Abstract
LATERAL ORGAN BOUNDARIES (LOB) DOMAIN (LBD) constitute a family of plant-specific transcription factors with key roles in the regulation of plant organ development, pollen development, plant regeneration, pathogen response, and anthocyanin and nitrogen metabolisms. However, the role of LBDs in fruit ripening and in grapevine (Vitis vinifera L.) development and stress responses is poorly documented. By performing a model curation of LBDs in the latest genome annotation 50 genes were identified. Phylogenetic analysis showed that LBD genes can be grouped into two classes mapping on 16 out of the 19 V. vinifera chromosomes. New gene subclasses were identified that have not been characterized in other species. Segmental and tandem duplications contributed significantly to the expansion and evolution of the LBD gene family in grapevine as noticed for other species. The analysis of cis-regulatory elements and transcription factor binding sites in the VviLBD promoter regions suggests the involvement of several hormones in the regulation of LBDs expression. Expression profiling suggest the involvement of LBD transcription factors in grapevine development, berry ripening and stress responses. Altogether this study provides valuable information and robust candidate genes for future functional analysis aiming to clarify mechanisms responsible for the onset of fruit ripening and fruit defense strategies.
Introduction
Transcription factors play an important role in the regulation of plant development and disease response. Among them, LATERAL ORGAN BOUNDARIES DOMAIN (LBD) proteins defined by a conserved N-terminal LATERAL ORGAN BOUNDARIES (LOB) domain is a family of plant-specific transcription factors with key roles in the regulation of plant organ development1,2. The heterodimeric interactions between the Arabidopsis AS1, AS2, and JLO proteins are involved in the establishment of organ boundaries3. AS2 (LBD6) interacts with AS1 in the process of leaf formation and are known to be required for repression of meristematic genes and establishment of leaf adaxial-abaxial polarity4. These proteins are also involved in the development of sepal and petal primordia of flowers by repressing boundary-specifying genes for normal development of the organ5. JLO/LBD30 is a general regulator of cell specification and organ patterning throughout plant development6. On the other hand, LBD16, LBD18, and LBD29 regulate lateral root organogenesis in Arabidopsis as direct targets of Aux/IAA–ARF modules in the auxin signalling pathway7.
Recent studies showed that these proteins are also involved in pollen development, plant regeneration, photomorphogenesis, pathogen response, and anthocyanin and nitrogen metabolisms2. In this way, LBD27/SIDECAR POLLEN (SCP) and LBD10 are Arabidopsis microspore-specific LBD proteins having cooperative and unique roles in male gametophyte development8. The Arabidopsis LBD proteins LBD16, LBD17, LBD18, and LBD29 are key regulators in the callus induction process associated with plant regeneration, and establish a molecular link between auxin signalling and the plant regeneration program9. Arabidopsis LBD20 is a Fusarium oxysporum susceptibility gene that appears to regulate components of jasmonic acid (JA) signalling required for full elicitation of F. oxysporum- and JA-dependent responses10. Arabidopsis LBD25/DDA1 is involved in the regulation of light/dark-dependent hypocotyl elongation11. LBD proteins have also been involved in developmental processes in non-model plants such as secondary phloem growth in Populus12 and pulvinus differentiation and petiole development in legumes13.
The characteristic LOB domain comprises a C-block containing four cysteine with spacing (CX2CX6CX3C) required for DNA-binding activity, a Gly-Ala-Ser (GAS) block and a leucine zipper- like coiled-coil motif (LX6LX3LX6L) responsible for protein dimerization1,2. Several LBD proteins are capable to form homo- and hetero- dimers2,3,8,14. Recently, it was demonstrated that the conserved proline residue in the GAS block is also crucial for the DNA-binding activity of Arabidopsis LBD16 and LBD18 proteins which have a role in lateral root formation15. The LBD gene family can be divided into two classes according to the structure of the LOB domain16,17. Class I LBD genes contain a perfectly conserved CX2CX6CX3C zinc finger-like domain and an LX6LX3LX6L leucine zipper-like coiled-coil motif, whereas class II LBD genes only have a conserved zinc finger-like domain17. The majority of LBD genes belong to class I. Class II LBD proteins have an incomplete, probably not functional, leucine zipper that cannot form a coiled–coil structure1.
Functional analysis, mainly in Arabidopsis, rice and maize, revealed that class I LBD genes are mostly involved in plant development such as lateral organ (leaf and flower) development1,2, and in auxin signal transduction cascade that leads to the formation of lateral roots7,18–20. By contrast, class II genes seem to be involved in metabolism, particularly as repressors of anthocyanin synthesis and N availability signals in the plant21,22.
LOB domain proteins are suggested to act as transcription factors based on their nuclear localization7,23, and their capacity to bind to DNA motifs24. The DNA-binding affinity of ASL4 (LOB) was reduced by interacting with bHLH048 proteins24. The variable C-terminal region of LBD proteins confers transcriptional control on downstream gene expression14.
In silico genome analyses predicted the presence of 43 LBD members in Arabidopsis thaliana and Zea mays, 35 in Oryza sativum and 58 in Malus domestica 17,25–27. As more species have their complete reference genome sequenced, additional LBD genes can be identified and the biological roles of this poorly studied gene family clarified.
Grapevine has been a widely-studied species during the last decade at the genomics level. The release of the whole grapevine genome sequence in 2007 represented a breakthrough to promote its molecular genetics analysis28. Based on the published sequence data, comprehensive analysis of a given gene family can be performed to uncover its molecular functions, evolution and gene expression profiles. These analyses can contribute to the understanding of how genes in gene families control traits at a genome-wide level.
Recent preliminary analyses predicted 40 LBD genes in the grapevine genome29 using an older version of the grapevine genome and without manual curation. In this work, we have identified 50 LBD genes and have performed a detailed structural analysis and mapping of these genes on the grapevine chromosomes. This gene family has been compared with similar families in thirty-three plant species. Finally, identification of cis-acting regulatory elements in promoter regions together with expression analyses based on microarray and RNAseq data suggest that LBD proteins are involved in the process of grape ripening and in the plant response to abiotic and biotic stresses.
Results
Structural annotation of LBD genes, phylogenetic analysis, and nomenclature
Genes that were previously identified as LATERAL ORGAN BOUNDARIES DOMAIN in the grapevine genome30 were used to performed sequence comparison analyses with BLASTX, either against the most up to date gene predictions from CRIBI V1 and V2, the NCBI refseq (remapped on the 12Xv2 of the genome assembly) and the VCOST (on the 12Xv2 of the genome assembly). Analyses were also performed directly against the reference genome sequence with TBLASTX to check whether any potential gene could have been missed by these predictions. By using these approaches, we identified 50 genome regions that shared homology with at least one of the genes.
Gene models were curated using the data collected from gene structure comparisons using different databases as well as the available inflorescence and flower RNAseq data from the laboratory (data not shown). RNAseq data allowed to evaluate whether newly detected genes, not represented in microarray data, showed expression, by redoing the bioinformatics analysis of original RNAseq data with an updated GFF file. A total of 50 LBD genes having a putatively functional structure were identified in the grapevine genome (Table 1), which is similar to the number of genes identified in Arabidopsis genome (43 genes)16,17. Data relative to the detection of LBD genes in previous genome annotations or gene-sets are summarized in Supplementary Table S1. The majority of the genes were identified in all the annotations. However, four genes were not detected in the automatic annotation CRIBIv1, three were not detected in the CRIBIv2, six were missing in the VIB annotation, and two in the NCBI refseq annotation. Representative sequences for each gene model were selected from the different annotations based on their quality (apparently full length gene when compared to other species, no chimera): 13 were selected from the CRIBI, 2 from the VIB annotation and the remaining 35 from the refseq annotation. These genes are integrated in the Grapevine annotation V3 recently published31.
Table 1.
Genome localization of the 50 grapevine VviLBD genes.
