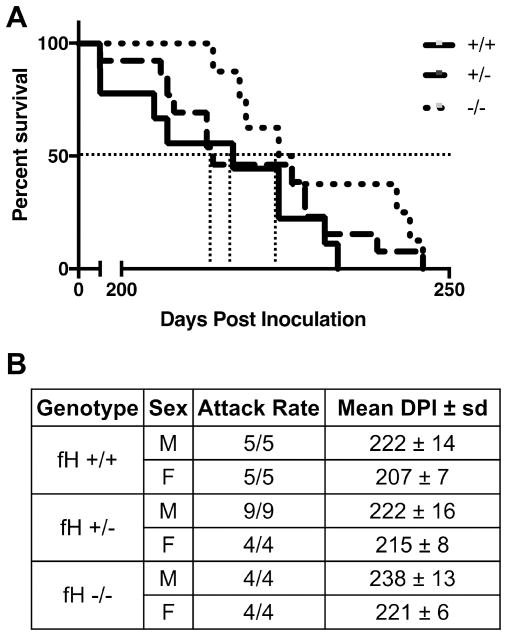
In a blind-to-genotype study, we challenged littermates with a single peritoneal dose of RML5 and monitored daily for onset of clinical disease (Figure 3). Aligning with early accumulation studies, fH−/− mice resisted the onset of terminal prion disease significantly longer than fH−/+ and wild type mice. We also noted a significant sex difference among the different genotypes, with male mice resisting terminal prion disease longer than females.
Figure 3. Factor H deficiency delays terminal prion disease.
Mice of blinded Cfh genotype received a single, intraperitoneal dose of RML5 scrapie prions and were euthanized at the onset of terminal disease, then genotype and prion disease confirmed. Statistical analysis (Mantel-Cox) accounting for sex and genotype revealed a significant difference between survival curves (A, p=0.0018) and mean dpi to terminal disease (B) for fH−/− and −/+ of both individual (p = 0.002) and combined (p = 0.047) sexes.
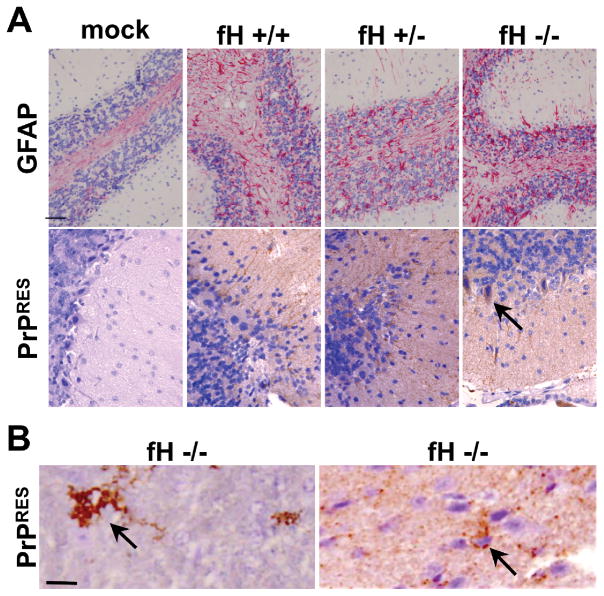
Histological examination of brains from these mice confirmed neuropathology consistent with terminal prion disease, including astrogliosis, vacuolation and PrPSc deposition (Figure 4). In mice expressing fH, we consistently observed typical diffuse PrPSc staining indicative of the RML5 mouse-adapted prion strain. However, in a subset of fH deficient mice we observed striking differences in PrPSc deposition patterns that included florid plaques, numerous puncta and even PrPSc contained in intracellular structures reminiscent of inclusion bodies.
Figure 4. Factor H knockout mice contain unique PrPSc deposition profiles.
