Abstract
Some microRNAs (miRNAs) are known to suppress breast cancer. However, whether the expressions of these tumor suppressive miRNAs translate to patient survival were not investigated in large cohort. Nine miRNAs (miR-30a, miR-30c, miR-31, miR-126, miR-140, miR-146b, miR-200c, miR-206, and miR-335) known to be tumor suppressive miRNAs in breast cancer were investigated in Genomic Data Common data portal miRNA-Seq dataset and The Cancer Genome Atlas (TCGA) (n = 1052). Of the 9 miRNAs, miR-30a, miR-30c, miR-126, miR-140, miR-206, and miR-335 were found to have significantly lower expression in breast cancer tissues compared to paired normal breast tissue. High expression of miR-30a or miR-200c was associated with significantly better overall survival (OS). Gene Set Enrichment Analysis (GSEA) demonstrated that low expression levels of miR-30a had the tendency to associate with gene enrichment of EMT, while miR-200c did not, in TCGA cohort, and our findings support the need of validation using large cohort to use miRNA as prognostic biomarker for patients with breast cancer.
Introduction
The “Central Dogma” of cell biology suggests that genetic information flows in a unidirectional manner from DNA to messenger RNA (mRNA) and then to protein, which determines the cell function and behavior. In the human genome, more than 90% of the DNA sequence can be transcribed to RNAs, but only about 2% of RNA encodes proteins. The remaining 98%, which does not, are termed non-coding RNAs (ncRNA). In the last decade, ncRNAs have garnered increased appreciation for their important roles in regulating various biological processes1–6. Small ncRNA, termed microRNA (miRNA), was first reported in 1993 regarding its role in regulating lin-4 mRNA translation in Caenorhabditis elegans 7,8.
Since then, miRNA research has expanded rapidly, and many important functions of miRNA in intra- or extra-cellular regulation have been discovered. It is now known that miRNAs have two major post-transcriptional epigenomic regulatory functions: 1) translational repression of mRNA, and 2) mRNA cleavage9. In 2002, the first role of miRNAs in cancer was discovered. Expressions of miR-15 and miR-16 were suppressed in patient samples and cell lines of chronic lymphocytic leukemia, leading to the discovery of tumor suppressive miRNAs10. In 2005, He et al. showed that a cluster of miRNAs, the miR-17–92 polycistron, can promote tumor formation as a potential human oncogene and coined the term oncogenic miRNA11.
Regarding breast cancer, Iorio et al. first reported cancer-related miRNAs with specific breast cancer features, such as estrogen and progesterone receptor expression, tumor stage, vascular invasion, and proliferation index12. The roles of miRNAs in breast cancer biology have been continuously investigated13–15. It has been demonstrated that oncogenic miRNAs inhibit tumor suppressive genes or activate some oncogenic pathways, whereas suppressive miRNAs inhibit oncogenic gene function through post-translational modification9,16,17. The target gene is not a one-to-one correspondence. In other words, one miRNA targets several genes or pathways through post-translational mechanisms. It has been clearly demonstrated both in vivo and in vitro that the low levels of tumor suppressive miRNA are associated with cancer aggressiveness, such as cancer proliferation or tumor metastasis, and high levels of tumor suppressive miRNA inhibit cancer growth17. However, many of these reports have not been validated in large cohorts, which limit the statistical power of these studies. For instance, Tavazoie et al. demonstrated that miR-126 and miR-335 showed tumor suppressive function, and high expression levels of the each of these miRNAs was associated with better survival. However, the number of patients in this analysis was only 20. Recently, miR-200c was also demonstrated to have tumor suppressive function through epithelial-mesenchymal transition (EMT) in breast cancer. Song et al. and Damiano et al. reported that breast cancer patients with high expression levels of miR-200c had better prognosis. However, both of these studies also had relatively small cohorts from single institutions (n = 134 and 51, respectively).
The Cancer Genome Atlas (TCGA) is a joint collaboration of the National Cancer Institute (NCI) and the National Human Genome Research Institute of the National Institute of Health that has collected treatment naïve primary cancer samples from over 10,000 patients on over 30 tumor types and provides genomic and epigenomic data obtained by high-throughput sequencing techniques18. Approximately 1,100 breast cancer samples were collected in TCGA cohort18, which enables investigators to utilize large sample sizes and thereby have sufficient statistical power for analyses. For example, our group has recently demonstrated that angiopoietin pathway associated with poor prognosis utilizing TCGA as a representative database providing mRNA expression data and survival outcomes in breast cancer19.
Although there have been several reports that demonstrated that some miRNAs have tumor suppressive functions in breast cancer, their general clinical relevance remains unclear because their effects have only been studied in small cohorts. Therefore, the aim of this study was to investigate the clinical significance of tumor suppressive miRNAs in breast cancer utilizing TCGA, which is a “big data” set that provides sufficient statistical power with proven high quality genetic samples.