| Locus ID | Short Name | Strand | Position | Locus ID | Short Name | Strand | Position |
|---|---|---|---|---|---|---|---|
| Vitvi08g00144 | LBDIa1 LBD20 | + | 2648151–2649186 | Vitvi13g00551 | LBDIf5 | − | 5061981–5063306 |
| Vitvi15g00736 | LBDIa2 LBD19 | + | 14992229–14994929 | Vitvi13g00552 | LBDIf6 | − | 5073630–5074839 |
| Vitvi15g00735 | LBDIa3 | − | 14983081–14985200 | Vitvi13g00549 | LBDIf7 | + | 5039284–5040392 |
| Vitvi07g00573 | LBDIa4 LBD16 | − | 6228524–6229895 | Vitvi13g00545 | LBDIf8 | + | 4985620–4986382 |
| Vitvi13g00333 | LBDIa5 LBD33 | + | 3457062–3457909 | Vitvi13g00546 | LBDIf9 | + | 5011144–5011863 |
| Vitvi07g00572 | LBDIa6 | + | 6220003–6220810 | Vitvi13g00556 | LBDIf10 | − | 5144789–5155693 |
| Vitvi17g00890 | LBDIc1 LOB | − | 10710469–10712731 | Vitvi13g00543 | LBDIf11 | + | 4954628–4956343 |
| Vitvi14g01707 | LBDIc2 | + | 27155871–27157919 | Vitvi06g00336 | LBDIf12 | − | 4180846–4182157 |
| Vitvi13g00085 | LBDIc3 LBD21 | − | 817035–817745 | Vitvi06g00338 | LBDIf13 | − | 4201430–4202390 |
| Vitvi00g00480 | LBDIc4 LBD6 | + | 11326893–11330106 | Vitvi16g01446 | LBDIg1 | + | 17405415–17406086 |
| Vitvi00g01060 | LBDIc5 | − | 22493178–22495116 | Vitvi15g01216 | LBDIi1 | + | 17253530–17254344 |
| Vitvi07g01328 | LBDIc6 | − | 18720228–18722222 | Vitvi15g01217 | LBDIi2 | + | 17259993–17261012 |
| Vitvi07g01326 | LBDIc7 | − | 18699491–18702938 | Vitvi04g01768 | LBDIi3 | + | 996279–997019 |
| Vitvi07g01327 | LBDIc8 | − | 18708083–18709964 | Vitvi14g01878 | LBDIi4 LBD27 | − | 28646091–28646927 |
| Vitvi16g00859 | LBDIc9 | − | 15931002–15932161 | Vitvi17g00520 | LBDIi5 | − | 6117662–6119015 |
| Vitvi19g01589 | LBDId1 LBD3 | + | 21536622–21539654 | Vitvi09g00188 | LBDIi6 LBD22 | − | 2066342–2067873 |
| Vitvi10g01237 | LBDId2 LBD4 | − | 17047348–17048794 | Vitvi12g00230 | LBDIi7 LBD2 | − | 3392281–3394534 |
| Vitvi06g00706 | LBDId3 | + | 7971884–7972497 | Vitvi11g00169 | LBDIi8 | + | 1720665–1722595 |
| Vitvi13g00109 | LBDId4 | + | 1022072–1022675 | Vitvi14g01193 | LBDIIa1 | + | 21211055–21212346 |
| Vitvi13g00144 | LBDId5 | − | 1309999–1311255 | Vitvi17g00325 | LBDIIa2 | − | 3791838–3793171 |
| Vitvi06g00772 | LBDId6 LBD13 | − | 8584220–8586134 | Vitvi01g00291 | LBDIIa3 | − | 3210258–3211492 |
| Vitvi13g01866 | LBDIf1 | − | 5100511–5101131 | Vitvi01g00290 | LBDIIa4 | + | 3204504–3205665 |
| Vitvi13g01867 | LBDIf2 | − | 5102158–5103523 | Vitvi18g00677 | LBDIIb1 | + | 7746353–7747276 |
| Vitvi13g00555 | LBDIf3 | − | 5130143–5136580 | Vitvi07g01610 | LBDIIb2 LBD39 | + | 21897655–21899042 |
| Vitvi13g00559 | LBDIf4 | − | 5173575–5179644 | Vitvi03g00628 | LBDIIc1 | − | 7098961–7099834 |
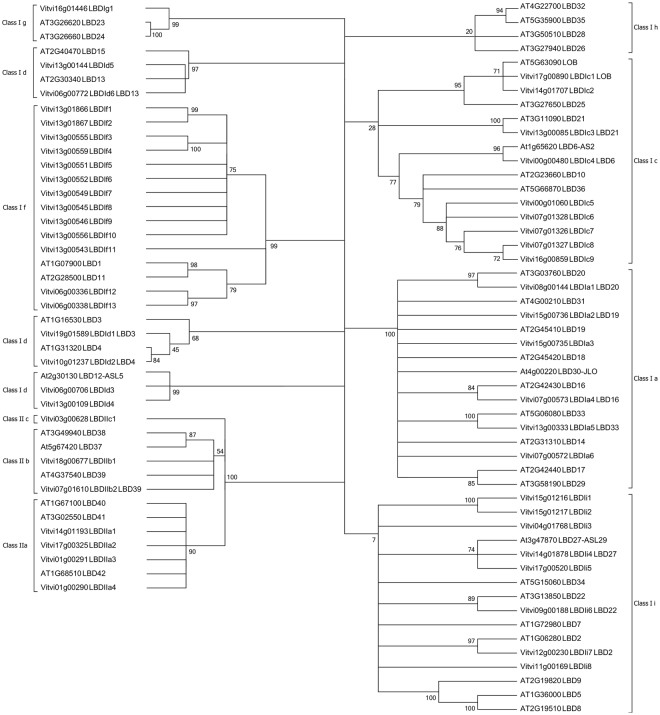
Regarding nomenclature, a phylogenetic tree of the LBD protein coding genes in V. vinifera and Arabidopsis was constructed (Fig. 1) as suggested by the Super-Nomenclature Committee for Grape Gene Annotation (sNCGGa)32. A bootstrap value of 70 as recommended by the Committee allowed to discriminate the genes within the majority of the classes but for some of them the phylogenetic analysis was complemented by motif analysis to detect conservation within classes and determine the affiliation of the genes inside some classes. The use of lower bootstrap values allowed to retrieve the same classes as in Arabidopsis. Class d is the only family where the genes are not all within the same branch. The genes are part of a subtree with the class f, but all the genes clustered with an Arabidopsis gene from class Id with a bootstrap value higher than 70. Class Ic is hardly conserved with a bootstrap of 28 necessary to maintain the tree architecture. However, clear consensus is found in the GAS motif and all the genes clustered with an Arabidopsis gene from class Ic with a bootstrap value higher than 70. Class IIb require a bootstrap of 54 to maintain the tree architecture but also clear conservation is observed in the LX6LX3LX6 motif. Class Ii requires a bootstrap of 7 to maintain the tree architecture, which is rather low and four genes were not clustering with an Arabidopsis gene from class Ic with a bootstrap value higher than 70. As in other species, VviLBD genes fall into two classes: Class I with 43 genes and Class II with 7 genes, relative to 37 and six in Arabidopsis16,17. Class I VviLBD genes were grouped into six subclasses (a, c, d, f, g, and i) and Class II genes into three subclasses (a–c). Arabidopsis LBD genes were not clustered in subclass IIc, which includes only the VviLBDIIc1 gene. Only two Arabidopsis LBD genes (LBD1 and LBD11) were grouped in subclass If with thirteen VviLBDIf (1–13) genes. For individual gene nomenclature, since both Arabidopsis nomenclature and previously named Vitis genes were named based on a poorly informative numeric code and few clear orthologs were identified, gene symbols/names were adapted to the class, the subclass and a distinctive number as proposed for Vitis genes nomenclature32. Correspondences among different nomenclatures are described in Supplementary Table S1.
Figure 1.
Phylogenetic analysis of grapevine and Arabidopsis LBD genes. Two classes were identified, Class I subdivided into six subclasses (a,c,d,f,g, and i) and Class II into three (a–c).
Regarding the exon/intron structure (Supplementary Fig. S1), the majority of the VviLBD genes presented two exons (37 genes), as it is commonly observed in other plant species25,26,33,34. Nevertheless, four of them have a non-coding exon (LBDIc1 LOB, LBDIc6, LBDIc8, LBDIi1), while LBDIc6 expression was not detected according to RNAseq data. Thirteen genes present a different exon/intron structure comparing to the other 37 genes: five of them did not have any intron (LBDIc2, LBDIc3, LBDIc9, LBDIi3, and LBDIi4), and seven of them contained three exons (LBDIc5, LBDId5, LBDId6, LBDIf3, LBDIf4, LBDIi7, and LBDIi8). However, LBDIc5 presented two non-sense exons. Finally, LBDIc4 presented five exons, although four of them were predicted as non-sense. Four of the five genes with predicted non-sense exons belong to Class Ic. The size of the LBD gene locus varied ten times, ranging from 603 nucleotides (VviLBDId4) to 6437 nucleotides (VviLBDIf3).
Motif analyses and orthologous relationships
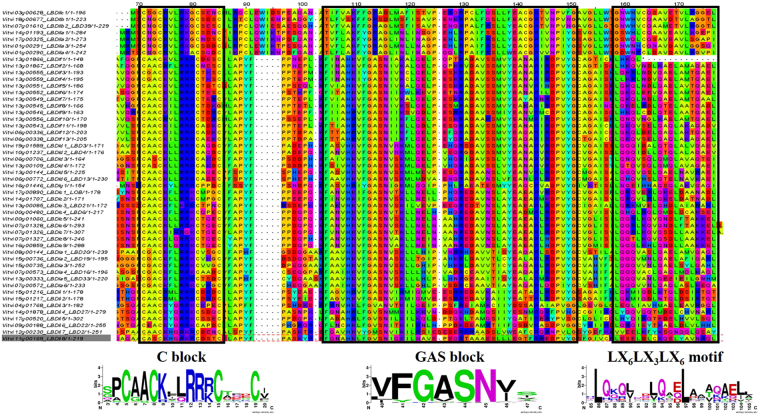
The LBD transcription factor family has a conserved LOB domain in the N terminus that comprises a C-block, a GAS block and a leucine-zipper-like coiled-coil motif16,17. Multiple sequence alignment within all of the VviLBD predicted proteins showed that the CX2CX6CX3C zinc finger-like domain was conserved in all 50 predicted protein sequences (Fig. 2, Supplementary Fig. S2). In addition, VviLBD proteins had a completely conserved G amino acid at the GAS block (Fig. 2). Class I LBD proteins presented a phenylalanine (F) and a histidine (H) completely conserved at the FX2(V/A)H motif, which represents the beginning of the GAS block. At the DP(V/I) YG motif of the Arabidopsis LBD proteins17, the proline (P) and the glycine (G) were completely conserved in all predicted grapevine proteins. The conserved proline residue in the GAS block was demonstrated in Arabidopsis to be essential in the biological function of the LBD proteins, since their replacement by leucine residues precludes LBD18-dependent control of the lateral root development via inhibition of the DNA-binding activity15. Valine (V) and leucine residues in the GAS block as well as a glutamine (Q) in the leucine zipper-like motif were found to be needed for motor organ specification in pea13. As observed for other plant species, leucine zipper-like motif (LX6LX3LX6L) was observed only in Class I VviLBD proteins and absent in Class II proteins, which suggests distinct functions of both classes. N and C terminals beyond the 3 blocks were not conserved at all among any sequences indicating that they probable play only a marginal role in protein function (Supplementary Fig. S2). It is however noteworthy that none of class II proteins presents a N terminal but this is not specific of the class; other class I proteins do not have it either. Class If protein present a longer, serine-enriched N terminal.
Figure 2.
VviLBD protein alignment and motif analysis. Conserved domains were highlighted with black boxes. CX2CX6CX3C zinc finger-like domain was conserved in all 50 predicted VviLBD protein sequences while the leucine zipper-like motif (LX6LX3LX6L) was observed only in the class I VviLBD proteins. Details on protein structure are shown in Supplementary Fig. S1.
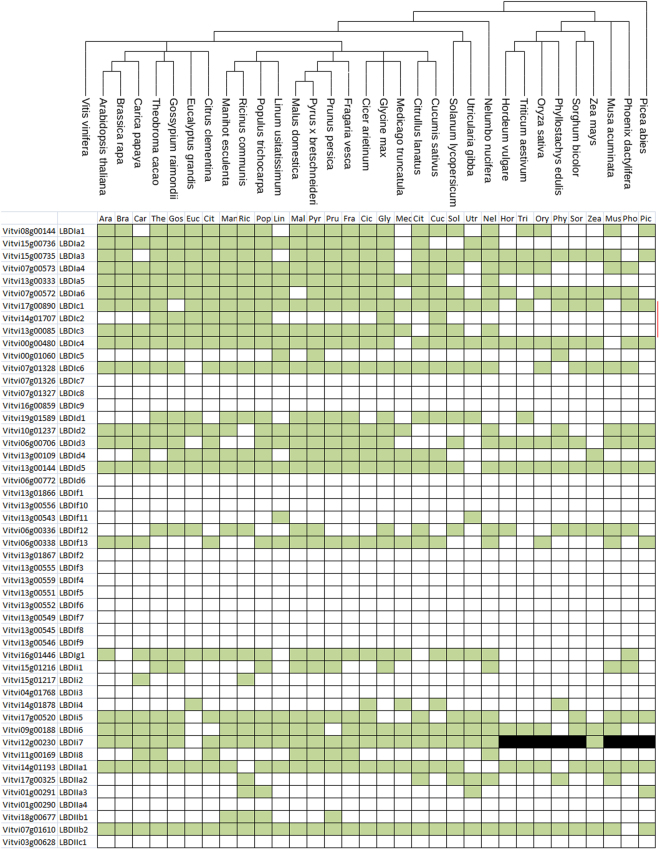
The orthologous relationship of LBD genes in V. vinifera and other plant species was analysed as previously described35 (Fig. 3). Orthologous relationships were classified into two categories depending on whether or not a one-to-one relationship with a given species gene was detected. Since the 3 blocks previously mentioned were highly conserved, homology between a grapevine gene and many LBD genes was systematically detected, except for VviLBDIi7 with most monocot species (in black in Fig. 3). Twenty genes showed a one-to-one orthologue relationship with an Arabidopsis gene when the comparison was carried out only with Arabidopsis. These genes likely correspond to well-conserved functions between both species.