(A) Infected mice of all genotypes displayed prion neuropathology, including astrogliosis and PrPSc deposition. No prion deposition was observed in mock infected mice. All infected fH wild type and hemizygous mice contained typical, diffuse RML5 deposition, whereas fH deficient mice contained cerebellar prion deposition suggestive of inclusion bodies at terminal disease (A, bottom panel). We observed florid plaques (B, left), and puncta (B, right) in a subset of fH knockout mice. Arrows point to prion deposition unique to fH knockout mice. Scale bars, 100 μm.
fH directly binds PrPC and prion amyloid
Previous reports indicate a molecular interaction between various conformations of recombinant PrP and fH, but whether fH bound in vivo-derived prion amyloid remained unknown. We therefore used surface plasmon resonance to test the interaction of fH with highly enriched prion rods isolated from hamsters infected with two distinct strains of TME prions, of which the HY strain is lymphotropic, while DY is not. Factor H bound the lymphotropic HY strain with slightly higher affinity (310 nM) than DY TME strain (343 nM, Figure 5).
Figure 5. Factor H directly interacts with prion amyloid enriched from infected brain.
(A) Prion rods were enriched from infected brains as previously described and (B) are protease K (PK) resistant. The line to the left of the blot marks 25 kD molecular weight. (C) Factor H biochemically interacts with prion rods of both HY and DY strains manually coupled to a surface plasmon resonance sensor chip. (D) Affinity analyses show that fH bound HY prions with slightly higher affinity than DY prions. Traces from the bottom to top of the sensograms show binding of 0 nM, 5 nM, 10 nM, 100 nM, 100 nM, 500 nM, 1000 nM, and 2000 nM fH analytes to prions coated on the chip.
Discussion
We investigated the influence of Complement regulatory protein Factor H in establishing prion infection after peripheral insult. Our data show fH directly binds prions and promotes splenic propagation, neuroinvasion, and terminal disease onset. While fH−/− mice eventually succumbed to prion disease, lymphotropism and subsequent neuroinvasion were impaired. Interestingly, splenic prion burdens did not differ between genotypes at 30 dpi. These data reveal no differences in initial trafficking of inocula to spleens, but impaired prion replication during the nonclinical phase of disease, suggesting fH may stabilize prions or promote propagation once captured in the lymphoreticular system (LRS). We previously showed that prions could traffic to lymphoid tissues cell-autonomously (25), independent of an intact Complement system. Perhaps soluble fH acts as a transport chaperone for such autonomous prions. Assessing the effects of pre-treating prion inocula with fH on early cell-mediated and -autonomous prion trafficking, as well as degradation could help test this hypothesis. We observed no differences in splenic prion accumulation or neuroinvasion by 150 dpi.
We can envision several roles for fH in directly promoting prion disease. As discussed above, fH could act as a soluble prion receptor and cell-autonomously shuttle prions to extraneural sites of prion replication. Alternatively, or even additionally, fH bound to PrPC-expressing cells could capture prions, bringing them to the vicinity of PrPC molecules, which then misfold and join the growing prion aggregate. Furthermore, fH may preferentially bind prions adopting distinct conformations or possessing certain post-translational modifications, thereby selecting for certain strains to replicate in the LRS.
Previous studies using mouse models of Scrapie (9, 13) and CWD (12) reveal that mice genetically or pharmacologically deficient in C3 accumulate less prions and resist terminal disease longer than their C3-sufficient controls. Prion opsonization likely promotes not only local degradation of prions by macrophages, but also trafficking to and subsequent propagation in lymphoid tissues, hallmark areas of early prion replication. fH deficiency results in increased C3 convertase formation and causes unregulated cleavage of C3 and downstream depletion of serum C3 (15). However, Klein et al. (2001) reported no difference in progression to terminal prion disease in C3 deficient and wild type mice upon a high-dose challenge. Here, we report a significantly delayed onset to terminal prion disease upon high dose challenge in mice lacking fH and, therefore C3, compared to wild type mice. Thus, fH deficiency likely plays a more substantial role in prolonging prion disease than simply eliminating C3.