Results
Literature search to identify tumor suppressive miRNAs in breast cancer
We conducted a literature search using PubMed Central to identify tumor suppressive miRNAs in breast cancer. We identified several tumor suppressive miRNAs that have been reported by multiple groups but that lack validation using a sufficiently large cohort. We selected 9 miRNAs for our analysis: miR-30a, miR-30c, miR-31, miR-126, miR-140, miR-146b, miR-200c, miR-206, and miR-335, which have been reported to be tumor suppressive miRNAs in breast cancer (Table 1)20–60.
Table 1.
Candidates of tumor suppressive miRNAs based on literature search in breast cancer.
| miRNA | Target or related gene/pathway | Significant function | Reference |
|---|---|---|---|
| miR-30a | VIM | Inhibits cell migration and invasion | 20–25 |
| Eya2 | Suppresse cell proliferation and migration | ||
| MTDH | Suppresse tumor growth and metastasis | ||
| Slug | Suppress epithelial mesenchymal transition (EMT) | ||
| ITGB3 | Suppress cell invasion | ||
| UBE3C | Suppress cell proliferation and migration | ||
| miR-30c | VIM, TWF1 | Suppress cell invasion | 25–27 |
| NF-kB, TRADD, CCNE1 | Negatively regulate cell cycle | ||
| miR-31 | RhoA | Inhibits several steps of the invasion-metastasis cascade | 28–32 |
| WAVE3, RhoA | Reduces cancer progression and metastasis | ||
| GNA13 | Reduce cell invasion | ||
| PRKCE | Sensitizes cells to apoptosis | ||
| miR-126 | IRS1 | Suppress cancer progression | 33–36 |
| IGFBP2, MERTK, PITPNC1 | Reduces metastasis and angiogenesis | ||
| No specific target | Reduces tumorigenesis and metastasis | ||
| miR-140 | ALDH1/SOX9 | Reduce stemm cell formation | 37–39 |
| SOX2/SOX9 | Inhibit stemm cell signaling | ||
| COL4A1, ITGA6, MARCKSL | Reduce cell proliferation and migration | ||
| miR-146b | NFkB, IL-6/STAT3 | Inhibit migration and invasion and metastasis | 40–43 |
| FOXP3 | Triggering apoptosis | ||
| BRMS1 | Suppress breast cancer metastasis | ||
| miR-200c | TGF-b, ZEB1/2, SNAIL1/2 | Suppress EMT | 44–50 |
| ELP1/HDAC2 | Suppress EMT | ||
| KRAS | Inhibit tumor growth | ||
| PRKAR1A, PRKACB | Reduce migration | ||
| miR-206 | Cx43 | Reduces migration, invasion and metastasis | 51–56 |
| MKL1/IL11 pathway | Inhibits cancer cell stemness and metastasis | ||
| VEGF, MAPK3, and SOX9 | Inhibit cell invasion and angiogenesis | ||
| Tbx3 | Inhibit cell proliferation, invasion, and maintenance of the cancer stem cell population | ||
| TGF-β, NRP1, SMAD2 | Suppresses EMT | ||
| CORO1C | Inhibit cell migration | ||
| ER-a | Suppress cell proliferation | ||
| miR-335 | SOX4, TNC | Suppresses metastasis and migration | 33, 57–59 |
| SOX4, TNC | Selective metastasis suppressor and tumor initiation | ||
| BRCA1 | Inhibit cell proliferation and activate apoptosis |
Briefly, miR-30a and miR-30c have been reported to inhibit cell migration and invasion through targeting vimentin or other epithelial mesenchymal transition (EMT)-related molecules such as Slug or TWIF120,23,27. MiR-30a also regulates UBE3C, a ubiquitin protein ligase family, resulting in suppression of cell proliferation and migration25. MiR-31 demonstrates cell proliferative and invasive properties through miR-31-mediated down-regulation of WAVE3, GNA13, PRKCE, or RhoA29–31,33,61. MiR-126 and miR-335 are tumor suppressive miRNAs that reduce bone- or lung-metastasis using cell-based comprehensive miRNA microarray analysis in human breast cancer34. MiR-335 also suppresses metastasis and migration through targeting of the progenitor cell transcription factor SOX4 and extracellular matrix component tenascin C (TNC)34. MiR-140 promotes cancer stem cell formation in basal-like early stage breast cancer through miR-140/ALDH1/SOX9 axis38. Another group also reported that miR-140/SOX2/SOX9 axis regulates cancer stem cells in early breast cancer39. miR-140 also contributes to tumor suppressive effect by targeting COL4A1, ITGA6 and MARCKSL1 in breast cancer40. MiR-146b shows tumor suppressive function through the regulation of NF-κB-IL-6/STAT3 signaling pathway in breast cancer41,43. FOXP3-miR-146 family-NF-κB axis provides tumor suppressor function such as inhibition of cell growth or tumor metastasis in vitro or in vivo assay44. MiR-200c has been reported to show significant tumor suppressive function in several solid tumors including breast cancer62. Specifically, miR-200c suppresses TGF-β signaling pathway and targets ZEB1/2 or SNAIL1/2, resulting in inhibition of EMT in breast cancer45,46,50,51. MiR-200c was also reported to regulate EMT through PELP1/HDAC247. MiR-200c inhibits breast cancer proliferation by targeting KRAS49. Lastly, miR-206/TWF1/MKL1-SRF/IL-11 signaling pathway inhibits breast cancer initiation and progression56.