Figure 3.
Grapevine LBD genes orthology against plant species with sequenced genomes. Green color represents one-to-one orthologs in the species (ortholog one-to-one = best match in the species that has the grapevine deduced protein as the best match in grapevine); white color represents no one-to-one homology match, and black color represents no match in the species.
In this context, a phylogenetic tree considering several mono and dicotyledonous species was constructed to identify genes with widely conserved functions among species (Fig. 3, Supplementary Fig. S3). VviLBDIa3, VviLBDIc1, VviLBDIc4, VviLBDId5, VviLBDIIa1 and VviLBDIIb2 presented orthologues at least in 88% of the species selected for comparison and could be involved in evolutionarily conserved functions.
This analysis did not detect orthologs for seventeen LBD genes while less than five orthologs were detected for six genes, mainly belonging to subclass If and Class II. This results may indicate that those proteins play a specific role in grapevine and in fact, Class IIc seems to be a Vitis vinifera species-specific subgroup. Regarding VviLBDIa2, VviLBDIa6 and VviLBDc3 they might have evolved after the monocot-dicot divergence since no orthologs were identified for them in the analysed monocot species. Supplementary Fig. S3 shows a cluster of the Vitis genes from the LBD1f subclass indicating a possible duplication event that appeared later and might be specific of the Vitis genus. Additionally, a Ka/Ks analysis was performed using the Ka/Ks calculation tool (http://services.cbu.uib.no/tools/kaks) on all the orthologs detected in the species for each grapevine gene, but no positive selection involving a grapevine gene in our gene set could be detected (no branch showed Ka/Ks ≫ 1, Supplementary Table S1).
Chromosomal location of the LBD genes
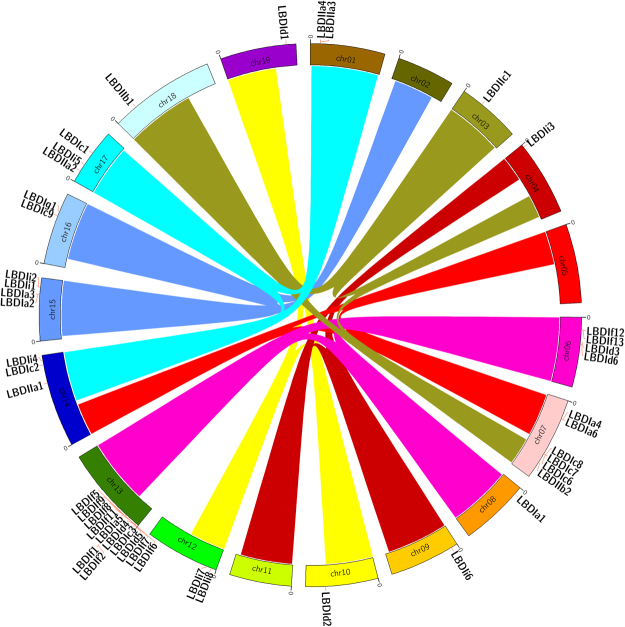
Grapevine LBD genes are unevenly distributed among the nineteen chromosomes. They are located in all chromosomes, except on chromosomes 2, 5 and 11 (Fig. 4). Two genes, LBDIc4 LBD6 and LBDIc5, were located on two scaffolds not assembled yet into any chromosome (they appear in the fictional chromosome “Unknown”). The highest number of VviLBD genes (15) was located on chromosome 13. The high number of LBD genes in this chromosome is mainly due to tandem repetition of genes belonging to the same subclass, in particular subclass LBD If genes. As highlighted by the orthology analysis, this duplication of class f probably occurred recently in Vitis since no ortholog was found in any other species. In contrast, chromosomes 3, 4, 8, 9, 10, 11, 12, 18, and 19 all carried a single LBD gene.
Figure 4.
Chromosomal localization of grapevine LBD genes. Links with the same colors in different chromosomes show previously described paralogous regions23. LBD genes from the same subclass were located in chromosomal regions that were previously identified as paralogous segments.
LBD genes belonging to the same subclass were located in chromosomal regions that were previously identified as paralogous segments resulting from ancestral polyploidization events28,36. In this way, LBD genes from subclass If are located in chromosomes 13 and 6, though LBD1f in chr 13 was located mostly just beside the presumed paralogous segment (Fig. 4). This is highly similar to what was obtained in a previous study for the GRAS sub-family LISCL35. The LISCL genes are also duplicated in the same area close to the paralogous region and have paralogs in Chr6. LBD1f and LISCL are at the same distance in chr13 and chr06 (1.7 Mb). It is possible that this area belongs actually to the paralogous region, since the paralog analysis was performed in the very original 8X version of the genome28 and might need an update. Class II genes are specific of two groups of paralogous segments, one group on chromosomes 1, 14, and 17 and another group on chromosomes 3, 7, 18. This indicates that all these subclasses predate the ancestral polyploidization events and likely play specific roles in grapevine since their functions were not redundant and were not discarded throughout evolution.
In addition, there are also tandem repetitions of genes belonging to different subclasses, like VviLBDIa5, VviLBDId4-5 and VviLBDIc3. These data revealed that segmental duplication and tandem duplications contributed significantly to the expansion and evolution of the LBD gene family.
Cis-acting regulatory elements in promoter regions
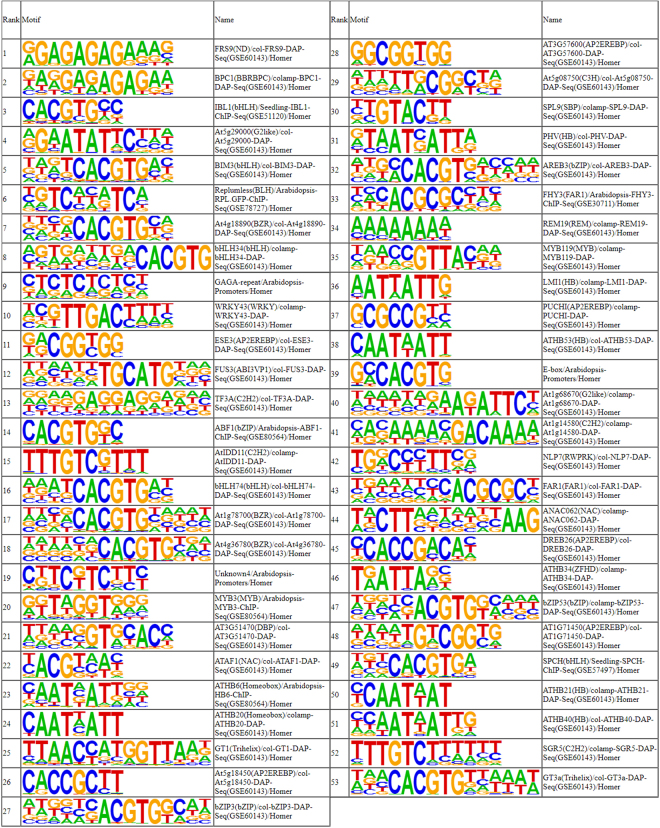
Analysis of cis-regulatory elements in the VviLBD promoter regions was performed using the PlantCARE (Supplementary Fig. S4; Supplementary Table S2) and PlantPAN databases (Supplementary Fig. S5; Supplementary Table S3). In addition to the core cis-elements, including the TATA box and CAAT box motifs presented in all promoter regions (data not shown), several regulatory motifs were identified and are associated with light regulation (BOX I, BOX 4, ACE, MRE), low temperature and heat stress responses (LTR, HSE), defence and stress responses (e.g. TC-rich repeats), hormonal regulation such as salicylic acid (e.g TCA-element, CA-element), methyl jasmonate (e.g CGTCA-motif), ethylene (e.g ERE), auxin (AuxRR-core, TGA-element), abscisic acid (ABRE, motif IIb, CE3), gibberellin (P-box, TATC-box, GARE-motif) and regulatory motifs related to tissue-specific expression (e.g Skn-1_motif, motif I, as1, GCN4_motif, RY-element) or developmental processes/ cell differentiation (HD-Zip 1, HD-Zip 2). Several transcription factor binding sites were also identified which have been widely associated with developmental processes and with biotic and abiotic stress responses namely AP2/ERF, NAC, C2H2, SBP, WRKY, Myb, bZIP and bHLH binding sites. Furthermore, these cis-regulatory elements were enriched in LBD promoter regions (Fig. 5; at P value < 0. 01). Interestingly, analyses of de novo motifs using Homer platform enabled the identification of a new motif GGTTGAATACAC as being enriched in VviLBD promoters (Supplementary Table S4). A similar known motif to this one was identified as GATGGAATAC (Supplementary Table S4).
Figure 5.
Enrichment of motifs on promoter regions of grapevine LBD genes. Several transcription factor binding sites were identified as enriched: FRS9 (ND); BPC1 (BBRBPC); IBL1 (bHLH); At5g29000 (G2like); BIM3 (bHLH); Replumless (BLH); At4g18890 (BZR); bHLH34 (bHLH); GAGA-repeat; WRKY43 (WRKY); ESE3 (AP2EREBP); FUS3 (ABI3VP1); TF3A (C2H2); ABF1 (bZIP); AtIDD11 (C2H2); bHLH74 (bHLH); At1g78700 (BZR); At4g36780 (BZR); Unknown4; MYB3 (MYB); AT3G51470 (DBP); ATAF1 (NAC); ATHB6 (Homeobox); ATHB20 (Homeobox); GT1 (Trihelix); At5g18450 (AP2EREBP); bZIP3 (bZIP); AT3G57600 (AP2EREBP); At5g08750 (C3H); SPL9 (SBP); PHV (HB); AREB3 (bZIP); FHY3 (FAR1); REM19 (REM); MYB119 (MYB); LMI1 (HB); PUCHI (AP2EREBP); ATHB53 (HB); E-box; At1g68670 (G2like); At1g14580 (C2H2); NLP7 (RWPRK); FAR1 (FAR1); ANAC062 (NAC); DREB26 (AP2EREBP); ATHB34 (ZFHD); bZIP53 (bZIP); AT1G71450 (AP2EREBP); SPCH (bHLH); ATHB21 (HB); ATHB40 (HB); SGR5 (C2H2); GT3a (Trihelix).
It should be highlighted that the enrichment in MYB binding sites is known to regulate the expression of genes involved in phenylpropanoid metabolism. Interestingly, VviLBDIf6 co-expressed with a gene coding for MYB divaricate and VviLBDIa1 with a gene coding for a bHLH family transcription factor (Table 2). The bHLH048 transcription factor was already found to interact with the AtASL4 from Arabidopsis, and its interaction reduces the affinity of LOB gene for its 6-bp GCGGCG consensus DNA motif24.