Complement control proteins contain highly homologous series of 60–70 amino acids termed short consensus repeats (SCRs). SCRs recognize certain C3/C4 cleavage products, and each Complement control protein elicits distinct downstream functions. For example, Complement receptor 2 (CR2 or CD21) recognize opsonized pathogens via its SCRs and reduce the threshold of B cell activation, whereas Factor H recognizes sialylated host products via its SCRs to prevent autoimmunity. Essentially, these proteins stimulate opposing effects using highly homologous SCRs. Domain mapping studies in our laboratory reveal CD21 containing the first two SCRs as sufficient to recognize prion amyloid (our unpublished data - manuscript in preparation). We therefore conclude CD21 is a cell-surface prion receptor which facilitates prion propagation in lymphoid tissues prior to neuroinvasion. We propose fH is a both a soluble and cell-associated prion receptor and assists in prion disease similarly to CD21. We therefore conclude SCRs on complement control proteins, such as CD21 and fH, bind prions either cell-mediated or cell-autonomously and promote lymphotropism and subsequent neuroinvasion.
Sjoberg, et al reported fH biochemically interacts with various confomers of PrP, but preferentially bound oligomeric species (19). Amyloid plaques, both in Alzheimer’s and prion diseases, appear to be relatively innocuous forms of the misfolded protein, whereas oligomeric species cause the cytotoxic features (26). Perhaps oligomeric species, due to their smaller size and less amyloidogenic status, are more sensitive to protease or cell-mediated degradation. Factor H may bind, thereby protect, and shuttle protease-sensitive oligomers to extraneural sites where oligomers could propagate into, perhaps even seed, amyloid production. We show fH binds the lymphotropic HY strain with slightly higher affinity than the non-lymphotropic DY prion strain. However, this slight affinity difference likely does not fully explain lymphotropic differences for these strains. Furthermore, these studies only test interactions with prion amyloid derived from hamster brain. Whether fH differentially binds HY- or DY- TME oligomeric species remains undetermined, as does whether fH differentially binds RML5 mouse prions, mostly due to the difficulty isolating sufficient RML5 prion rods from mouse brains to perform those analyses. HY and DY hamster prions likely form conformations distinct from RML5 mouse prions, another lymphotropic strain used in these infection studies. We predict, based on the in vivo infection studies reported here, that fH binds RML5 prions as well. We are currently investigating the role of fH in prion strain selection, stabilization, and trafficking in vivo.
Baskakov and colleagues convincingly demonstrated that sialylation status differs among prion strains (27,29), which could dictate fH binding. Katorcha et al. reported that PrPSc sialylation increased prion infectivity (28). PMCA-generated material derived from de-sialylated substrate did not infect wild type mice, whereas PMCA-generated material from fully sialylated substrate exhibited 100% attack rate. fH may bind sialylated PrPSc and facilitate disease via the shuttling mechanism described above. Hepatocarcinoma cells utilize sialic acids to selectively adhere to secondary lymphoid organs (reviewed in 29). Perhaps fH aids sialylated prions infect lymphoid organs via a similar mechanism. Accordingly, de-sialylated PMCA material was undetectable in spleen or brain after serial rounds of PMCA, indicative of either degradation or urinary or fecal secretion. Perhaps fH could explain both observations by binding sialic acid on both cancerous cells and prions, causing an evasion of cellular destruction and granting entry to lymphoid tissues.
Factor H could also be involved in selecting different prion strains to be propagated in lymphoid organs by preferentially binding sialylated prions. Although we show here that fH binds both lymphotropic and non-lymphotropic strains, their sialylation status is unknown. Factor H has also been shown to bind recombinant PrP free of post-translational modifications (19, 20), and therefore may dictate prion strain selection and disease outcomes independent of, or in addition to, sialylation status. Qualitative, histological examination revealed a higher proportion of fH−/− mice contained florid or intracellular prion deposition/accumulation versus the typical and extremely consistent diffuse RML5 deposition in wild type mice (Figure 4), supporting the hypothesis that fH is a key host factor in prion strain selection. Additionally, soluble fH may prevent certain prion strains like RML5 from aggregating into puncta, causing diffuse prion deposition.
Altogether, these data support a role for fH in peripheral prion disease pathogenesis by directly interacting with infectious prions and promoting lymphotropism and subsequent neuroinvasion. We propose fH selects for certain prion strains. Whether this prion selection occurs at the level of prion conformation, sialylation status, degradation evasion or a combination of these processes remains to be determined.