Expression levels of the 9 tumor-suppressive miRNAs in breast cancer tissue and paired normal breast tissue
The expression levels of each of the 9 tumor suppressive miRNAs in breast cancer tissue were compared with their paired normal breast tissue using TCGA dataset (n = 103 each group). Of the 9 tumor suppressive miRNAs, only miR-30a, miR-30c, miR-126, miR-140, miR-206, and miR-335 were found to show significantly lower expression levels in breast cancer tissue compared with paired normal breast tissue (p < 0.0001, p < 0.001, p < 0.0001, p < 0.0001, p < 0.0001, and p < 0.0001, respectively). Unexpectedly, miR-31, miR-146b, and miR-200c did not show any significant differences (Fig. 1). Interestingly, miR-200c showed higher expression levels in cancer tissue than in normal tissue, which was an opposite trend from previous reports (Fig. 1).
Figure 1.
Expression levels of the 9 tumor-suppressive miRNAs in breast cancer samples and their paired normal breast samples retrieved from TCGA dataset (n = 103). One-sided p < 0.05 was considered statistically significant for analysis of expression levels in cancer vs. normal tissue (tested normal greater than tumor).
Prognostic relevance of the tumor suppressive miRNA expression in breast cancer patients
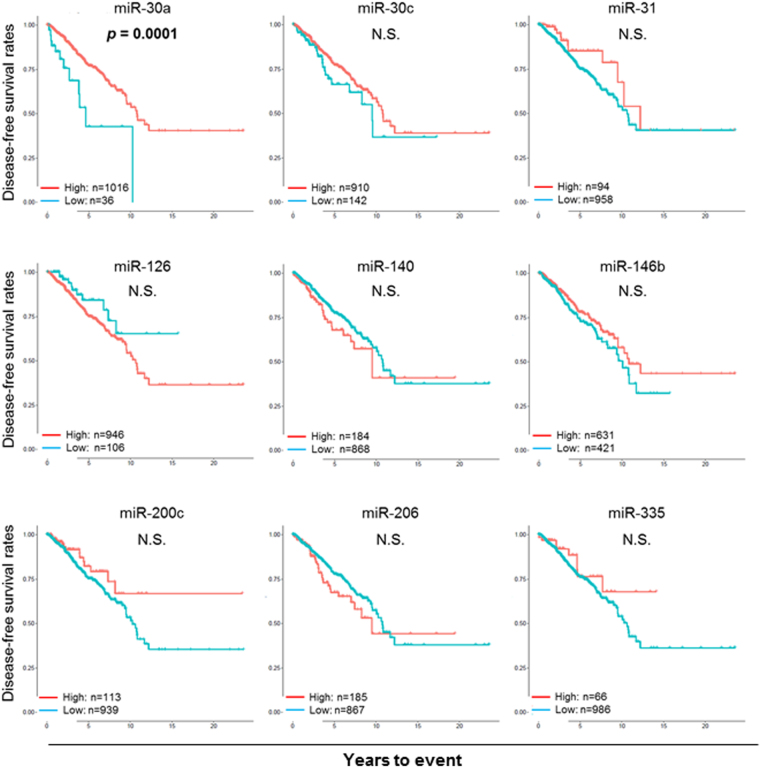
In order to determine the prognostic relevance of the 9 tumor suppressive miRNAs in breast cancer, OS was analyzed using the Kaplan-Meier curves and log rank test between the high and low expression groups of each miRNAs. The OS analysis demonstrated that high expression levels of miR-30a and miR-200c demonstrated significantly better survival (Log rank test, p = 0.0026 and p = 0.0266, respectively), while the other 6 miRNAs, miR-30c, miR-31, miR-140, miR-146b, miR-206, and miR-335, demonstrated no significant survival difference between high and low expression groups (Fig. 2). Surprisingly, high expression of miR-126 showed significantly worse prognosis (Log rank test, p = 0.0333) (Fig. 2), which was contrary to previous reports. As for the DFS analysis, high expression levels showed significantly better survival only for miR-30a (Log rank test, p = 0.0001), while high expression of miR-200c showed only a tendency toward better prognosis (Log rank test, p = 0.097) (Fig. 3).