Table 2.
Co-expression analysis of the VviLBD genes.
| Unique_ID/Nimblegen probeset | Functional annotation | Functional Categories |
|---|---|---|
| VIT_15s0046g00230 | VviLBDIi1 | LOB domain family transcription factor |
| VIT_15s0046g00240 | VviLBDIi2 | LOB domain family transcription factor |
| VIT_01s0011g03540 | VviLBDIIa3 | LOB domain family transcription factor |
| VIT_10s0003g03490 | GA 2-oxidase | Metabolism. Secondary metabolism. Terpenoid metabolism. Diterpenoid metabolism. Diterpenoid biosynthesis |
| VIT_14s0006g02950 | VviLBDIIa1 | LOB domain family transcription factor |
| VIT_12s0057g00170 | Wound-induced | Response to stimulus. Stress response. Abiotic stress response. wounding |
| VIT_07s0197g00040 | VviLBDIc7 | LOB domain family transcription factor |
| VIT_07s0031g02270 | Tonoplast monosaccharide transporter2 | Transport overview. Electrochemical Potential-driven Transporters. Porters. Major Facilitator Superfamily. Sugar Porter |
| VIT_14s0066g00680 | VviLBDIc2 | LOB domain family transcription factor |
| VIT_11s0016g05450 | Equilibrative nucleoside transporter ENT8 splice variant | Transport overview. Electrochemical Potential-driven Transporters. Porters. Equilibrative Nucleoside Transporter |
| VIT_01s0011g03530 | VviLBDIIa4 | LOB domain family transcription factor |
| VIT_08s0007g04480 | Pectinesterase family | Cellular process.Cellular component organization and biogenesis.Cell wall organization and biogenesis.Cell wall metabolism.Cell wall modification.Pectin modification |
| VIT_06s0004g07790 | VviLBDId6 LBD13 | LOB domain family transcription factor |
| VIT_02s0025g02940 | Caffeic acid O-3-methyltransferase | Metabolism. Secondary metabolism. Phenylpropanoid metabolism. Phenylpropanoid biosynthesis |
| VIT_12s0028g03580 | Lectin-receptor like protein kinase 3 | Signalling. Signalling pathway. Protein kinase |
| VIT_14s0068g01360 | GEM-like protein 5 | Cellular process. Cell growth and death |
| VIT_02s0025g02920 | Quercetin 3-O-methyltransferase 1 | Metabolism. Secondary metabolism. Phenylpropanoid metabolism. Flavonoid metabolism. Flavonoid biosynthesis |
| VIT_15s0048g00830 | VviLBDIa3 | LOB domain family transcription factor |
| VIT_18s0001g15390 | Gaiacol peroxidase | Metabolism. Primary metabolism. Amino acid metabolism. Aromatic amino acid metabolism. Phenylalanine metabolism. Phenylalanine biosynthesis |
| VIT_17s0000g09030 | Disease resistance protein (NBS-LRR class) | Diverse functions. Gene family with diverse functions. NBS-LRR superfamily |
| VIT_15s0048g00500 | Pectinesterase family | Cellular process. Cellular component organization and biogenesis. Cell wall organization and biogenesis. Cell wall metabolism. Cell wall modification. Pectin modification |
| VIT_13s0019g03780 | VviLBDIf6 | LOB domain family transcription factor |
| VIT_07s0031g02280 | MYB divaricata | Development. Reproductive development. Flower development |
| VIT_08s0056g01650 | VviLBDIa1 LBD20 | LOB domain family transcription factor |
| VIT_11s0103g00200 | Anthranilate N-benzoyltransferase | Metabolism. Primary metabolism. Amino acid metabolism. Aromatic amino acid metabolism. Aromatic amino acid biosynthesis |
| VIT_01s0127g00860 | Aborted microspores AMS | Regulation overview. Regulation of gene expression. Regulation of transcription. Transcription factor. bHLH family transcription factor |
| VIT_18s0001g15690 | Endo-1,4-beta-glucanase | Cellular process. Cellular component organization and biogenesis. Cell wall organization and biogenesis. Cell wall metabolism. Cell wall catabolism. Cellulose catabolism |
| VIT_18s0001g15680 | Cellulase | Cellular process. Cellular component organization and biogenesis. Cell wall organization and biogenesis. Cell wall metabolism. Cell wall catabolism. Cellulose catabolism |
| VIT_15s0021g02170 | Chalcone and stilbene synthase | Metabolism. Secondary metabolism. Phenylpropanoid metabolism. Flavonoid metabolism. Flavonoid biosynthesis |
| VIT_17s0000g05490 | VviLBDIi5 | LOB domain family transcription factor |
| VIT_09s0002g04380 | Plastidic glucose transporter 2 | Transport overview. Electrochemical Potential-driven Transporters. Porters. Major Facilitator Superfamily. Sugar Porter |
| VIT_12s0059g02500 | Constans-like 11 | Development. Reproductive development. Flower development |
| VIT_18s0001g13580 | Kinesin motor protein | Cellular process. Cellular component organization and biogenesis. Cytoskeleton organization and biogenesis. Microtubule organization and biogenesis. Microtubule-driven movement |
| VIT_03s0063g00510 | Leucine-rich repeat | Diverse functions. Gene family with diverse functions. NBS-LRR superfamily |
| VIT_06s0009g01830 | Invertase, neutral/alkaline | Metabolism. Primary metabolism. Carbohydrate metabolism. Monosaccharide metabolism. Galactose metabolism |
| VIT_07s0031g01870 | Zinc finger (CCCH-type) family protein | Regulation overview. Regulation of gene expression. Regulation of transcription. Transcription factor. C3H family transcription factor |
| VIT_00s2422g00010 | Hexokinase-2 | Metabolism. Primary metabolism. Carbohydrate metabolism. Glycolysis Gluconeogenesis |
| VIT_00s0288g00050 | V-type H+-transporting ATPase subunit G | Metabolism. Primary metabolism. Generation of metabolite precursors and energy. Electron transport. Respiratory-chain phosphorylation |
| VIT_19s0014g01240 | Morphogenesis of root hair 1 MRH1 | Development. Root development |
| VIT_18s0122g00910 | Mlo5 | Cellular process. Cell growth and death. Cell death |
| VIT_17s0000g07750 | Zinc finger protein 5 | Regulation overview. Regulation of gene expression. Regulation of transcription. Transcription factor. C2H2 family transcription factor |
| VIT_07s0005g01640 | feronia receptor-like kinase | Signalling. Signalling pathway. Protein kinase |
| VIT_00s0225g00170 | Peroxidase | Metabolism. Primary metabolism. Amino acid metabolism. Aromatic amino acid metabolism. Phenylalanine metabolism. Phenylalanine biosynthesis |
For some genes the list of co-expression is not complete. Further details are presented in Supplementary Table 2.
Expression analysis of grapevine LBD genes
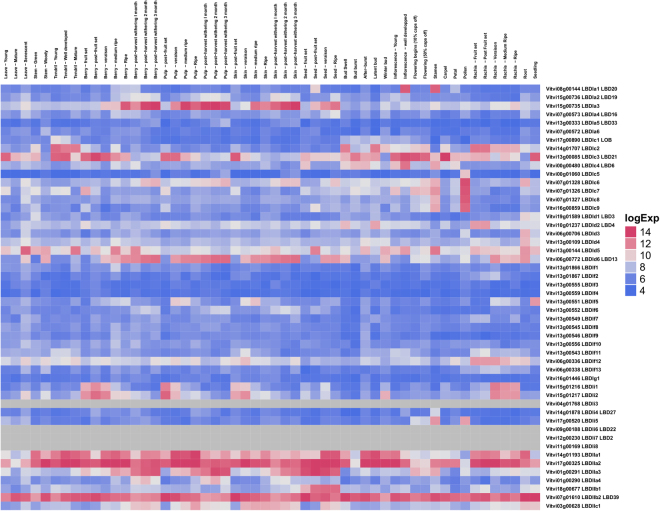
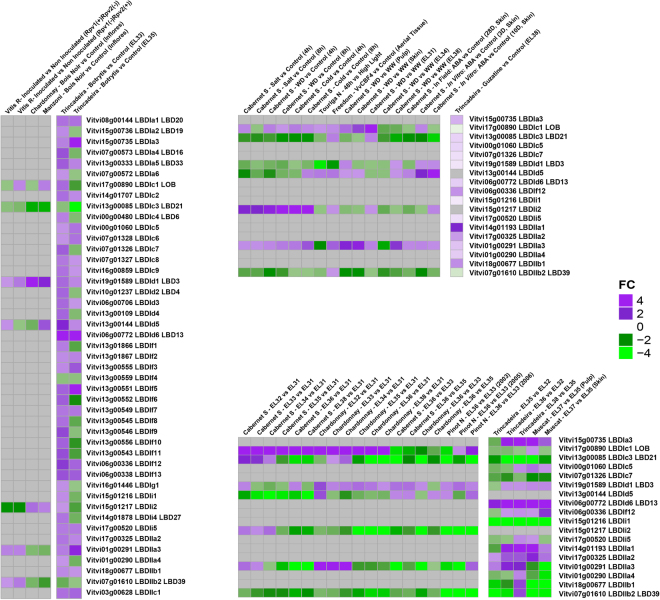
Three distinct approaches were performed to characterize LBD genes expression in grapevine: (i) an atlas of expression of the LBD genes was constructed based on the absolute value of gene expression published in the grapevine gene expression atlas37 (Fig. 6). Plant Ontology (PO) was attributed when a gene was clearly expressed in a given tissue. (ii) co-expression analysis was preformed based on the same original data using relative values of expression of all genes, centered on the average expression (Supplementary Table S5). The main objectives of this analysis were to determine expression patterns and to identify genes that were following the same pattern as the LBD genes and that could be under the same regulatory elements, or under the regulation of the LBD gene itself. The results presented in Table 2 revealed that twelve genes showed a correlation with other genes with a Pearson Correlation Coefficient (PCC) threshold of 0.2. Finding the optimal PCC threshold to retrieve functionally related genes was affected by the method of gene expression database construction and the target gene function38, but the PCC that was chosen was very stringent. (iii) public expression data was analysed in order to identify the behaviour of LBD genes during berry development and ripening and upon abiotic and biotic stress conditions (Fig. 7). Figure 7 presented the expression value among the experiments where difference in expression of LBD genes was detected.
Figure 6.
Expression of LBD genes in grapevine tissues. Gradient color is expressed in RMA-normalized intensity value on the Nimblegen microarray.
Figure 7.
Expression of LBD genes during grape berry development and ripening, and upon abiotic and biotic stresses. Left experiments of each heatmap were performed with GeneChip microarrays, and right experiments were performed with GrapeGen microarray. Grape berry development: developmental stages from EL31 to EL38; cultivars Cabernet Sauvignon, Chardonnay, Pinot Noir, Trincadeira and Muscat. Abiotic stress experiments: salt, cold, water deficit, high light, ABA. Biotic stress experiments: P. viticola, BoisNoir and Botrytis cinerea.