Acknowledgments
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke awards R01 NS056379 and F31NS087762 and National Institute of Allergy and Infectious Disease award R56AI122273, of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Dr. Hae-Eun Kang for aid and consultation in the SPR experiments, Joshua Thurman for breeding pairs of fH deficient mice, and Jason Bartz for hamster brains.
References
- 1.Somerville RA, Birkett CR, Farquhar CF, Hunter N, Goldmann W, Dornan J, Grover D, Hennion RM, Percy C, Foster J, Jeffrey M. Immunodetection of PrPSc in spleens of some scrapie-infected sheep but not BSE-infected cows. J Gen Virol. 1997;78:2389–2396. doi: 10.1099/0022-1317-78-9-2389. [DOI] [PubMed] [Google Scholar]
- 2.Bartz JC, Dejoia C, Tucker T, Kincaid AE, Bessen RA. Extraneural prion neuroinvasion without lymphoreticular system infection. J Virol. 2005;79:11858–11863. doi: 10.1128/JVI.79.18.11858-11863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Béringue V, Herzog L, Jaumain E, Reine F, Sibille P, Le Dur A, Vilotte JL, Laude H. Facilitated cross-species transmission of prions in extraneural tissue. Science. 2012 doi: 10.1126/science.1215659. [DOI] [PubMed] [Google Scholar]
- 5.Montrasio F, Frigg R, Glatzel M, Klein MA, Mackay F, Aguzzi A, Weissmann C. Impaired prion replication in spleens of mice lacking functional follicular dendritic cells. Science. 2000;288:1257–1259. doi: 10.1126/science.288.5469.1257. [DOI] [PubMed] [Google Scholar]
- 6.Brown K, Ritchie DL, Mabbott NA, Williams A, Fraser H, Morrison WI, Bruce ME. Scrapie replication in lymphoid tissues depends on prion protein-expressing follicular dendritic cells. Nat Med. 1999;5:1308–1312. doi: 10.1038/15264. [DOI] [PubMed] [Google Scholar]
- 7.Zabel MD, Heikenwalder M, Prinz M, Arrighi I, Schwarz P, Kranich J, von Teichman A, Haas KM, Zeller N, Tedder TF, Weis JH, Aguzzi A. Stromal complement receptor CD21/35 facilitates lymphoid prion colonization and pathogenesis. J Immunol. 2007;179:6144–6152. doi: 10.4049/jimmunol.179.9.6144. [DOI] [PubMed] [Google Scholar]
- 8.McCulloch L, Brown KL, Bradford BM, Hopkins J, Bailey M, Rajewsky K, Manson JC, Mabbott NA. Follicular dendritic cell-specific prion protein (PrP) expression alone is sufficient to sustain prion infection in the spleen. PLoS Pathog. 2011 doi: 10.1371/journal.ppat.1002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein MA, Kaeser PS, Schwarz P, Weyd H, Xenarios I, Zinkernagel RM, Carroll MC, Verbeek JS, Botto M, Walport MJ, Molina H, Kalinke U, Acha-Orbea H, Aguzzi A. Complement facilitates early prion pathogenesis. Nat Med. 2001;7:488–492. doi: 10.1038/86567. [DOI] [PubMed] [Google Scholar]
- 10.Klein MA, Frigg R, Raeber AJ, Flechsig E, Hegyi I, Zinkernagel RM, Weissmann C, Aguzzi A. PrP expression in B lymphocytes is not required for prion neuroinvasion. Nat Med. 1998;4:1429–1433. doi: 10.1038/4022. [DOI] [PubMed] [Google Scholar]
- 11.Michel B, Ferguson A, Johnson T, Bender H, Meyerett-Reid C, Pulford B, von Teichman A, Seelig D, Weis JH, Telling GC, Aguzzi A, Zabel MD. Genetic depletion of complement receptors CD21/35 prevents terminal prion disease in a mouse model of chronic wasting disease. J Immunol. 2012 doi: 10.4049/jimmunol.1201579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel B, Ferguson A, Johnson T, Bender H, Meyerett-Reid C, Wyckoff AC, Pulford B, Telling GC, Zabel MD. Complement protein C3 exacerbates prion disease in a mouse model of chronic wasting disease. Int Immunol. 2013 doi: 10.1093/intimm/dxt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mabbott NA, Bruce ME, Botto M, Walport MJ, Pepys MB. Temporary depletion of complement component C3 or genetic deficiency of C1q significantly delays onset of scrapie. Nat Med. 2001;7:485–487. doi: 10.1038/86562. [DOI] [PubMed] [Google Scholar]