Figure 2.
Expression of 9 selected tumor suppressive miRNAs in breast cancer was studied for their impact on overall survival (OS). OS was compared using the Kaplan-Meier curves and log rank test between the high (red line) and low (blue line) expression groups determined by each miRNA-specific thresholds. P value in bold type indicates statistical significance.
Figure 3.
Expression of 9 tumor suppressive miRNAs in breast cancer was studied for their impact on patient’s disease-free survival (DFS). DFS was compared using the Kaplan-Meier curves and log rank test between the high (red line) and low (blue line) expression groups determined by each miRNA-specific thresholds. P value in bold type indicates statistical significance.
Survival analyses of the suppressive miRNAs in different stages and subtypes
To further clarify which clinical stage and histology miR-30a and mir-200c demonstrate their tumor suppressive function, we conducted survival analyses on different TNM stages and subtypes for these 2 miRNAs. We found that high expression level of miR-30a was significantly associated with better survival in patients with stage II and IV cancer (p = 0.043 and 0.0053, respectively), and ER positive and non-triple negative subtypes (Log rank test, p = 0.0172, p = 0.0001, and p = 0.0168, respectively) (Fig. 4). Breast cancer is known to have two important subtypes, which have distinct signaling networks and drug targets63 and distinct prognostic signatures64. Therefore, we conducted survival analysis of luminal and basal-like subtypes of breast cancer based upon PAM50 classification. High expression level of miR-30a was significantly associated with better survival in both luminal and basal-like breast cancer subtypes (p = 0.0012 and p = 0.011, respectively) (Supplementary Figure S1). High expression of miR-200c was found to be significantly associated with better prognosis in patients with ER positive breast cancer. Patients with high expression of miR-200c demonstrated the trend towards better prognosis in both early and advanced stage breast cancer, but it was not statistically significant (Fig. 5).
Figure 4.
OS analyses of miR-30a in each stage and subtypes (ER positive and non-triple negative subgroups). OS was compared using the Kaplan-Meier curves and log rank test between the high (red line) and low (blue line) expression groups determined by the miRNA-30a-specific thresholds. P value in bold type indicates statistical significance.
Figure 5.
OS analyses of miR-200c in each stage and subtypes (ER positive and non-triple negative subgroups). OS was compared using the Kaplan-Meier curves and log rank test between the high and low expression groups determined by the miRNA-200c-specific thresholds. P value in bold type indicates statistical significance.
Univariate and multivariate Cox regression stratified analyses in TCGA dataset were performed on 2 selected miRNAs: miR-30a and miR-200c. When clinical stage or hormonal subtypes were defined as covariates in Cox regression analyses, the 2 miRNAs demonstrated no significant differences (Supplementary Table S1).
Association between EMT and TGF-β signaling related gene sets and miR-30a or miR-200c expression levels using GSEA
MiR-30a and miR-200c, which demonstrated significant survival associations in TCGA cohort, were previously demonstrated to have tumor suppressive function through EMT in breast cancer (Supplementary Figure S2). Therefore, we conducted GSEA to validate whether the miRNA expression levels were associated with EMT using TCGA cohort, as well as EMT-related gene set of transforming growth factor-β (TGF-β) signaling. Predefined gene sets of the HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION and HALLMARK_TGF_BETA_SIGNALING from GSEA, previously described to be involved in EMT and TGF-β, were used in this analysis. Interestingly, low expression levels of miR-30a showed a tendency with high enrichment score of EMT gene set (ES: -0.41, NES: -1.10, p = 0.393) and TGF-β signaling gene set (ES: -0.46, NES: -1.62, p = 0.053), while low expression levels of miR-200c did not show any association in TCGA cohort (EMT, ES: -0.27, NES: -0.71, p = 0.754; TGF-β signaling, ES: -0.28, NES: -1.02, p = 0.450) (Fig. 6).
Figure 6.
GSEA for expression levels of miR-30a or miR-200c. GSEA analyses were performed for HALLMARK EPITHELIAL MESENCHYMAL TRANSITION and HALLMARK TGF BETA SIGNALING using TCGA. ES, enrichment score; NES, normalized enrichment score.