Tissue specific gene expression
Based on the V. vinifera cv. Corvina gene expression atlas39, several LBD genes showed a strong tissue specificity of expression, with the majority of Class I genes being poorly expressed in the different tissues (Fig. 6). VviLBDIa3 and VviLBDId6 were highly expressed mainly in ripe berry tissues. VviLBDId6 was shown to be co-expressed (Table 2, Supplementary Table S5) with genes involved in phenylpropanoid metabolism, including two caffeic acid O-3-methyltransferase genes (VIT_02s0025g02940 and VIT_02s0025g02930) and a quercetin 3-O-metyltransferase gene (VIT_02s0025g02920), as well as with signaling and cell growth and death-related genes (VIT_12s0028g03580 and VIT_14s0068g01360, respectively). VviLBDIc3 had high expression in young leaves, young and well developed tendril, inflorescences, and in berry tissues mainly at the beginning of fruit development (green and véraison stages). In addition, transcripts corresponding to subclass Ic genes, VviLBDIc6-9, seemed to be more abundant in pollen and stamen.
Interestingly, VviLBDIa1 was only expressed in well-developed inflorescence and stamen and may have a specific function in the development of these tissues; it also co-expresses with genes involved in cell wall and secondary metabolism and transport (VIT_18s0001g15690, VIT_18s0001g15680, VIT_11s0103g00200, VIT_19s0015g00960). VviLBDIc7 was co-expressed with TONOPLAST MONOSACCHARIDE TRANSPORTER 2 (VIT_07s0031g02270). The class II genes VviLBDIIa1, VviLBDIIa2, and VviLBDIIb2 were abundantly expressed in almost all grapevine tissues, and, VviLBDIIa3 was more abundant in seeds and post-harvest berry tissues. Differential expression of LBD genes in diverse tissues were also observed for other plant species including Arabidopsis, rice, and maize17,26,40,41.
Gene expression during berry development and ripening
Expression studies regarding berry development and ripening revealed the involvement of LBD genes in different stages (Fig. 7). VviLBDIc1 gene was highly expressed after EL31 stage42, i.e. pea-size berries, both in cv. Cabernet Sauvignon and cv. Chardonnay. However, this expression profile can be cultivar and/or season dependent. In cv. Trincadeira, VviLBDId6 was expressed along berry ripening as previously mentioned for cv. Corvina. The same holds true for VviLBDIa3 which showed up-regulation at the onset of ripening and was co-expressed with genes involved in stress response (Table 2, Supplementary Table S5) namely a Disease Resistance protein (NBS-LRR class) (VIT_17s0000g09030) and a Gaiacol peroxidase (VIT_18s0001g15390). On the other hand, VviLBDIi1 was down-regulated in advanced ripening stages.
Concerning class II genes, VviLBDIIa1, VviLBDIIa2, VviLBDIIa3 seem to play a role in grape ripening. Furthermore, VviLBDIIa1 co-expressed with wound-induced genes involved in abiotic stress response and VviLBDIIa3 co-expressed with a gene coding for GA 2-oxidase (VIT_10s0003g03490).
Gene expression upon abiotic stress
Expression analysis concerning abiotic stress conditions (Fig. 7) revealed that VviLBDIi2 was up-regulated under salt, cold and water deficit conditions in shoot tips. VviLBDId5 expression in berry skin showed a positive response to in vitro ABA treatment. VviLBDIIa3 was also up-regulated in pulp and skin submitted to water deficit. Interestingly, VviLBDIIa1 responded to guazatine treatment, an inhibitor of polyamine oxidase involved in polyamine catabolism43.
On the other hand, some VviLBD genes presented mostly down-regulation upon abiotic stress such as VviLBDIc3 and VviLBDIIb2. VviLBDId1 was strongly down-regulated after 48 h of high light exposure.
Gene expression upon biotic stress
Regarding biotic stress conditions (Fig. 7), VviLBDIi2 was down-regulated in partially and completely resistant plants (resistance genes named Rpv1 and Rpv2) when inoculated with Plasmopara viticola. VviLBDId1 was up-regulated in inflorescences presenting Bois noir disease, with higher expression in cv. Chardonnay. On the other hand, VviLBDIc3 was down-regulated in the same conditions. This gene was also down-regulated in grape berries infected with Botrytis cinerea, with lower expression after long exposure (véraison stage). VviLBDId6 and VviLBDIIa1 were strongly up-regulated after Botrytis infection. VviLBDIIa1 co-expressed with six wound-induced genes as previously mentioned (Table 2, Supplementary Table S5). VviLBDIa3, VviLBDIf5 and VviLBDIIa3 were up-regulated upon Botrytis infection with higher expression at véraison stage. However, the majority of VviLBDs seemed to participate in the early response towards Botrytis attack.
Discussion
Prediction of putative biological functions for a given gene family can be approached based on genomic and transcriptomic available data with improved bioinformatics tools. In this study, we performed an extensive analysis of the LBD genes on the 12x Vitis vinifera genome sequence based on the isolation of the complete set of genes identified in PN40024. Characterization of LBD gene family and their putative functions was performed in grapevine based on detailed gene structure and expression analyses, chromosome localization, and comparative phylogenetic analyses with other sequenced genomes from different monocot and eudicot species.
LOB domain gene family in grapevine
The LOB domain gene family, also known as ASYMMETRIC LEAVES2-like (ASL) gene family, was firstly described in the past decade16,17 and several studies have been made to unveil their role in plant processes. Specific LBD proteins characterized in Arabidopsis and in crop plants including rice seem to display a wide functional diversity. They were found involved in the regulation of several developmental processes, namely meristem programming, leaf patterning, inflorescence development, embryogenesis, lateral root formation, vascular patterning, as well as metabolic processes such as anthocyanin and nitrogen metabolisms1,2. More recently, they have been also associated with biotic stress responses10,44–46.
The exhaustive analysis of the grapevine LBD genes performed in this study has led to the identification of 50 LBD genes. The N-terminal LOB domain that characterizes this gene family was identified in all predicted proteins. The VviLBD genes were located to sixteen of the nineteen grapevine chromosomes. Phylogenetic analysis and evolutionary relationships divided the LBD gene family into two classes, Class I and Class II, as previously observed to other plant species, characterized by the presence or absence of functional leucine-zipper-like domains, respectively16,17,25,26,33,40. The majority of VviLBD genes belongs to Class I. Grapevine LBD genes were further clustered into nine subclasses, six in Class I and three in Class II. Minor clades of the two major classes were also identified in other plant species: class I was divided in five subgroups in rice and maize, four in Arabidopsis, Lotus japonicus and Medicago truncatula, and seven in Malus domestica 25,26,40,47. Class II was divided in two subclasses in apple and maize25,40.
The gene structure analysis revealed that 74% (37 out of 50) of the VviLBD genes contained two exons, as the majority of LBD genes in other plant species25,26,47, indicating a conserved structure during evolution. On the other hand, the highly variable C-terminal domain in LBD proteins from grapevine and other plant species indicate functional diversity associated with this gene family47. Experiments of LOB domain swapping for the AS2 gene revealed that, despite the high similarity, the domain cannot be functionally replaced by a LOB domain of other family members indicating that dissimilar amino acid residues in the N-terminal are also important for the functional specificities of these transcription factors family members47. The highly-conserved C-block (CX2CX6CX3C) domain is present in all grapevine LBD proteins reinforcing its functional importance mainly associated with the DNA-binding process17,47. Class I proteins presented the leucine-zipper-like motif, which includes five hydrophobic amino acids (valine, isoleucine, leucine) separated by six variable amino acid residues and has been linked to protein-protein interaction.
The grapevine LBD gene family with 50 members is larger than the 43 LBD genes of Arabidopsis, the 24 in barley, the 31 in mulberry; the 35 in rice, 36 in Sorghum bicolor; the 38 in Lotus japonicus, and the 44 in maize16,17,26,33,34,40,48–50. Medicago truncatula and apple present a higher number of LBD genes, containing 57 and 58, respectively25,33. With approximately the same genome size, Vitis vinifera harbours more LBD genes than Lotus japonicus (approximately 487 and 470 Mb, respectively). Despite Malus domestica had almost the double genome size of Vitis vinifera, this fruit tree species only contains eight more genes25. Therefore, a large LBD density variation is observed among plant species.
Expansion of the grapevine LBD gene family likely took place by segmental/chromosomal duplication, as observed for other species from different taxonomic groups25,34,47. Duplicated genes like VviLBDIi1 and VviLBDIi2 might show functional redundancy, as suggested by their similar expression profile or co-expression in several grapevine tissues. Their functional study might unveil the evolutionary role of gene duplication and their contribution in plant processes. Duplication events are more likely to be retained for gene families involved in signal transduction and transcriptional regulation51. Nevertheless, no additional grapevine LBD genes co-expressed together which might indicate different functions or specialization in most cases. In fact other likely duplicated genes such as VviLBDIf12 and VviLBDIf13, showed a clear expression divergence in the expression analysis which might suggest their functional diversification. Tandem duplicated events were mainly associated with subclass If, which contained tandem repeated genes with high similarity. VviLBDIf12 and VviLBDIf13 presented several orthologs in other plant species, which might indicate conserved function. Nevertheless, for the remaining members of subclass If no ortholog was identified in the studied species, suggesting grapevine-specific functions. Moreover, some genes from specific subclasses were found in paralogous regions of the grapevine genome derived from polyploidization event28. Among them, subclass If had members in chromosomes 6 and 13, subclass IIa in chromosomes 1 and 17, and subclass IIb in chromosomes 7 and 18.
Expression patterns across a variety of tissues indicate roles of LBD genes in regulation of metabolism and organ differentiation
VviLBD genes showed different expression patterns across the grapevine tissues. No subclass-specific expression pattern was observed, as occurred in other species namely L. japonicus, M. truncatula and apple, suggesting gene-specific function or localization regardless the phylogenetic subclass25,33. For example, VviLBDIIb2 is highly expressed in almost all grapevine tissues whereas VviLBDIIb1 seemed to be expressed only in seeds. LBD38, the Arabidopsis ortholog of VviLBDIIb2, is involved in nitrogen and anthocyanin metabolism, as well as their close homologs LBD37 and LBD39 21. Moreover, the LBD37 rice ortholog, OsLBD37/ASL39, was also associated with nitrogen metabolism, particularly in nitrogen remobilization and senescence52. This could suggest a conserved function of these genes across plant species and therefore VviLBDIIb2 could regulate nitrogen and anthocyanin metabolism in a wide range of grapevines tissues. In fact, orthologs for this gene were found in 32 out of the 33 plants studied (Fig. 3).