- 14.Carroll MV, Sim RB. Complement in health and disease. Adv Drug Deliv Rev. 2011;63:965–975. doi: 10.1016/j.addr.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Pickering MC, Cook HT, Warren J, Bygrave AE, Moss J, Walport MJ, Botto M. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat Genet. 2002;31:424–428. doi: 10.1038/ng912. [DOI] [PubMed] [Google Scholar]
- 16.Zipfel PF, Edey M, Heinen S, Józsi M, Richter H, Misselwitz J, Hoppe B, Routledge D, Strain L, Hughes AE, Goodship JA, Licht C, Goodship TH, Skerka C. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3:e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava S, Makarava N, Katorcha E, Savtchenko R, Brossmer R, Baskakov IV. Post-conversion sialylation of prions in lymphoid tissues. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1517993112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjöberg AP, Nyström S, Hammarström P, Blom AM. Native, amyloid fibrils and beta-oligomers of the C-terminal domain of human prion protein display differential activation of complement and bind C1q, factor H and C4b-binding protein directly. Mol Immunol. 2008 doi: 10.1016/j.molimm.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell DA, Kirby L, Paulin SM, Villiers CL, Sim RB. Prion protein activates and fixes complement directly via the classical pathway: implications for the mechanism of scrapie agent propagation in lymphoid tissue. Mol Immunol. 2007;44:2997–3004. doi: 10.1016/j.molimm.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 21.Pulford B, Spraker TR, Wyckoff AC, Meyerett C, Bender H, Ferguson A, Wyatt B, Lockwood K, Powers J, Telling GC, Wild MA, Zabel MD. Detection of PrPCWD in feces from naturally exposed Rocky Mountain elk (Cervus elaphus nelsoni) using protein misfolding cyclic amplification. J Wildl Dis. 2012;48:425–434. doi: 10.7589/0090-3558-48.2.425. [DOI] [PubMed] [Google Scholar]
- 22.Meyerett C, Michel B, Pulford B, Spraker TR, Nichols TA, Johnson T, Kurt T, Hoover EA, Telling GC, Zabel MD. In vitro strain adaptation of CWD prions by serial protein misfolding cyclic amplification. Virology. 2008;382:267–276. doi: 10.1016/j.virol.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Safar J, Wang W, Padgett MP, Ceroni M, Piccardo P, Zopf D, Gajdusek DC, Gibbs CJ., Jr Molecular mass, biochemical composition, and physicochemical behavior of the infectious form of the scrapie precursor protein monomer. Proc Natl Acad Sci U S A. 1990;87:6373–6377. doi: 10.1073/pnas.87.16.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson TE, Michel BA, Meyerett C, Duffy A, Avery A, Dow S, Zabel MD. Monitoring immune cells trafficking fluorescent prion rods hours after intraperitoneal infection. J Vis Exp. 2010 doi: 10.3791/2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel B, Meyerett-Reid C, Johnson T, Ferguson A, Wyckoff AC, Pulford B, Bender H, Avery A, Telling G, Dow S, Zabel MD. Incunabular immunological events in prion trafficking. Sci Rep. 2012 doi: 10.1038/srep00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature. 437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katorcha E, Makarava N, Savtchenko R, Baskakov IV. Sialylation of the prion protein glycans controls prion replication rate and glycoform ratio. Sci Rep. 2015 doi: 10.1038/srep16912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katorcha E, Makarava N, Savtchenko R, D’Azzo A, Baskakov IV. Sialylation of prion protein controls the rate of prion amplification, the cross-species barrier, the ratio of PrPSc glycoform and prion infectivity. PLoS Pathog. 2014 doi: 10.1371/journal.ppat.1004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baskakov IV, Katorcha E. Multifaceted Role of Sialylation in Prion Diseases. Front Neurosci. 2016 doi: 10.3389/fnins.2016.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]