It is important to examine whether miR-30a or miR-200c targets are associated with Cancer Hallmark-associated gene sets; apoptosis, cell cycle, cell death, cell motility, DNA repair, immune response, and phosphorylation65,66. The GSEA of C5 demonstrated that some gene sets are significantly associated with miR-30a and miR-200c expression. Several gene sets related to cell cycle, DNA repair, or phosphorylation are associated with miR-200c expression. Some gene sets related to phosphorylation are associated with miR-30a expression (Supplementary Dataset S1). Although the evidence is not robust enough, the result of the GSEA suggests that miR-30a and miR-200c might play potential roles in cancer biology linked to Cancer Hallmark-associated gene sets.
Discussion
During the last decade, many studies have demonstrated the individual biodiversity of breast cancer such as the intrinsic subtypes based on genome wide analyses67–69, and other sources of breast cancer heterogeneity70–72. Clinical classifications have been used to estimate the efficacy of hormonal therapy or molecular targeted therapy in clinical settings based on these analyses68,69. For instance, the luminal subtypes were generated through coding-gene expression. On the other hand, ncRNA expression has not been as extensively studied. In 2005, Iorio et al. first demonstrated that several miRNAs were deregulated in human breast cancer using miRNA microarray12 and many reports have been published in that field since then. Although many tumor-related miRNAs, including oncogenic miRNAs and tumor suppressive miRNAs, have been elucidated as prognostic biomarkers in breast cancer, few of them have been validated using large cohorts.
TCGA dataset includes comprehensive genetic and epigenetic information, as well as clinical data such as age, gender, race, pathological diagnosis, survival, and tumor recurrence of over 1000 breast cancer patients. To date, few reports have utilized this public dataset as a validation cohort for tumor suppressive miRNA in breast cancer. Therefore, we first conducted a systematic literature search and determined promising candidates of tumor suppressive miRNA as prognostic biomarkers in breast cancer. We then evaluated their clinical relevance using TCGA cohort. We analyzed 9 tumor suppressive miRNAs (miR-30a, miR-30c, miR-31, miR-126, miR-140, miR-146b, miR-200c, miR-206, and miR-335) all previously reported in breast cancer. To our surprise, we found that only 2 out of the 9 selected miRNAs (miR-30a and miR-200c) demonstrated prognostic significance in TCGA cohort.
MiR-30a has been reported to target EMT-related molecules (such as vimentin or Slug) and to suppress tumor cell migration and invasion in breast cancer20,23 as well as other solid cancers73. MiR-30a has also been demonstrated to inhibit several critical oncogenes, such as Eya2, ITGB3, or UBE3C, and suppress tumor growth, cell migration and invasion21,22,24,25. MiR-200c is one of the most well-known tumor suppressive miRNAs in cancer. Hurteau et al. first demonstrated that overexpression of the miR-200c leads to reduced expression of ZEB1 and increased expression of E-cadherin in breast cancer cell lines45. Further, many reports have demonstrated that miR-200c has tumor suppressive functions related to EMT in breast cancer46,47,50,51 and other solid cancers74–77. However, the clinical relevance of miR-30a and miR-200c as prognostic biomarkers has never been investigated using large cohorts. In the present study, we found that high expression levels of miR-30a or miR-200c were associated with better OS and DFS in breast cancer using TCGA cohort. To our knowledge, this is the first report that demonstrates the prognostic relevance of tumor suppressive miRNAs in breast cancer patients using a sufficiently large cohort.
We recognize that there are limitations with our study. The TCGA dataset was collected from multiple institutes, which may introduce selection biases into our methods. There was also missing data such as some patients in TCGA cohort lacked clinical data. Further, although TCGA has a large sample size of patients with breast cancer, the number of patients with paired normal breast tissue was significantly smaller, which may have hindered the statistical power for that analysis.
In conclusion, we found that high expression of 2 tumor-suppressive miRNAs, miR-30a and miR-200c, was associated with better OS, whereas miR-30c, miR-31, miR-126, miR-140, miR-146b, miR-206, and miR-335 was not. To our knowledge, this is the first report that elucidated the feasibility of utilizing a publicly available database, such as TCGA, to validate the clinical relevance of tumor suppressive miRNA for patients with breast cancer.
Materials and Methods
Literature search to identify well established tumor suppressive miRNAs in breast cancer
We conducted a literature search using PubMed Central between 2005 and 2016 to identify well established tumor suppressive miRNAs in breast cancer. The criteria for selection were: 1) at least two research groups have demonstrated that the selected miRNA possesses only tumor suppressive function (and not oncogenic function) both in vitro and in vivo, 2) the target mRNA or signaling pathway of the miRNA have been identified in breast cancer, and 3) the clinical relevance of miRNA has yet to be elucidated using a large cohort of breast cancer patients.