Specific tissue expression patterns suggest the involvement of VviLBDIa1 and VviLBDIc3 in the development of floral organs. Interestingly, VviLBDIa1 co-expressed with cell wall-related genes (endo-1,4-beta-glucanase, cellulose), secondary metabolism genes (anthranilate N-benzoyltransferase, chalcone and stilbene synthase) and with a bHLH family transcription factor (aborted microspores AMS). In rice, down-regulation of a LBD gene (OsIG1) led to developmental abnormalities of various floral organs53 providing links between LBD proteins and floral organ development as it could be the case in grapevine. Furthermore, in Arabidopsis thaliana ASYMMETRIC LEAVES1 and ASYMMETRIC LEAVES1 2 (AS1 and AS2) and JAGGED (JAG) genes were shown to function in sepal and petal primordia to repress boundary-specifying genes for normal development of the organ5. In grapevine, the ortholog of the Arabidopsis LBD6/AS2 is VvLBDIc4 which is also expressed in flower organs but at a lower extent than VviLBDIa1 and VviLBDIc3. Also noteworthy is the pollen specific expression of VviLBDIc6, VviLBDIc7, VviLBDIc8 and VviLBDIc9 which may have redundant functions. VviLBDIc6 has orthologs in several species including AtLBD36 which was previously shown to be expressed in pollen54.
LBD genes may be involved in berry development and ripening through interaction with growth regulators
In grapevine, some LBD genes showed differential expression during fruit ripening, in particular the up-regulated genes VviLBDIa3, VviLBDId6, VviBDIIa1 and the down-regulated VviLBDIc3, VviLBDIi1 and VviLBDIi2. VviLBDIIa3 was up-regulated at the initial development stages in cv. Chardonnay suggesting an involvement in fruit set and early developmental stages characterized by intense cell division. Nevertheless, VviLBDIIa3 showed higher expression on the ripe stage of cv. Trincadeira, suggesting an expression pattern dependent on the variety. This gene is also co-expressed with a gene coding for gibberellin 2-oxidase (VIT_10s0003g03490) that inactivates endogenous bioactive gibberellins (GAs), suggesting an involvement of VviLBDIIa3 in GA metabolism during fruit-set. In cv. Corvina, VviLBDIIa3 was highly expressed in seed at fruit set and post-fruit set stages but also in ripe and post-harvest berries (Fig. 7). Little is known about the direct involvement of GAs on berry ripening, nevertheless, some evidences suggest a possible role in flowering and initial stages of berry development55. Additionally, differential accumulation of bioactive GAs was observed from flowering to fruit set, and this accumulation is finely regulated by the abundance and localization of GA oxidase transcripts56. Interestingly, AtLBD40, a close homolog of grapevine LBD subclass IIa genes, was reported to be a direct target of DELLA (growth-repressing transcription factor) in GA signalling pathway and to be down-regulated by gibberellin57.
VviLBDId6, expressed in ripe and post-harvest berry tissues, and also showed differential expression under biotic conditions. This gene was co-expressed with several genes involved in secondary metabolism (caffeic acid O-3-methyltransferase and quercetin 3-O-metyltransferase gene), signalling pathways (lectin-receptor like protein kinase 3) and cell growth and death (GEM-like protein 5). The expression of the close Arabidopsis homolog, AtLBD15/ALS11 leads to down-regulation of several cellulose synthase genes and is activated by a key regulator of secondary cell wall synthesis58.
In Vitis vinifera, LBD1d6 and LBDIa3 were identified as positive molecular markers of ripening stage in three Portuguese cultivars59. Analysis of cis-acting elements suggests modulation of these genes by several growth regulators (ABA, methyl jasmonate, auxin, ethylene) and in response to stress (Supplementary Table S2). VviLBDIa3 promoter showed a MYB binding site involved in flavonoid biosynthetic genes regulation. Interestingly, these two genes (VIT_06s0004g07790, VIT_15s0048g00830) were also identified as switch genes together with MYB transcription factors, cellulase, expansin B and caffeic acid 3-O-methyltransferase due to the fact that they are expressed at low levels in vegetative/green tissues and show a significant increase in mature/woody organs, suggesting a potential regulatory role during this developmental transition60. The putative participation of LBD genes in fruit ripening as suggested here is additionally supported by studies in banana where Ma LBD 1 3 was found to be ripening inducible61.
Interestingly, the promoter of VviLBDIa3 was one of the promoters of LBD genes with AuxRR-core motif involved in auxin responsiveness. The Arabidopsis ortholog of VviLBDIa3, LBD18/ASL20, together with LBD16 and LBD29, are key regulators of lateral root initiation/formation as direct targets of AUXIN RESPONSE FACTORs7,19,23. Arabidopsis LBD16, 17, 18 and 29 were also found to have an important role in in vitro callus formation induced by auxin9. Furthermore, both Arabidopsis and banana LBD genes were shown to directly regulate expression of EXPANSIN genes, encoding cell wall-loosening factors61–64 that are also modulated during grape ripening65.
Other LBD genes such as VviLBDIi1 were less expressed during grape ripening. In fact, this gene as well as VviLBD1c3 were identified as negative biomarkers of ripening stage in three Portuguese cultivars59. Additionally, VviLBDIi1 possesses a cis-acting element involved in ABA responsiveness, a growth regulator that increases during ripening55, suggesting that this LBD gene might be negatively regulated by ABA.
Brassinosteroids are steroidal plant hormones that have been proposed as ripening promoters in non-climacteric fruits, in particular grape berries55. The Arabidopsis LOB gene negatively regulates the accumulation of brassinosteroids in organ boundaries66. VviLBDIc1 is an ortholog of LOB and seems to be down-regulated during grape ripening as suggested here and in previous studies59, and possibly interacts with brassinosteroids. However, it should be noted that at pea-size stage of berry development VviLBDIc1 expression seems to be very low compared to the following ripening stages highlighting the importance of conducting detailed temporal studies of gene expression.
Altogether VviLBD1d6, VviLBDIa3 and VviLBD1c3 are robust candidates to participate in the regulation of the onset of the grape ripening program.
Expression of LBD genes upon abiotic and biotic stresses
In grapevine, some class I genes showed differential expression under abiotic stress conditions. Particularly, VviLBDIi2 is up-regulated under salt, cold and water deficit conditions, VviLBDId5 is up-regulated after in vitro ABA treatment, and VviLBDId1, down-regulated after 48 h of high light exposure. Interestingly, VviLBDIi2 presents in his promoter a MYB binding site involved in drought-inducibility, VviLBDId5 a cis-acting element involved in the abscisic acid responsiveness and VviLBDId1 many elements involved in light responsiveness, suggesting the involvement of LBDs in abiotic stress response.
Few studies focused on the role of LBD genes in abiotic stress. However, in Medicago truncatula, LBD1 gene was reported to have an important role in root architecture under salt stress67. Additionally, MtHB1, an ABA and salinity responsive transcription factor, was found to directly recognize a specific cis-acting element in the MtLBD1 promoter68. By contrast, several Sorghum bicolor LBD genes where highly induced under salt and drought stress conditions, suggesting a role in abiotic stress response50, whereas, in banana fruit, MaLBD5 expression was induced by cold and methyl jasmonate treatment69.
The majority of VviLBDs seem to participate in the early response towards Botrytis attack as previously mentioned. However, VviLBDId6 and VviLBDIIa1 were strongly up-regulated after Botrytis infection both at EL33 (green berries) and EL35 (véraison). VviLBDIIa1 was also found co-expressed with six wound-induced genes as previously referred (Table 2, Supplementary Table S5). Interestingly, the promoter of this gene presents several cis-elements related to abiotic stress and methyljasmonate responses. Jasmonates were previously proposed to be involved in grape response to Botrytis infection70. Expression profiles of several Class II AtLBD genes revealed induction by pathogens, including necrotrophic fungal pathogens Alternaria brassicicola and B. cinerea, root pathogen Phytophthora parasitica (oomycete) and the root-knot nematode Meloidogyne incognita, suggesting a role in plant defence mechanisms44. VviLBDIIa3 seems to be involved in GA regulation of fruit set and fruit ripening but this gene was also up-regulated upon Botrytis infection with higher expression at véraison stage. Although gibberellins are mainly associated with plant growth and development, they have been recently related to response to pathogen attack71. Still, further studies are required to elucidate the role of GA in plant defence that remains very complex and unclear. Genes coding for gibberellin 20 oxidase were up-regulated after B. cinerea infection70, suggesting activation of GA metabolism in defence response possibly with the involvement of VviLBDIIa3.
The involvement of the LBD genes in stress response has been poorly studied so far though some genes have been shown to play a role in disease susceptibility2. AtLBD20 was the first LBD gene associated with disease susceptibility10. In grapevine, AtLBD20 homolog/ortholog did not show a relevant expression level in berries upon B. cinerea fungal infection, however VviLBDIa3, belonging to the same clade, was up-regulated after long exposure to B. cinerea inoculation. In addition, VviLBDIa3 gene was found to be co-expressed with several genes including a NBS-LRR gene (VIT_17s0000g09030) related to defence and a pectinesterase gene (VIT_15s0048g00500), involved in cell wall modification processes. Nucleotide-binding site (NBS) leucine-rich repeats (LRR) proteins are involved in the recognition of pathogen effectors with virulence functions72.
Besides VviLBDIa3 and VviLBDIIa3, VviLBDIf5 was up-regulated upon Botrytis infection at véraison stage. Although no ortholog could be found for this gene, it belongs to the clade If that comprises the AtLBD1 and AtLBD11. The closest Citrus sinensis homolog of these Arabidopsis genes, CsLOB1, was found to function as disease susceptible gene in citrus bacterial canker, a disease caused by multiple Xanthomonas species45. Moreover, CsLOB2 and CsLOB3, belonging to the same clade as CsLOB1, were found to have a similar role as CsLOB1 in citrus bacterial canker73. Another putative disease susceptible gene might be VviLBDIi2 which was down-regulated in partially and completely resistant plants derived from Muscadinia rotundifolia when inoculated with Plasmopara viticola. Interestingly, VviLBDIi2 presented in its promoter cis-acting elements involved in salicylic acid responsiveness, a hormone known to be involved in response to biotrophic pathogens70.
The involvement of grapevine LBD genes in response to biotic stress was also noticed for Bois noir disease. VviLBDIc3 showed down- regulation in inflorescences presenting Bois noir disease, in grape berries after long exposure to B. cinerea, cold and ABA treatment, which could suggest that some LBD genes may be simultaneously modulated by abiotic and biotic stress conditions.
Conclusions
LOB domain (LBD) transcription factors families have been characterized in several plant species and shown to participate in the regulation of developmental programs and stress responses. Nevertheless, the role of LBDs in fruit ripening has been poorly documented. Modulation of LBD genes expression during grape berry development and ripening indicates that these processes may be under regulation of LBD transcription factors. In addition, several grapevine LBD genes bared cis-elements in their 5′ regulatory region associated with defence and hormonal regulation which together with expression and co-expression analyses supports their involvement in the abiotic and biotic stress response mechanisms. Candidate genes were identified that exhibit broad response to stress (e.g. VviLBDIc3) or could be involved in grape ripening and grape defence (e.g.VviLBDId6). Altogether this data may be used for functional characterization of genes and ulterior improvement of fruit quality traits and resilience to abiotic and biotic stresses.