Extraction of miRNA-Seq and clinical dataset from TCGA
All data including the expression levels of the miRNAs of interest (miRNA-Seq) and clinical data were obtained from TCGA breast cancer cohort through the Genomic data common (GDC) data portal. The survival data of the breast cancer patients in the TCGA was obtained as previously reported19. Among the 1,097 patient breast cancer samples logged in TCGA, 1,052 samples that had both miRNA-Seq data and survival information were used in this study. Since TCGA is a collection of de-identified publically accessible database, Institutional Review Board review was waived.
Comparison of miRNAs expression levels between breast cancer and paired normal breast tissue using TCGA cohort
To evaluate the expression level of each candidate tumor suppressive miRNA in breast cancer tissue, the miRNA-Seq expression quantification data of breast cancer tissue (n = 103) and the paired normal breast tissue (n = 103) were retrieved from the GDC data portal.
Prognostic analysis of the tumor suppressive miRNAs using TCGA cohort
Overall survival (OS) was defined as the time from the date of diagnosis to the date of death by any cause, and disease-free survival (DFS) was defined as the time from the date of diagnosis to the date of diagnosis of a recurrent breast cancer. Patients who did not have an event were censored at the last date of follow-up or after 10 years from clinical records. OS or DFS was compared using the Cox proportional hazard model between expression groups (high versus low) determined by each miRNA-specific thresholds. Namely, differences in the OS between the two groups were assessed at multiple candidate cutoff points within the range of observed expression value, and the optimal cut point was chosen based on the statistical significance of the Cox proportional hazard model. Stratified analyses were also performed. The covariates in the models included tumor TNM stage (American Joint Commission on Cancer Clinical Cancer Staging 7th edition), estrogen receptor (ER), progesterone receptor (PR), and HER2 status. In TCGA data set, the histological subtypes were determined using pathological molecular subtyping78,79. Univariate and multivariate Cox regression stratified analyses for OS were also conducted and the covariates in the models included tumor TNM stage, ER, PR, HER2 status, and the expression levels of each miRNA of interest.
Gene Set Enrichment Analysis (GSEA) for miRNA expression
To investigate whether the miRNAs of interest had significant associations with metastasis-related gene sets, GSEA was conducted with the miRNAs of interest and mRNA expression data from TCGA. GSEA was performed using software provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp)80. We performed GSEA for Hallmark gene sets, which summarized and represented specific well-defined biological states or processes and displayed coherent expression.
Statistical analysis
All statistical analyses were performed using R software (http:///www.r-project.org/) and Bioconductor (http://bioconductor.org/). Data of miRNA expression was normalized using DESeq. 2 package81 and log-transformed. Patients were dichotomized into low-expression group and high-expression groups based on the miRNA expression levels. A running Cox proportional hazard statistics was applied to determine the threshold of the dichotomization82. To compare the survival curves of individual groups, the Kaplan-Meier method with log-rank tests and Cox proportional hazard models were used when appropriate. To test the proportional Hazard assumption in Cox models, Schoenfeld residuals test was used. The reported results included hazard ratios (HR) and 95% confidence intervals (CI). One-sided p < 0.05 was considered statistically significant for analysis of expression levels in cancer vs. normal tissue (tested normal greater than tumor), and two-sided p < 0.05 was considered statistically significant for survival analysis.
Electronic supplementary material
Acknowledgements
Kazuaki Takabe is supported by NIH/NCI grant R01CA160688 and Susan G. Komen Investigator Initiated Research Grant IIR12222224, as well as Institutional Support Grant 71–4085–01. This work was also supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Cancer Institute’s Bioinformatics and Biostatistics Shared Resources. We would also appreciate TCGA consortium.
Author Contributions
Conception and design: T. Kawaguchi and K. Takabe. Development of methodology: T. Kawaguchi, L. Yan, and K. Takabe. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): T. Kawaguchi, Q. Qi, X. Peng, L. Yan, and S. Liu. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): T. Kawaguchi, Q. Qi, X. Peng, and L. Yan. Writing, review, and/or revision of the manuscript: T. Kawaguchi, E. Gabriel, Y. Jessica, and K. Takabe. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Q. Qi, X. Peng, L. Yan, and S. Liu, Study supervision: T. Kawaguchi, and K. Takabe
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16112-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science (New York, N.Y.) 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science (New York, N.Y.) 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (New York, N.Y.) 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes & development. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nature reviews. Drug discovery. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science (New York, N.Y.) 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 9.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Mulrane L, McGee SF, Gallagher WM, O’Connor D. P. miRNA dysregulation in breast cancer. Cancer research. 2013;73:6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 17.van Schooneveld E, et al. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast cancer research: BCR. 2015;17:21. doi: 10.1186/s13058-015-0526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nature medicine. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 19.Ramanathan, R. et al. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast cancer research and treatment10.1007/s10549-017-4102-2 (2017). [DOI] [PMC free article] [PubMed]
- 20.Cheng CW, et al. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast cancer research and treatment. 2012;134:1081–1093. doi: 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 21.Fu J, et al. miR-30a suppresses breast cancer cell proliferation and migration by targeting Eya2. Biochemical and biophysical research communications. 2014;445:314–319. doi: 10.1016/j.bbrc.2014.01.174. [DOI] [PubMed] [Google Scholar]
- 22.Zhang N, et al. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene. 2014;33:3119–3128. doi: 10.1038/onc.2013.286. [DOI] [PubMed] [Google Scholar]
- 23.Chang CW, et al. MicroRNA-30a increases tight junction protein expression to suppress the epithelial-mesenchymal transition and metastasis by targeting Slug in breast cancer. Oncotarget. 2016;7:16462–16478. doi: 10.18632/oncotarget.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Liu C, Zhao C, Zhai L, Lv S. Downregulation of beta3 integrin by miR-30a-5p modulates cell adhesion and invasion by interrupting Erk/Ets1 network in triple-negative breast cancer. International journal of oncology. 2016;48:1155–1164. doi: 10.3892/ijo.2016.3319. [DOI] [PubMed] [Google Scholar]
- 25.Xiong, J., Wei, B., Ye, Q. & Liu, W. MiR-30a-5p/UBE3C axis regulates breast cancer cell proliferation and migration. Biochemical and biophysical research communications10.1016/j.bbrc.2016.03.069 (2016). [DOI] [PubMed]
- 26.Rodriguez-Gonzalez FG, et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast cancer research and treatment. 2011;127:43–51. doi: 10.1007/s10549-010-0940-x. [DOI] [PubMed] [Google Scholar]
- 27.Bockhorn J, et al. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast cancer research and treatment. 2013;137:373–382. doi: 10.1007/s10549-012-2346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shukla K, et al. MicroRNA-30c-2-3p negatively regulates NF-kappaB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Molecular oncology. 2015;9:1106–1119. doi: 10.1016/j.molonc.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sossey-Alaoui K, et al. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. International journal of cancer. Journal international du cancer. 2011;129:1331–1343. doi: 10.1002/ijc.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Molecular cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korner C, et al. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon) The Journal of biological chemistry. 2013;288:8750–8761. doi: 10.1074/jbc.M112.414128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulrane L, Gallagher WM, O’Connor DP. A novel mechanism of regulation of the anti-metastatic miR-31 by EMSY in breast cancer. Breast cancer research: BCR. 2014;16:467. doi: 10.1186/s13058-014-0467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasheed SA, et al. MicroRNA-31 controls G protein alpha-13 (GNA13) expression and cell invasion in breast cancer cells. Molecular cancer. 2015;14:67. doi: 10.1186/s12943-015-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, et al. The cell growth suppressor, mir-126, targets IRS-1. Biochemical and biophysical research communications. 2008;377:136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 36.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 37.Zhu N, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Molecular and cellular biochemistry. 2011;351:157–164. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, et al. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene. 2014;33:2589–2600. doi: 10.1038/onc.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gernapudi R, et al. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast cancer research and treatment. 2015;150:685–695. doi: 10.1007/s10549-015-3326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salem O, et al. The highly expressed 5′isomiR of hsa-miR-140-3p contributes to the tumor-suppressive effects of miR-140 by reducing breast cancer proliferation and migration. BMC genomics. 2016;17:566. doi: 10.1186/s12864-016-2869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhaumik D, et al. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurst DR, et al. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer research. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang M, et al. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-kappaB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Science signaling. 2014;7:ra11. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu R, et al. FOXP3 Controls an miR-146/NF-kappaB Negative Feedback Loop That Inhibits Apoptosis in Breast Cancer Cells. Cancer research. 2015;75:1703–1713. doi: 10.1158/0008-5472.CAN-14-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer research. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 46.Bai WD, et al. MiR-200c suppresses TGF-beta signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. International journal of cancer. Journal international du cancer. 2014;135:1356–1368. doi: 10.1002/ijc.28782. [DOI] [PubMed] [Google Scholar]