Methods
Identification of LBD genes
Genes previously identified as encoding LOB domain proteins in Grimplet et al.30 were blasted (blastp and tblastn) against the grapevine genome 12Xv.2 (https://urgi.versailles.inra.fr/Species/Vitis/Data-Sequences/Genome-sequences), the non-redundant list of genes in Grimplet et al.30, the NCBI refseq (both remapped on the 12Xv2 of the genome assembly) and the COST annotation gene set available at the ORCAE website (http://bioinformatics.psb.ugent.be/orcae/). Results from different analyses were manually cross-checked to identify new potential loci corresponding to LBD genes in the grapevine genome. The UGene software74 was used to design the gene models on the grapevine genome and test their structure.
Gene structure analysis
The potential coding DNA sequences (CDS) were blasted (blastx) against the NCBI public database to compare the structures with other known LBD genes in other species and the NCBI Refseq predictions of the grapevine genes. When discrepancies were observed, gene models were corrected using the UGene software. Loci bearing non-functional genes were eliminated from the list. A GFF file with the LBD genes was designed, uploaded into the IGV software and the RNAseq data available on flowers in the laboratory were used to double-check the exon structure of the genes.
Promoter analysis
Promoter cis-acting regulatory elements within 1.5 kb of the upstream sequence from the ATG initial codon of each grapevine LBD gene were analyzed with PlantCARE software75 (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Analysis of transcription factor binding sites (TFBSs) of the 3 kb upstream sequence region of the initial codon was also performed using the Plant Promoter Analysis Navigator (PlantPAN) software76 (http://plantpan2.itps.ncku.edu.tw/).
Enrichment of cis-regulatory elements
Motif analysis of known and de novo motifs was performed using Homer v4.977 (http://homer.ucsd.edu/homer/motif/). With this end, grapevine promoter sequences (2.5 kb upstream of the coding sequence) of LBD genes were retrieved from Regulatory Sequence Analysis Tools (RSAT, http://floresta.eead.csic.es/rsat/). Additionally, in order to prevent overlapping between neighbouring genes, noorf option was performed.
Sequence alignment and phylogenetic analysis
Sequence information on previously reported LOB domain proteins of A. thaliana was retrieved from the Arabidopsis Information Resource (https://www.arabidopsis.org/). Evolutionary analyses were conducted in MEGA641. Multiple sequence alignment was inferred using MUSCLE78. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model79. The bootstrap consensus tree inferred from 100 replicates was taken to represent the evolutionary history of the taxa analyzed80. Branches corresponding to partitions reproduced in less than 30% of bootstrap replicates were collapsed. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The coding data was translated assuming a standard genetic code table. All positions with less than 95% site coverage were eliminated. The genes were named according to Grimplet and co-workers32 based on the distance homology with Arabidopsis genes.
The alignment file between Arabidopsis and grapevine sequences was uploaded to the Jalview and UGene software for manual adjustment of the alignment and manual motif editing. Motifs were flagged and labelled for the grapevine genes; additional motifs of high homology were also identified (at least 50% homology within the members of the subfamily on at least 10 amino acids) among grapevine sequences.
Expression analysis
Expression data were retrieved from 3 different microarray platforms (Affymetrix Genchip (16k probesets) GrapeGen (21k probesets), Vitis Nimblegen array (29k probesets) and from our in-house RNAseq projects. Data normalization was performed on all the array of each platform (RMA normalization). After retrieving the values for the probesets corresponding to each gene, the values for the 3 or 4 replicates of the same condition were averaged to obtain a total of up to 256 conditions (organ, cultivar, treatment, platform) for the genes present in all platform. Based on expression data of the grapevine gene expression atlas37, a plant ontology ID was attributed to each gene if expression intensity in a tissue was above a defined threshold of absolute log2 value of 8 or absolute value of 256. The same data were used for the co-expression analysis with the whole set of genes available on the Nimblegen platform. Hierarchical clustering with Pearson correlation as metric and average linkage cluster method was performed. Genes considered as having the same profile should present a distance threshold between each other lower than of 0.2.
For further evaluation of gene expression samples corresponding to several stages of grapevine development and ripening and several abiotic and biotic stress conditions were used37,39,65,70,81–96. Heat maps were performed with the ComplexHeatmap R package (https://github.com/jokergoo/ComplexHeatmap).
Sequence comparison among diverse plant species
We performed a sequence comparison using the LBD genes from 33 plant species (Arabidopsis thaliana, Brassica rapa, Carica papaya, Theobroma cacao, Gossypium raimondii, Eucalyptus grandis, Citrus clementina, Manihot esculenta, Ricinus communis, Populus trichocarpa, Linum usitatissimum, Malus domestica, Pyrus bretschneideri, Prunus persica, Fragaria vesca, Cicer arietinum, Glycine max, Medicago truncatula, Citrullus lanatus, Cucumis sativus, Solanum lycopersicum, Utricularia gibba, Nelumbo nucifera, Hordeum vulgare, Triticum aestivum, Oryza sativa subsp. indica, Phyllostachys heterocycla, Sorghum bicolor, Zea mays, Musa acuminata, Phoenix dactylifera, Picea abies) retrieved at http://planttfdb.cbi.pku.edu.cn. We identified orthologous genes in genomes from the thirty-three species following what was performed in Jaillon et al.28. Each pair of predicted gene sets was aligned with the BLASTp algorithm, and alignments with an e-value lower than 1e−20 and sequence homology higher than 40% were retained. If a comparison is above that value, the two genes were considered homologs. Two genes, A from Vitis genome (GV) and B from a given species genome (GX), were considered orthologs one-to-one if B was the best match for gene A in GX and A was the best match for B in GV. A phylogenetic tree was constructed with the LBD genes from these species with the same parameters as before.
A Ka/Ks analysis was performed using the Ka/Ks calculation tool (http://services.cbu.uib.no/tools/kaks) on all the orthologs detected in the species for each grapevine gene with the default parameters.
Data availability statement
All the data published in this article will be available for scientific community.
Electronic supplementary material
Acknowledgements
Funding was provided by the Portuguese Foundation for Science and Technology (PEst-OE/BIA/UI4046/2014, PD/BD/114385/2016, FCTInvestigator IF/00169/2015). JG was supported by the Ramon y Cajal program (RYC-2011-07791) and the AGL2014-59171-R project from the Spanish MINECO. PA-R was supported by a Portuguese fellowship (SFRH/BPD/72070/2010) and an Australian Research Council grant (LP130100347). Additionally, we would like to thank to Dr. Michael Considine for his support in the enrichment of cis-regulatory elements analysis.
Author Contributions
A.F. and J.G. designed the study. J.G., P.A.-R., D.P. and A.F. analysed the data. D.P. and A.F. wrote the manuscript with valuable input from J.G. and J.M. All the authors revised and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jérôme Grimplet and Diana Pimentel contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16240-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Xu C, Luo F, Hochholdinger F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016;21:159–167. doi: 10.1016/j.tplants.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Rast MI, Simon R. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell. 2012;24:2917–2933. doi: 10.1105/tpc.112.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semiarti E, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 5.Xu B, et al. Arabidopsis Genes AS1, AS2, and JAG Negatively Regulate Boundary-Specifying Genes to Promote Sepal and Petal Development. Plant Physiol. 2008;146:566–575. doi: 10.1104/pp.107.113787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borghi L, Bureau M, Simon R. Arabidopsis JAGGED LATERAL ORGANS Is Expressed in Boundaries and Coordinates KNOX and PIN Activity. Plant Cell. 2007;19:1795–1808. doi: 10.1105/tpc.106.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MJ, Kim M, Lee MR, Park SK, Kim J. Lateral Organ Boundaries Domain (Lbd)10 Interacts With Sidecar Pollen/Lbd27 To Control Pollen Development In Arabidopsis. Plant J. 2015;81:794–809. doi: 10.1111/tpj.12767. [DOI] [PubMed] [Google Scholar]
- 9.Fan M, Xu C, Xu K, Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012;22:1169–1180. doi: 10.1038/cr.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thatcher LF, Powell JJ, Aitken EA, Kazan K, Manners JM. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt Susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012;160:407–418. doi: 10.1104/pp.112.199067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangeon A, Bell EM, Lin WC, Jablonska B, Springer PS. Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J Exp Bot. 2011;62:221–233. doi: 10.1093/jxb/erq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yordanov YS, Regan S, Busov V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell. 2010;22:3662–3677. doi: 10.1105/tpc.110.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, et al. Conserved genetic determinant of motor organ identity in Medicago truncatula and related legumes. Proc Natl Acad Sci USA. 2012;109:11723–11728. doi: 10.1073/pnas.1204566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majer C, Xu C, Berendzen KW, Hochholdinger F. Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.) Philos Trans R Soc Lond B Biol Sci. 2012;367:1542–1551. doi: 10.1098/rstb.2011.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HW, Kim MJ, Park MY, Han KH, Kim J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol Plant. 2013;6:1722–1725. doi: 10.1093/mp/sst037. [DOI] [PubMed] [Google Scholar]
- 16.Iwakawa H, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 17.Shuai B, Reynaga-Pena CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, et al. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. [DOI] [PubMed] [Google Scholar]
- 19.Feng Z, Zhu J, Du X, Cui X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta. 2012;236:1227–1237. doi: 10.1007/s00425-012-1673-3. [DOI] [PubMed] [Google Scholar]
- 20.Lee HW, Cho C, Kim J. Lateral Organ Boundaries Domain16 and 18 Act Downstream of the AUXIN1 and LIKE-AUXIN3 Auxin Influx Carriers to Control Lateral Root Development in Arabidopsis. Plant Physiol. 2015;168:1792–1806. doi: 10.1104/pp.15.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin G, Tohge T, Matsuda F, Saito K, Scheible W-R. Members of the LBD Family of Transcription Factors Repress Anthocyanin Synthesis and Affect Additional Nitrogen Responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheible W-R, et al. Genome-Wide Reprogramming of Primary and Secondary Metabolism, Protein Synthesis, Cellular Growth Processes, and the Regulatory Infrastructure of Arabidopsis in Response to Nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009;151:1377–1389. doi: 10.1104/pp.109.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhang S, Su L, Liu X, Hao Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS One. 2013;8:e57044. doi: 10.1371/journal.pone.0057044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Yu X, Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006;39:248–262. doi: 10.1016/j.ympev.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Schnable PS, et al. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science. 2009;326:1112. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 28.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 29.Cao HUI, Liu C-Y, Liu C-X, Zhao Y-L, Xu R-R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J Genet. 2016;95:515–526. doi: 10.1007/s12041-016-0660-z. [DOI] [PubMed] [Google Scholar]