- 47.Roy SS, et al. Significance of PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of breast cancer. Oncogene. 2014;33:3707–3716. doi: 10.1038/onc.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie G, et al. Tumour-initiating capacity is independent of epithelial-mesenchymal transition status in breast cancer cell lines. Br J Cancer. 2014;110:2514–2523. doi: 10.1038/bjc.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song C, et al. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget. 2015;6:34968–34978. doi: 10.18632/oncotarget.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perdigao-Henriques R, et al. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene. 2016;35:158–172. doi: 10.1038/onc.2015.69. [DOI] [PubMed] [Google Scholar]
- 51.Damiano V, et al. Epigenetic silencing of miR-200c in breast cancer is associated with aggressiveness and is modulated by ZEB1. Genes, chromosomes & cancer. 2017;56:147–158. doi: 10.1002/gcc.22422. [DOI] [PubMed] [Google Scholar]
- 52.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer research. 2008;68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, et al. miR-206 inhibits cell migration through direct targeting of the actin-binding protein coronin 1C in triple-negative breast cancer. Molecular oncology. 2014;8:1690–1702. doi: 10.1016/j.molonc.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amir S, et al. Regulation of the T-box transcription factor Tbx3 by the tumour suppressor microRNA-206 in breast cancer. Br J Cancer. 2016;114:1125–1134. doi: 10.1038/bjc.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang Z, Bian X, Shim H. Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer. Biochemical and biophysical research communications. 2016;477:461–466. doi: 10.1016/j.bbrc.2016.06.076. [DOI] [PubMed] [Google Scholar]
- 56.Samaeekia, R. et al. MicroRNA-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clinical cancer research: an official journal of the American Association for Cancer Research10.1158/1078-0432.ccr-16-0943 (2016). [DOI] [PMC free article] [PubMed]
- 57.Yin K, et al. MiR-206 suppresses epithelial mesenchymal transition by targeting TGF-beta signaling in estrogen receptor positive breast cancer cells. Oncotarget. 2016;7:24537–24548. doi: 10.18632/oncotarget.8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heyn H, et al. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. International journal of cancer. Journal international du cancer. 2011;129:2797–2806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- 59.Png KJ, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes & development. 2011;25:226–231. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, et al. Aryl hydrocarbon receptor agonists induce microRNA-335 expression and inhibit lung metastasis of estrogen receptor negative breast cancer cells. Molecular cancer therapeutics. 2012;11:108–118. doi: 10.1158/1535-7163.MCT-11-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valastyan S, Weinberg RA. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell cycle (Georgetown, Tex.) 2010;9:2124–2129. doi: 10.4161/cc.9.11.11843. [DOI] [PubMed] [Google Scholar]
- 62.Koutsaki M, Spandidos DA, Zaravinos A. Epithelial-mesenchymal transition-associated miRNAs in ovarian carcinoma, with highlight on the miR-200 family: prognostic value and prospective role in ovarian cancer therapeutics. Cancer letters. 2014;351:173–181. doi: 10.1016/j.canlet.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 63.Zaman N, et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell reports. 2013;5:216–223. doi: 10.1016/j.celrep.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 64.Li J, et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nature communications. 2010;1:34. doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao S, et al. Identification and Construction of Combinatory Cancer Hallmark-Based Gene Signature Sets to Predict Recurrence and Chemotherapy Benefit in Stage II Colorectal Cancer. JAMA oncology. 2016;2:37–45. doi: 10.1001/jamaoncol.2015.3413. [DOI] [PubMed] [Google Scholar]
- 66.Wang E, et al. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Seminars in cancer biology. 2015;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 67.Sparano JA, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. The New England journal of medicine. 2015;373:2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 69.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gancberg D, et al. Comparison of HER-2 status between primary breast cancer and corresponding distant metastatic sites. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2002;13:1036–1043. doi: 10.1093/annonc/mdf252. [DOI] [PubMed] [Google Scholar]
- 71.Simon R, et al. Patterns of her-2/neu amplification and overexpression in primary and metastatic breast cancer. Journal of the National Cancer Institute. 2001;93:1141–1146. doi: 10.1093/jnci/93.15.1141. [DOI] [PubMed] [Google Scholar]
- 72.Bonsing BA, et al. Allelotype analysis of flow-sorted breast cancer cells demonstrates genetically related diploid and aneuploid subpopulations in primary tumors and lymph node metastases. Genes, chromosomes & cancer. 2000;28:173–183. doi: 10.1002/(SICI)1098-2264(200006)28:2<173::AID-GCC6>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 73.Kumarswamy R, et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. International journal of cancer. Journal international du cancer. 2012;130:2044–2053. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 74.Marchini S, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12:273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 75.Yu J, et al. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Molecular cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ceppi P, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Molecular cancer research: MCR. 2010;8:1207–1216. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 77.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature cell biology. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 78.Goldhirsch A, et al. Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sobin LH, G. M. & Wittekind, C. (ed International Union Against Cancer) Ch. 73–77, (Wiley-Blackwell, 2009).
- 80.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crowley, J, L. M., Jacobson, J. & Salmon, S. Proceedings of the First Seattle Symposium in Biostatistics Survival Analysis. Vol. 123 199-229 (Springer, 1997).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.