- 30.Grimplet J, et al. Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC research notes. 2012;5:213. doi: 10.1186/1756-0500-5-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canaguier A, et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3) Genomics data. 2017;14:56–62. doi: 10.1016/j.gdata.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimplet J, et al. The grapevine gene nomenclature system. BMC Genomics. 2014;15:1077. doi: 10.1186/1471-2164-15-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang T, Fang GY, He H, Chen J. Genome-Wide Identification, Evolutionary Analysis and Expression Profiles of LATERAL ORGAN BOUNDARIES DOMAIN Gene Family in Lotus japonicus and Medicago truncatula. PLoS One. 2016;11:e0161901. doi: 10.1371/journal.pone.0161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Ma B, Zeng Q, Xiang Z, He N. Identification and characterization of Lateral Organ Boundaries Domain genes in mulberry, Morus notabilis. Meta Gene. 2016;8:44–50. doi: 10.1016/j.mgene.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimplet J, Agudelo-Romero P, Teixeira RT, Martinez-Zapater JM, Fortes AM. Structural and Functional Analysis of the GRAS Gene Family in Grapevine Indicates a Role of GRAS Proteins in the Control of Development and Stress Responses. Front Plant Sci. 2016;7:353. doi: 10.3389/fpls.2016.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velasco R, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fasoli M, et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant cell. 2012;24:3489–3505. doi: 10.1105/tpc.112.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obayashi T, Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009;16:249–260. doi: 10.1093/dnares/dsp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tattersall EA, et al. Transcript abundance profiles reveal larger and more complex responses of grapevine to chilling compared to osmotic and salinity stress. Funct Integr Genomics. 2007;7:317–333. doi: 10.1007/s10142-007-0051-x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang YM, Zhang SZ, Zheng CC. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J Genet. 2014;93:79–91. doi: 10.1007/s12041-014-0342-7. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Bio Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coombe BG. Growth Stages of the Grapevine: Adoption of a system for identifying grapevine growth stages. Aust J Grape Wine Res. 1995;1:104–110. doi: 10.1111/j.1755-0238.1995.tb00086.x. [DOI] [Google Scholar]
- 43.Agudelo-Romero P, et al. Perturbation of polyamine catabolism affects grape ripening of Vitis vinifera cv. Trincadeira. Plant Physiol Biochem. 2014;74:141–155. doi: 10.1016/j.plaphy.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Thatcher LF, Kazan K, Manners JM. Lateral organ boundaries domain transcription factors: new roles in plant defense. Plant Signal Behav. 2012;7:1702–1704. doi: 10.4161/psb.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, et al. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci USA. 2014;111:E521–E529. doi: 10.1073/pnas.1313271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabrera J, et al. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 2014;203:632–645. doi: 10.1111/nph.12826. [DOI] [PubMed] [Google Scholar]
- 47.Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009;58:525–537. doi: 10.1111/j.1365-313X.2009.03797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo BJ, et al. A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.) J Zhejiang Univ Sci B. 2016;17:763–774. doi: 10.1631/jzus.B1500277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo JH, Weng L, Luo D. Isolation and expression patterns of LATERAL ORGAN BOUNDARIES-like genes in Lotus japonicus. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2006;32:202–208. [PubMed] [Google Scholar]
- 50.Wang S, et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics. 2010;10:533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- 51.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albinsky D, et al. Metabolomic Screening Applied to Rice FOX Arabidopsis Lines Leads to the Identification of a Gene-Changing Nitrogen Metabolism. Mol Plant. 2010;3:125–142. doi: 10.1093/mp/ssp069. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, et al. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J Exp Bot. 2015;66:99–112. doi: 10.1093/jxb/eru396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loraine AE, McCormick S, Estrada A, Patel K, Qin P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013;162:1092–1109. doi: 10.1104/pp.112.211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fortes AM, Teixeira R, Agudelo-Romero P. Complex Interplay of Hormonal Signals during Grape Berry Ripening. Molecules. 2015;20:9326–9343. doi: 10.3390/molecules20059326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giacomelli L, et al. Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set: functional characterization and evolution of grapevine gibberellin oxidases. J Exp Bot. 2013;64:4403–4419. doi: 10.1093/jxb/ert251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zentella R, et al. Global Analysis of DELLA Direct Targets in Early Gibberellin Signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu L, Guo JS, Zhou C, Zhu J. Ectopic expression of LBD15 affects lateral branch development and secondary cell wall synthesis. Arabidopsis thaliana. 2014;73:111–120. [Google Scholar]
- 59.Agudelo-Romero P, et al. Search for Transcriptional and Metabolic Markers of Grape Pre-Ripening and Ripening and Insights into Specific Aroma Development in Three Portuguese Cultivars. PLoS One. 2013;8:e60422. doi: 10.1371/journal.pone.0060422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palumbo MC, et al. Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell. 2014;26:4617–4635. doi: 10.1105/tpc.114.133710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ba, L.-j. et al. The Banana MaLBD (LATERAL ORGAN BOUNDARIES DOMAIN) Transcription Factors Regulate EXPANSIN Expression and Are Involved in Fruit Ripening. Plant Mol Biol Report32, 1103–1113 (2014).
- 62.Kim J, Lee HW. Direct activation of EXPANSIN14 by LBD18 in the gene regulatory network of lateral root formation in Arabidopsis. Plant Signal Behav. 2013;8:e22979. doi: 10.4161/psb.22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee HW, Kim J. EXPANSINA17 Up-Regulated by LBD18/ASL20 Promotes Lateral Root Formation During the Auxin Response. Plant Cell Physiol. 2013;54:1600–1611. doi: 10.1093/pcp/pct105. [DOI] [PubMed] [Google Scholar]
- 64.Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013;73:212–224. doi: 10.1111/tpj.12013. [DOI] [PubMed] [Google Scholar]
- 65.Fortes AM, et al. Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol. 2011;11:149. doi: 10.1186/1471-2229-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bell EM, et al. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci USA. 2012;109:21146–21151. doi: 10.1073/pnas.1210789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ariel FD, Diet A, Crespi M, Chan RL. The LOB-like transcription factor MtLBD1 controls Medicago truncatula root architecture under salt stress. Plant Signal Behav. 2014;5:1666–1668. doi: 10.4161/psb.5.12.14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ariel F, et al. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell. 2010;22:2171–2183. doi: 10.1105/tpc.110.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ba LJ, Kuang JF, Chen JY, Lu WJ. MaJAZ1 Attenuates the MaLBD5-Mediated Transcriptional Activation of Jasmonate Biosynthesis Gene MaAOC2 in Regulating Cold Tolerance of Banana Fruit. J Agric Food Chem. 2016;64:738–745. doi: 10.1021/acs.jafc.5b05005. [DOI] [PubMed] [Google Scholar]
- 70.Agudelo-Romero P, et al. Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J Exp Bot. 2015;66:1769–1785. doi: 10.1093/jxb/eru517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Bruyne L, Höfte M, De Vleesschauwer D. Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol Plant. 2014;7:943–959. doi: 10.1093/mp/ssu050. [DOI] [PubMed] [Google Scholar]
- 72.DeYoung BJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, et al. Homologues of CsLOB1 in citrus function as disease susceptibility genes in citrus canker. Mol Plant Pathol. 2017;18:798–810. doi: 10.1111/mpp.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okonechnikov K, Golosova O, Fursov M, team U. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 75.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang W-C, Lee T-Y, Huang H-D, Huang H-Y, Pan R-L. PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics. 2008;9:561–561. doi: 10.1186/1471-2164-9-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 80.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 81.Albertazzi G, et al. Gene expression in grapevine cultivars in response to Bois Noir phytoplasma infection. Plant Science. 2009;176:792–804. doi: 10.1016/j.plantsci.2009.03.001. [DOI] [Google Scholar]
- 82.Cramer GR, et al. Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics. 2007;7:111–134. doi: 10.1007/s10142-006-0039-y. [DOI] [PubMed] [Google Scholar]
- 83.Deluc LG, et al. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics. 2007;8:429. doi: 10.1186/1471-2164-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Espinoza C, et al. Gene expression associated with compatible viral diseases in grapevine cultivars. Functl Integr Genomics. 2007;7:95–110. doi: 10.1007/s10142-006-0031-6. [DOI] [PubMed] [Google Scholar]
- 85.Fung RWM, et al. Powdery Mildew Induces Defense-Oriented Reprogramming of the Transcriptome in a Susceptible But Not in a Resistant Grapevine. Plant Physiol. 2008;146:236–249. doi: 10.1104/pp.107.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grimplet J, et al. Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics. 2007;8:187. doi: 10.1186/1471-2164-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pilati S, et al. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at veraison. BMC Genomics. 2007;8:428. doi: 10.1186/1471-2164-8-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sreekantan L, et al. Differential floral development and gene expression in grapevines during long and short photoperiods suggests a role for floral genes in dormancy transitioning. Plant Mol Biol. 2010;73:191–205. doi: 10.1007/s11103-010-9611-x. [DOI] [PubMed] [Google Scholar]
- 89.Lund ST, Peng FY, Nayar T, Reid KE, Schlosser J. Gene expression analyses in individual grape (Vitis vinifera L.) berries during ripening initiation reveal that pigmentation intensity is a valid indicator of developmental staging within the cluster. Plant Mol Biol. 2008;68:301–315. doi: 10.1007/s11103-008-9371-z. [DOI] [PubMed] [Google Scholar]
- 90.Vega A, Gutierrez RA, Pena-Neira A, Cramer GR, Arce-Johnson P. Compatible GLRaV-3 viral infections affect berry ripening decreasing sugar accumulation and anthocyanin biosynthesis in Vitis vinifera. Plant Mol Biol. 2011;77:261–274. doi: 10.1007/s11103-011-9807-8. [DOI] [PubMed] [Google Scholar]
- 91.Carvalho LC, Vilela BJ, Mullineaux PM, Amancio S. Comparative transcriptomic profiling of Vitis vinifera under high light using a custom-made array and the Affymetrix GeneChip. Mol Plant. 2011;4:1038–1051. doi: 10.1093/mp/ssr027. [DOI] [PubMed] [Google Scholar]
- 92.Diaz-Riquelme J, Grimplet J, Martinez-Zapater JM, Carmona MJ. Transcriptome variation along bud development in grapevine (Vitis vinifera L.) BMC plant biology. 2012;12:181. doi: 10.1186/1471-2229-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tillett RL, et al. Identification of tissue-specific, abiotic stress-responsive gene expression patterns in wine grape (Vitis vinifera L.) based on curation and mining of large-scale EST data sets. BMC Plant Biol. 2011;11:86. doi: 10.1186/1471-2229-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lijavetzky D, et al. Berry flesh and skin ripening features in Vitis vinifera as assessed by transcriptional profiling. PLoS One. 2012;7:e39547. doi: 10.1371/journal.pone.0039547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pontin MA, et al. Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biol. 2010;10:224. doi: 10.1186/1471-2229-10-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carbonell-Bejerano P, et al. Thermotolerance responses in ripening berries of Vitis vinifera L. cv Muscat Hamburg. Plant Cell Physiol. 2013;54:1200–1216. doi: 10.1093/pcp/pct071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data published in this article will be available for scientific community.